Abstract
The US Food and Drug Administration-cleared ViroSeq HIV-1 Genotyping System (ViroSeq) and other population sequencing-based human immunodeficiency virus type 1 (HIV-1) genotyping methods detect antiretroviral drug resistance mutations present in the major viral population of a test sample. These assays also detect some mutations in viral variants that are present as mixtures. We compared detection of the K103N nevirapine resistance mutation using ViroSeq and a sensitive, quantitative point mutation assay, LigAmp. The LigAmp assay measured the percentage of K103N-containing variants in the viral population (percentage of K103N). We analyzed 305 samples with HIV-1 subtypes A, C, and D collected from African women after nevirapine administration. ViroSeq detected K103N in 100% of samples with >20% K103N, 77.8% of samples with 10 to 20% K103N, 71.4% of samples with 5 to 10% K103N, and 16.9% of samples with 1 to 5% K103N. The sensitivity of ViroSeq for detection of K103N was similar for subtypes A, C, and D. These data indicate that the ViroSeq system reliably detects the K103N mutation at levels above 20% and frequently detects the mutation at lower levels. Further studies are needed to compare the sensitivity of different assays for detection of HIV-1 drug resistance mutations and to determine the clinical relevance of HIV-1 minority variants.
The US Food and Drug Administration-cleared ViroSeq HIV-1 Genotyping System (ViroSeq) and other population sequencing-based genotyping methods detect mutations present in the major viral population in a test sample. These assays also detect some mutations that are present at lower levels. Using recombinant viral stocks, we previously demonstrated that ViroSeq reliably detects drug resistance mutations present in 40% of the viral population in samples with viral loads from 2000 to 5000 copies/ml1, lower level mixtures were not evaluated in that study. In another study, ViroSeq detected the K103N mutation in a recombinant human immunodeficiency virus type 1 (HIV-1) strain at a level of 10%.2
HIV-1 variants with the K103N mutation are often selected in women who receive a single dose of the antiretroviral drug nevirapine for prevention of HIV-1 mother-to-child transmission.3,4 We evaluated the sensitivity of ViroSeq for detection of K103N in 305 clinical plasma samples collected from African women 6 to 8 weeks after single dose nevirapine administration. Samples were collected from women in the HIV Network for Prevention Trial (HIVNET) 012 trial5,6 (Ugandan women, 146 subtype A samples and 95 subtype D samples) and the Nevirapine-zidovudine trial7 (Malawian women, 64 subtype C samples) and were analyzed with ViroSeq in previous studies.3,8 In this study, the level of K103N in test samples was quantified using a sensitive point mutation assay, LigAmp. The LigAmp assay involves mutation-specific ligation of two adjacent oligonucleotides hybridized to a DNA template. Ligated oligonucleotides are quantified in a second step using a real-time polymerase chain reaction (PCR)-based detection method.9,10
Materials and Methods
Human experimentation guidelines of the US Department of Health and Human Services and those of the authors’ institutions were followed in the conduct of this research.
HIV Genotyping with the ViroSeq System
HIV-1 genotyping was performed with the ViroSeq HIV-1 Genotyping System (Celera Diagnostics, Alameda, CA) according to the manufacturer’s instructions. Genotypes were analyzed only if bi-directional sequence data were obtained at all positions of nevirapine resistance mutations, including K103N.
Analysis of K103N with the LigAmp Assay
The LigAmp assay was performed using 50 pg of PCR products from the ViroSeq system. DNA concentrations were determined using a NanoDrop spectrophotometer (ND-1000; NanoDrop Technologies, Wilmington, DE). PCR products were incubated with 1 pmol of upstream oligonucleotide and 0.5 pmol of downstream oligonucleotide (gel-purified; Invitrogen Corp., Carlsbad, CA), with 2 U of Pfu DNA ligase in 1× Pfu Ligase Buffer (Stratagene, La Jolla, CA) in a 12.5-μl reaction volume. Ligation oligonucleotides used for detection of K103N (AAA→AAC) in subtypes A, C, and D include M13 tails (underlined). Upstream oligonucleotides also include a LacZ probe-binding site (italics) and an intentional mismatch at the third position from the 3′ end (G, bold). The upstream oligonucleotides were as follows: subtype A: 5′-ACTGTAAAACGACGGCCAGTGTTCCCCTCAAACTGGCAGATGCACGAGGAATACCACATCCAGCAGGTCTAAAAAAGGAC-3′; subtype C: 5′-ACTGTAAAACGACGGCCAGTGTTCCCCTCAAACTGGCAGATGCACGAGGAATACCACACCCAGCAGGGTTAAAAAAGGAC-3′; subtype D: 5′-ACTGTAAAACGACGGCCAGTGTTCCCCTCAAACTGGCAGATGCACGAGGAATACCACATCCTGCAGGGCTAAAAAAGGAC-3′. The downstream oligonucleotides were as follows: subtype A: 5′-AAATCAGTAACAGTACTAGATGTGGGGGTGGTCATAGCTGTTTCCTGCA-3′; subtype C: 5′-AAATCAGTGACAGTACTGGATGTGGGGGTGGTCATAGCTGTTTCCTGCA-3′; subtype D: 5′-AAATCAGTAACAGTACTGGATGTGGGTGTGGTCATAGCTGTTTCCTGCA-3′. Samples were denatured at 95°C for 1 minute, followed by 99 cycles alternating 95°C for 30 seconds with 50°C for 4 minutes. Ligated oligonucleo-tides were detected using the 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA). Each 25-μl reaction contained 5 pmol of M13 forward primer (5′-CTGTAAAACGACGGCCAGTG-3′), 5 pmol of M13 reverse primer (5′-TGCAGGAAACAGCTATGACCA-3′), 5 μl of the unpurified ligation reaction, 12.5 μl of TaqMan Universal PCR Master Mix (Applied Biosystems), and 5 pmol of the LacZ probe (FAM-5′-TCCCCTCAAACTGGCAGATGCACG-3′-BHQ-1, FAM = 6-carboxyfluoresceine, BHQ = black hole quencher; Integrated DNA Tech-nology, Coralville, IA). Reactions were incubated at 50°C for 2 minutes and 95°C for 10 minutes, followed by 50 cycles of 95°C for 10 seconds alternating with 64°C for 1 minute. The cycle threshold was set in the middle of the linear range of the amplification curve for each experiment (log scale). Paired plasmids with and with-out the K103N mutation were used as reference reagents for each subtype. The pol region of each plasmid was amplified, and mixtures were prepared at mutant DNA concentrations of 100, 10, 1, 0.1, 0.01, and 0%. The standard curve was analyzed in each experiment, and the percentage of K103N-containing variants was plotted against the cycle threshold value. The standard curve was used to determine the percentage of K103N-containing variants in each sample, as described.10
Statistical Methods
The (p) sensitivity of the ViroSeq assay for detection of different levels (X) of K103N was modeled using the logistic function p = exp(a + bX)/[1 + exp(a + bX)], where a and b were estimated from the data using Proc Logist (SAS version 9.1; SAS, Cary, NC).
Results
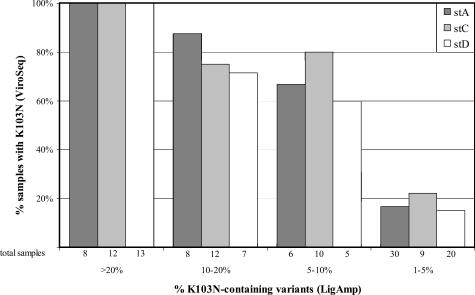
The set of 305 plasma samples was analyzed with the LigAmp assay to quantify the percentage of K103N-containing variants in each sample. Those results were compared with results obtained with the ViroSeq system in previous studies.3,4 ViroSeq detected K103N in 79 (25.9%) of the 305 samples and LigAmp detected K103N at >1% in 140 (45.9%) of the 305 samples. ViroSeq detected K103N in 33 (100%) of 33 samples with >20% K103N, in 21 (77.8%) of 27 samples with 10 to 20% K103N, in 15 (71.4%) of 21 samples with 5 to 10% K103N, and in 10 (16.9%) of 59 samples with 1 to 5% K103N. Similar results were obtained for samples with subtypes A, C, and D (Figure 1). Using a logistic regression function for detection, when K103N was present at or above X ≥17.3%, there was at least a 95% chance of detecting the mutation with the ViroSeq assay.
Figure 1.
Detection of the K103N mutation by the ViroSeq system in samples with different levels of K103N-containing HIV-1 variants. Samples with subtype A, C, and D were analyzed with the LigAmp assay to determine the percentage of HIV-1 variants in the viral population that had the K103N mutation. Samples with >1% K103N detected in the LigAmp assay were divided into categories for each subtype with >20% K103N, >10% but ≤20% K103N, >5% but ≤10% K103N, and >1% but ≤5% K103N. The graph indicates the percentage of samples in each category that had K103N detected using the ViroSeq system. The number of samples tested in each sample subset is indicated below each bar.
Discussion
Previous studies have documented high sensitivity and specificity of the ViroSeq system for detection of HIV-1 drug resistance mutations,1 and excellent performance of ViroSeq for analysis of diverse HIV-1 strains.10 This report demonstrates that ViroSeq consistently detects the K103N mutation in plasma samples with subtypes A, C, and D at levels over 20% of the viral population, and often detects the K103N mutation at lower levels. Detection of K103N at levels between 1 and 20% in 107 (35.1%) of the 305 samples was unlikely to represent false positives in the LigAmp assay, because K103N was detected in only 1 (0.4%) of 238 available samples from these women collected before nevirapine administration (data not shown). Peaks suggesting the presence of the K103N mutation were visible in ViroSeq electropherograms for all of the samples with 5 to 20% K103N and many of the samples with 1 to 5% K103N. Identification of K103N as present or absent during sequence editing was performed using guidelines provided with the ViroSeq system, recognizing that mutation identification at low levels may be influenced by a variety of factors.11
Sensitive point mutation assays, such as LigAmp, have been used recently to detect and quantify HIV-1 minority variants.9,12 However, the clinical significance of HIV-1 minority variants (eg, those below the level of detection of current US Food and Drug Adminstration-cleared HIV genotyping assays) is not known. Further studies are needed to determine the sensitivities of different assays for detection of HIV-1 drug resistance mutations, and to determine the clinical relevance of HIV-1 minority variants.
Acknowledgments
We thank the staffs in Uganda, Malawi, and at Johns Hopkins for assistance with sample processing. We also thank the clinical trials specialist Melissa Allen (Family Health International).
Footnotes
Supported by the HIV Network for Prevention Trials (sponsored by the US National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), and the Department of Health and Human Services (DHHS), through contract N01-AI-35173 with Family Health International, and contract N01-AI-45200 with Fred Hutchinson Cancer Research Center, and subcontracts with Makerere University, contract NOI-AI-35173–417); the HIV Prevention Trials Network (HPTN) sponsored by the NIAID, National Institutes of Child Health and Human Development (NICH/HD), National Institute on Drug Abuse, National Institute of Mental Health, and Office of AIDS Research, of the NIH, DHHS (contracts U01-AI-46745 and U01-AI-48054); the Adult AIDS Clinical Trials Groups (NIH, Division of AIDS, NIAID (grants U01-AI-38858); R01-HD042965–01); AIDS Fogarty International Research Collaborative Award (award 5R03TW01199) and supplement from the Fogarty International Center, NIH; The Doris Duke Charitable Foundation, New York; and the National Science Foundation (grant EIA 02–05116).
References
- Eshleman SH, Crutcher G, Petrauskene O, Kunstman K, Cunningham SP, Trevino C, Davis C, Kennedy J, Fairman J, Foley B, Kop J. Sensitivity and specificity of the ViroSeq HIV-1 Genotyping System for detection of HIV-1 drug resistance mutations using an ABI PRISM 3100 Genetic Analyzer. J Clin Micro. 2005;43:813–817. doi: 10.1128/JCM.43.2.813-817.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halvas E, Aldrovandi G, Balfe JP, Beck IA, Boltz V, Frenkel LM, Hazelwood J, Johnson V, Kearney M, Kovacs A, Kuritzkes DR, Metzner KJ, Nissley D, Nowicki M, Ziermann R, Zhao Y, Jennings C, Bremer J, Brambilla D, Mellors J. Updated, blinded, multicenter comparison of the sensitivity of different technologies to detect and quantify a minor drug-resistant HIV-1 variant. Antiviral Therapy. 2003;8:S102. [Google Scholar]
- Eshleman SH, Guay LA, Mwatha A, Brown E, Cunningham SP, Musoke P, Mmiro F, Jackson JB. Characterization of nevirapine (NVP) resistance mutations in women with subtype A vs. D HIV-1 6–8 weeks after single dose NVP (HIVNET 012). J Acquir Immune Defic Syndr. 2004;35:126–130. doi: 10.1097/00126334-200402010-00004. [DOI] [PubMed] [Google Scholar]
- Eshleman SH, Hoover DR, Chen S, Hudelson SE, Guay LA, Mwatha A, Fiscus SA, Mmiro F, Musoke P, Jackson JB, Kumwenda N, Taha T. Nevirapine (NVP) resistance in women with HIV-1 subtype C, compared with subtypes A and D, after the administration of single-dose NVP. J Infect Dis. 2005;192:30–36. doi: 10.1086/430764. [DOI] [PubMed] [Google Scholar]
- Guay LA, Musoke P, Fleming T, Bagenda D, Allen M, Nakabiito C, Sherman J, Bakaki P, Ducar C, Deseyve M, Emel L, Mirochnick M, Fowler MG, Mofenson L, Miotti P, Dransfield K, Bray D, Mmiro F, Jackson JB. Intrapartum and neonatal single-dose nevirapine compared with zidovudine for prevention of mother-to-infant transmission of HIV-1 in Kampala, Uganda: HIVNET-012 randomised trial. Lancet. 1999;354:795–802. doi: 10.1016/S0140-6736(99)80008-7. [DOI] [PubMed] [Google Scholar]
- Jackson JB, Musoke P, Fleming T, Guay L, Bagenda D, Allen M, Nakabiito C, Sherman J, Bakaki P, Owor M, Ducar C, Deseyve M, Emel L, Mirochnick M, Fowler MG, Mofenson L, Miotti P, Gigliotti M, Bray D, Mmiro F. Intrapartum and neonatal single-dose nevirapine compared with zidovudine for prevention of mother-to-child transmission of HIV-1 in Kampala, Uganda: 18 months follow-up of the HIVNET 012 randomised trial. Lancet. 2003;362:859–868. doi: 10.1016/S0140-6736(03)14341-3. [DOI] [PubMed] [Google Scholar]
- Taha TE, Kumwenda NI, Hoover DR, Fiscus SA, Kafulafula G, Nkhoma C, Nour S, Chen S, Liomba G, Miotti PG, Broadhead RL. Nevirapine and zidovudine at birth to reduce perinatal transmission of HIV in an African setting: a randomized controlled trial. JAMA. 2004;292:202–209. doi: 10.1001/jama.292.2.202. [DOI] [PubMed] [Google Scholar]
- Shi C, Eshleman SH, Jones D, Fukushima N, Hua L, Parker AR, Yeo CJ, Hruban RH, Goggins MG, Eshleman JR. LigAmp: sensitive detection of single nucleotide differences. Nat Methods. 2004;1:141–147. doi: 10.1038/nmeth713. [DOI] [PubMed] [Google Scholar]
- Flys T, Nissley DV, Claasen CW, Jones D, Shi C, Guay LA, Musoke P, Mmiro F, Strathern JN, Jackson JB, Eshleman JR, Eshleman SH. Sensitive drug resistance assays reveal long-term persistence of HIV-1 variants with the K103N nevirapine (NVP) resistance mutation in some women and infants after single dose NVP: HIVNET 012. J Infect Dis. 2005;192:24–29. doi: 10.1086/430742. [DOI] [PubMed] [Google Scholar]
- Eshleman SH, Hackett J, Swanson P, Cunningham SP, Drews B, Brennan C, Devare SG, Zekeng L, Kaptué L, Marlowe N. Performance of the Celera Diagnostics ViroSeq HIV-1 Genotyping System v2.0 for sequence-based analysis of diverse HIV-1 strains. J Clin Micro. 2004;42:2711–2717. doi: 10.1128/JCM.42.6.2711-2717.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Eshleman SH, Brambilla DJ, Palumbo PE, Bremer JW. Evaluation of the editing process in HIV-1 genotyping assays. J Clin Micro. 2003;35:614–622. doi: 10.1128/JCM.41.7.3265-3272.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JA, Li JF, Morris L, Martinson N, Gray G, McIntyre J, Heneine W. Emergence of drug-resistant HIV-1 after intrapartum administration of single-dose nevirapine is substantially underestimated. J Infect Dis. 2005;192:16–23. doi: 10.1086/430741. [DOI] [PubMed] [Google Scholar]