Abstract
Most gastrointestinal stromal tumors (GISTs) carry activating mutations of the KIT gene encoding the receptor tyrosine kinase KIT. In a previous study we were able to show an association between the lack of KIT mutations (wild-type GISTs) and the presence of a significant epithelioid tumor component. A very recent study described the occurrence of PDGFRα mutations in KIT wt GISTS. Therefore, we studied a panel of 87 GISTs for mutations in the hot spot regions of the PDGFRα gene with single strand conformation polymorphism analysis and sequencing and correlated the PDGFRα status with pathomorphological data. We detected 20 cases with exon 18 mutations but none with exon 12 mutations. The mutations were located in the second kinase domain of PDGFRα with 16 point mutations, and four larger deletions of 9 to 12 bp. All cases with mutations in the PDGFRα gene revealed wild-type KIT in common regions of mutation, ie, exons 9 and 11. Most interestingly, the occurrence of PDGFRα mutations was significantly associated with a higher frequency of epithelioid or mixed morphology (18 of 20 cases, P < 0.0001) and gastric location (all cases, P = 0.0008). Our data indicate that GISTs represent distinctive entities, differing in genetic, biological, and morphological features.
Gastrointestinal stromal tumors (GISTs) are the most frequent mesenchymal tumors in the digestive tract.1 They are characterized by the expression of the type III receptor tyrosine kinase KIT encoded by the KIT proto-oncogene.2 The latter carries gain-of-function mutations in the majority of cases leading to a ligand-independent autoactivation of the KIT receptor.3,4,5,6 However, a subset of GISTs is lacking any KIT mutations which is particularly critical as these tumors may be less sensitive to treatment with imatinib (Glivec), a tyrosine kinase inhibitor,7 than KIT mutation-positive tumors.
Very recently, activating mutations of the platelet-derived growth factor receptor α (PDGFRα) gene were described in a subset of KIT wild-type GISTs (wt GISTs).8,9 PDGFRα is a member of the subfamily of type III receptor tyrosine kinases, which includes KIT receptor, PDGF receptor β, FLK-3, and CSF-1 receptor. All members are characterized by high sequence homologies especially in the juxtamembranous (JM) and the tyrosine kinase (TK) domains. KIT mutations in GISTs are preferentially found in exon 11 encoding the JM domain, less often in exon 9 (extracellular domain) and rarely in exon 13 and 17 (TK domains).3,6,10,11 According to the first description of Heinrich et al8 PDGFRα mutations seem to cluster in analogous regions known for KIT mutations with exon 12 mutations in the JM domain and exon 18 mutations in the TK domain.
In the present study, we investigated the occurrence of PDGFRα mutations in 87 GISTs and compared 41 tumors with known KIT mutations with 46 GISTs without detectable KIT mutations in exons 9 or 11. We found PDGFRα mutations in 20 cases (43.5% in the group of wt GISTs), all of them without KIT mutations in the most frequently mutated exons 11 and 9. None of the GISTs with KIT mutations carried PDGFRα mutations. We evaluated clinicopathological data, histomorphological subtypes, and immunohistochemical expression patterns for PDGFRα and KIT receptor and compared PDGFRα-mutation-positive GISTs, KIT mutation-positive tumors and those without detectable KIT or PDGFRα mutations.
Materials and Methods
Tissues and Clinical Data
In 87 cases from the files of the Department of Pathology, University of Bonn Medical Center, including 43 cases sent from other institutions for reference opinion, DNA was extracted from formalin-fixed, paraffin-embedded tissue for mutational analysis. KIT mutational status has been published in part previously.3,12
Criteria for GIST Diagnosis and Classification
GIST diagnosis was confirmed by immunohistochemical analysis using antibodies against CD117 (KIT receptor), CD34, bcl-2, α-actin, desmin, S-100 protein, vimentin (all DAKO, Hamburg, Germany), and Ki-67 (MIB-1, Dianova, Hamburg, Germany) as previously described.3 Additionally, PDGFRα-expression was evaluated using a monoclonal antibody (Santa Cruz Biotechnologies, Santa Cruz, CA, USA; C-20, dilution 1:50). Specificity of the antibody against PDGFRα was controlled by peptide blocking (Santa Cruz Biotechnologies; blocking peptide, sc-338 P) and by Western blot analysis showing a specific band of approximately 185 kd (not shown). Immunohistochemical results were assessed in a semi-quantitative manner using the categories strong, intermediate, weak, or negative. The categories were defined as follows: strong, strong or intermediate positivity in more than 75% of tumor cells; intermediate, strong or intermediate positivity in more than 10% of tumor cells or weak positivity in more than 75% of tumor cells; weak, any positivity in less than 10% of tumor cells; and negative, no positivity. Proliferative activity was evaluated by counting mitoses per 50 high-power fields (HPFs). MIB1-index was determined by counting stained nuclei in 1000 tumor cells and is given in %.
Histomorphologically, GISTs were subtyped according to Fletcher et al13 into three categories: spindle cell type, epithelioid type, or mixed type. Potential risk for aggressive behavior was evaluated according to Fletcher et al (Table 1).13
Table 1.
Risk Assessment
| Tumor size | Mitotic count | |
|---|---|---|
| Very low risk | < 2 cm | <5/50 HPFs |
| Low risk | 2–5 cm | <5/50 HPFs |
| Intermediate risk | < 5 cm | 6–10/50 HPFs |
| 5–10 cm | <5/50 HPFs | |
| High risk | > 5 cm | >5/50 HPFs |
| >10 cm | any mitotic rate | |
| any size | >10/50 HPFs |
Data according to Fletcher et al.13
Cases without samples from the primary tumor, in which only metastases were evaluated, were excluded from risk assessment (three cases with KIT mutations, two cases without detectable mutation in KIT or the PDGFRα gene).
Analysis of PDGFRα Mutations in Exons 12 and 18 and KIT Mutations in Exons 9 and 11
For PDGFRα mutational analysis, tumor tissue for DNA extraction was marked on H&E-stained slides and microdissected from serial sections (10 μm). Tissue slides were deparaffinized by xylene. Total DNA was extracted after pretreatment with proteinase K and absorption on silica-gel-membranes (Qiagen, Hilden, Germany) and analyzed by single strand conformational polymorphism analysis (SSCP). Therefore, intronic PCR primers were designed to amplify exons 12 and 18. PDGFRα DNA was amplified by PCR using the following primers: exon 12A forward: 5′-ttcaccagttacctgtcctg-3′ and reverse: 3′-ccatctgggctgattgattc-5′, product size 84 bp; exon 12B forward: 5′-gaatcaatcagcccagatgg-3′ and reverse: 3′-accaagcactagtccatctc-5′, product size 102 bp; exon 18 forward: 5′-cttttccatgcagtgtgtcc-3′ and reverse: 3′-cactgcctttcgacacatag-‘5, product size 137 bp.
PCR was performed in 10-μl reactions containing 1.0 μl DNA, 10 mmol/L Tris-HCl (pH 8.3), 40 mmol/L KCl, 1.0 to 1.5 mmol/L MgCl2, 200 mmol/L of each dNTP, 20 pM of each primer, and 0.25 U Platinum Taq polymerase (Life Technologies, Invitrogen GmbH, Karlsruhe, Germany). PCR reaction was carried out on an Uno II Thermoblock (Biometra, Göttingen, Germany). Initial denaturation at 94°C for 3 minutes was followed by 41 cycles and a final extension step (5 minutes at 72°C). The cycles included denaturation at 94°C for 40 seconds, annealing at 55 to 57°C for 40 seconds, and extension at 72°C for 35 seconds. PCR products were diluted with formamide, denaturated at 94°C for 10 minutes, and single strands were separated on polyacrylamide gels under two different conditions. Single and double strands of the PCR products were visualized by silver staining as described previously.14 DNA single strand bands showing an altered mobility in comparison to reference products were excised from the wet gel. DNA was eluted in H2O for 2 hours at 50°C, precipitated by centrifugation at 12000 × g for 30 minutes and re-amplified. The products were purified using spin columns (QIAquick PCR Purification kit, Qiagen). Cycle sequencing (ABI PRISM Dye Terminator Sequencing Ready Reaction kit, Applied Biosystems, Weiterstadt, Germany) was done on a TC 9600 thermocycler (Perkin Elmer, Rodgau-Jügesheim, Germany) with 20 ng of PCR product as template according to the protocol of the manufacturer. The sequencing products were separated on an 6%, 1:19 bisacrylamide:acrylamide gel on an ABI 373A sequencer (Applied Biosystems). All sequence alterations were confirmed by an independent PCR amplification followed by SSCP, re-amplification, and sequencing to exclude PCR artifacts. Analysis of KIT mutations in exons 9 and 11 was performed as previously described.3,12
Sample Composition
We evaluated all available GISTs lacking KIT mutations in a larger series and compared them with a defined subset of previously described GISTs3,12 with known KIT mutational status. Therefore, the data do not represent the incidence of specific mutations in an unselected cohort of GISTs.
Statistics
Statistical analysis was carried out using a commercially available computer program (SAS for Windows, release 8.01, SAS Institute Inc., Cary, NC, USA). For comparison of frequency counts the Chi-square test or the Fisher’s exact test were used, where appropriate. Correlations of quantitative variables were assessed by the method of Spearman. Logistic regression was used to investigate a possible influence of covariates on binominal or ordinal parameters. In general, backward elimination with a covariate removal criteria of P = 0.05 was used.
Results
PDGFRα Mutations
In 20 of 46 (43.5%) GISTs without detectable KIT mutation in exons 9 or 11, shifts in the SSCP for exon 18 of the PDGFRα gene were observed in comparison with the control (blood of a healthy person), whereas no SSCP shifts were detected for exon 12. None of the tumors with known KIT mutation showed SSCP shifts in exon 12 or 18 of PDGFRα.
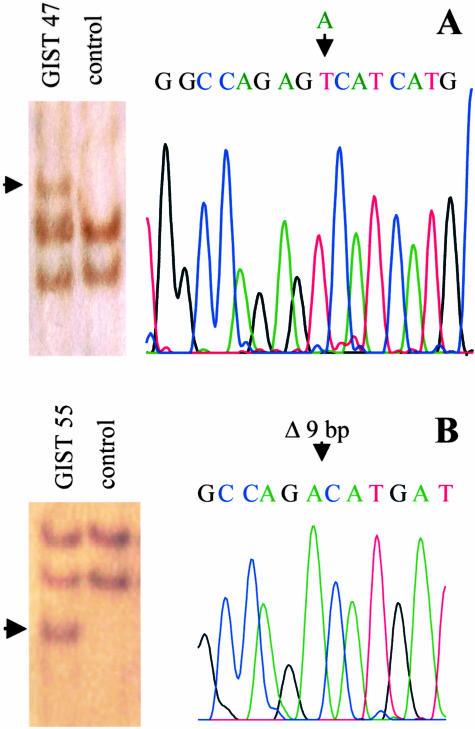
All tumors were sequenced on both strands of exon 12 and 18 PDGFRα gene independently of the detection of SSCP shifts. Whereas all cases with altered bands in the SSCP showed mutations in exon 18, none showed mutations in exon 12. Three cases (numbers 42, 46, and 57) carried a 12 bp-deletion in codons 843 to 846 resulting in the loss of the amino acids isoleucine, methionine, histidine, and asparagine. Sixteen cases (numbers 43–45, 47–54, 56, and 58–61) showed a point mutation in codon 842 leading to an amino acid exchange from asparagine to valine. One case (number 55) carried a 9-bp deletion leading to an amino acid change in codon 842 from asparagine to alanine and to a loss of codons 843 to 845 (isoleucine, methionine, and histidine). Examples for SSCP shifts and sequence analysis in exon 18 of PDGFRα are shown for GIST 47 (point mutation; Figure 1A) and GIST 55 (9-bp deletion; Figure 1B).
Figure 1.
SSCP and sequence analysis in exon 18 of PDGFRα. A: GIST 47: Point mutation (T -> A) leading to an amino acid exchange from asparagine to valine; DNA fragment with altered mobility marked by an arrow. B: GIST 55: Deletion of 9 bp leading to an amino acid change in codon 842 from asparagine to alanine and to a loss of codons 843 to 845 (isoleucine, methionine, and histidine; DNA fragment with altered mobility marked by an arrow).
Clinical Data
The series of 41 GISTs with known KIT mutation included 3 benign, 20 malignant, and 8 GISTs with uncertain malignant potential according to Miettinen et al.15 According to Fletcher,13 7 tumors were of very low risk, 10 of low risk, 5 of intermediate risk, and 16 of high risk. In three cases, risk assessment was not performed as the tumor tissue was taken from metastases and not from the primary lesion. Sixteen patients were male and 25 were female. Median age was 65 years (mean 62 years, SD 13.1 years, range, 34 to 86 years). The median tumor diameter was 6.5 cm (mean 7.6 cm, SD 6.1 cm, range, 0.6 cm to 29 cm). Twenty-four primary tumors were found in the stomach, 12 in the small bowel, and two in the rectum. In one GIST, a biopsy had been taken from the upper abdomen, one tumor tissue was taken from the peritoneum and in another case, a sample from a liver metastasis was evaluated. In the subgroup of 13 GISTs with KIT mutation in exon 9 tumors occurred preferentially in the small bowel (n = 8, 61.5% in the group of tumors with known primary location) whereas only two tumors were located primarily in the stomach (18.2%). One GIST was detected in the rectum and two other samples were from liver or peritoneal metastases, respectively. In the subgroup of 28 GISTs with KIT mutation in exon 11, primary location in the stomach predominated with 22 cases (78.6%) whereas only four tumors were located in the small bowel (14.3%) and one in the rectum (Table 2).
Table 2.
Clinicopathologic Data and Types of 41 KIT Mutation Positive, PDGFRα Mutation Negative GISTs
| No. | Sex | Age | Location | φ (cm) | Category | MC | Histomorphology | AA sequence in KIT |
|---|---|---|---|---|---|---|---|---|
| 1 | f | 69 | stomach | 1.1 | benign | 0 | spindle cell type | W557G |
| 2 | m | 79 | stomach | 0.6 | benign | 0 | spindle cell type | V559D |
| 3 | m | 68 | stomach | 4.0 | benign | 0 | spindle cell type | V556H, Q557_V560del |
| 4 | f | 52 | stomach | 4.0 | benign | 1 | spindle cell type | Y570_P576del |
| 5 | m | 66 | stomach | 3.5 | benign | 0 | spindle cell type | V555_Q556del |
| 6 | f | 61 | stomach | 3.3 | benign | 1 | spindle cell type | W557del |
| 7 | f | 69 | stomach | 0.7 | benign | 0 | spindle cell type | V560del |
| 8 | f | 75 | stomach | 1.6 | benign | 0 | spindle cell type | Q557H, W558T, K559del, I563M, N564I |
| 9 | f | 77 | stomach | 4.5 | benign | 0 | spindle cell type | S590_G602ins |
| 10 | f | 59 | stomach | 5.0 | benign | 3 | spindle cell type | V559D |
| 11 | m | 60 | stomach | 1.0 | benign | 0 | spindle cell type | A504_Y505ins* |
| 12 | m | 68 | small bowel | 1.0 | benign | 0 | mixed cell type | A504_Y505ins* |
| 13 | m | 71 | small bowel | 0.7 | benign | 0 | spindle cell type | A504_Y505ins* |
| 14 | m | 86 | stomach | 8.0 | uncertain | 0 | spindle cell type | L576P |
| 15 | f | 83 | stomach | 7.0 | uncertain | 1 | spindle cell type | V559D |
| 16 | f | 38 | small bowel | 4.5 | uncertain | 0 | mixed cell type | E561P,E562_D579del |
| 17 | f | 68 | stomach | 10.0 | uncertain | 1 | spindle cell type | D579del |
| 18 | m | 55 | small bowel | 3.0 | uncertain | 2 | spindle cell type | A504_Y505ins* |
| 19 | f | 86 | stomach | 7.0 | uncertain | 0 | spindle cell type | A504_Y505ins* |
| 20 | f | 52 | small bowel | 5.0 | uncertain | 0 | spindle cell type | A504_Y505ins* |
| 21 | m | 56 | rectum | 3.0 | uncertain | 0 | spindle cell type | A504_Y505ins* |
| 22 | m | 64 | small bowel | 9.5 | malignant | 10 | epithelioid cell type | A504_Y505ins* |
| 23 | f | 63 | stomach | 12.0 | malignant | 26 | spindle cell type | W557_K558del |
| 24 | f | 71 | stomach | 15.0 | malignant | 5 | spindle cell type | Q577_L578ins |
| 25 | f | 65 | stomach | 11.0 | malignant | 0 | spindle cell type | Q556H,W557T,K558_V559del |
| 26 | f | 35 | stomach | 7.7 | malignant | 48 | spindle cell type | W557_K558del |
| 27 | f | 46 | stomach | 9.5 | malignant | 22 | spindle cell type | W557_K558del |
| 28 | m | 50 | small bowel | 13.0 | malignant | 1 | mixed cell type | V559A |
| 29 | f | 49 | colon | 15.0 | malignant | 2 | spindle cell type | W557_K558del |
| 30 | m | 65 | stomach | 10.5 | malignant | 1 | spindle cell type | Y553N |
| 31 | f | 68 | stomach | 1.9 | malignant | 6 | spindle cell type | V559D |
| 32 | m | 42 | stomach | 29.0 | malignant | 40 | spindle cell type | W557_K558del |
| 33 | f | 74 | liver | ND | malignant | 18 | spindle cell type | A504_Y505ins* |
| 34 | m | 60 | small bowel | 6.0 | malignant | 8 | spindle cell type | V559_G565del |
| 35 | m | 57 | upper abd. | 14.0 | malignant | 16 | spindle cell type | W557_K558del |
| 36 | f | 75 | stomach | ND | malignant | 28 | epithelioid cell type | P577_L580 ins |
| 37 | f | 67 | small bowel | ND | malignant | 1 | spindle cell type | V560del |
| 38 | m | 53 | small bowel | 20.0 | malignant | 14 | spindle cell type | A504_Y505ins* |
| 39 | f | 39 | peritoneum | ND | malignant | 4 | spindle cell type | A504_Y505ins* |
| 40 | f | 68 | small bowel | 13.0 | malignant | 12 | spindle cell type | A504_Y505ins* |
| 41 | f | 34 | small bowel | 10.0 | malignant | 2 | mixed cell type | A504_Y505ins* |
Abbreviations: m, male; f, female; φ, maximum diameter in cm; MIB-1 index, percentage of cells showing nuclear staining for MIB-1; MC, mitotic count (mitoses/50 HPFs); histomorphology, histomorphological subtype according to Fletcher et al.,13; AA, amino acid; abd., abdomen; del, deletion; ins, insertion; ND, no data available;
, in exon 9, all others in exon 11.
The panel of 20 GISTs carrying PDGFRα mutations in exon 18 included nine benign, four malignant, and seven GISTs with uncertain malignant potential. One GIST belonged to the group of very low risk, eight were of low risk, seven of intermediate risk, and four of high risk. Fourteen patients were male and six were female. Median age was 63.5 years (mean 63 years, SD 13.1 years, range, 26 to 88 years). Median tumor size was 5.3 cm (mean 6.7 cm, SD 5.5 cm, range 1.1 to 23 cm). Interestingly, all 20 tumors were found in the stomach (Table 3).
Table 3.
Clinicopathologic Data and Types of 20 KIT Mutation Negative, PDGFRα Mutation Positive GISTs
| No. | Sex | Age | Location | φ (cm) | Category | MC | Histomorphology | AA sequence* |
|---|---|---|---|---|---|---|---|---|
| 42 | F | 88 | stomach | 4.5 | benign | 0 | spindle cell type | I843_D846del |
| 43 | m | 52 | stomach | 1.1 | benign | 0 | spindle cell type | D842V |
| 44 | m | 72 | stomach | 2.5 | benign | 2 | epithelioid cell type | D842V |
| 45 | m | 58 | stomach | 2.8 | benign | 0 | mixed cell type | D842V |
| 46 | f | 62 | stomach | 2.2 | benign | 0 | mixed cell type | I843_D846del |
| 47 | m | 61 | stomach | 2.5 | benign | 0 | mixed cell type | D842V |
| 48 | m | 26 | stomach | 3.0 | benign | 1 | mixed cell type | D842V |
| 49 | m | 65 | stomach | 4.2 | benign | 1 | mixed cell type | D842V |
| 50 | m | 62 | stomach | 3.3 | benign | 0 | epithelioid cell type | D842V |
| 51 | m | 73 | stomach | 5.3 | uncertain | 0 | mixed cell type | D842V |
| 52 | m | 49 | stomach | 6.5 | uncertain | 0 | epithelioid cell type | D842V |
| 53 | m | 57 | stomach | 6.0 | uncertain | 0 | epithelioid cell type | D842V |
| 54 | w | 81 | stomach | 8.0 | uncertain | 1 | mixed cell type | D842V |
| 55 | m | 70 | stomach | 8.0 | uncertain | 3 | mixed cell type | D842A, I843_H845del |
| 56 | m | 68 | stomach | 10.0 | uncertain | 2 | epithelioid cell type | D842V |
| 57 | f | 45 | stomach | 5.2 | uncertain | 1 | mixed cell type | I843_D846del |
| 58 | f | 68 | stomach | 7.0 | malignant | 8 | epithelioid cell type | D842V |
| 59 | f | 69 | stomach | 10.2 | malignant | 4 | mixed cell type | D842V |
| 60 | m | 73 | stomach | 23.0 | malignant | 0 | mixed cell type | D842V |
| 61 | m | 60 | stomach | 19.5 | malignant | ND | epithelioid cell type | D842V |
Abbreviations: m, male; f, female; φ, maximum diameter in cm; MC, mitotic count (mitoses/50 HPFs); histomorphology, histomorphological subtype according to Fletcher et al13;
, AA (amino acid) sequence in PDGFRα exon 18; ND, no data available.
The remaining 26 cases lacking KIT mutations in exons 9 or 11 and PDGFRα mutations consisted of 10 benign, 13 malignant, and 3 GISTs with uncertain malignant potential. Five GISTs were of very low risk, 6 of low risk, 4 of intermediate risk, and 9 of high risk. Fifteen patients were male and 11 were female. Median age was 61 years (mean 58.7 years, SD 14.1 years, range, 26 to 83 years). Median size of the tumors was 6.5 cm (mean 7.3 cm, SD 5.9 cm, range, 0.1 cm to 22.0 cm). Twelve tumors were located in the stomach, 11 in the small bowel, and one in the esophagus. In two cases without available tumor tissue from the primary tumor, samples from peritoneal metastases or an intra-abdominal recurrence was analyzed, respectively (Table 4).
Table 4.
Clinicopathologic Data and Types of 26 KIT and PDGFRα Mutation Negative GISTs
| No. | Sex | Age | Location | φ (cm) | Category | MC | Histomorphology | DNA sequence* |
|---|---|---|---|---|---|---|---|---|
| 62 | f | 83 | esophagus | 0.1 | benign | 0 | spindle cell type | wt KIT and PDGFRα |
| 63 | m | 55 | stomach | 0.3 | benign | 0 | spindle cell type | wt KIT and PDGFRα |
| 64 | m | 45 | stomach | 4.5 | benign | 0 | mixed cell type | wt KIT and PDGFRα |
| 65 | m | 76 | stomach | 3.2 | benign | 0 | spindle cell type | wt KIT and PDGFRα |
| 66 | m | 78 | stomach | 0.4 | benign | 0 | spindle cell type | wt KIT and PDGFRα |
| 67 | f | 58 | stomach | 2.0 | benign | 1 | mixed cell type | wt KIT and PDGFRα |
| 68 | m | 62 | stomach | 2.5 | benign | 1 | spindle cell type | wt KIT and PDGFRα |
| 69 | f | 38 | stomach | 3.0 | benign | 4 | spindle cell type | wt KIT and PDGFRα |
| 70 | m | 62 | small bowel | 2.0 | benign | 1 | spindle cell type | wt KIT and PDGFRα |
| 71 | f | 38 | stomach | 4.5 | benign | 0 | spindle cell type | wt KIT and PDGFRα |
| 72 | m | 67 | small bowel | 4.0 | uncertain | 0 | spindle cell type | wt KIT and PDGFRα |
| 73 | m | 54 | stomach | 6.5 | uncertain | 2 | mixed cell type | wt KIT and PDGFRα |
| 74 | m | 26 | stomach | 8.0 | uncertain | 0 | spindle cell type | wt KIT and PDGFRα |
| 75 | f | 67 | small bowel | 13.0 | malignant | 0 | mixed cell type | wt KIT and PDGFRα |
| 76 | f | 60 | small bowel | 8.0 | malignant | 0 | spindle cell type | wt KIT and PDGFRα |
| 77 | f | 65 | recurrence | ND | malignant | 12 | spindle cell type | wt KIT and PDGFRα |
| 78 | f | 68 | stomach | 22.0 | malignant | 7 | epithelioid cell type | wt KIT and PDGFRα |
| 79 | m | 50 | small bowel | 10.0 | malignant | 0 | spindle cell type | wt KIT and PDGFRα |
| 80 | f | 77 | stomach | 11.0 | malignant | 2 | spindle cell type | wt KIT and PDGFRα |
| 81 | m | 68 | small bowel | 15.0 | malignant | 3 | mixed cell type | wt KIT and PDGFRα |
| 82 | m | 50 | peritoneum | 2.5 | malignant | 90 | mixed cell type | wt KIT and PDGFRα |
| 83 | m | 68 | small bowel | 8.0 | malignant | 7 | spindle cell type | wt KIT and PDGFRα |
| 84 | f | 43 | small bowel | 20.0 | malignant | 2 | spindle cell type | wt KIT and PDGFRα |
| 85 | m | 57 | small bowel | 11.1 | malignant | 4 | spindle cell type | wt KIT and PDGFRα |
| 86 | m | 72 | small bowel | 8.0 | malignant | 10 | spindle cell type | wt KIT and PDGFRα |
| 87 | f | 39 | small bowel | 13.5 | malignant | 0 | mixed cell type | wt KIT and PDGFRα |
Abbreviations: m, male; f, female; φ, maximum diameter in cm; MIB-1 index, percentage of cells showing nuclear staining for MIB-1; MC, mitotic count (mitoses/50 HPFs); histomorphology; histomorphological subtype according to Fletcher et al13;
, DNA sequences in KIT exon 9 and 11, in PDGFRα exon 12 and 18; ND, no data available.
Gender and Location
In the series with PDGFRα mutation there was a predominance of male patients (14 of 20) that was inverse in the series with KIT mutation (16 of 41; P = 0.0231) but was also observed in patients with wild-type mutation pattern (15 of 26).
Interestingly, all GISTs with PDGFRα mutation were located in the stomach, whereas tumors with KIT mutation or with wild-type status in both genes were found also in the small bowel (P = 0.0008).
Risk Assessment
Comparing the risk of aggressive behavior according to Fletcher et al13 KIT mutation-positive GISTs and tumors lacking any mutations belonged more frequently to the high-risk group (42% and 38%) than those with PDGFRα mutation (20%; P = 0.0983, Table 5). However, the differences according to the Fletcher risk assessment did not reach statistical significance.
Table 5.
Risk Assessment According to Fletcher et al.13 in Relation to Mutational Status in KIT and PDGFRα Gene
| Risk | KIT mutation positive* (n = 38) | No detectable KIT or PDGFRα mutation** (n = 24) | PDGFRα mutation positive*** (n = 20) |
|---|---|---|---|
| Very low | 7 (18.4%) | 3 (12.5%) | 1 (5.0%) |
| Low | 10 (26.3%) | 8 (33.3%) | 8 (40.0%) |
| Intermediate | 5 (13.2%) | 4 (16.7%) | 7 (35.0%) |
| High | 16 (42.1%) | 9 (37.5%) | 4 (20.0%) |
KIT exon 9 or 11;
, KIT exon 9 and 11, PDGFRα exon 12 and 18;
, PDGFRα exon 18.
Histomorphological Subtypes
There was a statistically significant higher frequency of mixed (11 of 20) and epithelioid (7 of 20) tumor types in the series of PDGFRα mutation-positive GISTs as compared to KIT mutation-positive tumors (4 of 41 and 2 of 41, respectively, P < 0.0001) and as compared to wild-type tumors (7 of 26 and 1 of 26, respectively, P = 0.00016).
Immunohistochemistry of KIT Receptor and PDGFRα
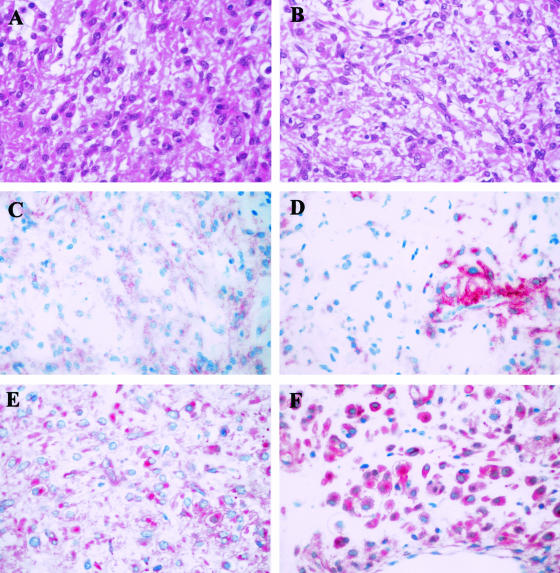
A stronger PDGFRα-expression was found in PDGFRα mutation-positive tumors compared to the lesions lacking mutations in both genes (OR 9.259, P = 0.0068) but was not significantly higher than in KIT mutation-positive GISTs (OR 3.194, P = 0.1586). Figure 2 shows two examples of GISTs (numbers 47 and 55) with PDGFRα mutation in exon 18 both exhibiting a strong PDGFRα expression and a rather weak or only focal KIT-receptor expression.
Figure 2.
Histomorphology and expression of KIT and PDGFRα receptors in two PDGFRα-mutated GISTs: Both tumors exhibited a mixed phenotype (GIST 55 in A, GIST 47 in B; H&E). GIST 55 showed a weak KIT receptor expression (C) and a strong membranous and dot-like cytoplasmatic PDGFRα receptor expression (E). GIST 47 showed a strong, but only focal KIT receptor expression (D) and a strong membranous and cytoplasmatic PDGFRα receptor expression (F). (Original magnification, ×400, A–F).
All 41 GISTs with known KIT mutation showed a strong or intermediate KIT receptor expression. In the series with KIT and PDGFRα wild-type sequence KIT receptor expression was less intensive but demonstrable in all but one case. In one case, GIST diagnosis was confirmed because of strong vimentin expression and lack of myogenic or neurogenic markers. The lowest KIT receptor expression was found in the series with PDGFRα mutation including three GISTs lacking KIT receptor expression. Lower KIT receptor expression of PDGFRα-mutated GISTs was found particularly in comparison with KIT-mutated tumors (OR = 0.045; P = 0.0003) and was still significantly different when compared with GISTs lacking mutations (OR = 0.229, P = 0.0244). A detailed summary of the results from all immunohistological stainings is depicted in Table 6.
Table 6.
Immunohistochemical Expression Profiles in Relation to Mutational Status in KIT and PDGFRα Gene
| Antibody
|
KIT mutation positive* (n = 41)
|
No detectable KIT or PDGFRα mutation** (n = 26)
|
PDGFRα mutation positive*** (n = 20)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Positivity (in %) | strong | intermediate | weak | none | strong | intermediate | weak | none | strong | intermediate | weak | none |
| KIT | 95.1 | 4.9 | — | — | 77.0 | 7.7 | 11.5 | 3.8 | 50.0 | 10.0 | 25.0 | 15.0 |
| PDGFRα | 70.3 | 16.2 | 5.4 | 8.1 | 46.2 | 26.9 | 15.4 | 11.5 | 90.0 | 5.0 | 5.0 | — |
| CD34 | 82.9 | 14.6 | 2.4 | — | 53.9 | 19.2 | 11.5 | 15.4 | 60.0 | 15.0 | 10.0 | 15.0 |
| bcl-2 | 48.8 | 14.6 | 22.0 | 14.6 | 42.3 | 23.1 | 3.8 | 30.8 | 75.9 | 10.0 | 10.0 | 5.0 |
| vimentin | 82.9 | 17.7 | — | — | 88.5 | 3.8 | 7.7 | — | 90.0 | 5.0 | 5.0 | — |
| sm-actin | 2.4 | 7.3 | 22.0 | 68.3 | — | 11.5 | 30.8 | 57.7 | — | 15.0 | 25.0 | 60.0 |
| desmin | — | 9.8 | 7.3 | 82.9 | — | 7.7 | 7.7 | 84.6 | — | 5.0 | 15.0 | 80.0 |
| S-100 | — | — | 9.8 | 90.2 | 3.8 | 3.8 | 11.6 | 80.8 | — | — | 5.0 | 95.0 |
, KIT exon 9 or 11;
, KIT exon 9 and 11, PDGFRα exon 12 and 18;
, PDGFRα exon 18.
Discussion
In the majority of GISTs, detection of mutations in the receptor tyrosine kinase KIT is regarded as an important step in their molecular pathogenesis. In most of these cases, a strong expression of KIT protein accompanies the mutated receptor. In contrast, KIT wild-type GISTs (wt GISTs) show a different phenotype with respect to KIT expression which is less prominent and also in some cases with respect to their cytologic composition.3
Recently, Heinrich et al8 described mutations in the PDGFRα gene in 14 of 40 GISTs (35%) with wild-type sequence in KIT. They could demonstrate that these mutations lead to autophosphorylation of the receptor protein in the same way as shown for the KIT receptor. Hirota et al9 could confirm these data in 5 of 8 KIT wt GISTs. PDGFRα belongs to the same type III receptor tyrosine kinase subfamily as KIT with a similarity of 35% of amino acids between both proteins. Both genes are located on the long arm of chromosome 4 in close vicinity and both are believed to be derived from a common ancestor during evolution.
In the present study we found mutations in exon 18 of the PDGFRα gene in 20 of 46 GISTs without detectable KIT mutation. In contrast to the results of Heinrich et al8 and Hirota et al9 we could not detect activating mutations in exon 12 of the PDGFRα gene despite direct sequencing of all cases independently of our SSCP results. This result suggests that exon 12 mutations might be less frequent than exon 18 mutations. Furthermore, it cannot be ruled out that different genetic backgrounds of the populations studied may exist. All 41 GISTs with known KIT mutation showed a wild-type sequence in exons 12 and 18 of the PDGFRα gene. Thus, PDGFRα mutations seem to be an alternative cause for GIST development. Clarification is necessary as to whether mutant PDGFRα transforms by itself or if it needs to form heterodimers with wt KIT to transform GIST precursors, as proposed by Hirota et al.9 In 26 tumors, mutations could not be detected in either exon 9 and 11 of KIT or in the PDGFRα gene, suggesting a third subgroup with a still unknown pathogenesis. It cannot be ruled out completely that single cases in this group may harbor a KIT mutation in exon 13 or 17. However, detection rate in these two exons encoding the tyrosine kinase domains I and II is extremely low in other series. Lasota et al11 found KIT exon 13 mutations in 2 of 200 tumors and Rubin et al.10 in 2 of 48 GISTs. The latter group described KIT exon 17 mutations in 2 of 48 GISTs, Heinrich et al16 found KIT exon 17 mutations in 2 of 127 GISTs.
Our data indicate that GISTs with PDGFRα mutation in exon 18 differ from tumors with KIT mutations and from those lacking mutations in both genes according to their location, their histomorphological features, their immunohistochemical expression pattern, and their proliferative activity.
Surprisingly, all tumors carrying a PDGFRα mutation were located in the stomach. In contrast, GISTs with KIT mutations occurred also in the small bowel (12 of 36) with tumors carrying exon 9 mutations even predominating in the small bowel (8 of 10). This suggests that GISTs are also genetically heterogeneous with respect to their site of origin. The progenitor cells giving rise to gastric GISTs seem to undergo different genetic hits compared to GISTs in other locations. There are different explanations for this finding. First, specific genotoxic events may only occur in the specific microenvironment of the stomach. However, the nature of such external factors is not known. Second, the progenitor cells leading to GISTs of the stomach may be different from those at other sites, or they may represent another stage of progenitor differentiation prone to be transformed by PDGFRα mutations.
Interestingly, the vast majority of GISTs with PDGFRα mutation (90%) displayed a mixed or epithelioid phenotype whereas KIT mutation-positive GISTs exhibited almost always a spindled histomorphology (85.4%) and also the majority (65.4%) of tumors lacking any mutations were composed of spindle cells. The occurrence of PDGFRα mutations may indicate an alternative activation mechanism that has similar, but not identical, functional consequences. Activated PDGFRα induces redistribution of cellular filaments, cell ruffling, and motility responses.17 Therefore, the epithelioid phenotype may be a direct consequence of the mutation. The fact that two cases had a spindle cell phenotype although they carry a PGRFRα mutation indicates that additional signals may influence the cellular architecture.
Comparing the levels of KIT receptor and PDGFRα expression, there was an association of mutational status and expression level. KIT receptor expression was lower in PDGFRα-mutation-positive GISTs than in tumors carrying KIT mutations and vice versa. Heinrich et al8 showed that activated KIT and PDGFRα receptors regulate a similar but not identical signaling cascade. They found phosphorylated STAT5 only in cells transfected with mutant KIT but not in cells transfected with mutant PDGFRα. This argues against the possibility that mutated PDGFRα simply dimerize wild-type KIT receptor and induce an identical activation response.
It is remarkable that differences between KIT and PDGFRα-mutated GISTs are observed not only on the level of location and morphology but also when comparing the mitotic count and the resulting classification and risk of aggressive behavior. These findings should be confirmed in longitudinal clinical studies.
In summary, our finding of PDGFRα mutations in KIT wild-type GISTs support the findings of Heinrich et al8 that these mutations may represent an alternative event in GIST pathogenesis. Furthermore, our analysis provides evidence that although these events are not equivalent, they define different genetic, phenotypic, and prognostic subsets of GISTs. Very recently, Heinrich et al16 demonstrated that PDGFRα mutations in GISTs may lead to resistance to tyrosine kinase inhibitors, such as imatinib mesylate, underlining the impact of mutational analysis in Kit and PDGFRα for therapeutic approaches. Further studies will be needed to dissect the functional consequences of these genetic events that are of particular interest in relation to GIST treatment with tyrosine kinase inhibitors.
Acknowledgments
We thank Susanne Steiner, Barbara Reddemann, Ellen Pagen, Dorota Denkhaus, and Gerrit Klemm for technical assistance.
Footnotes
E.W. and A.H. contributed equally to this paper.
References
- Miettinen M, Sarlomo-Rikala M, Lasota J. Gastrointestinal stromal tumours. Ann Chir Gynaecol. 1998;87:278–281. [PubMed] [Google Scholar]
- Sarlomo-Rikala M, Kovatich AJ, Barusevicius A, Miettinen M. CD117: a sensitive marker for gastrointestinal stromal tumors that is more specific than CD34. Mod Pathol. 1998;11:728–734. [PubMed] [Google Scholar]
- Wardelmann E, Neidt I, Bierhoff E, Speidel N, Manegold C, Fischer HP, Pfeifer U, Pietsch T. c-kit mutations in gastrointestinal stromal tumors occur preferentially in the spindle rather than in the epithelioid cell variant. Mod Pathol. 2002;15:125–136. doi: 10.1038/modpathol.3880504. [DOI] [PubMed] [Google Scholar]
- Hirota S, Isozaki K, Moriyama Y, Hashimoto K, Nishida T, Ishiguro S, Kawano K, Hanada M, Kurata A, Takeda M, Tunio GM, Matsuzawa Y, Kanakura Y, Shinomura Y, Kitamura Y. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 1998;279:577–580. doi: 10.1126/science.279.5350.577. [DOI] [PubMed] [Google Scholar]
- Miettinen M, Sarlomo-Rikala M, Lasota J. Gastrointestinal stromal tumors: recent advances in understanding of their biology. Hum Pathol. 1999;30:1213–1220. doi: 10.1016/s0046-8177(99)90040-0. [DOI] [PubMed] [Google Scholar]
- Taniguchi M, Nishida T, Hirota S, Isozaki K, Ito T, Nomura T, Matsuda H, Kitamura Y. Effect of c-kit mutation on prognosis of gastrointestinal stromal tumors. Cancer Res. 1999;59:4297–4300. [PubMed] [Google Scholar]
- van Osteroom AT, Judson I, Verweij J, Stroobants S, Donato di Paola E, Dimitrijevic S, Martens M, Webb A, Sciot R, van Glabbeke M, Silberman S, Nielsen OS. Safety and efficiacy of imatinib (STI571) in metastatic gastrointestinal stromal tumors: a phase I study. Lancet. 2001;358:1421–1423. doi: 10.1016/s0140-6736(01)06535-7. [DOI] [PubMed] [Google Scholar]
- Heinrich MC, Corless CL, Duensing A, Mc Greevey L, Chen CJ, Joseph N, Singer S, Griffith DJ, Haley A, Town A, Demetri GD, Fletcher CDM, Fletcher JA. PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 2003;299:708–710. doi: 10.1126/science.1079666. [DOI] [PubMed] [Google Scholar]
- Hirota S, Ohashi A, Nishida T, Isozaki K, Kinoshita K, Shinomura Y, Kitamura Y. Gain-of-function mutations in platelet-derived growth factor receptor a gene in gastrointestinal stromal tumors. Gastroenterology. 2003;125:660–667. doi: 10.1016/s0016-5085(03)01046-1. [DOI] [PubMed] [Google Scholar]
- Rubin BP, Singer S, Tsao C, Duensing A, Lux ML, Ruiz R, Hibbard MK, Chen CJ, Xiao S, Tuveson DA, Demetri GD, Fletcher CD, Fletcher JA. KIT activation is a ubiquitous feature of gastrointestinal stromal tumors. Cancer Res. 2001;61:8118–8121. [PubMed] [Google Scholar]
- Lasota J, Wozniak A, Sarlomo-Rikala M, Rys J, Kordek R, Nassar A, Sobin LH, Miettinen M. Mutations in exons 9 and 13 of KIT gene are rare events in gastrointestinal stromal tumors. Am J Pathol. 2000;157:1091–1095. doi: 10.1016/S0002-9440(10)64623-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardelmann E, Losen I, Hans V, Neidt I, Speidel N, Bierhoff E, Heinicke T, Pietsch T, Büttner R, Merkelbach-Bruse S. Deletion of Trp-557 and Lys-558 in the juxtamembrane domain of the c-kit protooncogene is associated with metastatic behavior of gastrointestinal stromal tumors. Int J Cancer. 2003;106:887–895. doi: 10.1002/ijc.11323. [DOI] [PubMed] [Google Scholar]
- Fletcher CD, Berman J, Corless CL, Gorstein F, Lasota J, Longley B, Miettinen M, O’Leary T, Remotti H, Rubin BP, Shmookler B, Sobin L, Weiss S. Diagnosis of gastrointestinal stromal tumors: a consensus approach. Hum Pathol. 2002;33:459–465. doi: 10.1053/hupa.2002.123545. [DOI] [PubMed] [Google Scholar]
- von Deimling A, Bender B, Louis DN, Wiestler OD. A rapid and non-radioactive PCR-based assay for the detection of allelic loss in human gliomas. Neuropathol Appl Neurobiol. 1993;19:524–529. doi: 10.1111/j.1365-2990.1993.tb00481.x. [DOI] [PubMed] [Google Scholar]
- Miettinen M, El-Rifai W, Sobin LH, Lasota J. Evaluation of malignancy and prognosis of gastrointestinal stromal tumors: a review. Hum Pathol. 2002;33:478–483. doi: 10.1053/hupa.2002.124123. [DOI] [PubMed] [Google Scholar]
- Heinrich MC, Corless CL, Demetri GD, Blancke CD, von Mehren M, Joensuu H, Mc Greevey LS, Chen CJ, Van den Abbeele AD, Druker BJ, Kiese B, Eisenberg B, Roberts PJ, Singer S, Fletcher CD, Silberman S, Dimitrijevic S, Fletcher JA. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumors. J Clin Oncol. 2003;21:4342–4349. doi: 10.1200/JCO.2003.04.190. [DOI] [PubMed] [Google Scholar]
- Eriksson A, Siegbahn A, Westermark B, Heldin C, Claesson-Welsh L. PDGF α- and β-receptors activate unique and common signal transduction pathways. EMBO J. 1992;11:543–550. doi: 10.1002/j.1460-2075.1992.tb05085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]