Abstract
We report on a series of 58 cases of angioimmunoblastic T-cell lymphoma (AILT) and 59 cases of peripheral T-cell lymphoma, unspecified (PTCL-NOS). Subsets of cases from both diagnostic groups were complicated by associated B-cell proliferations, and we performed B- and T-cell clonality studies and in situ hybridization for Epstein-Barr virus (EBV) to investigate the relationship between B-cell proliferation, B-cell clonality, and EBV. Using multiplex polymerase chain reaction assays based on the BIOMED-2 collaborative study, we detected TCRγ T-cell clones in 78 and 81% of AILT and PTCL-NOS cases, respectively, and IGH B-cell clones in 34 and 35% of AILT and PTCL-NOS cases, respectively. The majority of cases contained EBV-positive cells, including 50% of AILT and 57% of PTCL-NOS cases, and cases with B-cell proliferations were more often EBV-positive. Although a relatively high rate of B-cell clonality has been shown for AILT, our findings for PTCL-NOS differ from previous reports in that B-cell clonality was relatively frequent. Overall, a positive B-cell clone correlated, in part, with the presence of a B-cell proliferation but not with EBV. Our findings demonstrate that B-cell clonality is a common finding in AILT and PTCL-NOS, and its presence should not negate the diagnosis established by morphologic, immunophenotypic, and clinical findings.
Peripheral T-cell lymphoma (PTCL) is an uncommon malignancy, accounting for less than 10% of non-Hodgkin lymphomas worldwide. By current World Health Organization criteria, PTCLs are subclassified based on clinical, histologic, immunophenotypic, and genetic findings.1 However, the diagnosis is often difficult for pathologists because PTCLs are uncommon, and the histologic findings can be nonspecific and can overlap with reactive conditions. Perhaps the most challenging aspect is the lack of good immunophenotypic markers to establish clonality for T-cell lineage neoplasms, and pathologists often rely on DNA-based studies for evidence of clonality.
Given the difficulty in diagnosing PTCL, pathologists often perform both B- and T-cell clonality studies to clarify lineage and provide support for malignancy. Currently, the most commonly used method is the polymerase chain reaction (PCR), which has largely replaced Southern blot-based clonality assays. However, there are several caveats to this approach. In particular, studies evaluating large numbers of PTCLs by PCR show that simultaneous B- and T-cell clones occur relatively often (9 to 16%).2,3,4,5,6 These cases, with both B- and T-cell clones, present a diagnostic dilemma for the pathologist and treating oncologist.
There are numerous possible explanations for the detection of two clones in some cases, including those that are technical in nature, so-called lineage infidelity of a single clone, and the presence of two different clones in a sample.7 The phenomenon referred to as lineage infidelity, in this context, is the result of recombination of both T-cell receptor (TCR) and immunoglobulin (IG) genes in the same clone.7 This finding is more frequently seen in immature hematolymphoid neoplasms and is uncommon in mature B- and T-cell non-Hodgkin lymphomas.7,8,9,10 In recent years, cases of PTCL with associated B-cell proliferations have been described. Many of these cases are positive for both B- and T-cell clones, and in our series of peripheral T-cell lymphoma, unspecified (PTCL-NOS), a subset of B-cell proliferations were associated with Epstein-Barr virus (EBV).11 Others have reported similar B-cell proliferations in PCTL-NOS as well as angioimmunoblastic T-cell lymphoma (AILT), and many of these cases are EBV-positive and exhibit B-cell clones.12,13,14
We hypothesized that the relative frequency of B-cell clones in PTCL may be because of associated B-cell proliferations and/or EBV. To address this issue, we performed PCR-based B- and T-cell clonality studies on a total of 117 cases of PTCL. The series is composed of 58 cases of AILT, 12 of which show an associated B-cell proliferation, and 59 cases of PTCL-NOS, 11 of which show an associated B-cell proliferation. In addition, we performed in situ hybridization for EBV to characterize the relationship between the B-cell proliferation, EBV, and underlying T-cell lymphoma.
In this article, we describe the results of these studies, which include a high overall frequency of B-cell clones in both AILT (34%) and PTCL-NOS (35%) that correlates, in part, with an associated B-cell proliferation but not with EBV. Interestingly, cases without demonstrable B-cell proliferations also exhibited B-cell clonality, although with less frequency than cases with B-cell proliferations. This finding suggests that additional factors may contribute to B-cell clonality.
Materials and Methods
Cases
Fifty-eight cases of AILT and fifty-nine cases of PTCL-NOS were selected from the Laboratory of Hematopathology, Stanford University Medical Center, Stanford, CA. The cases were received between January 1, 2000, and April 4, 2005, and a diagnosis was rendered based on criteria established by the World Health Organization.1 AILT and PTCL-NOS cases were further subclassified as to whether they showed an associated-B-cell proliferation. Briefly, a B-cell proliferation was defined as a readily identifiable infiltrate of B cells complicating a PTCL as described by Higgins and colleagues.11 Cases were selected based on availability of paraffin-embedded tissue and/or residual DNA remaining from clonality studies performed for the original diagnosis. Use of tissue for this study was approved by the Stanford University Panel on Medical Human Subjects [protocol ID 79034; institutional review board number 348 (panel 1)].
Histology and Immunohistochemistry
Histologic sections were prepared from formalin-fixed, paraffin-embedded tissue by cutting 3- to 4-μm-thick sections and staining with hematoxylin and eosin. Sections were stained for immunohistochemistry on a Ventana BenchMark instrument (Ventana Medical Systems, Tucson, AZ) using the biotin-avidin technique in which diaminobenzidine was used as a chromogen.15 All cases were stained with at least one B- and one T-cell marker. The following monoclonal antibodies were used: 4C7 (anti-CD5; Vision Bio-System, Norwell, MA), 56C6 (anti-CD10; Vision Bio-System), L26 (anti-CD20; DAKO, Carpinteria, CA), IF8 (anti-CD21; DAKO), IB12 (anti-CD23; Vision Bio-System), JCV117 (anti-CD79a; DAKO), and B-B4 (anti-CD138; Serotec, Raleigh, NC). Polyclonal antibodies directed against the following antigens were used: CD3 (Cell Marque, Hot Springs, AR) and κ and λ immunoglobulin light chains (DAKO). Antigen retrieval was achieved through automated heat pretreatment for 4C7, 56C6, L26, IB12, JCV117, B-B4, and antibody against CD3 (Ventana Medical Systems). Antigen retrieval was achieved through automated protease pretreatment for IF8 and antibodies against immunoglobulin light chains (Ventana Medical Systems).
Isolation of DNA
DNA was obtained from formalin-fixed, paraffin-embedded tissue blocks by cutting four to eight 20-μm-thick sections followed by deparaffinization by extracting three times in 1.0 ml of either xylene or Histoclear (National Diagnostics, Atlanta, GA). The extracted tissue was washed two times in 1.0 ml of 100% ethanol and then dried at 65°C. The tissue was resuspended in 2 vol (30 to 250 μl) of a mixture of 200 mmol/L potassium chloride, 40 mmol/L Tris [Tris(hydroxymethyl)aminomethane]-hydrochloride (pH 8.5), 0.1% sodium dodecyl sulfate, and 0.6 mg/ml proteinase K and incubated at 55 to 65°C overnight. The sample was then boiled for 8 minutes and centrifuged at 15,000 × g for 10 minutes, and the DNA-containing supernatant was collected. To evaluate the yield of DNA, a 5-μl aliquot of supernatant was separated by electrophoresis on a 0.8% agarose gel (Invitrogen, Carlsbad, CA), stained with ethidium bromide, and visualized by ultraviolet illumination. Based on an estimate of DNA concentration, the crude extract was diluted with an appropriate volume of reagent-grade water (Teknova, Hollister, CA) and used directly in a PCR. Typically, a 1:10 to 1:20 dilution was prepared, and 5 μl was used for the PCR.
B-Cell and T-Cell Clonality Studies
Cases were evaluated for B- and T-cell clonality using two commercially available PCR-based kits that detect clonal rearrangements in IGH and TCRγ, respectively (InVivoScribe Technologies, San Diego, CA). These multiplex PCRs are based on a European collaborative study (BIOMED-2 Concerted Action).16 The PCRs were performed according to the manufacturers’ protocols. Briefly, a 5-μl aliquot of DNA sample was added to 45 μl of each reaction mixture and 1.25 U of AmpliTaq Gold (Applied Biosystems, Foster City, CA). Amplification was performed in a Perkin-Elmer 9700 thermocycler (Perkin-Elmer Applied Biosystems, Foster City, CA) by initially heating at 95°C for 7 minutes, followed by 35 cycles of 95°C for 45 seconds, 60°C for 45 seconds, and 72°C for 90 seconds. The final step was incubation at 72°C for 10 minutes.
After amplification, 1 μl of PCR product was added to 10 μl of HI-Deionized Formamide (Applied Biosystems) and 1 μl of ROX-250 internal size standard (Applied Biosystems). The mixture was then denatured at 95°C for 5 minutes, chilled on ice for 5 minutes, and resolved by capillary electrophoresis on an ABI 3100 instrument using performance-optimized polymer-4 (POP-4; Applied Biosystems). The data were stored electronically and analyzed using GeneScan analysis software (Applied Biosystems). Printed electropherograms were reviewed independently by two separate pathologists (B.T.T. and D.A.A.) who were blinded to the identity of specimens. Criteria for assigning a clone were similar to those used for clinical specimens at the Stanford molecular pathology laboratory. Briefly, a clone was required to yield one or two distinct peaks within the expected size range for a given primer set. In some cases, a polyclonal background was present in addition to the distinct peak(s). To meet criteria for a clone, the height of at least one distinct peak had to exceed that of the polyclonal background by at least twofold. When a primer set yielded three or more distinct peaks, the results were classified as oligoclonal, regardless of whether a polyclonal background was present or absent. Cases with a clonal peak demonstrated by one or multiple primer sets were scored as positive. Cases with only polyclonal and/or oligoclonal results were scored as negative.
Control specimens included clonal DNA isolated from cell lines (InVivoScribe), polyclonal DNA isolated from human tonsil specimens (InVivoScribe), a sensitivity control composed of clonal DNA diluted in polyclonal DNA, and a blank composed of reagent-grade water (Teknova). Multiple polyclonal DNA specimens, each isolated from different patients (InVivoScribe), consistently yielded polyclonal signals and no clonal signals. For a run of clonality studies to be accepted into the data set, a positive clone had to be detectable from the sensitivity control, which was prepared at 2 or 3% (expressed as μg of clonal DNA/μg of polyclonal DNA) for TCRγ or IGH assays, respectively. Individual DNA specimens had to yield detectable products of at least 300 nucleotides from the control amplification tube, which contained primers to amplify HLA-DQ. Samples yielding inadequate amplification were purified using a QIAamp DNA blood mini kit (Qiagen, Valencia, CA), and the purified DNA typically yielded adequate amplification. In our validation of the InVivoScribe TCRγ and IGH kits, we were able to detect clones in 80 to 90% of known B- and T-cell neoplasms, and amplification of the provided polyclonal control DNA (InVivoScribe) or DNA isolated from paraffin-embedded reactive tonsil specimens in our laboratory yielded only polyclonal results.
Sequencing of PCR Products
Eight cases yielding clonal Vγ10 rearrangements by GeneScan analysis were reamplified from the original DNA sample using a TCRγ clonality kit that uses primers of identical sequence to those used for GeneScan analysis, but the primers lack a 5′-fluorescent label, which would otherwise interfere with subcloning (InVivoScribe). The amplification products were separated by electrophoresis on a 2% low melting point agarose gel (Invitrogen), visualized by ethidium bromide staining, excised from the gel, and purified using a QIAquick gel extraction kit (Qiagen). The purified products were ligated to a plasmid vector using a TOPO TA cloning kit for sequencing (Invitrogen), and the ligation products were transformed into Escherichia coli. From each transformation, three bacterial colonies were picked, bacterial cultures were grown, and plasmid DNA was prepared using a PureLink quick plasmid miniprep kit (Invitrogen). The DNA was sequenced using a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems) with a bacteriophage T7 primer (Invitrogen). The resulting sequences were compared to those of Vγ10 listed on the international ImMunoGeneTics website (http://imgt.cines.fr/as accessed on January 23, 2006).
In Situ Hybridization for EBV
EBV-encoded small RNA (EBER) was detected from 3- to 4-μm-thick, formalin-fixed, paraffin-embedded tissue sections by in situ hybridization using a Ventana BenchMark instrument running a standardized program incorporating deparaffinization, hybridization to the Inform EBER probe cocktail, and staining with ISH iVIEW nitro blue tetrazolium (Ventana Medical Systems). Cases were evaluated for EBV-labeled cells under visible light microscopy, and the number of cells with blue-colored nuclei was visually estimated at 0, <1, 1 to 10, >10 to 100, or >100 per medium-power field using a ×15 ocular and ×20 objective lens on an Olympus BX50 microscope (Olympus, Tokyo, Japan). The minimum number of labeled nuclei required for a nonzero score was three per section, which is more than that typically seen in a lymph node or tonsil removed from a healthy individual without evidence of immunodeficiency (B.T.T. and R.A.W., unpublished observations). In approximately half of the cases studied for EBV (depending on availability of sample), a control hybridization was performed with an oligo-deoxythymidine probe to detect polyadenylated mRNA. In addition, an external positive control consisting of an EBV-positive malignancy (NK/T-cell lymphoma or nasopharyngeal carcinoma) was run with each set of hybridizations.
Statistical Analysis
The data were analyzed using the software program JMP-IN version 5.1 (SAS Institute, Cary, NC). Age was entered as a continuous numerical parameter, and all other data were entered as categorical parameters. Differences in age between categorically defined groups were evaluated by t-test and f-test, and the lower P value was recorded. Correlations between categorical parameters were evaluated by Pearson’s χ2 test and likelihood ratio test, and the lower P value was recorded. Only P values <0.15 were reported, except for some correlations with EBV, which were based on a lower number of cases.
Results
Cases
We collected 58 cases of AILT and 59 cases of PTCL-NOS from the Stanford hematopathology laboratory. Each case corresponded to a different patient, and all patients were adults. The mean ages of patients with AILT and PTCL-NOS were 64 and 62 years, respectively. Men slightly outnumbered women, accounting for 60 and 61% of patients with AILT and PTCL-NOS, respectively. Comparison of the AILT versus PTCL-NOS groups yielded no statistically significant difference in patient age or sex. The majority of specimens were from lymph nodes, and cases with involvement limited to skin, central nervous system, bone marrow, or body fluid/cavity were excluded. There were a total of 11 extranodal specimens, including five lung, two liver, one mediastinum, one spleen, one stomach, and one subcutaneous tissue specimen. The subcutaneous specimen was in the AILT group, and other extranodal specimens were in the PTCL-NOS group.
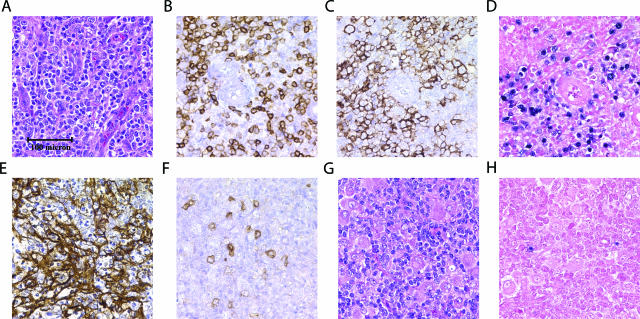
All cases of AILT showed the histologic features of an atypical mixed infiltrate within a background of prominent arborizing vessels. In Figure 1, A–F, representative histologic, immunohistochemical, and EBV in situ studies are shown for a case of AILT with an EBV-associated large B-cell proliferation. For 56 of the 58 total AILT cases, a network of extrafollicular dendritic cells was demonstrated by immunohistochemical staining for CD21 and/or CD23. In the remaining two cases, the diagnosis was rendered based on characteristic morphology and staining demonstrating T-cell lineage. A stain for CD10 was performed in 33 cases, demonstrating scattered CD10-positive cells unassociated with follicles in 20 cases, negative results in seven cases, and equivocal results in six cases. Eleven cases of AILT were complicated by an associated proliferation of B cells with seven composed of large B cells and four composed of small B cells. Two large B-cell proliferations and one small B-cell proliferation were extensive and met criteria for B-cell lymphoma based on the sheet-like architecture of the B-cell proliferation. One additional case showed an associated proliferation of plasma cells and was considered in the subgroup of cases with B-cell proliferations. The B-lineage proliferations were demonstrated by stains for various antigens including CD20, CD79a, CD138, and κ and λ immunoglobulin light chains.
Figure 1.
Histology of representative AILT and PTCL-NOS cases. All panels shown at identical magnification. A–F: AILT with EBV-associated large B-cell proliferation stained with H&E (A), immunohistochemistry with anti-CD5 (B), anti-CD20 (C), in situ hybridization for EBV (D), anti-CD21 (E), or anti-CD10 (F). G and H: PTCL-NOS stained with H&E (G) or examined by in situ hybridization for EBV (H). Images were captured with a Nikon Eclipse E1000 microscope (Nikon, Tokyo, Japan) using a ×20/0.75 objective lens (Nikon) and a Spot CCD camera (Diagnostic Instruments, McHenry, IL) with Spot software version 4.0.5 for image acquisition.
The PTCL-NOS group was composed of cases that met criteria for peripheral T-cell lymphoma, unspecified, by the current World Health Organization classification.1 These criteria are exclusionary in nature, and cases with morphologic, immunophenotypic, or clinical features diagnostic or suggestive of specific subtypes of PTCL were excluded from the PTCL-NOS group. In Figure 1, G and H, representative histologic and EBV in situ studies are shown for a case of PTCL-NOS. For 21 of the 59 total PTCL-NOS cases, an immunohistochemical stain for CD21 was performed and was negative in all 21 cases. A stain for CD10 was performed in 12 cases, demonstrating scattered CD10-positive cells in three cases, negative results in five cases, and equivocal results in four cases. Eleven cases of PTCL-NOS were complicated by an associated proliferation of B cells with seven composed of large B cells, three composed of small B cells, and one composed of mixed small and large B cells. One large B-cell proliferation and one small B-cell proliferation were classified as B-cell lymphoma based on the sheet-like architecture of the B-cell proliferation. The B-cell proliferations were demonstrated by stains for various antigens including CD20, CD79a, and κ and λ immunoglobulin light chains.
T-Cell Clonality Studies
We performed T-cell clonality studies using a multiplex PCR developed in a European BIOMED-2 collaborative study that detects clonal rearrangements in the T-cell receptor γ chain gene (TCRγ).16 Data from a representative TCRγ clonality study are shown in Figure 2, and the results for all cases are summarized in Table 1. The proportion of cases with a positive T-cell clone was 78 and 81% for total AILT and total PTCL-NOS cases, respectively. These figures included cases with associated B-cell proliferations, and when considered separately, 75% of AILT cases with B-cell proliferations and 73% of PTCL-NOS cases with B-cell proliferations were positive for T-cell clonality. There was no statistically significant correlation between T-cell clonality and diagnostic group, presence or absence of a B-cell proliferation, or the results of B-cell clonality or EBV studies. Interestingly, a positive T-cell clone correlated with older age (P = 0.045) and male sex (P = 0.009).
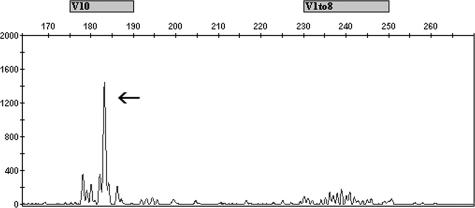
Figure 2.
Electropherogram of PCR for TCRγ rearrangements. The x axis indicates DNA length in nucleotides. The y axis indicates relative fluorescence units. Shown is electrophoresis of products from one multiplex tube, which contains primers to amplify Vγ1-8 and/or Vγ10 rearrangements, with expected product sizes above the x axis. The case was interpreted as positive with a clonal peak detected by Vγ10 primers (arrow) and polyclonal signals detected by Vγ1-8 primers.
Table 1.
T-Cell Clonality by PCR for TCRγ by Diagnosis
| Diagnosis | Number of cases studied | Number of positive cases (%) | Number of positive cases detected by specific primer sets (%)
|
||||
|---|---|---|---|---|---|---|---|
| Vγ1-8 | Vγ9 | Vγ10 | Vγ11 | Vγ9/11* | |||
| AILT | |||||||
| Without B-cell proliferation | 46 | 36 (78) | 23 (50) | 18 (39) | 22 (48) | 4 (9) | 6 (13) |
| With B-cell proliferation† | 12 | 9 (75) | 6 (50) | 2 (17) | 6 (50) | 0 (0) | 2 (17) |
| Total | 58 | 45 (78) | 29 (50) | 20 (34) | 28 (48) | 4 (7) | 8 (14) |
| PTCL-NOS | |||||||
| Without B-cell proliferation‡ | 48 | 40 (83) | 29 (60) | 20 (42) | 21 (44) | 4 (8) | 2 (4) |
| With B-cell proliferation | 11 | 8 (73) | 8 (73) | 4 (36) | 6 (55) | 0 (0) | 1 (9) |
| Total | 59 | 48 (81) | 37 (63) | 24 (41) | 27 (46) | 4 (7) | 3 (5) |
Vγ9/11 indicates a clonal PCR product within a range of sizes that is indeterminate between Vγ9 and Vγ11.
One case yielded oligoclonal Vγ10 and clonal Vγ9/11 products and was scored as positive.
One case yielded oligoclonal Vγ1-8 products only and was scored as negative.
Also listed in Table 1 are the frequencies of clonal detection for individual primer sets, which targeted rearrangements between specific variable genes (Vγ) and the joining segment (Jγ) of TCRγ. For a given case, clonality was often demonstrable by more than one primer set, as summarized in Table 2. There was no statistically significant difference in variable gene distribution between AILT and PTCL-NOS cases. Overall, rearrangements in Vγ1-8 were most common, detected in 50% and 63% of total AILT and PTCL-NOS cases, respectively. In addition, rearrangements in Vγ10 were observed frequently, occurring in 48% and 46% of total AILT and PTCL-NOS cases, respectively. Of the 93 total T-cell clones, 66 (71%) were detected by Vγ1-8 primers and 55 (59%) were detected by Vγ10 primers. The majority of Vγ10 rearrangements were seen in combination with Vγ1-8 and/or Vγ9 rearrangements, and cases with only Vγ10 rearrangements were uncommon, occurring in three AILT and three PTCL-NOS cases. Clonal rearrangements involving Vγ10 are reportedly uncommon in PTCL.17,18,19 Therefore, we sequenced eight PCR products corresponding to clonal Vγ10 rearrangements to confirm that the findings were not the result of nonspecific amplification or amplification of non-Vγ10 rearrangements by primers in the multiplex PCR. In all eight cases, the resulting sequence matched that of the Vγ10 segment.
Table 2.
T-Cell Clonality by PCR for TCRγ by Specific Variable Gene Rearrangement
| Rearrangement(s) | Number of cases
|
||
|---|---|---|---|
| AILT | PTCL-NOS | Total (%) | |
| Vγ1-8 | 8 | 12 | 20 (17.1) |
| Vγ1-8, Vγ10 | 9 | 7 | 16 (13.7) |
| Vγ1-8, Vγ9, Vγ10 | 7 | 7 | 14 (12.0) |
| Vγ1-8, Vγ9 | 2 | 8 | 10 (8.5) |
| Vγ1-8, Vγ9, Vγ10, Vγ11 | 2 | 1 | 3 (2.6) |
| Vγ1-8, Vγ10, Vγ11 | 0 | 2 | 2 (1.7) |
| Vγ1-8, Vγ11 | 1 | 0 | 1 (0.9) |
| Vγ10 | 3 | 3 | 6 (5.1) |
| Vγ9 | 2 | 0 | 2 (1.7) |
| Vγ11 | 1 | 0 | 1 (0.9) |
| Vγ9, Vγ10 | 7 | 7 | 14 (12.0) |
| Vγ9, Vγ11 | 0 | 1 | 1 (0.9) |
| Negative | 16 | 11 | 27 (23.1) |
B-Cell Clonality Studies
We performed B-cell clonality studies using a BIOMED-2-derived multiplex PCR that detects clonal rearrangements in the immunoglobulin heavy chain gene (IGH).16 Data from a representative IGH clonality study are shown in Figure 3, and the results for all cases are summarized in Table 3. The proportion of cases with a positive B-cell clone was 34 and 35% for total AILT and total PTCL-NOS cases, respectively. Of the 39 total cases with B-cell clones, 34 were positive for T-cell clonality and 5 were negative for T-cell clonality. Cases with associated B-cell proliferations showed a higher frequency of B-cell clonality than those without B-cell proliferations. For AILT cases, the frequencies of B-cell clonality were 50% for those with B-cell proliferations and 30% for those without B-cell proliferations, and for PTCL-NOS cases, the frequencies were 45% for those with B-cell proliferations and 33% for those without B-cell proliferations. When cases from both diagnostic groups were considered, B-cell clonality correlated, in part, with the presence of a B-cell proliferation (P = 0.13). There was no correlation between B-cell clonality and T-cell clonality or EBV.
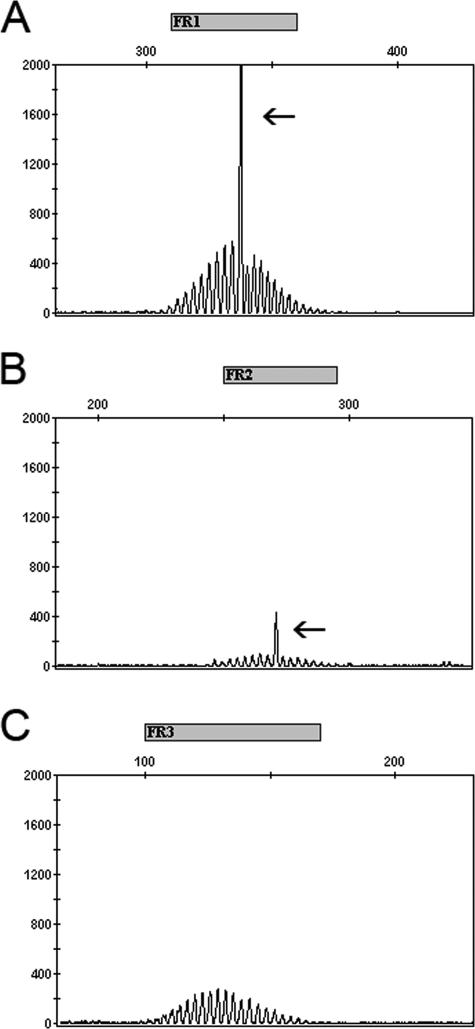
Figure 3.
Electropherograms of PCRs for IGH rearrangements. The x axis indicates DNA length in nucleotides. The y axis indicates relative fluorescence units. Shown are electrophoresis of products from three multiplex tubes, which contain primers targeting the FR1 (A), FR2 (B), and FR3 regions (C), with expected product sizes above the x axis. The case was interpreted as positive with clonal peaks detected by FR1 and FR2 primers (arrows) and polyclonal signals detected by FR3 primers.
Table 3.
B-Cell Clonality by PCR for IGH by Diagnosis
| Diagnosis | Number of cases studied | Number of positive cases (%) | Number of positive cases detected by specific primer sets (%)
|
||||
|---|---|---|---|---|---|---|---|
| FR1 | FR2 | FR3 | DH1-6 | DH7 | |||
| AILT | |||||||
| Without B-cell proliferation† | 44 | 13 (30) | 3 (7) | 2 (5) | 6 (14) | 4 (9) | 6 (14) |
| With B-cell proliferation | 12 | 6 (50) | 5 (42) | 3 (25) | 4 (33) | 1 (8) | 2 (17) |
| Total | 56* | 19 (34) | 8 (14) | 5 (9) | 10 (18) | 5 (9) | 8 (14) |
| PTCL-NOS | |||||||
| Without B-cell proliferation‡ | 46 | 15 (33) | 8 (17) | 3 (7) | 8 (17) | 10 (22) | 4 (9) |
| With B-cell proliferation | 11 | 5 (45) | 2 (18) | 1 (9) | 3 (27) | 2 (18) | 3 (27) |
| Total | 57* | 20 (35) | 10 (18) | 4 (7) | 11 (19) | 12 (21) | 7 (12) |
Framework region (FR) primer sets amplified rearrangements in IGH between variable segment (VH), diversity segment (DH), and joining segment (JH). DH primer sets amplified rearrangements between DH and JH.
Two cases of AILT and two cases of PTCL-NOS were not studied because of insufficient DNA.
One case yielded oligoclonal products with FR1 only and was scored as negative.
One case yielded clonal products with FR1 and oligoclonal products with DH1-6 and was scored as positive.
Also listed in Table 3 are the frequencies of clonal detection for different primer sets, amplifying either a complete VH-DH-JH rearrangement in the case of FR primers or an incomplete DH-JH rearrangement in the case of DH primers. Cases with both a complete (VH-DH-JH) rearrangement and incomplete (DH-JH) rearrangement can be explained by a productive rearrangement on one IGH allele and a nonproductive rearrangement on the other allele. Clonality was typically demonstrable by multiple primer sets, as summarized in Table 4. Among FR primers, the FR1 and FR3 primers were most sensitive, detecting 18 and 21 of the 39 total B-cell clones, respectively. The FR2 primers detected only nine clones, and the low sensitivity is likely technical in nature because these primers typically yielded weaker PCR products than the FR1 and FR3 primers in our laboratory (Figure 3). Among DH primers, the DH1-6 and DH7 primers detected 17 and 15 clones, respectively. Most clones detected by DH primers were also detected by FR primers. However, 14 of the 39 total B-cell clones were detected by DH primers only, consistent with the presence of an unamplifiable VH-DH-JH rearrangement on one allele and an amplifiable DH-JH rearrangement on the other allele. Of these 14 cases, 9 of 11 were EBV-positive, and for the remaining three cases, tissue was not readily available for in situ studies. Clonality of IGH demonstrable only by DH primers correlated with EBV (P = 0.015) but not with diagnosis, B-cell proliferation, or T-cell clonality.
Table 4.
B-Cell Clonality Detection by PCR for IGH by Specific Primer Sets
| Primer sets yielding IGH clones | Number of cases
|
||
|---|---|---|---|
| AILT | PTCL-NOS | Total (%) | |
| FR1 | 2 | 0 | 2 (1.8) |
| FR1, FR2 | 0 | 1 | 1 (0.9) |
| FR1, FR3 | 1 | 0 | 1 (0.9) |
| FR1, FR2, FR3 | 3 | 0 | 3 (2.7) |
| FR3 | 2 | 2 | 4 (3.5) |
| FR1, DH1-6 | 0 | 1 | 1 (0.9) |
| FR3, DH7 | 2 | 1 | 3 (2.7) |
| FR1, FR3, DH1-6 | 0 | 4 | 4 (3.5) |
| FR1, FR3, DH1-6, DH7 | 0 | 1 | 1 (0.9) |
| FR1, FR2, FR3, DH1-6 | 1 | 2 | 3 (2.7) |
| FR1, FR2, FR3, DH7 | 0 | 1 | 1 (0.9) |
| FR1, FR2, FR3, DH1-6, DH7 | 1 | 0 | 1 (0.9) |
| DH1-6 | 2 | 3 | 5 (4.4) |
| DH1-6, DH7 | 1 | 1 | 2 (1.8) |
| DH7 | 4 | 3 | 7 (6.2) |
| Negative | 37 | 37 | 74 (65.5) |
EBV Studies
We performed in situ hybridization for EBV EBER on approximately half of the cases. Representative in situ hybridizations for an AILT with an EBV-associated B-cell proliferation and a PTCL-NOS case are shown (Figure 1, D and H, respectively), and the results for all cases studied are summarized in Table 5. A positive result, which was defined as more than three EBV-labeled cells per section, was observed in 50% and 57% of total AILT and total PTCL-NOS cases, respectively. Cases with B-cell proliferations were more often EBV-positive and tended to show greater numbers of EBV-labeled cells. Among AILT cases, 45% without B-cell proliferations were EBV-positive, whereas 63% with B-cell proliferations were EBV-positive. For PTCL-NOS cases, 52% without B-cell proliferations were EBV-positive, and 75% with B-cell proliferations were EBV-positive. Overall, a positive EBV study correlated, in part, with the presence of a B-cell proliferation (P = 0.20) and male sex (P = 0.12). There was no correlation between EBV and diagnostic group, age, or the results of B- or T-cell clonality studies (P = 0.50 for correlation between EBV and B-cell clonality).
Table 5.
EBV Studies by in Situ Hybridization for EBER
| Diagnosis | Number of cases studied | Number of positive cases (%) | Number of EBV-labeled cells per medium-power field
|
||||
|---|---|---|---|---|---|---|---|
| 0 | Less than 1 | 1 to 10 | Greater than 10 to 100 | Greater than 100 | |||
| AILT | |||||||
| Without B-cell proliferation | 22 | 10 (45) | 12 | 2 | 3 | 3 | 2 |
| With B-cell proliferation | 8 | 5 (63) | 3 | 0 | 3 | 0 | 2 |
| Total | 30 | 15 (50) | 15 | 2 | 6 | 3 | 4 |
| PTCL-NOS | |||||||
| Without B-cell proliferation | 29 | 15 (52) | 14 | 2 | 4 | 7 | 2 |
| With B-cell proliferation | 8 | 6 (75) | 2 | 0 | 2 | 3 | 1 |
| Total | 37 | 21 (57) | 16 | 2 | 6 | 10 | 3 |
For each case with an associated B-cell proliferation, the results of the EBV and clonality studies along with the type of B-cell proliferation are listed in Table 6. Overall, 11 of 16 cases with B-cell proliferations were positive for EBV, and 11 of 23 had B-cell clones. In cases with high numbers of EBV-labeled cells, the EBV staining pattern correlated with that of CD79a or CD20, consistent with an EBV-associated B-cell proliferation (Figure 1, C and D). In cases with few numbers of labeled cells, the staining pattern for EBV was typically scattered and did not clearly correlate with that of B- or T-cell lineage markers. Essentially all combinations of type of B-cell proliferation and results of EBV and clonality studies were seen. For cases with small B-cell proliferations, three of five were EBV-positive, and five of seven had a B-cell clone. For cases with large B-cell proliferations, seven of nine were EBV-positive, and 5 of 14 had a B-cell clone. Among the EBV-positive cases, 4 of 11 had a B-cell clone, whereas for EBV-negative cases, three of five had a B-cell clone. Within the subset of cases with B-cell proliferations, there was no statistically significant correlation between EBV, type of B-cell proliferation, B-cell clonality, or T-cell clonality.
Table 6.
Summary of Results for Cases with Associated B-Cell Proliferations
| Diagnosis and type of B-cell proliferation | TCRγ clonality | IGH clonality | No. EBV-labeled cells/mpf | Lineage of EBV-labeled cells |
|---|---|---|---|---|
| AILT | ||||
| Large* | + | + | ND | NA |
| Large | + | − | ND | NA |
| Large | + | − | ND | NA |
| Small | + | + | 1 to 10 | Unclear |
| Small | − | + | >100 | Subset of B cells |
| Plasma cells | + | + | Negative | NA |
| Large | + | + | Negative | NA |
| Large* | + | − | Negative | NA |
| Large | − | − | 1 to 10 | Unclear |
| Large | + | − | >100 | Subset of B cells |
| Small | + | − | 1 to 10 | Unclear |
| Small* | − | + | ND | NA |
| PTCL-NOS | ||||
| Small | + | + | Negative | NA |
| Large | + | + | 1 to 10 | Unclear |
| Large | + | + | >10 to 100 | Large B cells |
| Large | + | + | ND | NA |
| Small* | + | + | ND | NA |
| Large | + | − | 1 to 10 | Large B cells |
| Small | + | − | Negative | NA |
| Small and Large | − | − | >10 to 100 | Large B cells |
| Large* | − | − | ND | NA |
| Large | − | − | >100 | Large B cells |
| Large | + | − | >10 to 100 | Subset of B cells |
ND, not done; NA, not applicable; mpf, medium-power field
These B-cell proliferations met criteria for B-cell lymphoma.
The lineage of EBV-labeled cells was determined by comparison of the in situ stain to that of B- and T-cell lineage markers. In some cases, the lineage of EBV-labeled cells could not be reliably determined, and the lineage is indicated as unclear.
Discussion
We detected T-cell clones in 78 and 81% of AILT and PTCL-NOS cases, respectively. These figures are consistent with those of previous reports,2,20,21,22,23,24 including a recent study on the BIOMED-2 PCR,25 which used the same TCRγ primers to demonstrate clonality in 86% of T-cell lymphoproliferative disorders. An important aspect of our study is the variable gene usage data, which highlight the advantage of multiplex PCR assays that target a wide range of variable genes over those that target only Vγ1-8. Although 71% of T-cell clones were detected by Vγ1-8 primers, the remaining 29% were only detectable by a combination of Vγ9, Vγ10, and Vγ11 primers. Lawnicki and colleagues19 reported similar findings with 25% of T-cell clones undetectable using Vγ1-8 primers alone. Interestingly, we also observed rearrangements in Vγ10 relatively frequently, occurring in 59% of T-cell clones. However, previous studies on variable gene usage in PTCL report rearrangements in Vγ10 as uncommon, occurring in 2 to 17% of cases.17,18,19 Because of this apparent discrepancy, we considered the possibility of nonspecific amplification or amplification of non-Vγ10 rearrangements by Vγ10 primers or other primers in the multiplex PCR. Therefore, we sequenced eight PCR products corresponding to Vγ10 rearrangements. In all eight cases, the sequence matched that of Vγ10, confirming the specificity of the BIOMED-2 primers for this region. Our findings suggest a higher sensitivity of the BIOMED-2 PCR for Vγ10 than methods used in previous reports, which was also observed in the initial BIOMED-2 study.16
Increased numbers of EBV-positive cells were observed in 50 and 57% of AILT and PTCL-NOS cases, respectively. These frequencies are comparable to those of previous reports on PTCL26,27,28,29 and more recent studies on AILT.20,30,31 In both AILT and PTCL-NOS groups, cases with associated B-cell proliferations were more often EBV-positive with higher numbers of labeled cells. In many cases, the majority of the B-cell proliferation was EBV-positive (Figure 1, C and D), suggesting that the virus stimulates B-cell proliferation. However, some B-cell proliferations were EBV-negative, indicating additional mechanisms for B-cell proliferation in some T-cell lymphomas.
The B-cell clonality studies provided the most interesting results, with B-cell clones detected in 34 and 35% of AILT and PTCL-NOS cases, respectively. Although previously reported frequencies of B-cell clonality in AILT are similar,20,23,32,33 those for PTCL range from 9 to 16%.2,4,5,22,34,35 We considered that some cases of PTCL-NOS were misclassified and actually represent AILT. However, PTCL-NOS cases did not have classic morphologic features of AILT. Furthermore, of the 20 cases of PTCL-NOS with a B-cell clone, an immunohistochemical stain for CD21 was performed in 12 cases, yielding negative results in all 12. In contrast, 56 of 56 AILT cases were positive for CD21 expansions, consistent with previous reports that CD21 staining is a virtually constant finding in AILT.36 Nevertheless, we cannot exclude that a morphologic spectrum exists between AILT and PTCL-NOS.
Overall, B-cell clonality correlated, in part, with the presence of a B-cell proliferation but not with EBV. However, cases without demonstrable B-cell proliferations also exhibited B-cell clones relatively frequently, suggesting other contributing factors. One factor contributing to the relatively high frequency of B-cell clones detected in the current study may be the inherent sensitivity of the BIOMED-2 IGH PCR, which uses primers targeting all three FRs. Most methods used in previous studies targeted only a single FR,2,5,22 a strategy shown to be less sensitive.34 If our data are compared by basing clonality on a single FR primer set (Table 3), 15 to 20% of cases would be considered positive for B-cell clonality. Another possibility is so-called lineage infidelity, referring to simultaneous TCRγ and IGH rearrangements occurring in the same cell.7 Although uncommon in mature B- and T-cell non-Hodgkin lymphomas,7 this phenomenon may have contributed more than in previous studies because of the sensitivity of our assays.
In contrast to previous studies,11,20 we did not observe the frequent presence of B-cell oligoclones in cases with EBV. This difference is likely related to methodology, with the BIOMED-2 IGH PCR yielding either positive or negative results rather than oligoclones. However, EBV was associated with B-cell clones detectable only by DH primers, a finding likely caused by the presence of an incomplete DH-JH rearrangement on one IGH allele and a complete VH-DH-JH rearrangement on the other allele that is unamplifiable. It is possible that the previous methods, which used FR primers only and showed frequent oligoclones, would have yielded oligoclonal results in these cases. Alternatively, our findings may represent B-cell clones with mutations that interfere with FR primer detection. In either case, understanding the clonal or oligoclonal nature underlying these findings will require correlation with nongenotypic methods such as immunophenotyping.
Collectively, the B-cell clonality and EBV results suggest a model whereby altered immune function in the locale of AILT or PTCL-NOS results in uncontrolled B-cell proliferation. The frequent presence of EBV is evidence of immune dysfunction, and in EBV-positive cases, the virus may further stimulate B-cell proliferation, which is supported by studies localizing EBV to B cells in PTCL.11,29,37 Uncontrolled B-cell proliferation may result in the emergence of a dominant clone over time. Evidence for this evolution is provided by PTCL patients who initially lack an associated B-cell proliferation but who later have specimens showing a B-cell proliferation, B-cell clone, or even an associated B-cell lymphoma.11,14 Overall, the model is similar to that of post-transplant lymphoproliferative disorder (PTLD), but it does not explain the occurrence of B-cell clones in PTCL lacking B-cell proliferations. It is possible that B-cell proliferations are not always readily demonstrable by morphology and immunohistochemistry, and because of the sensitivity and lack of quantitation of PCR, a B-cell clone may reflect processes ranging from an incipient or focal B-cell proliferation to overt B-cell lymphoma. Regardless of the mechanisms involved in B-cell clonality, the clinical significance remains unclear. Feller and colleagues32 reported that clonal IGH rearrangements in AILT were associated with increased overall survival. However, this finding was not statistically significant, possibly because of the low number of cases. Thus, until the clinical manifestations are better understood, the most important aspect of B-cell clonality in PTCL is recognizing that it occurs and that its presence should not negate the diagnosis of PTCL.
Acknowledgments
We thank Marla Lay, Linda Gojenola, Tarek Hajar, and Kyunga Seo for excellent technical assistance.
Footnotes
Supported by the Stanford Department of Pathology.
Related Commentary on page 426
References
- Jaffe ES, Harris NL, Stein H, Vardiman JW. Kleihues P, Sobin LH, editors. Lyon: IARC Press,; Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. 2001 [Google Scholar]
- Diss TC, Watts M, Pan LX, Burke M, Linch D, Isaacson PG. The polymerase chain reaction in the demonstration of monoclonality in T cell lymphomas. J Clin Pathol. 1995;48:1045–1050. doi: 10.1136/jcp.48.11.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriault C, Galoin S, Valmary S, Selves J, Lamant L, Roda D, Rigal-Huguet F, Brousset P, Delsol G, Al Saati T. PCR analysis of immunoglobulin heavy chain (IgH) and TcR-gamma chain gene rearrangements in the diagnosis of lymphoproliferative disorders: results of a study of 525 cases. Mod Pathol. 2000;13:1269–1279. doi: 10.1038/modpathol.3880232. [DOI] [PubMed] [Google Scholar]
- Garcia MJ, Martinez-Delgado B, Granizo JJ, Benitez J, Rivas C. IgH, TCR-gamma, and TCR-beta gene rearrangement in 80 B- and T-cell non-Hodgkin’s lymphomas: study of the association between proliferation and the so-called “aberrant” patterns. Diagn Mol Pathol. 2001;10:69–77. doi: 10.1097/00019606-200106000-00001. [DOI] [PubMed] [Google Scholar]
- Vergier B, Dubus P, Kutschmar A, Parrens M, Ferrer J, de Mascarel A, Merlio JP. Combined analysis of T cell receptor gamma and immunoglobulin heavy chain gene rearrangements at the single-cell level in lymphomas with dual genotype. J Pathol. 2002;198:171–180. doi: 10.1002/path.1192. [DOI] [PubMed] [Google Scholar]
- Noorali S, Pervez S, Moatter T, Soomro IN, Kazmi SU, Nasir MI, Smith JL. Characterization of T-cell non-Hodgkin’s lymphoma and its association with Epstein-Barr virus in Pakistani patients. Leuk Lymphoma. 2003;44:807–813. doi: 10.1080/1042819031000067747. [DOI] [PubMed] [Google Scholar]
- van Dongen JJ, Wolvers-Tettero IL. Analysis of immunoglobulin and T cell receptor genes. Part II: possibilities and limitations in the diagnosis and management of lymphoproliferative diseases and related disorders. Clin Chim Acta. 1991;198:93–174. doi: 10.1016/0009-8981(91)90247-a. [DOI] [PubMed] [Google Scholar]
- Smith LJ, Curtis JE, Messner HA, Senn JS, Furthmayr H, McCulloch EA. Lineage infidelity in acute leukemia. Blood. 1983;61:1138–1145. [PubMed] [Google Scholar]
- Greaves MF, Chan LC, Furley AJ, Watt SM, Molgaard HV. Lineage promiscuity in hemopoietic differentiation and leukemia. Blood. 1986;67:1–11. [PubMed] [Google Scholar]
- McCulloch EA. Lineage infidelity or lineage promiscuity? Leukemia. 1987;1:235. [PubMed] [Google Scholar]
- Higgins JP, van de Rijn M, Jones CD, Zehnder JL, Warnke RA. Peripheral T-cell lymphoma complicated by a proliferation of large B cells. Am J Clin Pathol. 2000;114:236–247. doi: 10.1309/72CM-KAXF-66DE-4XVA. [DOI] [PubMed] [Google Scholar]
- Lome-Maldonado C, Canioni D, Hermine O, Delabesse E, Damotte D, Raffoux E, Gaulard P, Macintyre E, Brousse N. Angio-immunoblastic T cell lymphoma (AILD-TL) rich in large B cells and associated with Epstein-Barr virus infection. A different subtype of AILD-TL? Leukemia. 2002;16:2134–2141. doi: 10.1038/sj.leu.2402642. [DOI] [PubMed] [Google Scholar]
- Xu Y, McKenna RW, Hoang MP, Collins RH, Kroft SH. Composite angioimmunoblastic T-cell lymphoma and diffuse large B cell lymphoma: a case report and review of the literature. Am J Clin Pathol. 2002;118:848–854. doi: 10.1309/VD2D-98ME-MB3F-WH34. [DOI] [PubMed] [Google Scholar]
- Zettl A, Lee SS, Rudiger T, Starostik P, Marino M, Kirchner T, Ott M, Muller-Hermelink HK, Ott G. Epstein-Barr virus-associated B-cell lymphoproliferative disorders in angioimmunoblastic T-cell lymphoma and peripheral T-cell lymphoma, unspecified. Am J Clin Pathol. 2002;117:368–379. doi: 10.1309/6UTX-GVC0-12ND-JJEU. [DOI] [PubMed] [Google Scholar]
- Bindl JM, Warnke RA. Advantages of detecting monoclonal antibody binding to tissue sections with biotin and avidin reagents in Coplin jars. Am J Clin Pathol. 1986;85:490–493. doi: 10.1093/ajcp/85.4.490. [DOI] [PubMed] [Google Scholar]
- van Dongen JJ, Langerak AW, Bruggemann M, Evans PA, Hummel M, Lavender FL, Delabesse E, Davi F, Schuuring E, Garcia-Sanz R, van Krieken JH, Droese J, Gonzalez D, Bastard C, White HE, Spaargaren M, Gonzalez M, Parreira A, Smith JL, Morgan GJ, Kneba M, Macintyre EA. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17:2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- Theodorou I, Raphael M, Bigorgne C, Fourcade C, Lahet C, Cochet G, Lefranc MP, Gaulard P, Farcet JP. Recombination pattern of the TCR gamma locus in human peripheral T-cell lymphomas. J Pathol. 1994;174:233–242. doi: 10.1002/path.1711740402. [DOI] [PubMed] [Google Scholar]
- Kneba M, Bolz I, Linke B, Bertram J, Rothaupt D, Hiddemann W. Characterization of clone-specific rearrangement T-cell receptor gamma-chain genes in lymphomas and leukemias by the polymerase chain reaction and DNA sequencing. Blood. 1994;84:574–581. [PubMed] [Google Scholar]
- Lawnicki LC, Rubocki RJ, Chan WC, Lytle DM, Greiner TC. The distribution of gene segments in T-cell receptor gamma gene rearrangements demonstrates the need for multiple primer sets. J Mol Diagn. 2003;5:82–87. doi: 10.1016/s1525-1578(10)60456-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JL, Hodges E, Quin CT, McCarthy KP, Wright DH. Frequent T and B cell oligoclones in histologically and immunophenotypically characterized angioimmunoblastic lymphadenopathy. Am J Pathol. 2000;156:661–669. doi: 10.1016/S0002-9440(10)64770-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber DA, Braziel RM, Bagg A, Bijwaard KE. Evaluation of T cell receptor testing in lymphoid neoplasms: results of a multicenter study of 29 extracted DNA and paraffin-embedded samples. J Mol Diagn. 2001;3:133–140. doi: 10.1016/S1525-1578(10)60664-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krafft AE, Taubenberger JK, Sheng ZM, Bijwaard KE, Abbondanzo SL, Aguilera NS, Lichy JH. Enhanced sensitivity with a novel TCRgamma PCR assay for clonality studies in 569 formalin-fixed, paraffin-embedded (FFPE) cases. Mol Diagn. 1999;4:119–133. doi: 10.1016/s1084-8592(99)80036-8. [DOI] [PubMed] [Google Scholar]
- Attygalle A, Al-Jehani R, Diss TC, Munson P, Liu H, Du MQ, Isaacson PG, Dogan A. Neoplastic T cells in angioimmunoblastic T-cell lymphoma express CD10. Blood. 2002;99:627–633. doi: 10.1182/blood.v99.2.627. [DOI] [PubMed] [Google Scholar]
- Lee SS, Rudiger T, Odenwald T, Roth S, Starostik P, Muller-Hermelink HK. Angioimmunoblastic T cell lymphoma is derived from mature T-helper cells with varying expression and loss of detectable CD4. Int J Cancer. 2003;103:12–20. doi: 10.1002/ijc.10758. [DOI] [PubMed] [Google Scholar]
- Sandberg Y, van Gastel-Mol EJ, Verhaaf B, Lam KH, van Dongen JJ, Langerak AW. BIOMED-2 multiplex immunoglobulin/T-cell receptor polymerase chain reaction protocols can reliably replace southern blot analysis in routine clonality diagnostics. J Mol Diagn. 2005;7:495–503. doi: 10.1016/S1525-1578(10)60580-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbjuhn P, Anagnostopoulos I, Hummel M, Tiemann M, Dallenbach F, Parwaresch MR, Stein H. Frequent latent Epstein-Barr virus infection of neoplastic T cells and bystander B cells in human immunodeficiency virus-negative European peripheral pleomorphic T-cell lymphomas. Blood. 1993;82:217–223. [PubMed] [Google Scholar]
- Zhou XG, Hamilton-Dutoit SJ, Yan QH, Pallesen G. High frequency of Epstein-Barr virus in Chinese peripheral T-cell lymphoma. Histopathology. 1994;24:115–122. doi: 10.1111/j.1365-2559.1994.tb01289.x. [DOI] [PubMed] [Google Scholar]
- Anagnostopoulos I, Hummel M, Stein H. Frequent presence of latent Epstein-Barr virus infection in peripheral T cell lymphomas. A review. Leuk Lymphoma. 1995;19:1–12. doi: 10.3109/10428199509059657. [DOI] [PubMed] [Google Scholar]
- Ho JW, Ho FC, Chan AC, Liang RH, Srivastava G. Frequent detection of Epstein-Barr virus-infected B cells in peripheral T-cell lymphomas. J Pathol. 1998;185:79–85. doi: 10.1002/(SICI)1096-9896(199805)185:1<79::AID-PATH52>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Khan G, Norton AJ, Slavin G. Epstein-Barr virus in angioimmunoblastic T-cell lymphomas. Histopathology. 1993;22:145–149. doi: 10.1111/j.1365-2559.1993.tb00093.x. [DOI] [PubMed] [Google Scholar]
- Vrsalovic MM, Korac P, Dominis M, Ostojic S, Mannhalter C, Kusec R. T- and B-cell clonality and frequency of human herpes viruses-6, -8 and Epstein Barr virus in angioimmunoblastic T-cell lymphoma. Hematol Oncol. 2004;22:169–177. doi: 10.1002/hon.740. [DOI] [PubMed] [Google Scholar]
- Feller AC, Griesser H, Schilling CV, Wacker HH, Dallenbach F, Bartels H, Kuse R, Mak TW, Lennert K. Clonal gene rearrangement patterns correlate with immunophenotype and clinical parameters in patients with angioimmunoblastic lymphadenopathy. Am J Pathol. 1988;133:549–556. [PMC free article] [PubMed] [Google Scholar]
- Lorenzen J, Li G, Zhao-Hohn M, Wintzer C, Fischer R, Hansmann ML. Angioimmunoblastic lymphadenopathy type of T-cell lymphoma and angioimmunoblastic lymphadenopathy: a clinicopathological and molecular biological study of 13 Chinese patients using polymerase chain reaction and paraffin-embedded tissues. Virchows Arch. 1994;424:593–600. doi: 10.1007/BF01069738. [DOI] [PubMed] [Google Scholar]
- Bagg A, Braziel RM, Arber DA, Bijwaard KE, Chu AY. Immunoglobulin heavy chain gene analysis in lymphomas: a multi-center study demonstrating the heterogeneity of performance of polymerase chain reaction assays. J Mol Diagn. 2002;4:81–89. doi: 10.1016/S1525-1578(10)60685-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang SS, Lin CN, Shen FC, Liao PJ, Liao YL, Chang JH, Tsai YC, Cho CY, Huang W. Detecting clonal rearrangement in non-Hodgkin’s lymphomas in Taiwan by polymerase chain reaction. Leuk Lymphoma. 2003;44:117–121. doi: 10.1080/1042819021000040314. [DOI] [PubMed] [Google Scholar]
- Jones D, Jorgensen JL, Shahsafaei A, Dorfman DM. Characteristic proliferations of reticular and dendritic cells in angioimmunoblastic lymphoma. Am J Surg Pathol. 1998;22:956–964. doi: 10.1097/00000478-199808000-00005. [DOI] [PubMed] [Google Scholar]
- Weiss LM, Jaffe ES, Liu XF, Chen YY, Shibata D, Medeiros LJ. Detection and localization of Epstein-Barr viral genomes in angioimmunoblastic lymphadenopathy and angioimmunoblastic lymphadenopathy-like lymphoma. Blood. 1992;79:1789–1795. [PubMed] [Google Scholar]