Abstract
Genetic aberrations in tumors are predictive for chemosensitivity and survival. A test is needed that allows simultaneous detection of multiple changes and that is widely applicable in a routine diagnostic setting. Multiplex ligation-dependent probe amplification (MLPA) allows detection of DNA copy number changes of up to 45 loci in one relatively simple, semiquantitative polymerase chain reaction-based assay. To assess the applicability of MLPA, we performed MLPA analysis to detect relevant genetic markers in a spectrum of 88 gliomas. The vast majority of these tumors (n = 79) were previously characterized by comparative genomic hybridization. With MLPA kit P088 (78 cases), complete and partial loss of 1p and 19q were reliably identified, even in samples containing only 50% tumor DNA. Distinct 1p deletions exist with different clinically prognostic consequences, and in contrast to the commonly used diagnostic strategies (loss of heterozygosity or fluorescent in situ hybridization 1p36), P088 allows detection of such distinct 1p losses. Combining P088 with P105 will further increase the accurate prediction of clinical behavior because this kit identified markers (EGFR, PTEN, and CDKN2A) of high-grade malignancy in 41 cases analyzed. We conclude that MLPA is a reliable diagnostic tool for simultaneous identification of different region-specific genetic aberrations of tumors.
The majority of gliomas can be classified as astrocytic (As) or oligodendroglial tumors (OTs), the latter including pure oligodendroglial (Os) or mixed oligo-astrocytic (OAs) tumors. An accurate distinction between OTs and As is important because of prognostic and therapeutic implications.1,2,3 Unfortunately, unequivocal histopathological criteria are lacking and differences in clinical behavior within a specific histopathological group have been reported (eg, two thirds of the anaplastic Os respond to PCV chemotherapy [procarbazine, lomustine (CCNU), and vincristine]).1,3,4 Fortunately, loss of 1p and 19q have been identified as diagnostic molecular markers in gliomas predicting response to chemotherapy and long survival.1,5,6,7,8,9,10 Identification of molecular alterations involved in the malignant progression of gliomas such as EGFR amplification and PTEN and CDKN2A loss next to loss of heterozygosity (LOH) 1p36 was reported to predict a less favorable prognosis and a less durable response to chemotherapy.10,11,12,13,14,15 A combined molecular diagnostic approach would therefore be of clinical relevance.
The most commonly applied techniques, LOH,10,16 fluorescent in situ hybridization (FISH),17,18 or quantitative microsatellite analysis,19,20 specifically analyze the chromosomal region 1p36 to identify OTs with a favorable prognosis and therapy response. The clinical value of such tests has been clearly proven. However, not all chemosensitive OTs were identified, and vice versa, some of the tumors with a loss of 1p36 proved to be chemoresistant.1,16 Furthermore, it is becoming increasingly clear that distinct types of 1p deletions (complete versus partial) exist in (oligodendro)gliomas, sometimes with opposite clinical and biological consequences.21,22 Unfortunately, however, the above-mentioned diagnostic strategies do not discriminate between these distinct types. Until the exact combination of genes on 1p/19q responsible for the favorable clinical behavior of these gliomas have been identified, analysis of multiple regions on 1p and 19q seems preferable, thereby enabling an even more accurate identification of these clinically favorable gliomas. Furthermore, it is noteworthy that techniques identifying copy number changes such as (array) comparative genomic hybridization (CGH) identified 1p and 19q gains in gliomas.23,24,25,26,27 These gains could be easily misinterpreted as loss of heterozygosity using microsatellite approach.23,24,25,26,27
Next to analysis of multiple 1p/19q loci, analysis of additional genes, for example those reported to be involved in malignant progression, will prove to be of additional diagnostic value. Unfortunately, with the above-mentioned techniques only one or a few loci can be analyzed per experiment. Array CGH or, at a lower resolution, conventional CGH provides an overview of copy number changes through the entire tumor genome.28,29,30 Because such experiments are rather specialized, they are not available in most standard molecular biology laboratories and may therefore be less suitable for a routine diagnostic setting. Multiplex ligation-dependent probe amplification (MLPA) is a technique by which up to 45 different sequences can be targeted in a single, semiquantitative polymerase chain reaction (PCR)-based experiment (see Figure 1).31 The sequences detected can be small (∼60 nucleotides), enabling analysis of fragmented DNA. Furthermore, the MLPA reaction is fast, relatively inexpensive, and easy to perform, and the equipment needed for MLPA analysis is present in most molecular biology laboratories.
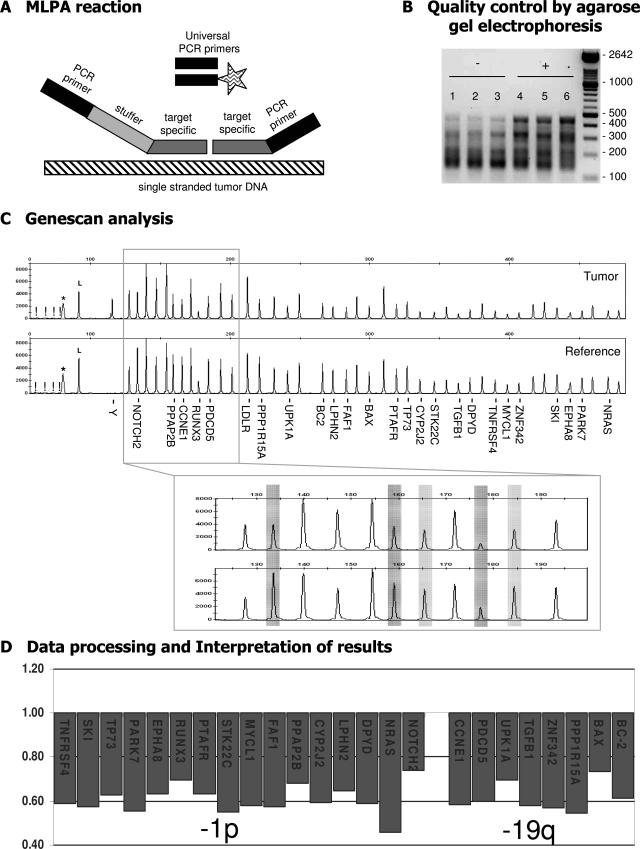
Figure 1.
Schematic outline of the MLPA procedure. During MLPA analysis up to 45 target-specific probes are used in one reaction. A: The unique target-specific sequences (gray) of the probes are hybridized on single-stranded genomic DNA (hatched) after which the two adjacent parts of the probe are joined through ligation and PCR amplification is performed with a fluorescently (FAM) labeled primer (indicated by the asterisk). Because identical PCR primers (black) are used for all probes, there are no primer-specific advantages in this procedure. Each probe has a unique length [attributable to variation in the length of the stuffer sequence (light gray)], and therefore gel electrophoresis can be used to separate the individual probe fragments. B: Agarose gel electrophoresis is performed as an early (and extra) control step for identification of suboptimal MLPA reactions (P088 is shown as an example). DNA fragments are smaller than 500 bp. In suboptimal samples (−; lanes 1 to 3) the large DNA fragments are underrepresented compared to adequate experiments (+; lanes 4 to 6). C: Capillary electrophoresis is performed to identify and quantify the amplification products. Probe names are provided for those located on 1p and 19q. The MLPA mix includes an internal control for DNA quantity and quality. If these are insufficient, fragments of 64, 70, 76, and 82 bp will appear (indicated by an exclamation mark in C) whereas a band of 94 bp indicates successful ligation and hybridization (indicated by an L in C). A nonspecific broad peak can be present, with the size depending on the electrophoresis apparatus and fluorescence used (indicated by an asterisk in C). In this example, MLPA analysis with kit P088 of an oligodendroglial tumor (N293) with −1p/−19q (as detected by CGH) is shown. Subsequently, ratio analysis is performed, and results are visualized showing the probes in their chromosomal order (described in Materials and Methods; threshold used for losses and gains set at 0.8 and 1.2, respectively).
Two MLPA kits were designed for molecular analysis of (oligodendro)gliomas, one to detect copy number changes on 1p and 19q (kit P088; MRC-Holland, Amsterdam, The Netherlands) and one to detect aberrations of EGFR, TP53, PTEN, CDKN2A, and ERBB2 (kit P105; MRC-Holland). To establish the potential of MLPA in a routine diagnostic setting, we analyzed a spectrum of 88 glial tumors using P088 (n = 78) and P105 (n = 41). The vast majority (79 of 88) of these tumors were previously characterized genetically by conventional CGH.
Materials and Methods
Samples
Eighty-eight specimens obtained from glioma patients treated in the Department of Neurosurgery of the Radboud University Nijmegen Medical Centre, The Netherlands, were selected. The use of brain tumor tissue after completing histopathological diagnosis for research purposes was approved by the ethics committee of the Radboud University Nijmegen Medical Centre, and informed consent was given by the patients. Tumors were classified according to the World Health Organization–2000 classification32 and included three pilocytic astrocytomas (A-I), six low-grade diffuse astrocytomas (A-II), one anaplastic astrocytoma (A-III), 24 glioblastomas multiforme (GBM), nine low-grade oligodendrogliomas (O-II), 16 anaplastic oligodendrogliomas (O-III), seven low-grade oligo-astrocytomas (OA-II), 17 anaplastic oligo-astrocytomas (OA-III), three low-grade ependymomas (E-II), and two anaplastic ependymomas (E-III). Most samples used in this study (79 of 88) were previously analyzed by conventional CGH.25,26,27
DNA was isolated from snap-frozen tumor tissue with the DNeasy tissue kit, as described by the manufacturer (Qiagen, Venlo, The Netherlands), supplemented with an additional wash step using the supplied wash buffer AW2 before elution. In case of paraffin-embedded tissue, 50-μm paraffin sections were cut and incubated in P-buffer (50 mmol/L Tris-HCl, pH 8.2, 100 mmol/L NaCl, 1 mmol/L ethylenediaminetetraacetic acid, 0.5% Tween 20, 0.5% Nonidet P-40, 20 mmol/L dithiothreitol) at 90°C for 15 minutes, after which a protein digestion was performed using 0.5 mg/ml of proteinase K (Roche Diagnostics GmbH, Mannheim, Germany) at 55°C overnight. Subsequently, DNA was isolated using the DNeasy tissue kit. Furthermore, DNA previously isolated using a salting out procedure25,33 was purified using the DNeasy tissue kit.
MLPA Procedure
P088 contains 15 1p probes, eight 19q probes, and 21 control probes specific to other chromosomes (MRC Holland). During our study a minor change (replacement of two 1p probe by a new 1p and control probe) was introduced by the manufacturer, and lot-nr 0804 was replaced by lot-nr 0305. SALSA P105 (lot-nr 0804) contains nine PTEN probes, five CDKN2A probes, eight TP53 probes, three EGFR probes, two ERBB2 probes, and 15 control probes. All MLPA probe pairs code for unique human single copy DNA sequences and were designed and prepared as described by Schouten and colleagues.31 Probe sequences and genes detected by the control probes are available on request by the manufacturer (MRC Holland). Probes used to detect imbalances are listed in Figures 2 to 5.
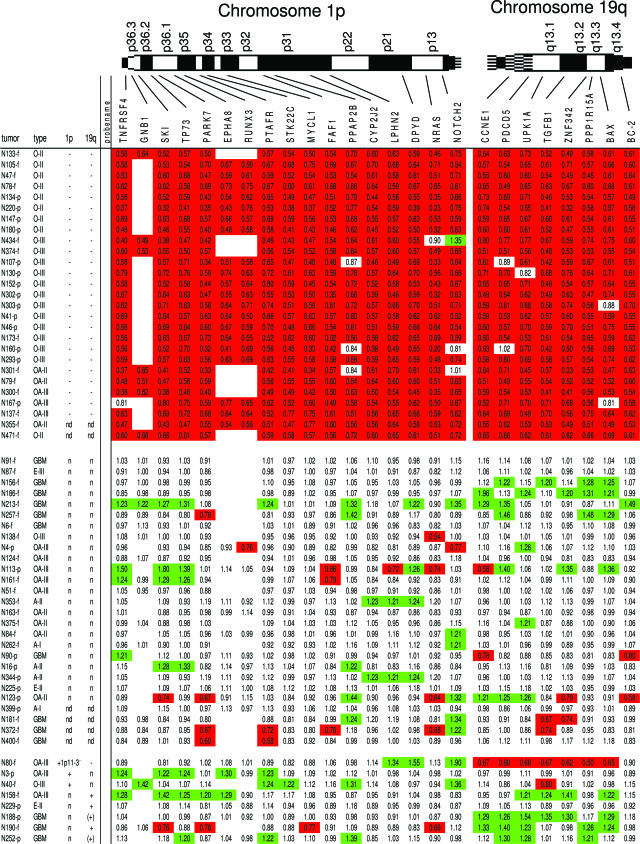
Figure 2.
Overview of the results of MLPA analysis of gliomas using kit P088 for detection of losses on chromosome 1p and 19q. In the top row the probe names analyzed are listed in chromosomal order. Red boxes represent ratios surpassing the threshold set to detect a loss (0.8), whereas green boxes show a ratio greater than 1.2. White boxes without a ratio are present as during our study 2 probes in kit P088 were replaced by others (EPHA8 and RUNX3 were only present in lot 0408). The left column contains tumor identification numbers (-f and -p: tumor DNA from snap-frozen or paraffin-embedded tissue, respectively). Tumors are grouped based on the CGH results, containing a complete loss of 1p and 19q, no losses or gains on 1p or 19q, or gains involving 1p and 19q, respectively. Tumors containing partial deletions are shown in Figure 4. In the next column the histopathological diagnosis is given (for abbreviations see Materials and Methods) followed by columns with the results of previous CGH analysis with regard to chromosome 1p and 19q (n, no aberration detected; −, loss of chromosome arm; +, gain of chromosome arm; nd, not done; (), CGH ratio clearly deviating from the normal ratio but not crossing the threshold that was set to detect a loss (0.8) or gain (1.2) by CGH).
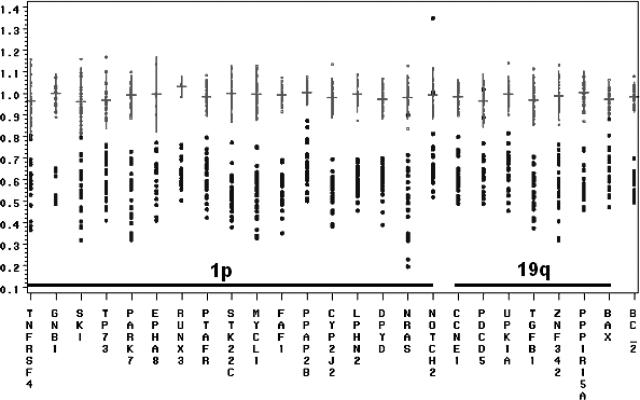
Figure 3.
Overview of the distribution of individual probe ratios in control and −1p/−19q-containing samples. The x axis shows probe names in chromosomal order whereas the y axis shows ratios. Probe ratios for the reference DNAs are shown in gray, and ratios in tumors with a complete loss of 1p and 19q as detected by CGH are shown in black. For reference DNA, horizontal lines represent mean probe values; vertical lines represent mean values ± 2 times SD; dot represents individual probe ratio. The increased variation in probe ratios among the −1p/−19q tumors compared to the control samples is caused by the fact that the amount of tumor cells within a tumor sample directly affecting the probe ratios varies among the different tumors. Mean probe ratios and standard deviations were therefore not calculated for tumor DNA.
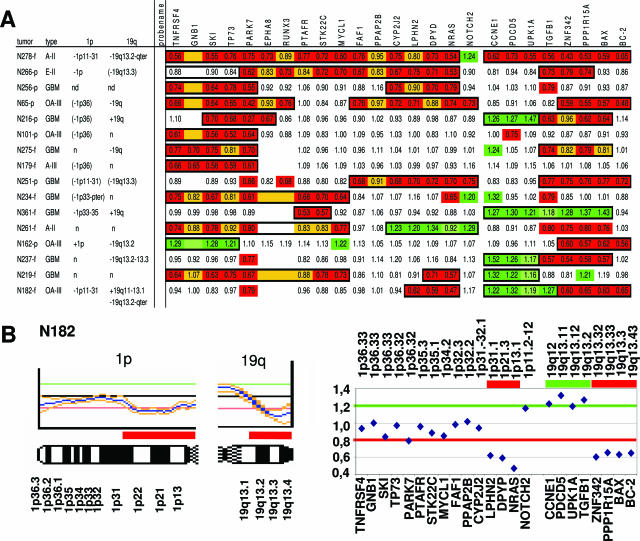
Figure 4.
Overview of the detection of partial 1p and 19q losses using MLPA kit P088. A: Partial deletions as detected by MLPA kit P088. Legends are as described for Figure 2. Partial deletions as detected by CGH are listed on the left. Red and green boxes represent probe ratios <0.8 and >1.2. Additionally, yellow and light green boxes represent regions that, based on the relative low/high ratios and ratios of adjacent probes, were considered to be lost or gained even though the threshold (0.8 and 1.2) was not reached. The total deleted or gained regions as detected by this method are boxed in black. B: An example of comparison of conventional CGH analysis (left) and MLPA analysis using kit P088 (right) both identifying a partial deletion on 1p and 19q (case N182). The partial deletions detected in case N182 by CGH involve 1p11-31 and 19q13.2-qter, whereas MLPA detects a loss on 1p from NRAS (1p13.1) to LPHN2 (1p31.1) and a loss on 19q from ZNF342 (19q13.32) to BC2 (19q13.43); in addition, MLPA analysis shows a partial gain on 19q, and this gain is also indicated by CGH, but here the threshold is not crossed.
Figure 5.
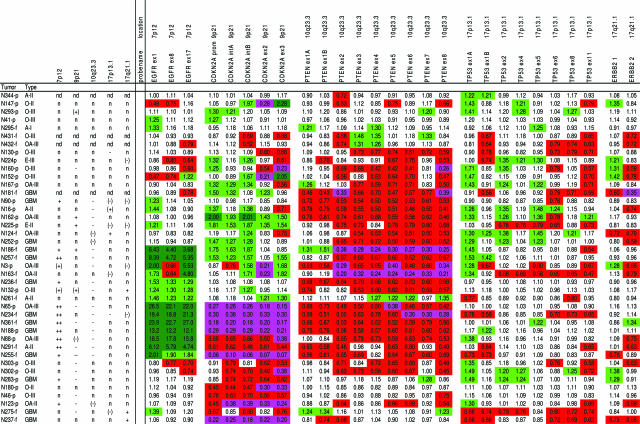
Results of MLPA analysis of gliomas using kit P105 (for EGFR, CDKN2A, PTEN, TP53, and ERBB2). See also legend in Figure 2. The probe names (gene name and exon analyzed are indicated) are listed; probe CDKN2A prom is located in the promotor region, CDKN2A intA is located 0.5 kb before the start of p14ARF exon 1, CDKN2A intB is located between p16 exon 1 and p14 exon 1, whereas ERBB2 1 and ERBB2 2 represent the 142-bp and 409-bp fragment of ERBB2 2, respectively (exact location not provided). CGH results are provided for chromosomal regions on which the genes are located [7p12 (EGFR), 9p21 (CDKN2A), 10q23.3 (PTEN), 17p13.1 (TP53) and 17q21.1 (ERBB2)]. Abbreviations used for CGH imbalances are as in Figure 2; the following symbols are also used: −, CGH ratio of ∼0.6 suggesting the presence of a homozygous deletion, ++, high copy amplification as indicated by a CGH ratio greater than 1.4. Red and green boxes represent probe ratios less than 0.8 and more than 1.2. Additionally, dark green boxes represent MLPA ratios more than 2.0 indicating high copy number amplifications, whereas pink boxes represent ratios less than 0.4, which may indicate homozygous deletions.
MLPA was performed as described by the manufacturer with minor modifications. Briefly, DNA (250 to 450 ng) was dissolved in 5 μl of TE-buffer (10 mmol/L Tris, pH 8.2, 1 mmol/L ethylenediaminetetraacetic acid, pH 8.0) or Milli-Q water, denatured, and subsequently cooled to 25°C. After adding the probe mix, the sample was denatured, and the probes were allowed to hybridize (16 hours at 60°C). After ligation of both probe pairs and inactivation of ligase, PCR was performed in a volume of 50 μl containing 10 μl of the ligation reaction mixture using the PTC 200 thermal cycler (MJ Research Inc., Waltham, MA) (33 cycles of denaturation at 95°C for 20 seconds, annealing at 60°C for 30 seconds, and extension at 72°C for 1 minute with a final extension of 20 minutes at 72°C). We included an additional agarose gel electrophoresis to examine MLPA efficiency (Figure 1). Fragments were separated and quantified by electrophoresis on an ABI 3730 capillary sequencer (Applied Biosystems, Foster City, CA) and Genemapper analysis (Applied Biosystems).34
In each set of MLPA experiments, in addition to the tumor samples to be analyzed, we included at least four normal control DNA samples for data processing. Reference DNAs were isolated either from blood of healthy volunteers or from normal (nontumorous) paraffin-embedded samples for use in MLPA analysis of tumor DNAs from frozen or paraffin-embedded tissue, respectively.34
MLPA Data Processing
Data analysis was performed in Excel as described by the manufacturer (MRC-Holland). First, the fraction of each peak was calculated by dividing the peak value (peak height or area) of each probe amplification product by the combined value of the control probes within the sample, to compensate for PCR efficiency of the individual samples. Subsequently this relative peak value or so-called probe fraction is divided by the mean probe fraction of this fragment within the included reference DNAs, generating the normalized peak value or the so-called probe ratio.
For quality control, five control peaks were included in each reaction: four control fragments generating amplification products of 64, 70, 76, and 82 bp if the DNA quality (amount or purity) is insufficient, and a fifth peak showing an amplification product of 94 bp that is indicative for hybridization and ligation efficiency. For these quality control fragments, the relative peak values are calculated (peak value/average peak value of the control probes within a sample) to identify unreliable experiments (Table 1). Based on our previous experience with MLPA kit P00534 and the current data set described in this study, the threshold was set at 0.8 for identification of losses and at 1.2 for gains. Ratios of the control probes were not further analyzed because these probes were targeting dispersed regions of different chromosomes, and for reliable detection of aberrations by MLPA, multiple probes for the gene or region of interest need to be analyzed.34
Table 1.
Guidelines for Reliable Detection of Loss of 1p and 19q Using MLPA Kit P088
| • Percentage of tumor cells in tumor sample should be at least 50%. |
| • DNA should be of high quality and preferably isolated by spin columns (eg, using the DNeasy tissue kit); DNA isolated differently should be purified. |
| • Frozen tissue is preferable over formalin-fixed, paraffin-embedded tissue, but the latter can also be used. |
| • 250 to 450 ng of DNA should be used. |
| • Agarose gel electrophoresis should be performed as a quality control step after the PCR is performed (in insufficient experiments larger DNA fragments are underrepresented). |
| • Evaluation of the MLPA DNA control peaks and ligation peak (included in each MLPA reaction) should be performed after capillary gel electrophoresis; relative peak values should be <0.1 for the DNA control peaks (64, 70, 76, and 82 bp) and >1.0 for the ligation peak (94 bp) (peak value divided by the average peak value of all control probes). |
| • DNAs used as a reference during data processing should show normal ratios; otherwise, they should be excluded from the reference pool. |
| • Threshold to detect losses and gains in tumor samples should be set at 0.8 and 1.2, respectively. |
| • Ratios of adjacent probes should be taken into consideration for the assessment of the presence of gains or losses. |
Results
Detection of −1p/−19q
MLPA experiments were performed using 250 to 450 ng (depending on concentration and availability) of control and tumor DNA isolated from snap-frozen tissue or formalin-fixed, paraffin-embedded tissue. Overall, tumors showing loss of the complete chromosome arm 1p and/or 19q as detected by CGH also showed loss of nearly all 1p and/or 19q probes (Figure 2). Vice versa, only occasional probes showed a loss in tumors without loss involving 1p or 19q as identified by CGH analysis. Figure 3 shows an overview of mean probe values ± 2× standard deviations in the control DNAs used in combination with the individual ratios for tumors with a complete loss of 1p/19q as detected by CGH. Partial deletions as detected by CGH were also detected by MLPA and could be better mapped with this latter technique (Figure 4). Although a loss is identified by a ratio less than 0.8, ratios greater than 1.2 indicate gained regions. Although CGH only detected a gain in a small number of cases (Figure 2), probes showing a ratio >1.2 were relatively frequently identified (as an isolated event or involving adjacent probes). So far the diagnostic value of 1p/19q imbalances encompasses detection of losses, and further evaluation of gains was considered beyond the scope of the present study.
Quality Control
To test the amount of normal DNA that can be present in a DNA-tumor sample without affecting MLPA reliability, a titration experiment was performed using DNA isolated from a subcutaneous human glioma xenograft line (E34) containing loss of 1p and 19q.35 As expected, MLPA probe ratios increased with increasing amounts of normal DNA (0, 20, 40, 50, 60, 80, and 100%) (data not shown). We found that loss of 1p and 19q can still reliably be detected in a sample containing 50% tumor DNA. The theoretically expected ratios for a loss of ∼0.5 (one of two alleles lost) were indeed detected in the 100% tumor DNA sample whereas in the sample with only 50% tumor cells, derived DNA-detected ratios were ∼0.75 (ie, the theoretical expected value).
Evaluating the MLPA quality control fragments in the P088 kit in our hands reliably showed relative peak values (peak value/average peak value of all control probes within the sample) of less than 0.1 for the DNA control peaks (64, 70, 76, and 82 bp), whereas the relative ligation peak (94 bp) value was at least 1, but usually greater than 1.2. An additional quality control step using agarose gel electrophoresis before capillary electrophoresis enabled us to reliably identify experiments that, after data processing, appeared to provide unreliable detection of −1p and −19q. In such inefficient experiments, agarose gel electrophoresis demonstrated under-representation of larger DNA fragments (see also Figure 1B).
Duplicate experiments were performed on 13 tumor DNAs, and probe ratios showed only minor differences. Furthermore, of 11 cases (6 without a loss on 1p or 19q as detected by CGH) analyzed using DNA isolated from paraffin-embedded tissue (p-DNA), MLPA was repeated using DNA isolated from frozen tissue (f-DNA) because several probes showed unexpected ratios in comparison to the CGH analysis or adjacent probes. In nine of these cases, the results improved using f-DNA; however, the same conclusions could be drawn from both the f-DNA and p-DNA analysis. In the remaining two cases, the results for f-DNA were similar to those obtained with p-DNA. Based on our experience guidelines were established for reliable detection of (partial or complete) losses on chromosome arms 1p and 19q in tumors (Table 1).
Detection of Other Genetic Markers
With MLPA kit P105 (testing EGFR, CDKN2A, PTEN, TP53, and ERBB2, located on 7p12, 9p21, 10q23.3, 17p13.1, and 17q21.1, respectively), we investigated 41 gliomas (Figure 5). Aberrant probe ratios for EGFR, CDKN2A, and PTEN were detected in the majority of the cases in which CGH identified an imbalance of the corresponding region (11 of 13, 10 of 12, and 16 of 17 cases, respectively). Furthermore, several aberrations were detected by MLPA that were not identified by CGH; this is most likely a result of the detection limit of CGH (2 to 10 Mb). Interestingly, all seven cases that contained high copy amplification of the 7p12 region as identified by CGH were also identified as such by MLPA, with the EGFR ratios primarily surpassing the 1.2 threshold (ranging from 3.6 to 26.5). Ratios for the CDKN2A probes ranged from 0.06 to 0.10 in N255, suggesting a homozygous deletion in the majority of the cells analyzed. Only in one case (N124) was an imbalance involving 17p13.1 clearly detected by CGH, demonstrating the need for evaluation of the reliability of these TP53 probes using gene-specific information. Furthermore, both probes included in P105 to analyze ERBB2 frequently showed opposite results, and it remains unclear which one is reliable. This finding underscores our previous observations that three or more probes should be analyzed for reliable detection of imbalances in the gene or chromosomal region of interest.34 Overall, aberrations previously identified by CGH, including the EGFR, CDKN2A, and PTEN region (7p12, 9p21, and 10q23, respectively), were correctly identified by P105 in the majority of cases.
Combining the P105 and P088 data of the 31 tumors that were analyzed with both kits, we found that a gain of EGFR usually does not coincide with −1p/−19q. Deletions of the CDKN2A gene were present in approximately one third of the tumors, irrespectively of their 1p/19q status. PTEN deletions were detected at a somewhat higher frequency in tumors without a 1p/19q loss [∼72% (18 of 25) versus 57% (8 of 14) of the cases, respectively].
Discussion
An increasing number of molecular markers with prognostic value in predicting better survival or response to therapy have been identified for different tumor types. Implementation in a routine diagnostic setting is warranted, preferably using techniques that enable simultaneous detection of multiple markers or loci and are not dependent on highly specialized equipment or personnel. MLPA is a very promising technique for routine diagnostics because it allows detection of DNA or RNA copy number changes of up to 45 loci in one relatively simple, semiquantitative PCR-based experiment.31 Most MLPA kits available are designed for oncological purposes and address either hereditary or sporadic tumors. Germline aberrations are easily detected, but analysis of (sporadic) tumor samples is much more complex because a whole spectrum of different chromosomes can be affected. Therefore, the included control probes need to be carefully selected and located on different chromosomes that are infrequently affected in the tumor type of interest. Furthermore, aberrations are usually only present in a subpopulation of the cells analyzed because of tumor heterogeneity or contaminating DNA from normal cells in the tumor tissue. Less stringent thresholds are therefore required to enable detection of such aberrations. Two recently designed MLPA kits are of special interest for glial tumors: one to identify loss of 1p and 19q (P088), which are indicative for chemosensitivity and long survival, and another to detect amplification or loss of EGFR, CDKN2A, and PTEN (indicating malignant progression) as well as amplification or loss of TP53 and ERBB2 (P105). In the present study we evaluated these kits using genetically characterized gliomas to establish the potential of MLPA in a routine diagnostic setting.
Detection of −1p/−19q
Early in 2003 the first MLPA kit was released that was specifically designed for detection of loss of 1p and 19q. In our pilot study we analyzed 14 (oligodendro)glial tumors with (n = 8) or without (n = 6) losses on 1p/19q as identified by CGH (unpublished data). Because we noticed that the design of this original kit was suboptimal, suggestions for improvement were provided to the manufacturer.36 Recently, a study was published by Natté and colleagues37 describing the use of this original kit on 19 tumors. An adapted way of data processing was proposed, analyzing only the 22 reliable probes (22 of 40) but enabling identification of their −1p/−19q gliomas as established by FISH. Tumors were considered to have a loss when at least four of the seven 1p probes or two of the four 19q probes showed a ratio less than 0.75. Following the procedure proposed by Natté and colleagues,37 we could indeed classify the seven tumors in our pilot study showing a complete 1p loss and five of the six tumors without this loss. However, a tumor containing a partial deletion of 1p11-31 was classified as a tumor without 1p loss. Identification of complete versus partial losses of 1p/19q in a single and relatively simple experiment is one of the advantages of MLPA over LOH or FISH and might prove to be of additional diagnostic value. Using this original kit the potential of MLPA is thus not fully exploited. A new kit has been designed for the detection of losses on 1p and 19q and is now commercially available (P088).
In the present study we show that, using this improved MLPA kit (P088), complete as well as partial losses of 1p and 19q can easily be identified. Vice versa, tumors without these losses at the CGH level were classified as such. Tumors not previously characterized by CGH could also easily be classified as having complete loss of 1p and 19q (N471 and N355) or as tumors without such losses (N181, N372, N399, and N400). Only in one case (N256) the MLPA results were difficult to interpret because in this tumor dispersed, nonadjacent 1p probes showed a ratio <0.8. Because pre-existent brain tissue was reported to be present in this case, MLPA analysis for diagnostic purposes using a tissue sample with a higher tumor load is warranted to provide more accurate results.
In cases without a clear complete loss of 1p or 19q, but with several adjacent probes on (one of) these chromosome arms showing a ratio <0.8, true partial deletions are likely to be present. Indeed, in some of these cases CGH ratios of 0.9 were detected (0.8 also being the threshold to detect losses at the CGH level) for the corresponding region. The scattered loss of individual 1p or 19q probes in tumors in which CGH did not detect a (partial) loss may represent small deletions that are undetectable by CGH. Alternatively, scattered losses for only part of the 1p or 19q probes may occur when these aberrations are present in a subpopulation of the cells. In several cases, ratios greater than 1.2 were detected for 1p and 19q probes while CGH did not indicate a gain. Because a ratio >1.2 often occurred in adjacent probes, this likely represents a partial gain that is too small to be detected by CGH. Furthermore, although this kit was designed to detect losses on 1p and 19q, probes for two genes on 19p (19p13.3; LDLR and SMARCA4) are included to distinguish losses restricted to 19q from loss of the complete chromosome 19. In contrast to gains (14 cases), losses of both 19p probes were not detected (data not shown). If preferred these 19p probe might be used to calculate relative copy number losses as performed during FISH analysis (19q/19p). However, this approach might be complex when partial losses are involved.
Quality Control
Our titration experiment revealed that MLPA analysis for −1p and −19q is rather robust and that a sample containing 50% of tumor cell-derived DNA can still be correctly identified. However, in our experience DNA used for MLPA analysis should be of high quality and 250 to 450 ng of DNA per tumor enables reliable identification of aberrations. Two control steps are instrumental in establishing the efficiency of MLPA: agarose gel electrophoresis after PCR (Figure 1) and subsequent evaluation of the MLPA quality control peaks after capillary electrophoresis (Table 1). Using agarose gel electrophoresis, some additional inefficient experiments were identified that were not recognized by evaluation of the quality control peaks. Furthermore, in some tumors a relative ligation peak less than 1.0 occurred because of a deletion including the ligation-specific control probe sequence (2q14). This occurred in combination with adequate results for DNA control peaks (< 0.1) and in this case is therefore not indicative for an inefficient experiment.
Detection of Other Genetic Markers
Of the tumors included in this part of the study, information about chromosomal imbalances (based on previous CGH investigations) was available for 37 of 41 cases. Because the P105 kit analyzes specific exons of genes, using the CGH information for validation of the results obtained by MLPA kit P105 analysis is less straightforward than for the MLPA kit P088, and further validation of P105 is warranted using gene-specific information for comparison. Nevertheless, in the majority of CGH and MLPA tumors analyzed showed comparable results for the chromosomal regions of EGFR, CDKN2A, PTEN, and TP53. Moreover, probe ratios enabled identification of high copy amplifications of EGFR or homozygous deletions of CDKN2A. MLPA detected several aberrations that were not identified by CGH, probably because of the detection limit of CGH (ie, 2 to 10 Mb), including a high copy gain for EGFR in one tumor.
Comparison of the results obtained with P088 and P105 revealed that differences in ratios between adjacent probes more often occurred in P105. As in 31 cases the same DNA stock was analyzed using P105 as well as P088, these differences are most likely not caused by DNA quality, quantity, or the presence of nontumorous cells. Although −1p and −19q are early events in the oncogenesis of (oligodendro)glial tumors, PTEN, EGFR, and CDKN2A aberrations are considered to be involved in malignant progression (late events).38 Such progression-associated aberrations may only be present in part of the tumor cells (tumor heterogeneity) and, as discussed above, may increase the chance of obtaining inconclusive results. Alternatively, as different regions in the genes are analyzed, these results may reflect the fact that some exons are more prone to aberrations than others.
The data processing for the results obtained with the P105 kit was performed in the same way as for the P088 kit. An alternative data processing procedure was suggested by the manufacturer [calculating the fractions using the sum of the nearest (fragments length) two to four control peaks instead of all control peaks]. However, this alternative approach did not improve the results. Because multiple chromosomal aberrations are usually present in tumors, part of the control probes may target regions containing an imbalance. Evaluation of too few control probes for data processing may in this situation primarily affect the calculation of the ratio of the probes of interest and lead to unreliable results.
Diagnostic Potential of MLPA Using Kit P088 and P105
The demand for implementation of molecular diagnostics in glioma patients is increasing. Usually LOH or FISH methods are used to detect loss of the 1p36 region, generally identifying a group of OTs with better chemoresponse and (progression-free) survival. In this study we established the clinical potential of MLPA for glioma diagnostics. Rather than evaluating an MLPA kit (P147) designed to analyze 1p36, we validated MLPA kit P088, which is designed to detect complete as well as partial deletion on 1p and 19q because it is becoming increasingly clear that there are distinct types of 1p/19q losses with specific clinical characteristics. For example, complete 1p loss associated with a complete 19q loss (without 19p loss) predicts longer survival and a better chemoresponse compared to partial telomeric or interstitial 1p losses not associated with a (complete)19q loss.8,21,23 In contrast, it has been reported that gliomas with partial LOH 1p and LOH 19q have a better chemoresponse.22 Concordantly, one of the patients in our series with an OA harboring −1pcen-p31 and −19q13.2-qter (Figure 4) showed a long survival (16 years since first surgery for low-grade glioma, which 9 years later appeared to have progressed to anaplastic OA at the histopathological level; unpublished data). Based on our results we conclude that using MLPA kit P088 reliably identifies not only complete but also partial losses on 1p and 19q in gliomas from both snap-frozen and paraffin-embedded tumor tissue containing at least 50% tumor cells. Using another MLPA kit (P105), genetic markers such as EGFR, PTEN, and CDKN2A [indicating high-grade malignancy in (glial) tumors] can be identified. Combining data of P088 and P105 thus enables identification of those −1p/−19q tumors that harbor these additional aberrations associated with malignant progression and reported to be predictive for shorter overall survival and duration of chemoresponse.10 Consequently, identification of these latter aberrations (P105) in gliomas without 1p/19q losses will also be predictive for a less favorable outcome.
Because MLPA is rather robust and relatively easy to perform, we expect that implementation of this technique will be very helpful in making genetic information that is relevant for estimation of prognosis and therapy available for individual glioma patients and will thereby facilitate customized therapeutic decision making. Furthermore, MLPA can be easily used to screen large sets of gliomas and will therefore enable further evaluation of the therapeutic and prognostic implications of complete versus specific partial 1p/19q losses (P088) as well as the additional diagnostic value of a gain of EGFR and loss of CDKN2A and PTEN in cases with or without 1p/19q losses (P105).
Acknowledgments
We thank Jan Schouten from MRC-Holland for kindly supplying the P088 and P105 kits and helpful discussions, Marcory van Dijk for sharing her MLPA expertise, Sandra Nooijen for technical assistance in optimizing the P105 MLPA assay, and the neurosurgeons of the Radboud University Nijmegen Medical Centre for their continuous collaboration.
Footnotes
Supported by the Dutch Cancer Society (Konigin Wihelmina Fonds: Katholic University Nijmegen 2004-3143) and the Pauline van Everdingen Foundation.
References
- Cairncross JG, Ueki K, Zlatescu MC, Lisle DK, Finkelstein D, Hammond RR, Silver JS, Stark PC, Macdonald DR, Ino Y, Ramsay DA, Louis DN. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst. 1998;90:1473–1479. doi: 10.1093/jnci/90.19.1473. [DOI] [PubMed] [Google Scholar]
- Schmid JS, Perry A, Borell TJ, Lee HK, O’Fallon J, Hosek SM, Kimmel D, Yates A, Burger PC, Scheithauer BW, Jenkins RB. Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. J Clin Oncol. 2000;18:636–645. doi: 10.1200/JCO.2000.18.3.636. [DOI] [PubMed] [Google Scholar]
- Cairncross JG. Aggressive oligodendroglioma: a chemosensitive tumor. Recent Results Cancer Res. 1994;135:127–133. doi: 10.1007/978-3-642-85039-4_13. [DOI] [PubMed] [Google Scholar]
- Kyritsis AP, Yung WKA, Bruner J, Gealson M, Levin VA. The treatment of anaplastic oligodendrogliomas and mixed gliomas. Neurosurgery. 1993;32:365–371. doi: 10.1227/00006123-199303000-00005. [DOI] [PubMed] [Google Scholar]
- Nutt CL, Mani DR, Betensky RA, Tamayo P, Cairncross JG, Ladd C, Pohl U, Hartmann C, McLaughlin ME, Batchelor TT, Black PM, von Deimling A, Pomeroy SL, Golub TR, Louis DN. Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Cancer Res. 2003;63:1602–1607. [PubMed] [Google Scholar]
- Smith JS, Perry A, Borell TJ, Lee HK, Fallon JO, Hosek SM, Kimmel D, Yates A, Burger PC, Scheithauer BW, Jenkins RB. Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, mixed oligoastrocytomas. J Clin Oncol. 2000;18:636–645. doi: 10.1200/JCO.2000.18.3.636. [DOI] [PubMed] [Google Scholar]
- van den Bent MJ, Looijenga LHJ, Langberg K, Dinjens W, Gravenland W, Uytdewilligen L, Sillevis Smitt PA, Jenkins RB, Kros JM. Chromosomal anomalies in oligodendroglial tumors are correlated with clinical features. Cancer. 2003;97:1276–1284. doi: 10.1002/cncr.11187. [DOI] [PubMed] [Google Scholar]
- Iuchi T, Namba H, Iwadate Y, Shishikura T, Kageyama H, Nakamura Y, Ohira M, Yamaura A, Osato K, Sakiyama S, Nakagawara A. Identification of the small interstitial deletion at chromosome band 1p34–p35 and its association with poor outcome in oligodendroglial tumors. Gen Chromosom Cancer. 2002;35:170–175. doi: 10.1002/gcc.10080. [DOI] [PubMed] [Google Scholar]
- Bauman GS, Ino Y, Ueki K, Zlatescu MC, Fisher BJ, Macdonald DR, Stitt L, Louis DN, Cairncross JG. Allelic loss of chromosome 1p and radiotherapy plus chemotherapy in patients with oligodendrogliomas. Int J Radiat Oncol Biol Phys. 2000;48:825–830. doi: 10.1016/s0360-3016(00)00703-3. [DOI] [PubMed] [Google Scholar]
- Ino Y, Betensky RA, Zlatescu MC, Sasaki H, Macdonald DR, Stemmer-Rachamimov AO, Ramsay DA, Cairncross JG, Louis DN. Molecular subtypes of anaplastic oligodendroglioma: implications for patient management at diagnosis. Clin Cancer Res. 2001;7:839–845. [PubMed] [Google Scholar]
- Maintz D, Fiedler K, Koopmann J, Rollbrocker B, Nechev S, Lenartz D, Stangl AP, Louis DN, Schramm J, Wiestler OD, von Deimling A. Molecular genetic evidence for subtypes of oligoastrocytomas. J Neuropathol Exp Neurol. 1997;56:1098–1104. doi: 10.1097/00005072-199710000-00003. [DOI] [PubMed] [Google Scholar]
- Wolter M, Reifenberger J, Blaschke B, Ichimura K, Schmidt EE, Collins VP, Reifenberger G. Oligodendroglial tumors frequently demonstrate hypermethylation of the CDKN2A (MTS1, p16INK4a), p14ARF, and CDKN2B (MTS2, p15INK4b) tumor suppressor genes. J Neuropathol Exp Neurol. 2001;60:1170–1180. doi: 10.1093/jnen/60.12.1170. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Nakamura M, Kros JM, Burkhard C, Yonekawa Y, Kleihues P, Ohgaki H. Phenotype versus genotype correlation in oligodendrogliomas and low-grade diffuse astrocytomas. Acta Neuropathol (Berl) 2002;103:267–275. doi: 10.1007/s004010100464. [DOI] [PubMed] [Google Scholar]
- von Deimling A, Fimmers R, Schmidt MC, Bender B, Fassbender F, Nagel J, Jahnke R, Kaskel P, Duerr E, Koopmann J, Maintz D, Steinbeck S, Wick W, Platten M, Muller DJ, Przkora R, Waha A, Blumcke B, Wellenreuther R, Meyer-Puttlitz B, Schmidt O, Mollenhauer J, Poustka A, Stangl AP, Lenartz D, von Ammon K, Henson JW, Schramm J, Louis DN, Wiestler OD. Comprehensive allelotype and genetic analysis of 466 human nervous system tumors. J Neuropathol Exp Neurol. 2000;56:544–558. doi: 10.1093/jnen/59.6.544. [DOI] [PubMed] [Google Scholar]
- Reifenberger G, Kros JM, Burger PC, Louis DN, Collins VP. Kleihues P, Cavenee WK, editors. Lyon: International Agency for Research on Cancer (IARC) Press,; Oligodendroglial tumors and mixed gliomas. WHO classification. Tumours of the Nervous System. 1999:55–70. [Google Scholar]
- Reifenberger G, Louis DN. Oligodendroglioma: toward molecular definitions in diagnostic neuro-oncology. J Neuropathol Exp Neurol. 2003;62:111–126. doi: 10.1093/jnen/62.2.111. [DOI] [PubMed] [Google Scholar]
- Smith JS, Alderete BE, Minn Y, Borell T, Perry A, Mohapatra G, Smith SM, Kimmel D, Fallon JO, Tates A, Feuerstein BG, Burger PC, Scheithauer BW. Localization of common deletion regions on 1p and 19q in human gliomas and their association with histological subtype. Oncogene. 1999;18:4144–4152. doi: 10.1038/sj.onc.1202759. [DOI] [PubMed] [Google Scholar]
- Hatanpaa KJ, Burger PC, Eshleman JR, Murphy KM, Berg KD. Molecular diagnosis of oligodendroglioma in paraffin sections. Lab Invest. 2003;83:419–428. doi: 10.1097/01.lab.0000059948.67795.ef. [DOI] [PubMed] [Google Scholar]
- Ginzinger DG, Godfrey TE, Nigro J, Moore DH, Suzuki S, Pallavicini MG, Gray JW, Jensen RH. Measurement of DNA copy number at microsatellite loci using quantitative PCR analysis. Cancer Res. 2000;60:5405–5409. [PubMed] [Google Scholar]
- Nigro JM, Takahashi MA, Ginzinger DG, Law M, Passe S, Jenkins RB, Aldape KD. Detection of 1p and 19q loss in oligodendroglioma by quantitative microsatellite analysis, a real-time quantitative polymerase chain reaction assay. Am J Pathol. 2001;158:1253–1262. doi: 10.1016/S0002-9440(10)64076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsberg J, Erkwoh A, Sabel MC, Kirsch L, Fimmers R, Blaschke B, Schlegel U, Schramm J, Wiestler OD, Reifenberger G. Oligodendroglial tumors: refinement of candidate regions on chromosome arm 1p and correlation of 1p/19q status with survival. Brain Pathol. 2004;14:121–130. doi: 10.1111/j.1750-3639.2004.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLendon RE, Herndon JE, West B, Reardon D, Wiltshire R, Rasheed BK, Quinn J, Friedman HS, Friedman AH, Bigner DD. Survival analysis of presumptive prognostic markers among oligodendrogliomas. Cancer. 2005;104:1693–1699. doi: 10.1002/cncr.21362. [DOI] [PubMed] [Google Scholar]
- Idbaih A, Marie Y, Pierron G, Brennetot C, Hoang-Xuan K, Kujas M, Mokhtari K, Sanson M, Lejeune J, Aurias A, Delattre O, Delattre JY. Two types of chromosome 1p losses with opposite significance in gliomas. Ann Neurol. 2005;58:483–487. doi: 10.1002/ana.20607. [DOI] [PubMed] [Google Scholar]
- Jeuken JWM, Boots-Sprenger SHE, Wesseling P. Zhang W, Fuller GN, editors. Boston: Jones and Bartlett Publishers,; Chromosomal imbalances in oligodendroglial tumors as detected by comparative genomic hybridization (CGH). Genomic and Molecular Neuro-Oncology. 2004:185–198. [Google Scholar]
- Jeuken JWM, Sprenger SHE, Wesseling P, Macville MVE, von Deimling A, Teepen HLJM, van Overbeeke JJ, Boerman RH. Identification of subgroups of high-grade oligodendroglial tumors by comparative genomic hybridization. J Neuropathol Exp Neurol. 1999;58:606–612. doi: 10.1097/00005072-199906000-00005. [DOI] [PubMed] [Google Scholar]
- Jeuken JWM, Sprenger SHE, Boerman RH, von Deimling A, Teepen HLJM, van Overbeeke JJ, Wesseling P. Subtyping of oligo-astrocytic tumours by comparative genomic hybridisation. J Pathol. 2001;194:81–87. doi: 10.1002/path.837. [DOI] [PubMed] [Google Scholar]
- Jeuken JWM, Sprenger SHE, Vermeer H, Kappelle AC, Boerman RH, Wesseling P. Chromosomal imbalances in primary oligodendroglial tumors and their recurrences; clues for malignant progression as detected by CGH. J Neurosurg. 2002;96:559–564. doi: 10.3171/jns.2002.96.3.0559. [DOI] [PubMed] [Google Scholar]
- Jeuken JWM, Sprenger SHE, Wesseling P. Comparative genomic hybridization: practical guidelines. Diagn Mol Pathol. 2002;11:193–203. doi: 10.1097/00019606-200212000-00002. [DOI] [PubMed] [Google Scholar]
- Cowell JK, Barnett GH, Nowak NJ. Characterization of the 1p/19q chromosomal loss in oligodendrogliomas using comparative genomic hybridization arrays (CGHa). J Neuropathol Exp Neurol. 2004;63:151–158. doi: 10.1093/jnen/63.2.151. [DOI] [PubMed] [Google Scholar]
- Kitange G, Misra A, Law M, Passe S, Kollmeyer TM, Maurer M, Ballman K, Feuerstein BG, Jenkins RB. Chromosomal imbalances detected by array comparative genomic hybridization in human oligodendrogliomas and mixed oligoastrocytomas. Gene Chromosom Cancer. 2005;42:68–77. doi: 10.1002/gcc.20108. [DOI] [PubMed] [Google Scholar]
- Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleihues P, Cavenee WK. Lyon, France: International Agency for Research on Cancer (IARC) Press,; Pathology and genetics. Tumours of the nervous system. World Health Organization Classification of Tumours. 2000 [Google Scholar]
- Miller SA, Dykes DD, Polsky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk MC, Rombout PD, Boots-Sprenger SH, Straatman H, Bernsen MR, Ruiter DJ, Jeuken JW. Multiplex ligation-dependent probe amplification for the detection of chromosomal gains and losses in formalin-fixed tissue. Diagn Mol Pathol. 2005;14:9–16. doi: 10.1097/01.pas.0000146701.98954.47. [DOI] [PubMed] [Google Scholar]
- Jeuken JWM, Sprenger SHE, Wesseling P, Bernsen HJJA, Suijkerbuijk RF, Roelofs F, Macville MVE, Gilhuis HJ, van Overbeeke JJ, Boerman RH. Xenografts genetically reflect glioblastoma biopsies: characterization of 11 glioblastoma xenograft lines by comparative genomic hybridization. J Neurosurg. 1999;92:652–658. doi: 10.3171/jns.2000.92.4.0652. [DOI] [PubMed] [Google Scholar]
- Jeuken JWM, Boots-Sprenger SHE, Wesseling P. Multiplex ligation dependent probe amplification for the detection of 1p and 19q loss in oligodendroglial tumors. Brain Pathol. 2005;15:364–365. doi: 10.1111/j.1750-3639.2005.tb00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natté R, van Eijk R, Eilers P, Cleton-Jansen AM, Oosting J, Kouwehove M, Kros JM, van Duinen S. Multiplex ligation dependent probe amplification for the detection of 1p and 19q loss in oligodendroglial tumors. Brain Pathol. 2005;15:192–197. doi: 10.1111/j.1750-3639.2005.tb00520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeuken JW, von Deimling A, Wesseling P. Molecular pathogenesis of oligodendroglial tumors. J Neurooncol. 2004;70:161–181. doi: 10.1007/s11060-004-2748-1. [DOI] [PubMed] [Google Scholar]