Abstract
We have developed a model system for studying differentiation in the mammary gland, by using two clonal cultures deriving from a rat breast adenocarcinoma. They differ in the ability to form domes, structures the significance of which is unknown. By using the subtractive cDNA library approach, we isolated a cDNA that is highly expressed in the dome-forming cells, and identical to the rat8 gene and highly homologous to the human 9–27 gene. Antisense treatment of the dome-forming cells specifically and reproducibly abolishes dome formation, while forced expression of the gene in non-dome-forming cells causes morphological changes suggestive of “flat” domes. In situ hybridization on rat tissues shows that the gene is expressed in epithelia, especially in those forming tubular structures, suggesting a relatedness between these structures and domes. Cytokeratin 8 and E cadherin are strongly expressed in the domes but not outside them, suggesting that the rat8 gene triggers the cells to express molecules that tighten the lateral connections between the cells; the process is likely to parallel that occurring during the differentiation of the mammary gland.
The genetic control of differentiation of specific organs or tissues is of great interest in itself, and also for understanding their pathology. It is, however, difficult to study the development of many organs because the stages are ill-defined; this applies especially to the mammary gland, as, outside pregnancy and lactation, cells in various stages of development are rare and immersed in a vast number of stromal cells.
This difficulty can be overcome by using cultures of cells undergoing differentiation in vitro; however, the relationship of the observed changes to the differentiation stages occurring in vivo is generally unknown. To overcome this difficulty we have taken the approach of identifying genes responsible for in vitro differentiation and to determine their site of action in vivo by in situ hybridization as a means to determine the correspondence.
The cells used are clonal derivatives of the Rama-25 line of rat mammary cells isolated by Bennett et al. (1) from a dimethylbenzanthracene-induced adenocarcinoma in Sprague–Dawley rats. Two sublines were used: LA7 and 106A10 (2). The LA7 subline grown on plastic dishes spontaneously undergoes differentiation, forming two kinds of structures, called “domes” and “ridges” (3), which are not formed by the other line.
The tendency of the LA7 line toward differentiation is noticeable in nude mice where it produces solid epithelial nodules with interspersed ducts, whereas the 106A10 line produces nodules of spinocellular cells without any recognizable differentiation (unpublished observations).
In this work we concentrated on the formation of domes, blister-formed by the detachment of the cell layer from the plastic, through a transcellular transport of water and ions. This has been shown for the kidney cell line MDCK, which undergoes the same differentiation (4). The formation of domes is highly reproducible, can be studied quantitatively, and can be markedly accelerated by inducers of differentiation such as dimethyl sulfoxide (DMSO) or 8-Br-cAMP and others (3). The general strategy we used was to prepare cDNAs from the LA7 in the process of forming domes under the action of DMSO (to maximize the effect), and from the uninduced 106A10 line, and to carry out subtraction experiments by using one or the other mRNA as the driver. In this way, genes that are expressed preferentially in dome-forming cells or nondome forming cells were isolated; and because the two sublines are closely related, it is likely that the genes expressed only, or most highly, in one cell line are related to its state of differentiation.
Here we report the isolation of a gene, whose activity is required for the differentiation into domes, from induced LA7 cells by using 106A10 mRNA as a driver. The gene is identical to the previously known rat8 gene (5) that is closely related (70% homology) to the 1–8 gene family; members of this family are part of the larger human multigene family (7) that has been shown to be interferon inducible (7, 24, 25). The rat8 gene is highly homologous to the 9–27 human gene (6), which is a member of this family. We find that this gene is widely expressed in animals, especially in several epithelia, including the tubular structures of the mammary gland.
MATERIALS AND METHODS
Cells.
The cell lines LA7 and 106A10 were cultured as described in Dulbecco (2). To study dome formation, sets of three identical 35-mm plates with 3 × 105 cells per cm2 were grown at confluence (for 48 hr) and differentiated in the presence of DMSO 1.5% as inducer (3). As the dome density varies in different parts of a culture, the domes were counted in square areas of 10.8 mm per side located at the center of each plate.
Construction of cDNA Libraries.
The mRNA was extracted from DMSO-induced LA7 cells; from 106A10 cells it was extracted without induction. Total RNA was isolated by the single-step acid/guanidinium isothiocyanate/phenol/chloroform extraction method (8) by using 50 × 106 cells. Poly(A)RNA was purified by affinity chromatography through three passages over oligo(dT)12–18-cellulose column as described in Aviv and Leder (9). Poly(A)+ RNA was then treated with RNase-free DNase I (GIBCO/BRL) 1 unit/μg at room temperature for 15 min.
To prepare cDNA tester from LA7 cells, 1 μg of purified mRNA was converted to first strand cDNA essentially as described in Takahashi and Ko (10). Synthesis of second strand cDNA was carried out by RNase H replacement according to Maggi et al. (11) with minor modifications. Following phenol/chloroform extraction, the DNA was purified by G50 spin column (Boehringer Mannheim). After ligation of the 5′ end phosphorylated synthetic adapters to the blunt DNA, excess adapters were removed by gel filtration chromatography by using Centricon-100 (Amicon). For each strand of cDNA synthesis, parallel reactions incorporating [32P]dATP were carried out to determine the efficiency of synthesis.
The oligomer-adapters ligated to blunt end cDNAs were LL-SalIA and LL-SalIB as reported by Ko et al. (12). All the cDNAs ligated to adapters were amplified by using the LL-SalIA oligomer as a primer and the PCR conditions were as recommended by Takahashi and Ko (10).
The mRNA driver from the 106A10 cell line was prepared according to the method outlined above, and biotinylated by UV crosslinking by using CLONTECH’s PhotoActivatable Biotin according to the manufacturer’s recommendations.
Differential Cloning by Subtracted Library Construction.
Twenty-five micrograms of biotinylated mRNA (the driver, in a 40-fold excess) from the 106A10 cell line was combined in 120 μl of TE (10 mM Tris/1 mM EDTA) with 600 ng of the LA7 double stranded cDNA (the tester), and denatured at 70°C for 5 min. Then 120 μl of 2× hybridization buffer (80% formamide/100 mM Hepes, pH 7.5/2 mM EDTA/0.2% SDS) was added before carrying out the hybridization at 65°C for 48 hr (13).
Streptavidin (25 μg; GIBCO/BRL) were then added to the mixture and incubated at room temperature for 25 min. Single-stranded cDNAs were separated from the cDNA·mRNA heteroduplex by removal of the biotin-streptavidin-cDNA·mRNA complex with phenol/chloroform extraction (14). After three cycles of subtraction the unhybridized cDNAs remaining in the inorganic phase were ethanol precipitated with 10 μg of glycogen carrier (Boehringer Mannheim). The efficiency of the method was checked by using an alternative protocol: the unhybridized cDNAs were recovered by using streptavidin conjugated to magnetic beads (Dynabeads M-280, Dynal, Great Neck, NY), and by removing the beads with the biotinylated cDNA-streptavidin by using a magnet (15). The unhybridized cDNA strands were PCR amplified by using the LL-SalIA primer. The PCR-amplified products were then cloned by direct ligation into the pCR II vector (Invitrogen) and transformed by using competent DH5α cells.
Clone Analysis and Sequencing.
To check for clone inserts, selected white colonies were amplified by PCR by using the “lone linker” LLSalIA oligomer. For sequencing we used the universal primer M13 from the pCR II vector. Nucleic acid sequence homology searches were analyzed by using fasta and wordsearch programs by searching the complete combined GenBank/EMBL databanks. Amino acid sequence homology searches were also analyzed by searching the complete SwissProt database. Selected amino acid sequences were analyzed by using blocks (16), while prosite was used to identify potential motifs within translated sequences.
Northern Blot Hybridization.
RNA extraction and Northern blot analysis were performed according to standard procedures (17) by using 10 μg of total RNA from each cell line (LA7 and 106A10). Probes were prepared from PCR-amplified clone inserts and [32P]dCTP-labeled by random priming by using Ready Prime (Amersham). Prehybridization, hybridization and washing conditions were carried out at 45°C according to standard procedures (17). A loading control was performed by using [32P]dCTP-labeled β-actin or 36B4 labeled cDNA probe representing a gene whose level of expression is independent from the action of inducers (18).
Northern blot analysis was also performed by using a mouse RNA blot (CLONTECH), and lab-produced rat RNA blots.
RNA in Situ Hybridization.
In situ hybridization was performed on rat mammary tissues as described (19) with the following modifications. Sections of 12 μm were cryostatically prepared, fixed with 4% paraformaldehyde, and hybridized overnight at 50°C with 35S-UTP-labeled sense and antisense p41 RNA probes or 35S-ATP-labeled sense and antisense oligoprobes designed from the sequence (accession no. X61381). The p41 sense and antisense RNA probes were obtained by in vitro transcription of the full-length p41 cDNA by using T7 and Sp6 primers, respectively. After hybridization and washing according to the conditions reported in ref. 20, autoradiography on x-ray films was for 14 to 33 days at 4°C. Bright-field photographs were performed directly on the x-ray films. Following development, the tissue sections were autoradiographed for 3–5 weeks by using the NTB2 emulsion (Kodak) and then photographed in dark-field. The specificity of the reaction was assessed by pretreating the adjacent section with 20 μg/ml RNase A, following paraformaldehyde fixation and hybridization in identical conditions. In situ hybridization experiments were also performed on LA7 and 106A10 cell cultures. The cells were paraformaldehyde fixed as described in the Immunofluorescence Microscopy section of the present paper, autoradiographed for 3–5 weeks by using NTB2 emulsion, and photographed in dark-field.
Antisense Oligodeoxynucleotide Methodology.
For inhibition studies of the p41 cDNA expression, three synthetic oligodeoxynucleotides of 20 bases were synthesized: antisense oligomer (5′-TTTCGTAGTTTGGGGGTTGT-3′) designed as a complementary sequence at the 5′ of the p41 coding region, starting at position 72; sense oligomer (5′-ACAACCCCCAAACTACGAAA-3′) from the same coding region; and scrambled oligomer (5′-GTCGTATTTGTGTGCAACCC-3′) a scrambled sequence with the same nucleotides used for the antisense oligomer. Phosphorothioate oligomers were also used at the concentration of 4 μg/ml (phosphorothioate modification was designed every two nucleotides). Thirty thousand cells per cm2 were placed into each of four flasks. After 24 hr, the medium was replaced with a fresh one containing inactivated serum. The flasks were further incubated for 24 hr upon addition to each flask of 40 μg/ml (1 mg/ml = 0.35 mM) (21) of one of the three oligomers, and a control culture flask was left untreated. Twenty-four hours later, DMSO at a final concentration of 1.5%, and an additional 20 μg/ml of each oligomer were added to the corresponding flasks.
Cells were maintained in culture for 72 hr, inspected for dome formation, photographed, and harvested for RNA extraction.
Immunofluorescence Microscopy.
Detection of cytokeratin 8 and E and P cadherin was performed on cells grown for 3 days on Permanox chamber slides with coverslips (Nunc). Cells were induced with 1.5% DMSO, fixed in a paraformaldehyde/PBS gradient from 0.5% to 4% for 20 min, and then incubated with commercial mAb raised against mouse cytokeratin 8 (Sigma) and E or P cadherin (Sigma) according the manufacturer’s protocol. The secondary antibody used was anti-mouse IgG fluorescein isothiocyanate-labeled (Vector Laboratories). Cells were microscopically examined and photographed by using a ×40 objective.
106A10 Cell Transformation with p41 cDNA.
The mouse mammary tumor virus (MMTV)-p41 expression vector was constructed replacing the neu gene fragment from the MMTV-neu plasmid (22) with the EcoRI-HindII segment made blunt by gap-filling with T4 DNA polymerase. Restriction analysis and sequencing were used to confirm that the insert was correctly oriented. The p41 MMTV vector carries the MMTV promoter (23), polyadenylation signal sequences and the neomycin phosphotransferase gene, which confers resistance to the antibiotic G418 (geneticin, GIBCO/BRL).
The 106A10 cells were transfected by electroporation by using 10 μg of the linearized MMTV-p41 recombinant vector. After 24 hr, selection was initiated by adding G418 (determined as being 0.8 mg/ml) to the culture medium. After one week, resistant clones were isolated by capillary duct aspiration, trypsinized and transferred one by one into 96-well plates, and then individually amplified. Active transcription of the 41cDNA was verified by RNA isolation and Northern blot analysis of the transfected cells.
RESULTS
cDNAs Identified Through Differential Cloning.
The construction of the subtracted cDNA library allowed the isolation of 97 cDNAs. In each clone the size of the insert was determined by PCR by using a primer complementary to the ligated adapters. The fragments were partially sequenced at both ends, and the sequences were compared with the combined GenBank/EMBL DNA and SwissProt databanks. By sequence analysis we were able to organize the cDNA clones into the following classes: 44 cDNAs were homologous to known genes (18 were mitochondrial and ribosomal mRNAs, 26 were different mRNAs); 32 represented unknown genes, of which 19 were homologous to various expressed sequence tags. The remaining 21 clones were either multiple copies of the same gene, or did not contain inserts.
Expression Analysis of Subtracted mRNAs.
The expression specificity of the isolated clones was checked by hybridizing each of them to filters containing total RNA from LA7 (induced and uninduced) and 106A10 cell lines.
The highest difference was found in two clones that were completely sequenced. One (clone p41) was found to be identical to the rat8 gene (5); the other (clone p144) was found to be identical to the rat metallothionein II mRNA (EMBL GenBank accession no. M11794).
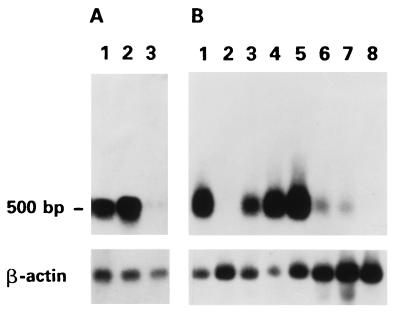
We focused our attention on the p41 cDNA defined from here on as rat8. This gene was preferentially expressed in LA7 cells both induced by DMSO and uninduced (Fig. 1A). Its expression, in contrast to that of the homologous gene present in lymphocytes (25), was not enhanced by interferons of classes I or II (data not shown). The expression of the gene in various tissues was determined by Northern blot analysis by using a commercially available (CLONTECH) mouse RNA blot. As shown in Fig. 1B, rat8 was expressed at high levels in lung, liver, heart, spleen, at low levels in kidney and muscle, but not in brain and testis. Significant expression was also seen in rat lung, heart, and kidney.
Figure 1.
Northern blot analysis of rat8 gene. (A) Lane 1: uninduced LA7, lane 2: DMSO-induced LA7; lane 3: 106A10 cell line. (B) Mouse tissues: heart (lane 1), brain (lane 2), spleen (lane 3), lung (lane 4), liver (lane 5), skeletal muscle (lane 6), kidney (lane 7), and testis (lane 8). Loading control with β-actin was performed (Bottom).
Effect of Antisense Oligonucleotides.
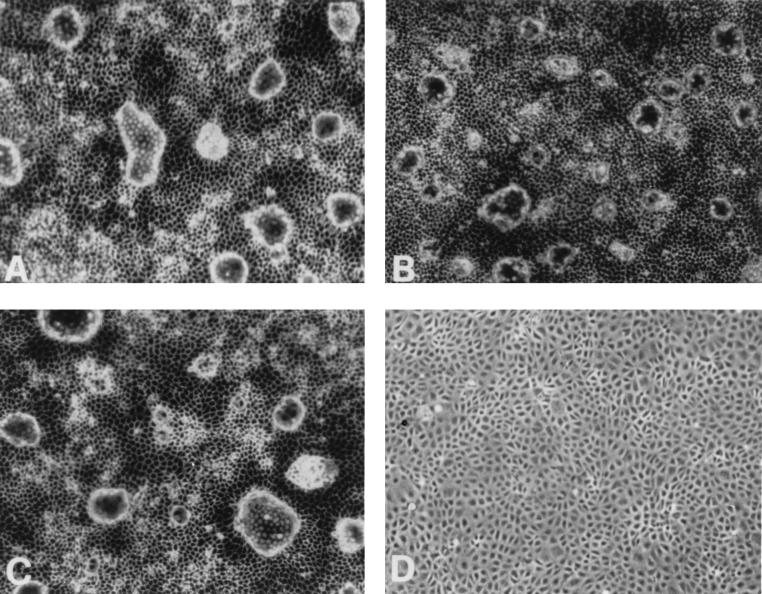
To determine whether rat8 was directly involved in the dome formation in LA7 cells, we tested the effect of an oligomer complementary to the mRNA sequence, both in phosphate and phosphorothioate form. As described in Materials and Methods, three synthetic oligomers, each 20 nucleotides long, were synthesized and used at the concentration of 40 μg/ml (phosphate) and 4 μg/ml (phosphorothioate): an antisense, a sense, and a scrambled one. Their effects were determined on LA7 cultures of comparable numbers of cells either induced by DMSO or uninduced. In either case, domes were formed in comparable numbers in cultures containing only medium, as well as in those containing the sense or the scrambled oligomer, but were completely absent in the cultures containing the antisense oligonucleotides (Fig. 2). This was true for induced and uninduced cultures. There was also a marked effect of the antisense on the appearance of the cell layer outside the domes: on microscopic examination of induced cultures treated with the antisense oligomer the layer appeared to be formed by separate although adjacent cells, with no structure between them, yet in the sense and scrambled controls, as in the untreated cultures, the boundaries between cells were very prominent, suggesting the existence of a conjoining structure. The antisense treatment had no appreciable effect on the growth of the culture.
Figure 2.
Antisense anti-rat8 oligonucleotide effect on dome formation of LA7 induced with DMSO. In all four panels, LA7 cells were induced to differentiate with 1.5% DMSO. Individual oligonucleotides were added in the following way: (A) None; (B) sense oligonucleotide (control); (C) scrambled oligonucleotide; (D) antisense oligonucleotide. See Materials and Methods for further details.
The antisense experiments were repeated three times with the phosphate oligomers and twice with the phosphorothioate oligomers, using three different preparations of the oligomers. The results were identical in all cases.
In Situ Hybridization with rat8 cDNA.
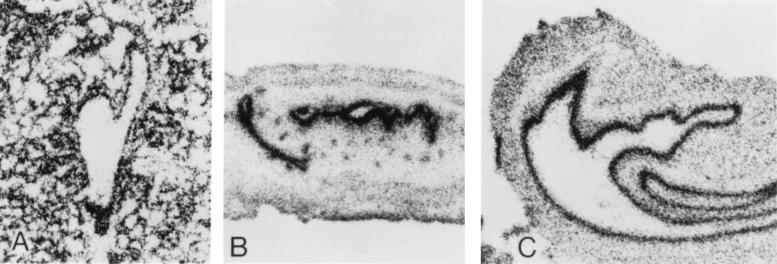
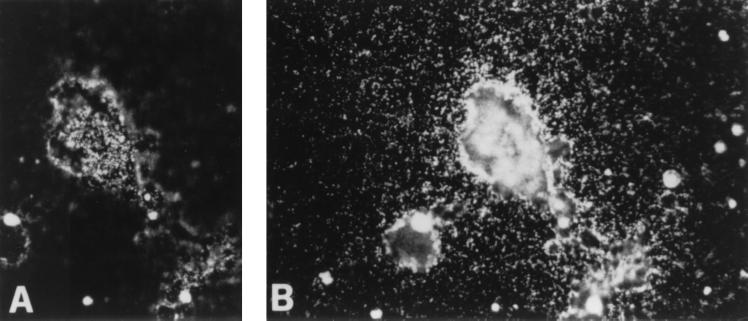
The expression of the gene was determined in the rat mammary gland, ovary, and lung. In the lactating mammary gland there is strong expression in the ducts and alveolar epithelia (Fig. 3). In the fallopian tube expression is strong in the epithelium lining the lumen. In the lung there is expression in the alveoli, but especially in the bronchial epithelium. It appears, therefore, that in these organs the gene is widely expressed in epithelia, with its main expression in epithelia that differentiate into tubular structures. In situ hybridization was also carried out by using induced LA7 cultures; the expression of the gene was seen in areas containing domes, being very strong in the domes, and quite evident around each dome, for a short distance (Fig. 4). Areas of the culture lacking domes showed much weaker expression.
Figure 3.
In situ hybridization of rat8 gene on rat tissue sections. Cryostatically prepared rat tissue sections were hybridized to 35S-labeled rat8 antisense RNA as described in Materials and Methods. (A) Lactating breast. (B) Fallopian tube. (C) Bronchus.
Figure 4.
In situ hybridization of rat8 mRNA on LA7 cells. Dark-field photographs of LA7 cells in the process of forming domes, hybridized with 35S-labeled rat8 antisense mRNA. Rat8 is markedly expressed in cells forming domes and cells surrounding the domes. (A) Indication of radioactivity above the dome cells; (B) Indication of the radioactivity of the cells outside the domes.
Epithelial Marker Expression.
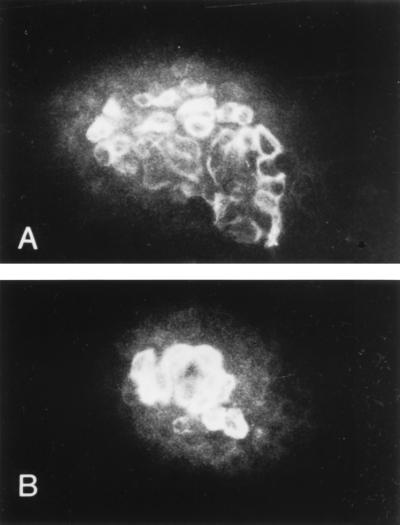
Three markers were used: antibodies specific for E and P cadherin, and antibody specific for cytokeratin 8. In dome-forming cultures the cells of the domes showed strong surface reactivity to E cadherin; the same cells also showed strong reactivity to cytokeratin 8 (Fig. 5), localized in the outer layer of the cytoplasm. The effect was stronger in induced cultures, in which the domes are larger. Cells outside domes showed weak or no reactivity to either antibody.
Figure 5.
Epithelial marker expression. Immunofluorescence microscopy of DMSO-induced LA7 cultures incubated with mAbs anti-cytokeratin 8 (A) and anti-E cadherin (B). Cells reacting with both the antibodies (A and B) are those that form the domes. The cells outside the domes showed weak or no reactivity to either antibody. The secondary antibody used was fluorescein-conjugated anti-mouse IgG.
Analysis of Transfected Clones.
Rat adenocarcinoma 106A10 cells were transfected with the MMTV-p41 recombinant vector designed to produce the full-length message from the rat8 cDNA under the control of MMTV long-terminal repeat promoter. One week after selection with G418 a total of 78 clones were isolated and transferred to 96-well plates, and individually amplified. We found that 32 clones could be further amplified. The expression of the mRNA41 was determined by mRNA isolation and Northern analysis. Among the 32 transfected clones, 10 exhibited high to medium levels of expression of rat8 mRNA. In the expressing clones at confluence we observed areas of aggregate cells, where the cell layer had a different organization, reminiscent of a flat dome (data not shown). No true domes were observed.
DISCUSSION
We have identified a gene, rat8, whose function is required for the differentiation of the LA7 cell line derived from a rat mammary adenocarcinoma. Cultures of this line undergo spontaneous differentiation into characteristic structures, called “domes.” The differentiation can be markedly increased by exposing the cells to inducers, such as DMSO, 8Br cAMP (3).
The rat8 gene is more highly expressed in LA7 cells than in the related line 106A10 that does not produce domes either spontaneously or after induction; rat8 expression is slightly enhanced by treatment with DMSO, which promotes the formation of domes. The fact that the expression of rat8 is required for the formation of domes, either spontaneously or under DMSO induction, is shown by the effect of antisense oligonucleotides to the gene, which abolish dome formation; it is also confirmed by the association of domes with cells expressing the gene in in situ experiments. In the cultures, the gene is expressed most strongly in the cells of the domes, and to a lesser extent, in those surrounding the domes; at a certain distance from the domes, the expression is weak. The production of domes requires, in addition, the expression of additional genes, i.e., genes involved with pump function (4).
The role of the rat8 gene is most likely that of tightening the lateral connection between the cells, because in the presence of the antisense oligonucleotide specific for the gene, the LA7 cultures show marked differences at the boundaries between cells; whereas in the presence of control oligonucleotides there appear to be structures conjoining the cells; these such structures seem absent in the presence of the antisense. This possibility is confirmed by the strong expression of E cadherin in the cells of the domes. In contrast, the basal adherence of the cells to the plastic seems to have been weakened in the domes, favoring their detachment. It is therefore likely that the formation of domes requires the simultaneous expression of several functions: the presence of tight connections between cells, a loosening of the connection with the plastic, and a pumping function causing the accumulation of liquid between the cell layer and the plastic of the dish. The lack of dome formation in cultures of 106A10 line transfected with the rat8 cDNA can be attributed to the presence of additional genetic differences in these cells: the patches of changed appearance of the cell layer seems to be the effect of the rat8 gene expression.
The observations with in situ hybridization that the rat8 gene is not expressed in all cells of an induced LA7 culture, raise the question as to what determines the localization of the expression. One possible interpretation is that initially, the rat8 gene is activated only in sparse individual cells, with the activity then spreading to surrounding cells; a dome would then begin to form as soon as enough cells have undergone the changes caused by the rat8 activation, altering the connections among themselves and between them and the plastic. The spreading of the rat8-positive areas may occur under the influence of signals generated by the positive cells themselves; such a requirement would explain why domes are formed only after the cell layer reaches a certain density, and, within the same cultures, in the areas where the cell density is greater; it would also explain the dome-inducing effect of medium conditioned by confluent LA7 cultures (3).
The in situ observations show that the rat8 gene is normally expressed in many epithelia that surround tubular structures. The localization of the rat8 gene expression, both in the domes of the LA7 cultures and in the ducts of mammary glands, and its effects on the cells, strongly suggests a correspondence between the two structures. The correspondence between the domes and mammary ducts is strengthened by the expression of E cadherin and cytokeratin 8 in the cells of domes. In fact, in various systems the activation of the expression of E cadherin accompanies their differentiation (26–29) and therefore is an index of differentiation of LA7 cells in the domes. Therefore, the demonstration of the requirement for the rat8 gene activity for the development of the domes suggests that it is responsible for one of the steps in tubular development. It may play an especially important role in developmental processes that generate epithelial structures from an undifferentiated matrix, as occurs in the formation of tubules in the end buds of mammary glands in rats (30), which must involve changes in the adhesion among cells. The fact that cells outside the domes, although expressing E cadherin very weakly, form an epithelial layer shows that its expression is not required for adhesion between cells; the function can be provided by other adhesion molecules. The presence of E cadherin in the domes probably indicates that it allows the formation of junctions of a special nature, enabling the cell layer to be lifted by the underlying hydrostatic pressure produced by the pumping function. The expression of a cytokeratin is also a sign of a differentiation process, because cytokeratins of various types are expressed in the cells of mammary ducts at various stages of differentiation (31). Cytokeratin 8, which is expressed in the domes, is frequently associated with tubular structures in other organs (32). Its expression in the domes may be related to that of E cadherin, which interacts with cytoplasmic molecules through catenins (33–35) and may participate in signaling, as has been shown for N cadherin in Xenopus mesoderm (33).
Although it is clear that the function of the rat8 gene is required in dome differentiation, it is not clear how it performs that function. Studies in lymphocytes have shown that the human homolog of this gene (known as 9–27, a member of the 1–8 family, in humans) is expressed at their surface where its protein forms a complex with several other proteins (36). It is thought that the complex is a receptor for unknown ligands, and that it controls cellular adhesion as well as cellular growth (37). In the LA7 cells the main effect of the activation of rat8 is in triggering cell adhesion, whereas its effect on cell growth is negligible. The expression of E cadherin may be part of its effect on adhesion. Yet this expression is localized to the cells forming domes, not to all cells expressing rat8, suggesting that the gene may be involved in a signaling pathway that activates E cadherin expression, and involves other genes.
The expression of cytokeratin may well be controlled by the same pathway, as it occurs only in cells expressing E cadherin. It seems possible, therefore, that the rat8 protein is an important intermediary in a pathway that controls the differentiation leading to dome formation in LA7 cultures, and of that leading to the development of tubules in the mammary gland and probably also in other epithelial organs. Dome formation appears, therefore, to be a good in vitro model, although probably simplified, for investigating developmental processes leading to the formation of tubular structures of various kinds within the mammary gland. The identification of the rat8 gene as being part of this process may be a starting point for characterizing the molecular events involved.
Acknowledgments
We would especially like to acknowledge Dr. Elena Cattaneo for her help and contribution to immunofluorescence microscopy. We thank Drs. Mineko Terao, Enrico Garattini, and Mauro Cimino for protocols and advice on in situ hybridization techniques. We also thank Drs. Sharon Evans, Mary Murray, and Eugenio Scanziani for their help and suggestions; Dr. M.G. Sacco for providing us with the MMTV-neu plasmid DNA; and Sophie Bevan for typing the manuscript. This manuscript was partially funded by Fondazione Italiana per la Ricerca sul Cancro. This is manuscript no. 15 of the Genoma 2000/ITBA Project funded by Cariplo.
ABBREVIATIONS
- DMSO
dimethyl sulfoxide
- MMTV
mouse mammary tumor virus
References
- 1.Bennett D C, Peachey L A, Durbin H, Rudland P S. Cell. 1978;15:283–298. doi: 10.1016/0092-8674(78)90104-6. [DOI] [PubMed] [Google Scholar]
- 2.Dulbecco R. Proc Natl Acad Sci USA. 1979;76:1256–1260. doi: 10.1073/pnas.76.3.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dulbecco R, Okada S. Proc R Soc London B. 1980;208:399–408. doi: 10.1098/rspb.1980.0058. [DOI] [PubMed] [Google Scholar]
- 4.Misfeldt D S, Hamamoto S T, Pitelka D R. Proc Natl Acad Sci USA. 1976;73:1212–1216. doi: 10.1073/pnas.73.4.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayzer D J, Brinson E, Runge M S. Gene. 1992;117:277–278. doi: 10.1016/0378-1119(92)90739-c. [DOI] [PubMed] [Google Scholar]
- 6.Deblandre G A, Marinx O P, Evans S S, Majjaj S, Leo O, Caput D, Huez G A, Wathelet M G. J Biol Chem. 1995;270:23860–23866. doi: 10.1074/jbc.270.40.23860. [DOI] [PubMed] [Google Scholar]
- 7.Lewin A R, Reid L E, McMahon M, Stark G R, Kerr I M. Eur J Biochem. 1991;199:417–423. doi: 10.1111/j.1432-1033.1991.tb16139.x. [DOI] [PubMed] [Google Scholar]
- 8.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 9.Aviv H, Leder P. Proc Natl Acad Sci USA. 1972;69:1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi N, Ko M S H. Genomics. 1994;23:202–210. doi: 10.1006/geno.1994.1478. [DOI] [PubMed] [Google Scholar]
- 11.Maggi A, Susanna L, Bettini E, Mantero G, Zucchi I. Mol Endocrinol. 1989;3:1165–1170. doi: 10.1210/mend-3-7-1165. [DOI] [PubMed] [Google Scholar]
- 12.Ko M S H, Ko S B H, Takahashi N, Nishiguchi K, Abe K. Nucleic Acids Res. 1990;18:4293–4294. doi: 10.1093/nar/18.14.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhyner T A, Biguet N F, Berrard S, Borbely A A, Mallet J. J Neurosci Res. 1986;16:167–181. doi: 10.1002/jnr.490160116. [DOI] [PubMed] [Google Scholar]
- 14.Sive H L, St. John T. Nucleic Acids Res. 1988;16:10937. doi: 10.1093/nar/16.22.10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez R I, Chader G J. Nucleic Acids Res. 1992;20:3528. doi: 10.1093/nar/20.13.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henikoff J G, Pietrokovski S, Henikoff S. Nucleic Acids Res. 1997;25:222–225. doi: 10.1093/nar/25.1.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 18.Rio M C, Bellocq J P, Gairard B, Rasmussen U B, Krust A, Koehl C, Calderoli H, Schiff V, Renaud R, Chambon P. Proc Natl Acad Sci USA. 1987;84:9243–9247. doi: 10.1073/pnas.84.24.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basset P, Wolf C, Rouyer N, Bellocq J P, Rio M C, Chambon P. Cancer (Philadelphia) 1994;74:1045–1049. doi: 10.1002/1097-0142(19940801)74:3+<1045::aid-cncr2820741511>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 20.Rasmussen U B, Wolf C, Mattei M G, Chenard M P, Bellocq J P, Chambon P, Rio M C, Basset P. Cancer Res. 1993;53:4096–4101. [PubMed] [Google Scholar]
- 21.Szczylik C, Skorski T, Nicolaides N C, Manzella L, Malaguarnera L, Venturelli D, Gewirtz A M, Calabretta B. Science. 1991;253:562–565. doi: 10.1126/science.1857987. [DOI] [PubMed] [Google Scholar]
- 22.Sacco M G, Mangiarini L, Villa A, Macchi P, Barbieri O, Sacchi M C, Monteggia E, Fasolo V, Vezzoni P, Clerici L. Gene Ther. 1995;2:493–497. [PubMed] [Google Scholar]
- 23.Fasel N, Pearson K, Buetti E, Diggelmann H. EMBO J. 1982;1:3–7. doi: 10.1002/j.1460-2075.1982.tb01115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelly J M, Gilbert C S, Stark G R, Kerr I M. Eur J Biochem. 1985;153:367–371. doi: 10.1111/j.1432-1033.1985.tb09312.x. [DOI] [PubMed] [Google Scholar]
- 25.Reid L E, Brasnett A H, Gilbert C S, Porter A C, Gewert D R, Stark G R, Kerr I M. Proc Natl Acad Sci USA. 1989;86:840–844. doi: 10.1073/pnas.86.3.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graff J R, Herman J G, Lapidus R G, Chopra H, Xu R, Jarrard D F, Isaacs W B, Pitha P M, Davidson N E, Baylin S B. Nucleic Acids Res. 1995;55:5195–5199. [PubMed] [Google Scholar]
- 27.Vleminckx K, Vakaet L, Mareel M, Fiers W, Van Roy F. Cell. 1991;66:107–119. doi: 10.1016/0092-8674(91)90143-m. [DOI] [PubMed] [Google Scholar]
- 28.Frixen U H, Behrens J, Sachs M, Eberle G, Voss B, Warda A, Lochner D, Birchmeier W. J Cell Biol. 1991;113:173–185. doi: 10.1083/jcb.113.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Navarro P, Gomez M, Pizarro A, Gamallo C, Quintanilla M, Cano A A. J Cell Biol. 1991;115:517–533. doi: 10.1083/jcb.115.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dulbecco R, Allen W R, Bowman M. Proc Natl Acad Sci USA. 1984;81:5763–5766. doi: 10.1073/pnas.81.18.5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allen R, Dulbecco R, Syka P, Bowman M, Armstrong B. Proc Natl Acad Sci USA. 1984;81:1203–1207. doi: 10.1073/pnas.81.4.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kasper M, von Dorsche H, Stosiek P. Histochemistry. 1991;96:271–277. doi: 10.1007/BF00271547. [DOI] [PubMed] [Google Scholar]
- 33.Holt C H, Lemaire P, Gurdon J B. Proc Natl Acad Sci USA. 1994;91:10844–10848. doi: 10.1073/pnas.91.23.10844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takeichi M. Science. 1991;251:1451–1455. doi: 10.1126/science.2006419. [DOI] [PubMed] [Google Scholar]
- 35.Ozawa M, Barbault H, Kemler R. EMBO J. 1989;8:1711–1717. doi: 10.1002/j.1460-2075.1989.tb03563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi S, Doss C, Levy S, Levy R. J Immunol. 1990;145:2207–2213. [PubMed] [Google Scholar]
- 37.Evans S S, Lee D B, Han T, Tomsi T B, Evans R L. Blood. 1990;76:2583. [PubMed] [Google Scholar]