Abstract
Background
The long bone abnormality (lbab) mouse is a new autosomal recessive mutant characterized by overall smaller body size with proportionate dwarfing of all organs and shorter long bones. Previous linkage analysis has located the lbab mutation on chromosome 1 between the markers D1Mit9 and D1Mit488.
Results
A genome-based positional approach was used to identify a mutation associated with lbab disease. A total of 122 genes and expressed sequence tags at the lbab region were screened for possible mutation by using genomic DNA from lbabl/lbab, lbab/+, and +/+ B6 mice and high throughput temperature gradient capillary electrophoresis. A sequence difference was identified in one of the amplicons of gene Nppc between lbab/lbab and +/+ mice. One-step reverse transcriptase polymerase chain reaction was performed to validate the difference of Nppc in different types of mice at the mRNA level. The mutation of Nppc was unique in lbab/lbab mice among multiple mouse inbred strains. The mutation of Nppc is co-segregated with lbab disease in 200 progenies produced from heterozygous lbab/+ parents.
Conclusion
A single nucleotide mutation of Nppc is associated with dwarfism in lbab/lbab mice. Current genome information and technology allow us to efficiently identify single nucleotide mutations from roughly mapped disease loci. The lbab mouse is a useful model for hereditary human achondroplasia.
Background
Annually, approximately 2.3% of infants in the United States are born short (small) for gestational age (SGA) [1]. Achondroplasia (short-limbed dwarfism) is the most common cause of human dwarfism [2], and it accounts for 70% of dwarfism cases [3] To explore causative genetic factor(s) for the development of achondroplasia in animal models, we focused on a novel spontaneous model of long bone abnormality called lbab mouse, which was originally found in the PL/J strain at The Jackson Laboratory (TJL)[4]. Homozygous mutants exhibit proportionate dwarfing of all organs and shorter long bones. The mutation has been transferred to the C57BL/6J strain to improve reproduction. The genetic locus responsible for the phenotype has previously been mapped to chromosome 1 (Chr 1) between markers D1Mit9 and D1Mit488 (53.5 cM) at TJL [4], but the responsible gene and the nature of mutation remained unclear.
Disease gene hunting has always been time-consuming and labor-intensive. For successful map-based cloning, a complicated fine mapping of a major locus is generally essential [5]. Recently, we developed an alternative, sequence-based, positional candidate cloning approach to bypass this bottleneck of cloning, and we have successfully identified several mutated genes in different mouse spontaneous mutants by applying this strategy [6,7]. Our strategy takes advantage of the availability of comprehensive murine sequence databases, polymerase chain reaction (PCR), high throughout PCR product analysis, and sequencing technologies to speed up the process of disease-related gene hunting. Interestingly, we identified a nucleotide mutation (C→G transversion) in gene Nppc in the lbab mice. Herein, we describe the detailed process of our cloning and validation.
Results
Body growth of lbab/lbab mice
All lbab/lbab mice housed at the University of Tennessee Health Science Center died before 7 days of age with remarkable changes in organ weight and body size (Fig. 1, Table 1). No detectable differences in survival rate or body size were noted between the lbab/+ and +/+ mice.
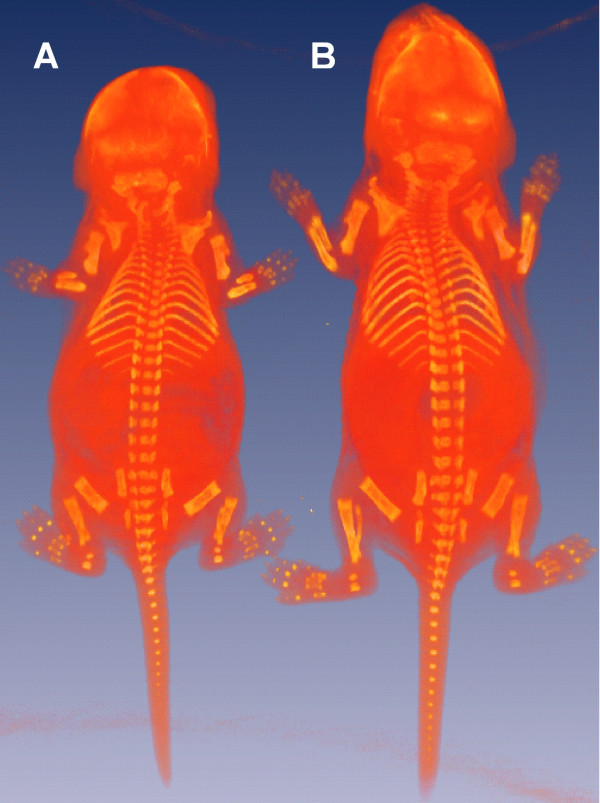
Figure 1.
Phenotypic assay of normal and lbab mice. A whole-body microCT scanned image of a pair of lbab/lbab and +/+ B6 mice in the same litter at the age of 6 days. A: lbab/lbab mouse, B: +/+ mouse.
Table 1.
Comparison of phenotypic parameters between mice with different genotypes at the lbab locus
| Parameter | +/+ (n = 10) | lbab/+ (n = 10) | lbab/lbab (n = 12) | lbab/lbab as % of +/+ |
| Body weight (g) | 3.45 ± 0.4221 | 3.61 ± 0.351 | 1.54 ± 0.255* | 44.1% |
| Body length (mm) | 55.9 ± 0.36 | 56.7 ± 0.34 | 34.0 ± 0.35* | 61.6% |
1 Values were mean ± SD. Mice of different gender were pooled as they were too young to show detectable differences; +/+ mice included 5 females and 5 males, 10 lbab/+ mice included 5 females and 5 males; 12 lbab/lbab mice included 6 females and 6 males. As shown, the body weight and body length from the lbab/lbab group were significantly smaller than in the +/+ and lbab/+ groups (* p < 0.05). There were no significant differences between +/+ and lbab/+ mice.
Target region of the mutation in lbab locus
Previous genetic analysis showed that the lbab mutation is located on mouse Chr 1 and is flanked by molecular markers D1Mit9 and D1Mit488 [4]. According to the Ensembl database, D1Mit488 is located between 91902920–91903043 bps. However, there is no physical position for D1Mit9 in the database, although we know from TJL database that it is positioned at 53.5 cM. From TJL's mouse genome informatics database, we found 13 molecular markers at the 53.5 cM position. The positions of most of these [8] markers are near 84 Mb [9]. Accordingly, we decided to start our investigation in the region between 83 and 90 Mb (Fig. 2A). Genomic sequences within this region are complete in the Ensembl database. There are a total of 122 transcripts, 70 of which represent genes and 52 represent expressed sequence tags (ESTs) (Table 2).
Figure 2.
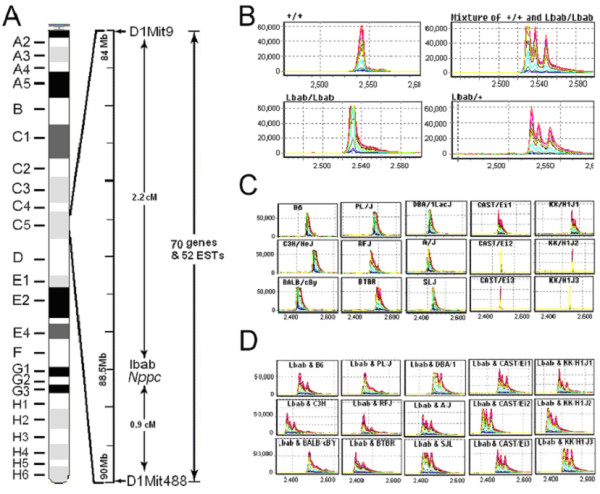
Schematic of the mutation identification in lbab mice. A: A genetic map of the lbab locus showing the relative locations of microsatellite markers and the total number of candidate transcripts within the lbab locus. B: PCR product analyses using the SpectruMedix system. The PCR products from different species of mice were pair mixed to check the possible sequence difference between normal B6 and PL/J mice (+/+), normal and homozygous lbab mice (mixture of +/+ and lbab/lbab), intrahomozygous lbab mice (lbab/lbab), and intraheterozygous mice (+/lbab). C: The PCR amplification products of exon 2 of the Nppc gene from 11 strains of mice (C57BL/6J, SJL/J, CAST/Ei/J, C3H/HeJ, PL/J, BALB/cJ, RF/J, KK/H1J, A/J, DBA/1J, and BTBR/J). D: PCR products from each of those strains were mixed separately with that of lbab/lbab mice. Each mixture showed multiple bands of signal, indicating the difference in their DNA sequences. The X-axis represents relative size of the PCR products. The Y-axis represents relative strength of signal or the amount of the PCR products.
Table 2.
Distribution of candidate transcripts around the lbab locus
| Region | Gene | EST |
| 83–84 | 14 | 4 |
| 84–85 | 3 | 5 |
| 85–86 | 1 | 6 |
| 86–87 | 22 | 13 |
| 87–88 | 20 | 11 |
| 88–89 | 10 | 6 |
| 89–90 | 0 | 7 |
| Total | 70 | 52 |
Initial screening of the targeted region
Because lbab mice are bred on a C57BL/6J (B6) background, we assumed that the majority of the background genome carrying the lbab mutation was from the B6 strain. Therefore, we isolated genomic DNA from both B6 and lbab/lbab mice. We designed 528 pairs of primers flanking first and last exons of 122 candidate transcripts by using Primer3 software[10]. We obtained the primers commercially (Illumina, San Diego, CA) and conducted PCR amplification with genomic DNAs from lbab/lbab and B6 mice, which were regarded as normal controls. The PCR products were then analyzed for the presence of sequence differences between lbab/lbab and normal mice by using the RevealTM system (SpectruMedix LLC, PA). We found variations in 25 PCR products between normal B6 and lbab/lbab mice (Table 3).
Table 3.
Primers flanking polymorphism sites between C57BL/6J and lbab/lbab mice
| Gene ID (Ensembl) | Genomec Position | Forward Primer's Sequence | Reverse Primer's Sequence |
| ENSMUSG00000036707 | 86006371–86006887 | GATGGGATGCAGTGTGTCAG | AAACTGCAGGCGCACTAAAC |
| ENSMUSESTG00000001751 | 83966699–83966785 | CAGCAGAAGTGAAACCAGGA | TGACTCTGAGCTCACCATGC |
| ENSMUSG00000026163 | 84087634–84088853 | GGGATGAACAGCATTTGAGA | TGCTCCCTGGGAAGTAACAG |
| ENSMUSESTG00000001946 | 85602128–85646987 | TGATTCCCAACTCCTTTTCAG | GGCAATACACCTTGCTACTCC |
| ENSMUSG00000026220 | 85713718–85742232 | ACATGCCTGGCTTGAAAGTT | GGATGGATCTTTGACATCACC |
| ENSMUSG00000026200 | 85713718–85742232 | GAAGTGGGTGAGTTGCAGTG | GCAAGAGGGTCAGAGGTTCA |
| ENSMUSESTG00000001946 | 85877547–85896383 | ACTGGAGCTCTGGTGTCCAT | GGAAGGGACAAACTCTTCCA |
| ENSMUSESTG00000001946 | 85877547–85896383 | TCCCTCACTTCATCCCAAAC | CCAACAACTTGCTCCATTGTAA |
| ENSMUSESTG00000001946 | 85877547–85896383 | GCATGGAGTCAGCTGTGCTA | GCTGTGCAGTGATCAGCAGT |
| ENSMUSG00000026228 | 86273443–86274025 | TGTTCAGTGGCAGGAACAAG | GAGTCACCAAAAGGCGAATG |
| ENSMUSESTG00000004625 | 86885787–86885885 | GCTTCGCTCTTGGCTCAAC | TCCTCTCGCACTGAACTGAA |
| ENSMUSESTG00000004625 | 87005019–87005248 | GGCAGTTAAACCAGAGAGCA | GCCCAAGACTCACCTCTGTC |
| ENSMUSG00000036511 | 87288388–87288749 | AGTTGCAAGGACAGCAGGAC | GCCTAGCTTGAGGTGCTGAC |
| ENSMUSG00000026253 | 87397283–87403022 | AGGCTGACACATGACACTCC | CCCCAGGTCTCAAATCCCTA |
| ENSMUSG00000026254 | 87405306–87405408 | ATCCACGGGGAGGTTAAAGT | ACGCGTCGAACTTGTTGTTC |
| ENSMUSG00000026254 | 87411304–87411418 | CTTGGGCTATGACCTTGTGG | CCTGTATCATCACCCCAAAC |
| ENSMUSG00000026254 | 87417820–87417956 | TCACTCTGTTCCTCCCTTAGGA | GGCCCAACTATGGAGCTACT |
| ENSMUSG00000026255 | 87484369–87484561 | TCCTTGGACCATGCAGGTAT | TATCCAACCAGGGAGCATTC |
| ENSMUSG00000026255 | 87504768–87505947 | CGACCCCATTCAAATCAGAG | TCAGAAGCCCAATCCTGAAC |
| ENSMUSG00000044104 | 87581463–87582114 | GCTGCATAGCTGGCTGCATA | AGAAGGCACTGGGGAAATCT |
| ENSMUSG00000048000 | 87641410–87641794 | ACAAGGCTGGAGTTGCTCTC | AGCCTCAAGAGAGGCGAGTA |
| ENSMUSG00000026259 | 87686025–87686185 | GTGCTTACCGCTCACTGACA | GCTTCAGGACATCTGCCTCT |
| ENSMUSG00000026288 | 87914088–87914491 | CTGAGGGCAGAATGACCAGT | CCCAGGTTTAGCCCTTCTTC |
| ENSMUSG00000026289 | 87985895–87986828 | GGGAAAGCATTCTCTGTCCT | GACAATCAAGACGCAAGGAG |
| ENSMUSG00000026289 | 87985895–87987101 | AGGCTTTGTACGCAGACTCC | GCACAAGTTGGTTTGGGACT |
We speculated that the identified sequence variations were likely to arise from the close linkage between the mutated gene and nearby sequences in the parental PL/J strain, even though the background of lbab mice was mainly on the B6 strain. To determine if this were the case, we isolated DNA from PL/J mice obtained from TJL and amplified the 25 variable fragments by using the same panel of primers. By comparing those DNA fragments with those from lbab/lbab mice, we did not find sequence differences between DNA products from lbab/lbab and PL/J mice (data not shown), suggesting that those 25 variations represent polymorphisms between PL/J and B6.
Detecting the mutation in lbab locus in a more focused region
Because of the recognition of sequence polymorphisms between PL/J and B6 mice in the targeted region, we made two changes in our follow-up screening. First, we switched our controls from B6 to PL/J mice. Using another panel of primers (n = 240 pairs) for PCR amplification of every exon of all candidate genes, we identified only one DNA fragment from lbab/lbab mice that was different from PL/J mice DNA. This fragment was from exon 2 of the gene ENSMUSG00000026241, representing the gene for natriuretic peptide precursor C (Nppc). The same pair of primers for this fragment was used for further genotyping as indicated in material and methods.
Because we used PL/J mice as controls for this cycle of screening, there was another concern that the sequence variation might be a polymorphism derived from the B6 strain. To address this issue, we used PCR amplification of genomic DNA from B6 mice and compared results to the PCR products from PL/J and lbab/lbab mice. The data showed that the amplified DNA fragment from lbab/lbab mice was different from both B6 and PL/J mice, while the fragments from PL/J and B6 were the same (Fig. 2B). To find out which nucleotide(s) was different between lbab/lbab and PL/J controls, we sequenced genomic DNA fragments from PL/J and lbab/lbab mice. The data revealed a C→G change from PL/J to lbab/lbab mice (Fig. 3A, left panel).
Figure 3.
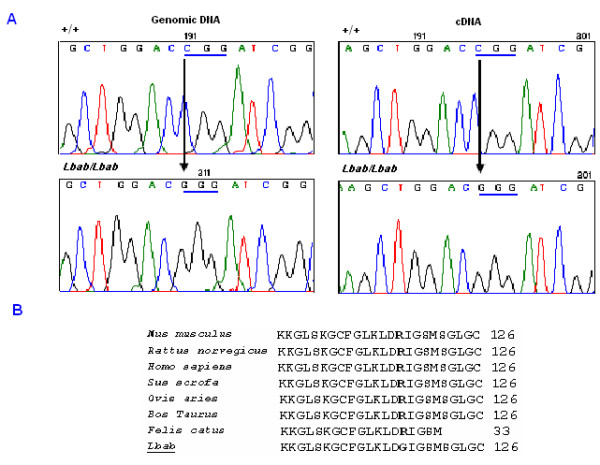
Schematic of the sequence changes of Nppc gene in lbab mice. A: A stretch of nucleotide sequence of Nppc in lbab/lbab and +/+ mice. A C→G transversion was found in both genomic DNA and cDNA sequences of Nppc in lbab/lbab mice compared with the sequence from PL/J mice. B: Alignment of the predicted amino acid sequence in the conserved domain of Nppc in different species. The C→G substitution leads Arg (R) to Gly (G) substitution at codon 117 on the conserved domain of Nppc.
To confirm the same difference at the cDNA level, we performed reverse transcriptase PCR (RT-PCR) on total RNAs from lbab/lbab, lbab/+, and +/+ mice by using primers that covered the mRNA sequence from exon 2 to exon 3 of the Nppc. The resultant RT-PCR products were sequenced using the SpectruMedix system, and the same C→G transversion from PL/J to lbab/lbab mice was found (Fig. 3A, right panel)
To evaluate the potential consequence of this point mutation in Nppc, we examined the translated amino acid sequence of Nppc. We found that this transversion predicts the substitution of arginine (R) for glycine (G) in a conserved domain of Nppc protein (Fig. 3B).
Confirmation of mutation
We conducted further experiments to confirm the single nucleotide change in the Nppc gene. Based on our initial screening, we think that it is very likely that the C→G change in exon 2 of the Nppc gene is causally related to the phenotypes in lbab/lbab mice. Because the lbab mutation arose from PL/J, theoretically, there should be no difference in the Nppc sequence between lbab and PL/J mice except the mutation. In addition, this single nucleotide replacement is the only change between homozygous lbab/lbab and homozygous normal PL/J mice in 122 transcripts. However, we could not completely rule out the possibility that we may have missed other mutations/polymorphisms. Moreover, one could question whether this is a spontaneous polymorphism in mouse strains or a random mutation that arose after the lbab was separated from the PL/J. Therefore, we carried out two more experiments to further ensure the association between the mutation and the disease. First, we examined sequence polymorphism in exon 2 of Nppc from nine other inbred strains (Fig. 2C). As shown in Fig. 2D, each such mixture showed multiple bands, indicating the difference between lbab/lbab and those strains. Second, we bred 200 mice from heterozygous lbab/+ parents to evaluate allele frequency in relation to the phenotype. We genotyped every offspring by using the original pair of primers that flank exon 2 of the Nppc gene. From those progeny, we found 14 lbab/lbab mice that exhibited a genotype of homozygous G/G nucleotide and a phenotype of lbab mice, while the remaining progeny had 74 homozygous C/C and 112 heterozygous C/G genotypes, all with a normal phenotype. Taken together, our data indicate that the C→G transversion in exon 2 of the Nppc gene is associated with the phenotypes observed in lbab mice.
Discussion
For the first time, we have identified a single nucleotide mutation by high throughput screening of a large genome region without fine mapping. The initial mapping at TJL was conducted with only 27 F2 animals. Linkage of lbab was first detected on Chr 1 with D1Mit231 and D1Mit9 by using the pooled sample. DNA samples were then typed for the individual 96 animals with these two markers and three additional Chr 1 markers [4]. By the standard strategy of classical positional cloning, the lbab locus could be further mapped. However, with the availability of mouse genome information and a tested protocol for high throughput screening of mutations [6,7], we directly searched genes based on the map from TJL. With the success of finding this mutation and others [6,7], we feel confident that we no longer need fine mapping for most mutations.
In this study, several lines of evidence indicate that a single nucleotide mutation of Nppc is associated with the lbab phenotype. First, Nppc is located within the genetic region of the lbab locus. Second, the Nppc mutation was the only defect detected among candidate genes and ESTs within the lbab locus from lbab mice. Because the lbab mutation was transferred from the PL/J strain to the B6 inbred strain, we evaluated the possibility of close linkages of nearby sequences from the PL/J mice by screening any sequence difference near the mutation area and later crossing with PL/J mice. There were no other differences between lbab/lbab mice and their two parental strains, so the possibility of other mutation involvement was ruled out. Third, cDNA sequence results agreed with the genomic DNA data. Last, we showed that the Nppc genotype is unique in lbab/lbab mice compared with nine other inbred strains, and the G/G Nppc genotype was closely associated with the phenotype in lbab mice.
Recently, several transgenic and knockout mouse studies have demonstrated that Nppc is critical in the prevention and rescue of achondroplasia [11,12]. A recent gene knockout study done by Chusho et al. [11] indicated that Nppc null mice of 129/Sv background showed severe dwarfism and early death. The lbab mice have a phenotype similar to Nppc knockout mice with two exceptions. First of all, the lbab mice develop an overall smaller body size. Second, the mutants exhibit proportionate dwarfing of all organs with the possible exception of the male reproductive tract, which appears extremely small [4]. However, much precise information may be obtained by a direct comparison between Nppc null mice and lbab mice. There may still be some difference between them because of the difference not only in the nature of the mutations but also in the genome backgrounds of those two models. Accordingly, we speculate that the identified point mutation of Nppc in lbab mice belongs to a loss-of-function mutation. As a key positive regulator of endochondral bone formation, Nppc seems to express its activity mainly through natriuretic peptide receptor 2 (Npr2) [11,13-15]. On the other hand, a recent study indicated that Nppc counteracts the activities of fibroblast growth factor signalling, which is a major negative regulatory pathway for long bone development, in both direct and indirect ways [16].
Murine Nppc is structurally similar to that of other species. The affected Arg at codon 117 on the Nppc domain is highly conserved among all members of the natriuretic peptide system and different species (Fig. 3B). The mechanisms for regulating Nppc expression are currently unknown. Importantly, the mutated nucleotide is also located in the common biologically active COOH-terminal 22 amino acid residue area, suggesting the critical significance of this amino acid residue in the functioning of these ligands during skeletal development.
Nppc was not in the list of candidate genes for the allelism test. According to the information on TJL webpage [4], allelisms tested were brachymorphic (bm) [17] with a ratio of disease/total of 0/42 progeny born, achondroplasia (cn) [18] with a ratio of 0/61 progeny born, osteochondrodystrophy (ocd) [19] with a ratio of 0/19 (4 unclassifiable) progeny born, and small (sml) [20] with a ratio of 0/59 progeny born. The first three loci are known to be located on Chr 19 [17], 4 [18], and 19 [19], respectively. The last one, sml, is either on Chr 6 or unknown [20]. Had Nppc been considered as a candidate gene, our initial screening would have been simpler, although we feel that some work is needed to exclude mutations in the nearby genes. In addition to the known function of Nppc, the fact that there is no other mutation in nearby genes in the lbab region is supporting evidence for the potential cause of the lbab phenotype by the single nucleotide mutation in Nppc.
Identifying the Nppc mutation in lbab mice provides useful information for human achondroplasia studies. It also demonstrates that, while candidate genes should be carefully examined based on gene function, it is feasible to identify mutated genes that are roughly mapped by linkage analysis by sequence-based positional candidate cloning strategy. We speculate that this strategy will be particularly useful for familial human diseases with small numbers of patients; in those cases, researchers usually have either a rough map or name/number of the chromosome of the disease locus. Furthermore, by using functional genomics and rodent models with spontaneous mutations yielding measurable phenotypes, we can rapidly identify mutational events in a cost-effective manner.
Conclusion
In the present study, a sequence-based positional candidate cloning approach was applied to identify a gene mutation in lbab mice with abnormal endochondral ossification. Our results suggest that a single nucleotide mutation in gene Nppc is likely to be the causative factor and that the lbab mouse may be a useful model for human achondroplasia studies.
Methods
Mice
A heterozygous (lbab/+) breeding pair of mice was purchased from TJL, and a breeding colony was established at the research animal facility of the University of Tennessee Health Science Center (UTHSC). Experimental animal procedures and mouse husbandry were performed in accordance with the National Institutes of Health's Guide for the Care and Use of Laboratory Animals and approved by the UTHSC Institutional Animal Care and Use Committee.
Genomic sequence information
Information about microsatellite markers and their locations were obtained from the Mouse Genome Informatics website [8] and the Ensembl mouse genome database [9]. The sequence data used in the paper were based on the information as of March 20, 2005.
DNA and RNA
Genomic DNAs (gDNA) were extracted from the livers of phenotypically normal +/+ and lbab/+ mice, as well as from mutant lbab/lbab mice, by using a Qiagen Genomic-tip20/G (Qiagen, Alameda, CA) following the manufacturer's instructions. After determining the quality and quantity of the DNA in an Eppendorf photometer (Eppendorf Scientific, Westbury, NY), we used DNAs for large-scale mutational screening. Total RNAs were extracted from femurs by using Trizol reagent (Invitrogen, Carlsbad, CA), and the quality of the total RNA was checked by electrophoresis on a Spectronic Genesys spectrophotometer (Spectronic Instruments, Rochester, NY).
High throughput screening
The gDNAs from lbab/lbab, lbab/+, and +/+ mice were used for temperature gradient capillary electrophoresis (TGCE) and sequence analysis using Reveal™ system (SpectruMedix; State College, PA). Based on the Ensembl and National Center for Biotechnology Information (NCBI) databases, primer pairs flanking the exons of known and predicted genes (including ESTs) within the lbab locus were designed using Primer3 software [10]. Primers were located approximately 100 bp 5' or 3' to each exon and, in general, produced 300–400 bp amplicons. For large exons that required multiple pairs of primers, primers were designed to allow neighboring DNA fragments to overlap each other by at least 50 bps. Primer pairs were synthesized from Illumina (San Diego, CA). PCR amplification of gDNA was performed in a 96-well plate format and consisted of 30–35 cycles at three temperatures: strand denaturation at 96°C for 30 sec, primer annealing at 54–60°C for 60 sec, and primer extension at 72°C for 120 sec. A TGCE device made by SpectruMedix was used to analyze amplicons from +/+, +/lbab, and lbab/lbab mice. The SpectruMedix system includes a high-throughput capillary electrophoresis instrument capable of analyzing 96 samples every 140 min. Heteroduplex analysis was subsequently performed using SpectruMedix software. Amplicons from lbab mice were sequenced if they differed from normal ones.
Genotyping
A pair of primers flanking the position of a single nucleotide mutation of the Nppc gene (forward primer: CTCTTGGGTGCAGAGCTAGG; reverse primer: AGCTGGTGGCAATCAGAAAA) was used to genotype +/+, lbab/+, and lbab/lbab mice. The PCR products from heterozygous and homozygous mice were mixed with those from normal mice. The individual and mixed PCR products were then run on TGCE to detect possible sequence variations. PCR amplification was performed in a total volume of 25 μl at a final concentration of 1.5 mM MgCl2 concentration and 0.2 mmol/L each dNTP, 0.2 μmol/L oligonucleotide primer, 100 ng template DNA, and 0.7 units Taq polymerase (Fisher Scientific, Pittsburgh, PA) for 35 cycles of 94°C for 1 min, 50–55°C for 1 min, and 72°C for 1 min.
RT-PCR amplification of Nppc
One-step RT-PCR kit (Invitrogen, Carlsbad, CA) was used to detect the expression of mutated Nppc at the mRNA level. Reactions were performed in a total volume of 50 μl with 8 ng/μl of total RNA, and 0.2 μM forward (AGCTGGTGGCAATCAGAAAA) and reverse (TCAGTGCACAGAGCAGTTCC) primers were used to amplify exons 2 to 3 of Nppc. First, cDNA synthesis and pre-denaturation were performed in single cycles at 50°C for 40 min and 94°C for 2 min. Next, PCR amplification was performed for 35 cycles: 94°C for 30 sec, 54–58°C for 36 sec, and 72°C for 2 min.
DNA sequencing
DNA sequencing was conducted to verify the mutation in the gDNA and cDNA of Nppc from different species of mice. PCR products from both gDNA and cDNA were purified using an AMPure PCR Purification Kit (Agencourt, Beverly, MA), and the resultant products were sequenced using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA). A total volume of 5 μl sequencing reactions including 2 μl Big Dye (plus Half-BD), 10–23 ng of purified DNA template, and 1–3 ρmoles of either forward or reverse universal sequencing primers was incubated for 37 cycles at 96°C for 180 sec, 50°C for 30 sec, and 60°C for 180 sec. Unreacted primers were removed by ethanol-acetate precipitation (3.75% 3 M NaOAc, 87.5% nondenatured 100% EtOH, and 8.75% dH2O, pH 4.6). The labeled products were dissolved in 0.02 mM EDTA in HiDi formamide prior to electrophoretically loading onto the SpectruMedix 96 capillary sequencing system. The same primers in the amplification of DNA fragments from either genomic DNA or mRNA were also used in the sequencing. Sequencing was conducted twice to verify the results for both gDNA and cDNA.
Statistical analysis
The body weight and body length for individual mice were measured at an average age of 5.5 days after birth. The weight and length differences between normal and lbab mice were analyzed for their statistical significance by using two-tail t-test with a cutoff p value of < 0.05.
Authors' contributions
YJ carried out the majority of the molecular work including DNA and RNA isolation, primer design, RT-PCR, genotyping, sequencing, and data analysis.
JY drafted the paper and provided helpful comments on experimental execution and data analysis.
FJ provided animal samples and performed TGCE running.
HY did the original screening.
LRD, XL, BAR, and JS provided valuable input and help in drafting the paper.
WG initiated and mentored the study, as well as provided the valuable framework to draft the paper.
All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
The authors thank Dr. Wesley Beamer and Cliff Rosen at The Jackson Laboratory for their reading and helpful comments for this manuscript. We thank Dr. David Armbruster, author's editor at UTHSC, for editing this manuscript. This work was supported by the Center of Genomics and Bioinformatics (WG) and Center in Connective Tissue Research (WG) at the University of Tennessee Health Science Center; Veterans Administration Medical Center in Memphis (WG); National Institute of Arthritis and Musculoskeletal and Skin Diseases, National Institutes of Health (R01 AR51190 to WG; R01 AR50785 to JS); National Eye Institute, National Institutes of Health (R01 EYO15073 to LRD); and National Center for Research Resources, National Institutes of Health (RR01183 to Muriel Davisson, LRD).
Contributor Information
Yan Jiao, Email: yjiao2@utmem.edu.
Jian Yan, Email: jyan1@utmem.edu.
Feng Jiao, Email: fjiao@utmem.edu.
HongBin Yang, Email: yanghb12@hotmail.com.
Leah Rae Donahue, Email: lrd@jax.org.
Xinmin Li, Email: lix@uchicago.edu.
Bruce A Roe, Email: broe@ou.edu.
John Stuart, Email: jstuart@utmem.edu.
Weikuan Gu, Email: wgu@utmem.edu.
References
- Lee PA, Chernausek SD, Hokken-Koelega AC, Czernichow P. International Small for Gestational Age Advisory Board consensus development conference statement: management of short children born small for gestational age, April 24-October 1, 2001. Pediatrics. 2003;111:1253–1261. doi: 10.1542/peds.111.6.1253. [DOI] [PubMed] [Google Scholar]
- Francomano CA. The genetic basis of dwarfism. N Engl J Med. 1995;332:58–59. doi: 10.1056/NEJM199501053320113. [DOI] [PubMed] [Google Scholar]
- Wikipedia http://en.wikipedia.org/wiki/Dwarfism
- TJL http://www.jax.org/mmr/lbab.html
- Karayiorgou M, Gogos JA. Schizophrenia genetics: uncovering positional candidate genes. Eur J Hum Genet. 2006;14:512–519. doi: 10.1038/sj.ejhg.5201587. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Li X, Beamer WG, Yan J, Tong Y, Goldowitz D, Roe B, Gu W. A deletion causing spontaneous fracture identified from a candidate region of mouse Chromosome 14. Mamm Genome. 2005;16:20–31. doi: 10.1007/s00335-004-2414-0. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Yan J, Zhao Y, Donahue LR, Beamer WG, Li X, Roe BA, Ledoux MS, Gu W. Carbonic anhydrase-related protein VIII deficiency is associated with a distinctive lifelong gait disorder in waddles mice. Genetics. 2005;171:1239–1246. doi: 10.1534/genetics.105.044487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MGI http://www.informatics.jax.org/
- Ensembl http://www.ensembl.org/Mus_musculus/mapview
- Pimer3 http://frodo.wi.mit.edu/
- Chusho H, Tamura N, Ogawa Y, Yasoda A, Suda M, Miyazawa T, Nakamura K, Nakao K, Kurihara T, Komatsu Y, Itoh H, Tanaka K, Saito Y, Katsuki M. Dwarfism and early death in mice lacking C-type natriuretic peptide. Proc Natl Acad Sci U S A. 2001;98:4016–4021. doi: 10.1073/pnas.071389098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasoda A, Komatsu Y, Chusho H, Miyazawa T, Ozasa A, Miura M, Kurihara T, Rogi T, Tanaka S, Suda M, Tamura N, Ogawa Y, Nakao K. Overexpression of CNP in chondrocytes rescues achondroplasia through a MAPK-dependent pathway. Nat Med. 2004;10:80–86. doi: 10.1038/nm971. [DOI] [PubMed] [Google Scholar]
- Yasoda A, Ogawa Y, Suda M, Tamura N, Mori K, Sakuma Y, Chusho H, Shiota K, Tanaka K, Nakao K. Natriuretic peptide regulation of endochondral ossification. Evidence for possible roles of the C-type natriuretic peptide/guanylyl cyclase-B pathway. J Biol Chem. 1998;273:11695–11700. doi: 10.1074/jbc.273.19.11695. [DOI] [PubMed] [Google Scholar]
- Koller KJ, Lowe DG, Bennett GL, Minamino N, Kangawa K, Matsuo H, Goeddel DV. Selective activation of the B natriuretic peptide receptor by C-type natriuretic peptide (CNP) Science. 1991;252:120–123. doi: 10.1126/science.1672777. [DOI] [PubMed] [Google Scholar]
- Schulz S. C-type natriuretic peptide and guanylyl cyclase B receptor. Peptides. 2005;26:1024–1034. doi: 10.1016/j.peptides.2004.08.027. [DOI] [PubMed] [Google Scholar]
- Krejci P, Masri B, Fontaine V, Mekikian PB, Weis M, Prats H, Wilcox WR. Interaction of fibroblast growth factor and C-natriuretic peptide signaling in regulation of chondrocyte proliferation and extracellular matrix homeostasis. J Cell Sci. 2005;118:5089–5100. doi: 10.1242/jcs.02618. [DOI] [PubMed] [Google Scholar]
- O'Brien EP, Novak EK, Keller SA, Poirier C, Guenet JL, Swank RT. Molecular map of chromosome 19 including three genes affecting bleeding time: ep, ru, and bm. Mamm Genome. 1994;5:356–360. doi: 10.1007/BF00356554. [DOI] [PubMed] [Google Scholar]
- Velinov M, Slaugenhaupt SA, Stoilov I, Scott CI, Jr., Gusella JF, Tsipouras P. The gene for achondroplasia maps to the telomeric region of chromosome 4p. Nat Genet. 1994;6:314–317. doi: 10.1038/ng0394-314. [DOI] [PubMed] [Google Scholar]
- Poirier C, Blot S, Fernandes M, Carle GF, Stanescu V, Stanescu R, Guenet JL. A high-resolution genetic map of mouse chromosome 19 encompassing the muscle-deficient osteochondrodystrophy (mdf-ocd) region. Mamm Genome. 1998;9:390–391. doi: 10.1007/s003359900777. [DOI] [PubMed] [Google Scholar]
- Lundberg P, Welander P, Openshaw H, Nalbandian C, Edwards C, Moldawer L, Cantin E. A locus on mouse chromosome 6 that determines resistance to herpes simplex virus also influences reactivation, while an unlinked locus augments resistance of female mice. J Virol. 2003;77:11661–11673. doi: 10.1128/JVI.77.21.11661-11673.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]