Abstract
Previously we developed an oligonucleotide sequencing microarray (MitoChip) as an array-based sequencing platform for rapid and high-throughput analysis of mitochondrial DNA. The first generation MitoChip, however, was not tiled with probes for the noncoding D-loop region, a site frequently mutated in human cancers. Here we report the development of a second-generation MitoChip (v2.0) with oligonucleotide probes to sequence the entire mitochondrial genome. In addition, the MitoChip v2.0 contains redundant tiling of sequences for 500 of the most common haplotypes including single-nucleotide changes, insertions, and deletions. Sequencing results from 14 primary head and neck tumor tissues demonstrated that the v2.0 MitoChips detected a larger number of variants than the original version. Multiple coding region variants detected only in the second generation MitoChips, but not the earlier chip version, were further confirmed with conventional sequencing. Moreover, 31 variations in noncoding region were identified using MitoChips v2.0. Replicate experiments demonstrated >99.99% reproducibility in the second generation MitoChip. In seven head and neck cancer samples with matched lymphocyte DNA, the MitoChip v2.0 detected at least one cancer-associated mitochondrial mutation in four (57%) samples. These results indicate that the second generation MitoChip is a high-throughput platform for identification of mitochondrial DNA mutations in primary tumors.
Identification of sensitive and specific molecular markers for early diagnosis of cancer is an area of active research, with immense potential benefits to patients. For example, patients with head and neck squamous cell carcinoma demonstrate a 90% 5-year survival rate if detected at stage I laryngeal cancer compared to a <50% 5-year survival rate for stage IV laryngeal cancer.1 Similarly, early detection of lung cancer is necessary to improve the dismal prognosis of this lethal tumor because most patients present with advanced inoperable disease.2 A number of studies have been focused on identifying tumor-specific genetic or epigenetic alterations for diagnosis of cancer in clinical samples such as serum or other body fluids.
Analysis of mitochondrial genome alterations has recently emerged as a new molecular approach to cancer diagnosis, receiving increasing attention in the field of cancer research. The potential use and advantage of mitochondrial (mt)DNA mutations over nuclear genome-based methods for similar detection approaches is attributable to the large numbers of mutated, often homoplasmic, mitochondria found in single cancer cells.3,4,5
Mitochondrial DNA mutations have been documented to exist in a wide range of human cancers including lung, colon, breast, prostate, and head and neck cancers and so forth.3,6,7 This commonality of mitochondrial mutations in human cancers suggests that analysis of mitochondrial DNA mutation patterns may serve as a unique molecular approach to detection of all types of cancers. In addition, mitochondrial mutations are found in premalignant lesions8,9,10 and therefore offer an opportunity for early detection and screening of human cancer. Indeed, detection of mitochondrial DNA mutations, widely reported in clinical samples including body fluids and serum, possesses potential for early detection and screening of a variety of human tumors.3,11,12 A high-throughput strategy for sequencing the mitochondrial genome is therefore required to translate the potential of mitochondrial mutations as a cancer biomarker to clinical practice.
Several techniques are currently available to detect genomic variations including enzymatic cleavage reaction, denaturing high-performance liquid chromatography, and automated capillary sequencers13,14,15; however, insufficient sensitivity or cumbersome methodology has primarily limited the routine use of these assays for sequencing the entire mitochondrial genome. We have recently developed a human mitochondrial array-based sequencing platform for rapid and high-throughput analysis of mitochondrial DNA.16 This first generation of MitoChip (MitoChip v1.0) was able to sequence both strands of the entire mitochondrial coding region in a single assay, demonstrating >99.99% reproducibility. However, a major limitation of the earlier version of MitoChip was its lack of tiling oligonucleotide probes for sequencing the mitochondrial displacement loop (D-loop). The D-loop is a noncoding region in the mitochondrial DNA molecule that controls both replication and transcription. Many of the somatic mitochondrial DNA mutations of primary tumors are found in the D-loop region.17,18 Additional conventional sequencing of the D-loop region is therefore required to provide a complete mitochondrial genomic profiling of cancer samples while using the MitoChip v1.0 as a screening tool.
To realize fully a high-throughput strategy for analysis of mtDNA, we present here a new second-generation version of MitoChip, referred to as MitoChip v2.0. The major difference is that the v2.0 chips are tilled with oligonucleotide probes for the entire 16.5-kb sequence of mtDNA (Table 1). In addition, to increase sensitivity of mutation detection, redundant probes are tilled in the MitoChip v2.0 for 500 of the most common haplotypes as observed in the MitoMap public database (http://www.mitomap.org/; accessed April, 2004). The reduced feature size of 8 μmol/L confers another advantage of the v2.0 MitoChips compared to the MitoChips v1.0 because it reduces oligonucleotide synthesis costs and decreases bleed through between adjacent tiled sequences. To evaluate the second generation MitoChip, we performed a comparison study between the v1.0 and v2.0 chips. The sequencing results of 14 primary tumor tissues demonstrated comparable sensitivity in the v2.0 MitoChips with >99.99% reproducibility, as reported previously for the v1.0 array. In addition, 31 variations in the D-loop region were successfully detected in the v2.0 chips. With sequencing of paired lymphocytes, somatic mtDNA mutations were detected in four of seven head and neck tumors with the MitoChip v2.0. The second generation MitoChip has thus proved to be a high-throughput sequencing microarray for sequencing the entire mitochondrial genome for early detection and clinical screening of human tumors.
Table 1.
Design of v2.0 MitoChip
| Total double-stranded DNA sequenced per Mitochip | 16,569 bp |
|---|---|
| Control (plasmid) DNA | 980 bp |
| Mitochondrial coding sequence | 15,451 bp |
| (Includes RCRS 573 through 16,024) | |
| Mitochondrial D-loop sequence | 1118 bp |
| (Includes RCRS 16,025 through 572) |
Materials and Methods
Design of Human Mitochondrial v2.0 Oligonucleotide Microarray
The MitoChip v2.0 was obtained from Affymetrix (commercially available GeneChip Human Mitochondrial Resequencing Array 2.0; Santa Clara, CA). Sequences comprising both strands of the entire 16,568-bp human mitochondrial genome were synthesized as overlapping 25-mers on high-density oligonucleotide arrays with 8 × 8-μm features. The Cambridge Reference Sequence was tiled as well as sequences representing 500 of the most common haplotypes observed in the MitoMap public database (http://www.mitomap.org/), which include single-nucleotide changes, insertions, and deletions. The MitoChip was fabricated using standard photolithography and solid-phase DNA synthesis by Affymetrix, as described previously.19,20 To query any given site from the human mitochondrial reference sequence, four features are tiled on the MitoChip. The four features differ only by the central or 13th base, which consists of each of the four possible nucleotides.
DNA Sample Source and Preparation
The performance of the v1.0 and v2.0 MitoChips was evaluated using DNA from 14 head and neck cancer samples from 14 individual patients. Of the 14 samples, seven had matched normal (lymphocyte DNA) samples to identify cancer-associated mutations. All tumor and normal specimens were collected before surgical resections with prior consent from patients. Tumor specimens were frozen and microdissected on a cryostat so that the tumor samples contained greater than 70% neoplastic cells. DNA from tumor sections was digested with 1% sodium dodecyl sulfate/proteinase K, extracted by phenol-chloroform, and ethanol precipitated. Control DNAs from peripheral lymphocytes were processed in the same manner as described previously.21
Polymerase Chain Reaction (PCR) Amplification
The entire mitochondrial DNA sequence was amplified in three overlapping long PCR fragments, with each reaction containing 50 ng of genomic DNA. The primers for PCR amplification were the same as in the previous report for MitoChip v1.016 and were selected using the Amplify 1.2 program as described.22 Amplification was accomplished in 50-μl PCRs performed in thin-walled polypropylene plates using the high-fidelity TaKaRa LA Taq (TaKaRa Mirus Bio, Madison, WI), as described in the previous report22 with slight modifications. Specifically, all PCR reactions were optimized to perform under standard conditions without addition of 5% dimethyl sulfoxide to aid in the amplification of GC-rich regions as reported previously.22 The cycling conditions for all reactions were as follows: 1) 95°C for 2 minutes, 2) 95°C for 15 seconds, 3) 68°C for 7 minutes, 4) repeat step 2 for 29 times, 5) final extension for 12 minutes. As a control for PCR amplification and subsequent hybridization, a 7.5-kb plasmid DNA (Tag IQ-EX template) was amplified concomitantly with the test samples, using forward and reverse primers included in the CustomSeq control kit (Affymetrix, Inc.). The specificity of the reactions was confirmed by agarose gel electrophoresis. The PCR products were purified using QIAQuick PCR Clean up kit (Qiagen, Inc., Valencia, CA), and the resultant purified DNA was resuspended in 30 to 40 μl vol of EB buffer (Affymetrix, Inc.). The concentration of each purified PCR product was determined spectrophotometrically, and a yield of 100 to 200 ng/μl of PCR reactions was routinely obtained.
Fragment Pooling, DNA Fragmentation, Labeling, and Chip Hybridization
The procedures on sample pooling, DNA fragmentation, and labeling were identical for both v1.0 and v2.0 MitoChips. Specifically, to obtain optimal performance across the microarray, we pooled equimolar amounts from the three amplified fragments to ensure that an equal number of targets existed for each probe. The pooled DNA fragments were digested with DNase I for 15 minutes in a 50-μl reaction containing Affymetrix fragmentation reagent (0.2 U of DNase I/μg DNA), 5 μl of OnePhorAll buffer (Amersham Life Sciences, Arlington Heights, IL), and EB buffer. Samples were then incubated at 95°C for 15 minutes to inactivate DNase I. Fragmented DNA was labeled by adding 2.0 μl of GeneChip DNA labeling reagent and 3.4 μl of 30 U/μl terminal deoxynucleotidyl transferase (both from Affymetrix).
Prehybridization, hybridization, washing, and scanning of the MitoChip were performed as described in the Affymetrix CustomSeq Resequencing protocol. The prehybridizations were performed for 15 minutes in 80-μl (for v2.0 chips) or 200-μl (for v1.0 chips) solution containing 3 mol/L tetramethylammonium chloride, 0.1% Tween 20, and 10 mmol/L Tris, pH 7.8. The chips were hybridized for 16 hours at 48°C with 60 rpm rotation in a hybridization solution containing 3 mol/L tetramethylammonium chloride, 100 μg/ml herring sperm DNA, 500 μg/ml bovine serum albumin, 10 mmol/L Tris, pH 7.8, 0.01% Tween 20, and 200 pmol/L control oligo. The chips were then washed on the Affymetrix fluidics station using the preprogrammed CustomSeq Resequencing wash protocols.
Automated Batch Analysis of Microarray Data
The analysis of microarray data for both v1.0 and v2.0 MitoChips was done using RA tools, a modified version of the previously described adaptive background genotype-calling scheme (ABACUS);22 the open source software is available at http://www.dpgp.org/. Briefly, RA tools uses an objective statistical framework to assign each genotype call a quality score, which is the difference between the log (base 10) likelihood of the best fitting and the second best fitting statistical model for assigning a genotype at any position on the sequencing array. The total quality score threshold (totThresh) is the quality score that a given base has to exceed to be called. Increasing this value requires increased support for base calls and, as a consequence, fewer bases are called. Bases that fail to reach this threshold are called “N.” The optimum total threshold quality score was determined empirically to be 12, which yields the highest base call rate with the lowest discrepancy between genotypes for replicate samples (see below). The original ABACUS algorithm and the RA tools version has been successfully applied for high-throughput variation detection in human and pathogenic organisms,22,23 as well as for detection of mitochondrial sequence variations in human embryonic stem cells.24
Conventional Dye Terminator Sequencing of Mitochondrial DNA
To confirm a subset of variations identified in coding region by the v2.0 MitoChip, six pairs of primers were designed to amplify mitochondrial segments. The forward and reverse primers were respectively 5′-CGATCAAAAGGGACAAGCAT-3′, and 5′-GGTTTGGGGCTAGGTTTAG-3′, for verifying site 921; 5′-GCTAAGACCCAAACTGGGATT-3, and 5′-GGCCCTGTTCAACTAAGCAC-3′, for sites 1189 and 1393; 5′-TGACCGCTCTGAGCTAAACC, and 5′-CTACCTTTGCACGGTTAGGG-3′ for site 1811; 5′-AACATCACCTCTAGCATCACCA-3′, and 5′-CGTCAGCGAAGGGTTGTAGT-3′ for site 3196; 5′-AGCATTCCCCCTCAAACCTA-3′, and 5′-GAGAGGAGGGTGGATGGAAT-3′ for site 4792; and 5′-CCATCCCTACGCATCCTTTA-3′, and 5′-TCCGAGGAGGTTAGTTGTGG-3′ for site 8392. To confirm variations identified in D-loop region, two overlapping fragments of 613 and 679 bp were PCR amplified and sequenced using primers reported previously.25 The purified DNA products were then sequenced with dye terminator platform using the ABI BigDye cycle sequencing kit (Applied Biosystems, Foster City, CA).
Results
Total Mitochondrial Base Calls
Samples used in the present study included 14 head and neck tumor DNA and seven matched lymphocyte DNA samples. The entire mitochondrial genome was amplified with three PCR reactions. To simplify the experiments, we modified the PCR conditions used previously16 and obtained an optimal PCR program for all of the reactions as described in the Materials and Methods. In addition, we reduced the total amount of DNA to 150 ng for the amplification of the entire mitochondrial genome. With this DNA we performed 47 MitoChip assays. Of the 47 assays, 14 tumor DNAs were sequenced with v1.0 MitoChip assays, the same DNA was also applied to v2.0 MitoChips for the purpose of comparison. Four samples selected from the original 14 samples were repeated with v2.0 MitoChips to examine the reproducibility of v2.0 chips. Each sample was repeated three times, thus a total of 12 comparisons was made to estimate the reproducibility of the v2.0 MitoChips. Finally, seven matched lymphocyte DNA samples (serving as normal control) were sequenced with the v2.0 MitoChips to identify the mutations. The same procedures for DNA fragmentation and hybridization were used for both v1.0 and 2.0 MitoChips. A different washing program was applied to the two versions of the MitoChips because of the different designs. Using a totThresh of 12, RA tools assigned a mean base call 96.8% (ranging from 95.8 to 97.8%) in v1.0 chips. For v2.0 chips the overall call rate across all of the chips was 94.6% (ranging from 89.2 to 96.8%) (Table 2). Although most chips exhibited a call rate ∼95%, one v2.0 MitoChip recalled 89.2%. Although the recall rate was slightly lower in v2.0 MitoChips compared with v1.0 chips, no significant differences were found in the mitochondrial base call rates between the v1.0 and v2.0 MitoChips.
Table 2.
Summary of v1.0 and v2.0 MitoChip Assays
| Total MitoChips assays | 47 |
| Total v1.0 MitoChip assays | 14 |
| Total mitochondrial DNA sequenced by v1.0 chips | 15,451*14 = 216,314 bp |
| Total percentage bases called in v1.0 chips (%) | 96.8 |
| Total v2.0 MitoChips assays | 33 |
| Total mitochondrial DNA sequenced by v2.0 chips | 16,569*33 = 546,777 bp |
| Total percentage bases called in v2.0 chips (%) | 94.6 |
Sensitivity of mtDNA Variation Detection between the v1.0 and v2.0 MitoChips
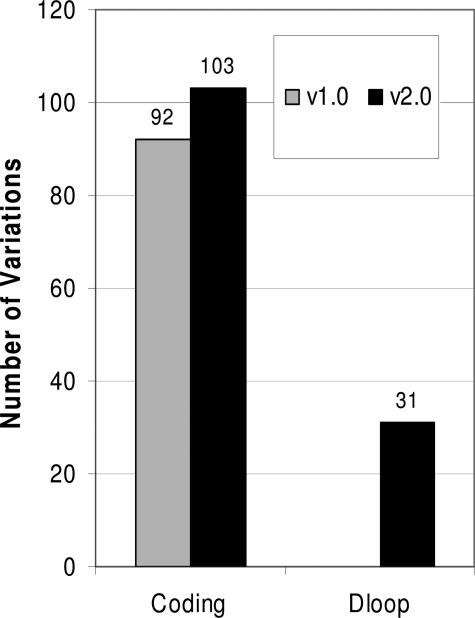
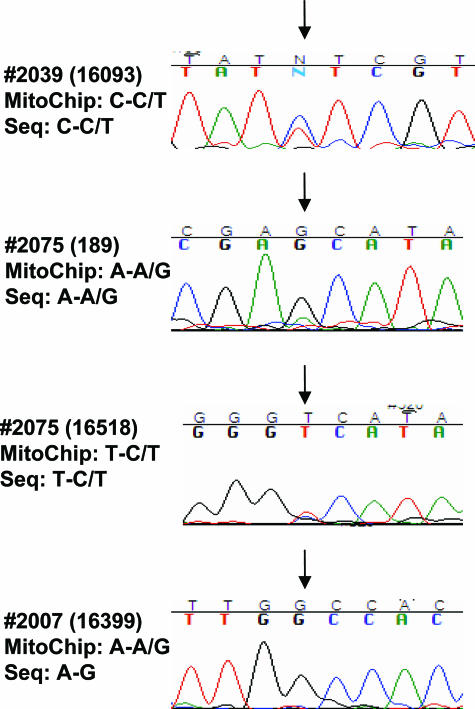
To evaluate the application of the v2.0 MitoChips, we first compared the sensitivity of variation detection between the v1.0 and v2.0 chips. Independent aliquots of the same PCR products were used for hybridization to both v1.0 and v2.0 MitoChips. For the comparison purposes, the use of the same PCR products was necessary to rule out any possibility of PCR-induced artifacts, although the polymerase used is known to have extremely high fidelity. Sequencing variants were identified by comparing to mitochondrial reference sequence (http://www.mitomap.org). A difference in sequence when compared to the reference sequence was referred to as variation. When a variation was present in matched normal tissues but absent in tumors, we termed these variations somatic mutations. Of the 120 variations, 75 were detected by both v1.0 and v2.0 MitoChips, and 17 variations were detected only in MitoChip v1.0, and the other 28 detected by MitoChip v2.0 only (Figure 1). From these comparison experiments the v2.0 MitoChips seem to exhibit a higher detection sensitivity. A question arises whether the variations detected in the v2.0 only were truly present or were an artifact of array-based sequencing. Since the MitoChip v1.0 has been evaluated before,16 the 75 variations detected by both types of MitoChips should be real. Therefore, we randomly selected eight variations of the 28 variations only detected by v2.0 MitoChips and sequenced these by conventional dye terminator platform. All of the eight variations were confirmed in the conventional sequencing with 100% accuracy (Table 3). It is worthy to note that 17 variations identified in v1.0 went undetected in v2.0 chips. This was mainly attributable to lack of ability to call some nucleotide positions in v2.0 chips (call rate of 94.2). As mentioned above, a new feature of the v2.0 MitoChip is its tiling of probes for detecting variations in the mitochondrial D-loop region. Indeed, 31 variations were detected in the v2.0 MitoChips (Figure 1). Of the 31 variations, 20 were homoplasmic and 11 were heteroplasmic. The criteria for the identification of heteroplasmic variations was based on the ability of the RA sequencing software to make a reliable call for heteroplasmy, using preset threshold values. Thus, this MitoChip may be able to detect even low-level heteroplasmy. We sequenced all these 31 variations detected in D-loop region. Nineteen of twenty homoplasmic variations were confirmed by conventional dye terminator platform. Five of eleven heteroplasmic variations were confirmed, and the remaining six appeared to be homoplasmic variations. It has been known that the MitoChip possesses higher sensitivity compared to the conventional sequencing assay. As showed in Figure 2, the heteroplasmic variation (C-C/T) in sample 2039 is clearly displayed in sequencing chromatogram. By examining the chromatograms two heteroplasmic variations were found to exist in sample 2075. However, in sample 2007, conventional sequencing detected a homoplasmic variation (A-G) rather than heteroplasmic variation (A-A/G) identified in MitoChip (Figure 2). It is likely that this heteroplasmic variation is real but cannot be detected by conventional sequencing assay because of its low sensitivity. Further studies including dilution experiments may be needed to fully evaluate the ability of MitoChip v2.0 to detect heteroplasmic variations.
Figure 1.
Comparison of detection of mitochondrial DNA variations by v1.0 and v2.0 MitoChips. A total of 14 tumor samples were sequenced with v1.0 and v2.0 chips, respectively. Variations were obtained by comparing the sequencing results to mitochondrial DNA reference from MitoMap database. The variations for coding region and D-loop part are presented separately.
Table 3.
Confirmation of Mitochondrial DNA Variations by Conventional Sequencing
| Sample code | Variation site | Variation (normal-variation) | Confirmation |
|---|---|---|---|
| 1759 | 4791 | A-G | Yes |
| 1759 | 1393 | G-A | Yes |
| 2007 | 3197 | T-C | Yes |
| 2051 | 921 | T-C | Yes |
| 2039 | 1189 | T-C | Yes |
| 2039 | 1811 | A-G | Yes |
| 2188 | 3197 | T-C | Yes |
| 2195 | 8393 | C-T | Yes |
Figure 2.
Chromatograms showing conventional sequencing of D-loop in representative samples. Arrows indicate the variation sites identified in MitoChips. The corresponding sequencing results of MitoChip and conventional assay are presented to the left of the corresponding chromatograms. Nucleotide sites are specified in parentheses.
Reproducibility of Array-Based Sequencing in v2.0 MitoChips
We used replicate experiments, consisting of independent amplifications of identical samples followed by hybridization to distinct microarrays of the same design, to determine the reproducibility of RA tools genotype calls at the totThresh of 12. To evaluate the quality we compared the number of bases called in common between the replicates and the fraction of bases called discordantly. Among the 182,997 bp called in the replicate experiments, 12 base sites were called discordantly (Table 4). This represents a between chip error rate of 0.00328%. The extremely low-level genotyping error in the replicate experiments represents >99.99% reproducibility of base calls between chips using array-based sequencing, which indicates the reliability of the second generation MitoChips.
Table 4.
v2.0 MitoChip Repeatability in Replicate Experiments
| Total comparisons in the replicate experiment | 12 |
| Total number of sites assigned in the replicate experiment | 182,997 bp |
| Total number of sites assigned differently | 12 bp |
| The overall call rate for the replicate chips | 94.6% |
| Percentage of sites assigned identically | 99.997% |
| (12/2*182,997 = 0.0000328) |
Identification of Mitochondrial Mutations in Both Coding and D-Loop Regions
The purpose of this comparison study is to evaluate the v2.0 MitoChip for its use in identifying mutations in clinical samples. Therefore, we analyzed a subset of seven paired primary tumor and lymphocyte DNA (as normal control) to determine whether the MitoChip v2.0 could identify somatic mutations. The seven primary samples were randomly selected from those head and neck tumor DNAs used in the comparison experiments. The final average base call rates for the DNA obtained from the matched lymphocytes were essentially identical to that of the primary tumor DNA (between 94 to 95% call rate), demonstrating that DNA obtained from these clinical samples is feasible for use in chip-based assays. Overall, four of seven (57%) head and neck tumor DNAs demonstrated multiple mutations. An important improvement of the v2.0 MitoChips over the v1.0 MitoChips is the inclusion of tiling probe for mitochondrial D-loop. Indeed, three of the four samples with mutations were found to carry mutations in the D-loop. In tumor sample no. 2007, 15 mutations were detected, four of which were in the D-loop. In addition, two were heteroplasmic mutations (Table 5). To exclude cross-contamination of specimens, we examined mutant sequences in a cohort of 83 head and neck tumors samples and noted that all mutant sequences were unique (unpublished data), indicating lack of cross contamination.
Table 5.
MtDNA Mutations in Tumor Sample No. 2007 Detected with v2.0 Chip
| Nucleotide position | Gene | DNA (N→T) | Protein |
|---|---|---|---|
| 73 | D-loop | A-G | |
| 16,192 | D-loop | C-T | |
| 16,270 | D-loop | C-T | |
| 16,399 | D-loop | A-A+G | |
| 2706 | 16sRNA | A-G | |
| 3197 | 16sRNA | T-C | |
| 7028 | COI | C-T | G-G |
| 9342 | COIII | G-G+A | G-S |
| 9477 | COIII | G-A | V-I |
| 11,467 | ND4 | A-G | L-L |
| 12,308 | tRNA | A-G | |
| 12,372 | ND5 | G-A | L-L |
| 13,617 | ND5 | T-C | I-I |
| 14,793 | Cyt B | A-G | H-R |
| 15,218 | Cyt B | A-G | T-A |
Discussion
Mitochondrial alterations have long been suspected as contributors to carcinogenesis.26 This hypothesis has been supported by recent studies that demonstrate positive contribution of mitochondrial mutations in the tumor development.27,28 More importantly, accumulating data on mitochondria suggest that mitochondrial DNA mutations play a wide range of roles in cancer development and can be used as biomarkers to both diagnosis and prognosis of human cancers. Thus, development of rapid and reliable approaches to sequence mitochondrial DNA has become an important tool. Microarray-based resequencing technology is one of the current existing approaches for identifying unknown DNA variations, and has displayed higher sensitivity and automation capabilities compared to conventional sequencing technologies.21,29 We have previously developed oligonucleotide-based sequencing microarray and used them to successfully identify mtDNA mutations in human samples.16 The purpose of this study was to develop a microarray platform for high-throughput analysis of the entire mitochondrial DNA sequence for identification of mutations in clinical samples. An important improvement of the redesigned second generation MitoChips is the tiling of the entire mitochondrial DNA (16,568 bp), thus allowing the sequencing of the entire mitochondrial genome on a single chip. In addition, redundant probes are tiled in the v2.0 MitoChip for 500 of the most common haplotypes in the mitochondrial sequence, which markedly increases the capacity for mutation detection. Moreover, we have made several modifications to optimize the whole procedure in the preparation of samples. These include 1) using same PCR program for all three reactions to amplify entire mtDNA, 2) using 150 ng of genomic DNA rather than 300 ng in the PCR reactions, and 3) reducing filling volume of chips. These improvements have greatly simplified the procedure and helped save samples and reagents.
Previous studies demonstrated that the MitoChips v1.0 exhibit higher sensitivity compared to other sequencing assays.16,30 All of the 14 tumor samples were found to contain mtDNA variations with both v1.0 and v2.0 MitoChips. Analysis of coding region showed that the v2.0 MitoChips detected 11 more variations than the first generation MitoChips in the coding region (Table 3). The increased sensitivity might be attributable to the redundant tiling of probes in the highly variable region of mitochondrial sequence. This increased sensitivity is of particular importance for the use of the MitoChips to the clinical purpose of diagnosis and prognosis of cancers. A limitation of the MitoChip v1.0 is its inability to detect variations in mitochondrial D-loop region. The D-loop region functions as a promoter for mitochondrial DNA, and alterations in this region may affect the transcription of mitochondrial genes. Accumulating data indicate the D-loop is a region frequently harboring mtDNA mutations in cancers.3 A recent report demonstrates that mitochondrial D-loop mutations are associated with low survival rate of colon cancers.31 Therefore, sequencing the D-loop part is necessary to explore biomarkers for early detection and prognosis of human tumors. To realize fully the high-throughput purpose, the redesigned MitoChip v2.0 includes probes for mitochondrial D-loop. Although preliminary, the data presented in the present study is promising in identification of variations in D-loop with v2.0 MitoChips. Further dilution experiments may prove the MitoChip v2.0 to be an excellent platform for identification of mtDNA variations present as a minority of analyzed DNA sample.
Like other sequencing technologies, accuracy is an important concern in mitochondrial DNA resequencing. With the MitoChip v1.0 we achieved a high reproducibility of >99.99%. In the present study an equivalent reproducibility was obtained with the second generation MitoChips when compared to the v1.0 chips (Table 4). Another important improvement in the present study is the reduction of total DNA amount to 150 ng for PCR amplification. This is particularly important in applications in which the amount of available DNA is limited.
Using the MitoChip v2.0 arrays we sequenced seven head and neck tumors. Four of the seven tumors were found to contain at least one somatic mutation. More importantly, three of the four tumors were found to contain D-loop mutations. With its high sensitivity and reproducibility, the v2.0 MitoChip may also be applied to screening of population for tumor and other genetic diseases. In addition, short amplicon primers for application to paraffin tissues are being developed. This will have application to forensic samples fixed in formalin or embedded in paraffin, thus expand the use of MitoChips.
Although extensive studies report on the identification of mitochondrial DNA mutations in a wide range of human cancers, the exact role of mitochondrial DNA mutations in tumor development and progression has not been established. Development of array-based technologies to detect mitochondrial genetic alterations may provide an opportunity for large-scale analysis of mitochondrial mutations in human cancers.
In conclusion, the redesigned MitoChip v2.0 arrays are able to resequence the entire mitochondrial genome. More importantly, the MitoChip v2.0 arrays express a higher sensitivity compared to the first generation arrays. Our study thus shows that the MitoChip v2.0 can be a high-throughput tool for sequencing the entire mitochondrial genome for early detection and clinical screening of human tumors.
Footnotes
Supported by the National Institute of Dental and Craniofacial Research (grants 1R01DE015939-01 to J.C. and 1R21CA107858-01 to A.M.), the Damon Runyon Cancer Research Foundation (Lilly clinical investigator CI-#9 to J.C.), the Flight Attendant Medical Research Institute (clinical innovator award to J.C.), the Maryland Cigarette Restitution Fund (to A.M.), the Sol Goldman Pancreatic Cancer Research Center (to A.M.), and the Michael Rolfe Foundation for Pancreatic Cancer Research (to A.M.).
References
- Forastiere A, Koch W, Trotti A, Sidransky D. Head and neck cancer. N Engl J Med. 2001;345:1890–1900. doi: 10.1056/NEJMra001375. [DOI] [PubMed] [Google Scholar]
- Frisch SM, Screaton RA. Anoikis mechanisms. Curr Opin Cell Biol. 2001;13:555–562. doi: 10.1016/s0955-0674(00)00251-9. [DOI] [PubMed] [Google Scholar]
- Fliss MS, Usadel H, Caballero OL, Wu L, Buta MR, Eleff SM, Jen J, Sidransky D. Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science. 2000;287:2017–2019. doi: 10.1126/science.287.5460.2017. [DOI] [PubMed] [Google Scholar]
- Polyak K, Li Y, Zhu H, Lengauer C, Willson JK, Markowitz SD, Trush MA, Kinzler KW, Vogelstein B. Somatic mutations of the mitochondrial genome in human colorectal tumours. Nat Genet. 1998;20:291–293. doi: 10.1038/3108. [DOI] [PubMed] [Google Scholar]
- Copeland WC, Wachsman JT, Johnson FM, Penta JS. Mitochondrial DNA alterations in cancer. Cancer Invest. 2002;20:557–569. doi: 10.1081/cnv-120002155. [DOI] [PubMed] [Google Scholar]
- Mydlo JH, Gerstein M. Patients with urologic cancer and other nonurologic malignancies: analysis of a sample and review of the literature. Urology. 2001;58:864–869. doi: 10.1016/s0090-4295(01)01394-2. [DOI] [PubMed] [Google Scholar]
- Herbst RS, Onn A, Sandler A. Angiogenesis and lung cancer: prognostic and therapeutic implications. J Clin Oncol. 2005;23:3243–3256. doi: 10.1200/JCO.2005.18.853. [DOI] [PubMed] [Google Scholar]
- Ha PK, Tong BC, Westra WH, Sanchez-Cespedes M, Parrella P, Zahurak M, Sidransky D, Califano JA. Mitochondrial C-tract alteration in premalignant lesions of the head and neck: a marker for progression and clonal proliferation. Clin Cancer Res. 2002;8:2260–2265. [PubMed] [Google Scholar]
- Jeronimo C, Nomoto S, Caballero OL, Usadel H, Henrique R, Varzim G, Oliveira J, Lopes C, Fliss MS, Sidransky D. Mitochondrial mutations in early stage prostate cancer and bodily fluids. Oncogene. 2001;20:5195–5198. doi: 10.1038/sj.onc.1204646. [DOI] [PubMed] [Google Scholar]
- Parrella P, Xiao Y, Fliss M, Sanchez-Cespedes M, Mazzarelli P, Rinaldi M, Nicol T, Gabrielson E, Cuomo C, Cohen D, Pandit S, Spencer M, Rabitti C, Fazio VM, Sidransky D. Detection of mitochondrial DNA mutations in primary breast cancer and fine-needle aspirates. Cancer Res. 2001;61:7623–7626. [PubMed] [Google Scholar]
- Nomoto S, Yamashita K, Koshikawa K, Nakao A, Sidransky D. Mitochondrial D-loop mutations as clonal markers in multicentric hepatocellular carcinoma and plasma. Clin Cancer Res. 2002;8:481–487. [PubMed] [Google Scholar]
- Okochi O, Hibi K, Uemura T, Inoue S, Takeda S, Kaneko T, Nakao A. Detection of mitochondrial DNA alterations in the serum of hepatocellular carcinoma patients. Clin Cancer Res. 2002;8:2875–2878. [PubMed] [Google Scholar]
- Liu MR, Pan KF, Li ZF, Wang Y, Deng DJ, Zhang L, Lu YY. Rapid screening mitochondrial DNA mutation by using denaturing high-performance liquid chromatography. World J Gastroenterol. 2002;8:426–430. doi: 10.3748/wjg.v8.i3.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medintz IL, Paegel BM, Blazej RG, Emrich CA, Berti L, Scherer JR, Mathies RA. High-performance genetic analysis using microfabricated capillary array electrophoresis microplates. Electrophoresis. 2001;22:3845–3856. doi: 10.1002/1522-2683(200110)22:18<3845::AID-ELPS3845>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Otway R, Tetlow N, Hornby J, Kohonen-Corish M. Evaluation of enzymatic mutation detection trade mark in hereditary nonpolyposis colorectal cancer. Hum Mutat. 2000;16:61–67. doi: 10.1002/1098-1004(200007)16:1<61::AID-HUMU11>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Maitra A, Cohen Y, Gillespie SE, Mambo E, Fukushima N, Hoque MO, Shah N, Goggins M, Califano J, Sidransky D, Chakravarti A. The human MitoChip: a high-throughput sequencing microarray for mitochondrial mutation detection. Genome Res. 2004;14:812–819. doi: 10.1101/gr.2228504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Cespedes M, Parrella P, Nomoto S, Cohen D, Xiao Y, Esteller M, Jeronimo C, Jordan RC, Nicol T, Koch WM, Schoenberg M, Mazzarelli P, Fazio VM, Sidransky D. Identification of a mononucleotide repeat as a major target for mitochondrial DNA alterations in human tumors. Cancer Res. 2001;61:7015–7019. [PubMed] [Google Scholar]
- Tamori A, Nishiguchi S, Nishikawa M, Kubo S, Koh N, Hirohashi K, Shiomi S, Inoue M. Correlation between clinical characteristics and mitochondrial D-loop DNA mutations in hepatocellular carcinoma. J Gastroenterol. 2004;39:1063–1068. doi: 10.1007/s00535-004-1445-3. [DOI] [PubMed] [Google Scholar]
- Pease AC, Solas D, Sullivan EJ, Cronin MT, Holmes CP, Fodor SP. Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proc Natl Acad Sci USA. 1994;91:5022–5026. doi: 10.1073/pnas.91.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipshutz RJ, Fodor SP, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nat Genet. 1999;21:20–24. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- Ahrendt SA, Halachmi S, Chow JT, Wu L, Halachmi N, Yang SC, Wehage S, Jen J, Sidransky D. Rapid p53 sequence analysis in primary lung cancer using an oligonucleotide probe array. Proc Natl Acad Sci USA. 1999;96:7382–7387. doi: 10.1073/pnas.96.13.7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler DJ, Zwick ME, Carrasquillo MM, Yohn CT, Tobin KP, Kashuk C, Mathews DJ, Shah NA, Eichler EE, Warrington JA, Chakravarti A. High-throughput variation detection and genotyping using microarrays. Genome Res. 2001;11:1913–1925. doi: 10.1101/gr.197201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CW, Albert TJ, Vega VB, Norton JE, Cutler DJ, Richmond TA, Stanton LW, Liu ET, Miller LD. Tracking the evolution of the SARS coronavirus using high-throughput, high-density resequencing arrays. Genome Res. 2004;14:398–405. doi: 10.1101/gr.2141004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra A, Arking DE, Shivapurkar N, Ikeda M, Stastny V, Kassauei K, Sui G, Cutler DJ, Liu Y, Brimble SN, Noaksson K, Hyllner J, Schulz TC, Zeng X, Freed WJ, Crook J, Abraham S, Colman A, Sartipy P, Matsui S, Carpenter M, Gazdar AF, Rao M, Chakravarti A. Genomic alterations in cultured human embryonic stem cells. Nat Genet. 2005;37:1099–1103. doi: 10.1038/ng1631. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Toyooka S, Miyajima K, Iizasa T, Fujisawa T, Bekele NB, Gazdar AF. Alterations in the mitochondrial displacement loop in lung cancers. Clin Cancer Res. 2003;9:5636–5641. [PubMed] [Google Scholar]
- Schumacher HR, Szekely IE, Patel SB, Fisher DR. Mitochondria: a clue to oncogenesis? Lancet. 1973;2:327. doi: 10.1016/s0140-6736(73)90836-2. [DOI] [PubMed] [Google Scholar]
- Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, Hall J, Lim S, Issa MM, Flanders WD, Hosseini SH, Marshall FF, Wallace DC. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci USA. 2005;102:719–724. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shidara Y, Yamagata K, Kanamori T, Nakano K, Kwong JQ, Manfredi G, Oda H, Ohta S. Positive contribution of pathogenic mutations in the mitochondrial genome to the promotion of cancer by prevention from apoptosis. Cancer Res. 2005;65:1655–1663. doi: 10.1158/0008-5472.CAN-04-2012. [DOI] [PubMed] [Google Scholar]
- Hacia JG, Sun B, Hunt N, Edgemon K, Mosbrook D, Robbins C, Fodor SP, Tagle DA, Collins FS. Strategies for mutational analysis of the large multiexon ATM gene using high-density oligonucleotide arrays. Genome Res. 1998;8:1245–1258. doi: 10.1101/gr.8.12.1245. [DOI] [PubMed] [Google Scholar]
- Jakupciak JP, Wang W, Markowitz ME, Ally D, Coble M, Srivastava S, Maitra A, Barker PE, Sidransky D, O’Connell CD. Mitochondrial DNA as a cancer biomarker. J Mol Diagn. 2005;7:258–267. doi: 10.1016/S1525-1578(10)60553-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lievre A, Chapusot C, Bouvier AM, Zinzindohoue F, Piard F, Roignot P, Arnould L, Beaune P, Faivre J, Laurent-Puig P. Clinical value of mitochondrial mutations in colorectal cancer. J Clin Oncol. 2005;23:3517–3525. doi: 10.1200/JCO.2005.07.044. [DOI] [PubMed] [Google Scholar]