Abstract
Background
Pseudomonas aeruginosa frequently colonizes and is responsible for severe ventilator-associated pneumonia in intubated patients. A quorum-sensing (QS) circuit, depending on the production of the two QS-signaling molecules (autoinducers, AIs) 3-oxo-C12-HSL and C4-HSL, regulates the production by P. aeruginosa of several virulence factors and is required for biofilm formation. Therefore QS-inhibition has been suggested as a new target for preventive and/or therapeutic strategies. However the precise role of QS during colonization and subsequent infections of intubated patients remains unclear.
Results
We wondered whether QS is active during colonization of intubated patients, and whether P. aeruginosa isolates growing inside the biofilm covering the intubation devices and those resident in the lungs of colonized patients differ in their QS-dependent phenotypes. We collected the intubation devices of eight patients colonized by P. aeruginosa. We detected 3-oxo-C12-HSL on eight, and C4-HSL on six of these devices. In three of these patients we also obtained P. aeruginosa isolates from tracheal aspirates at the time of extubation (n = 18), as well as isolates from the intubation devices (n = 25). We genotyped these isolates, quantified their AIs production, and determined three QS-dependent phenotypes (adherence capacity, biofilm and elastase production). The production of 3-oxo-C12-HSL was consistently increased for isolates from the intubation devices, whereas the production of C4-HSL was significantly higher for isolates from tracheal aspirates. Isolates from tracheal aspirates produced significantly higher amounts of elastase but less biofilm, and had a marginally reduced adhesion capacity than isolates from the intubation devices. Levels of 3-oxo-C12-HSL and elastase production correlated statistically for tracheal intubation isolates, whereas levels of 3-oxo-C12-HSL production and adhesion ability, as well as biofilm production, correlated weakly amongst intubation device isolates.
Conclusion
Our findings demonstrate that autoinducers are produced during the colonization of intubated patients by P. aeruginosa. The microenvironment, in which P. aeruginosa grows, may select for bacteria with different capacities to produce autoinducers and certain QS-dependent phenotypes. QS-inhibition might therefore affect differently isolates growing inside the biofilm covering intubation devices and those resident in the lungs.
Background
Pseudomonas aeruginosa is an opportunistic pathogen implicated in a wide variety of infections, particularly in burn victims, cancer patients and cystic fibrosis (CF) patients. In addition P. aeruginosa is a major cause of severe pneumonia in mechanically ventilated patients [1]. Such ventilator-associated pneumonia (VAP) are almost always preceded by the colonization of the upper respiratory tract. The presence of the intubation device is a major risk factor for this colonization [2]. As such the capacities of P. aeruginosa to adhere to the inert surface and grow inside a biofilm on the intubation device are thought to be essential for the subsequent colonization and infection of the lower respiratory tract [3].
After colonizing the respiratory tract of intubated patients, P. aeruginosa causes infections and extensive damage to host tissues via the production of a number of extracellular virulence factors [4]. The expression of several of these factors is controlled by a quorum-sensing (QS) circuitry involving at least two autoinducers (AIs): N-(3-oxododecanoyl)-Homoserine Lactone (3-oxo-C12-HSL) and N-butyryl-L-Homoserine Lactone (C4-HSL) [4]. The production of 3-oxo-C12-HSL and C4-HSL requires the autoinducer synthases encoded by the lasI and rhlI genes, respectively. These QS-systems were first described for their role in the regulation of extracellular virulence factors [5,6] but have recently been shown to regulate many other bacterial functions including biofilm formation [7-10].
The importance of a functional QS-circuit for the virulence of P. aeruginosa has been clearly established in vivo in animal models [11]. As a consequence QS-inhibition has been suggested as a potential target for new preventive and/or therapeutic strategies of P. aeruginosa infections.
However the precise role during human infections remains speculative. Most studies so far have focused on the capacity of clinical isolates to produce the two autoinducers and on the detection of the two AIs directly in clinical samples. P. aeruginosa isolates from CF patients produce AIs in vitro [12], and sputum of CF patients colonized by P. aeruginosa produces the same signals ex vivo [13]. AIs have also been detected in situ in CF lung tissue [14], in the sputum of CF patients [15,16], and in bronchoalveolar lavage fluid after lung transplantation [17]. Recently we observed that most strains initially colonizing intubated patients are QS-proficient [18]. Both 3-oxo-C12-HSL and C4-HSL have also been detected in biofilms, both in vivo and in situ [15,16,19]; moreover their relative abundance has been suggested as a marker for the biofilm mode of growth in CF patients [13].
Because much remains to be understood about the role of QS during human infections, and the potential of QS-inhibition in their prevention and/or treatment, we examined the behavior of QS during the colonization of intubated patients. To determine whether QS is active during the colonization of intubated patients, we first extracted and quantified AIs directly from the intubation devices of patients colonized with P.aeruginosa. Because of the multitude of phenotypes depending on QS we then wondered whether P. aeruginosa isolates growing in different microenvironments might vary in their capacity to produce AIs and differ in their expression of QS-dependent phenotypes. We therefore collected P. aeruginosa isolates from the surface of intubation devices and from tracheal aspirates obtained from the same patients on the day of extubation. For each patient, we grouped strains according to their genotype, and characterized them in terms of production of autoinducers, and capacity to adhere to PVC, and produce elastase and biofilm. Our results suggest that the relative production of autoinducers, and the QS-dependent phenotypes of P. aeruginosa vary according to their site of isolation.
Results
Study population
Patients' clinical characteristics are presented in Table 1. Underlying diseases included polytrauma, intra-cerebral hemorrhage, congestive heart failure and non-pseudomonal sepsis. Duration of mechanical ventilation ranged from 9 to 24 days. Detailed clinical characteristics are provided for the three patients from whom P. aeruginosa isolates were collected both from the intubation device and from a tracheal aspirate on the day of extubation:
Table 1.
Patient characteristics and intubation devices
| Patient | Underlying disease | Duration of intubation (days) | Tracheal aspirate on day of extubation | 3-oxo-C12-HSL (nmoles/cuff) | C4-HSL (nmoles/cuff) |
| 2 | Polytrauma | 9 | no | 0.4 | 100 |
| 5 | Nonpseudomonal sepsis | 13 | no | 0.5 | 17 |
| 7 | Intra-cerebral hemorrhage | 24 | no | > 10 | 22 |
| 8 | Polytrauma | 15 | yes | > 10 | 22 |
| 11 | Polytrauma | 18 | no | > 10 | 300 |
| 12 | Polytrauma | 9 | yes | > 10 | 2 |
| 13 | Congestive heart failure | 17 | yes | > 10 | 2 |
| 14 | Polytrauma | 16 | no | 6 | 24 |
Patient 8 was a 27-year-old male who suffered a severe polytrauma including cerebral contusion and hemorrhage. P. aeruginosa was cultured for the first time from his tracheal aspirates 7 days after his intubation. Therapeutic retreat was decided after 15 days of mechanical ventilation and he died without having received any antibiotics. The presence of P. aeruginosa in his tracheal aspirates was considered as colonization. The tracheal aspirate used in the present study was collected shortly before his death and extubation.
Patient 12 was a 21-year-old female who suffered a severe polytrauma including a cranio-cerebral trauma with diffuse axonal lesions and multiple rib fractures. P. aeruginosa was first cultured in bronchoalveolar lavage fluid four days after her intubation. P. aeruginosa pneumonia was diagnosed two days later and she was treated with an imipenem – tobramycin combination therapy. The clinical response was satisfying and she was extubated 3 days later still on antimicrobial therapy. A tracheal aspirate was obtained on the day of extubation.
Patient 13 suffered severe congestive heart failure with a cardiac arrest. P. aeruginosa was first cultured from a tracheal aspirate three days after her intubation. P. aeruginosa pneumonia was treated with cefepime during 10 days starting 6 days after intubation. She remained colonized by P. aeruginosa and was extubated after 17 days of mechanical ventilation.
In situ quantification of autoinducers on intubation devices
The cuff of each endotracheal intubation device was removed and autoinducers were extracted from the biofilm as described in Materials and Methods. 3-oxo-C12-HSL was detected on all cuffs, the amounts varying between 0.4 and more than 10 nmoles per cuff (Table 1). C4-HSL was detected on six of the eight devices tested and its amount varied between 17 and 300 nmoles per cuff (Table 1). The in situ isolation of autoinducers from biofilms of the majority of the intubation devices suggests that QS might play a role during colonization by P. aeruginosa of such devices. The fact that we detected C4-HSL only from 6 devices could result from the lower sensitivity of our C4-HSL bioassay (2 nmoles/cuff) compared to the one used to quantify 3-oxo-C12-HSL (0.01 nmoles/cuff).
Collection of P. aeruginosa isolates colonizing the intubation devices and the lungs of intubated patients
For the three patients in whom we obtained tracheal aspirates (TA) at the time of extubation we also collected P. aeruginosa isolates directly from the cuff of the intubation device (ID) as described in Material and Methods. The complete collection consisted of 18 TA isolates and 25 ID isolates partitioned as shown in Table 2. Genotyping of these strains by RAPD identified 4 different genotypes (B, C, D and P). Genotypes C (3 isolates from patient 12 and 1 isolate from patient 13) and P (3 isolates from patient 13) were only found in tracheal aspirates and were therefore excluded from the analyses described in the following paragraphs, when not otherwise stated. The final collection therefore consisted of 11 TA and 25 ID isolates representing genotype B (8 TA and 21 ID isolates) and genotype D (3 TA and 4 ID isolates) (Table 2).
Table 2.
Characterization of P. aeruginosa isolates from intubated patients
| Isolates | Patients | Genotype | Site of Isolation | Productionc of | Elastasea(OD495/OD660) | Productiona of | Adhesiona (%) | Biofilma (%) | ||
| Proteases | Rhamnolipids | 3-oxo-C12-HSL (μM) | C4-HSL (μM) | |||||||
| 61G3d | 8 | D | TA | + + + | BDb | 0.5 | 0.51 | 15 | 88 | 31 |
| 61E8 | 8 | D | TA | + + + | BD | 0.59 | 0.41 | 24 | 88 | 11 |
| 60A3d | 8 | B | TA | + + + | + | 0.68 | 0.33 | 7.8 | 100 | 13 |
| 60B5 | 8 | D | TA | + + + | BD | 0.65 | 0.36 | 5 | 139 | 33 |
| 63E8 | 8 | B | ID | + + + | + | 0.3 | 1.3 | 2.4 | 116 | 83 |
| 63E9 | 8 | B | ID | + + + | + + | 0.34 | 2.5 | 1.1 | 111 | 105 |
| 63E11d | 8 | B | ID | + + + | + + + | 0.29 | 0.19 | 2.9 | 105 | 89 |
| 63G3 | 8 | B | ID | + + + | + | 0.24 | 0.63 | 15 | 115 | 59 |
| 63G6 | 8 | D | ID | + + + | BD | 0.3 | 0.36 | 3 | 120 | 98 |
| 63G8 | 8 | B | ID | + + + | + + | 0.2 | 0.21 | 3.3 | 106 | 125 |
| 63H2 | 8 | B | ID | + + + | + + + | 0.12 | 2.7 | 2.9 | 137 | 74 |
| 62F2 | 8 | D | ID | + + + | BD | 0.32 | 0.37 | 3.1 | 99 | 10 |
| 62F3d | 8 | D | ID | + + + | + | 0.22 | 0.17 | 2.4 | 101 | 20 |
| 62G3 | 8 | D | ID | + + + | BD | 0.3 | 0.29 | 3.3 | 110 | 25 |
| 54A7 | 12 | C | TA | + + + | + + | 0.5 | 0.36 | 30 | 104 | 59 |
| 54A8 | 12 | B | TA | + + + | + | 0.63 | 0.41 | 19 | 115 | 49 |
| 54A9 | 12 | C | TA | + + + | + + + | 0.5 | 0.38 | 12 | 105 | 49 |
| 54B1 | 12 | C | TA | + + + | + | 0.64 | 0.77 | 30 | 99 | 60 |
| 54D5d | 12 | B | TA | + + + | + | 0.48 | 0.35 | 14 | 116 | 50 |
| 54D6d | 12 | B | TA | + + + | + + + | 0.43 | 0.31 | 18 | 80 | 53 |
| 54E1 | 12 | B | TA | + + + | + + | 0.72 | 1 | 7.9 | 121 | 58 |
| 55A1d | 12 | B | ID | + + | BD | 0.19 | 2.6 | 2.1 | 116 | 78 |
| 55A2 | 12 | B | ID | + + | + | 0.27 | 1.2 | 1.3 | 122 | 75 |
| 55A5 | 12 | B | ID | + + | + + | 0.26 | 2.5 | 2.8 | 137 | 102 |
| 55D2 | 12 | B | ID | + + | + | 0.27 | 2 | 3.4 | 118 | 68 |
| 55E4 | 12 | B | ID | + + | + + | 0.31 | 2.5 | 2.7 | 115 | 88 |
| 55F3d | 12 | B | ID | + + | + + + | 0.22 | 2.5 | 2.2 | 101 | 59 |
| 55G2 | 12 | B | ID | + + | + + | 0.17 | 4 | 2 | 117 | 84 |
| 55G4 | 12 | B | ID | + + | + | 0.19 | 1.3 | 1.6 | 112 | 103 |
| 55A6 | 12 | B | ID | + + | + + + | 0.25 | 2.1 | 3.4 | 126 | 109 |
| 56A1 | 13 | B | TA | + + + | + + | 0.64 | 0.44 | 6.4 | 85 | 46 |
| 59B3 | 13 | C | TA | + + + | + + | 0.45 | 0.3 | 7.9 | 88 | 39 |
| 59B4d | 13 | B | TA | + + + | + + | 0.47 | 0.16 | 9.5 | 83 | 48 |
| 59A1d | 13 | B | TA | + + + | + + + | 0.52 | 0.13 | 9.3 | 99 | 38 |
| 59B6 | 13 | P | TA | + + + | + + | 0.45 | 0.28 | 13.5 | 83 | 41 |
| 59C1 | 13 | P | TA | + + + | + + + | 0.52 | 0.27 | 11.3 | 106 | 77 |
| 59C2 | 13 | P | TA | + + + | + | 0.47 | 0.33 | 13.3 | 101 | 59 |
| 63A1 | 13 | B | ID | + + + | + + | 0.23 | 1.4 | 1.8 | 114 | 91 |
| 63A3d | 13 | B | ID | + + + | + + + | 0.3 | 0.33 | 3.1 | 137 | 77 |
| 61A12d | 13 | B | ID | + + + | + + + | 0.15 | 2.6 | 2.2 | 129 | 90 |
| 61G12 | 13 | B | ID | + + + | + + | 0.11 | 2.7 | 2.4 | 121 | 87 |
| 62C2 | 13 | B | ID | + + + | + + | 0.15 | 2.6 | 3.1 | 121 | 97 |
| 62C3 | 13 | B | ID | + + + | + + + | 0.08 | 1.3 | 3.3 | 143 | 86 |
a Values are mean of three independent experiments with a standard deviation of < 10%.
b BD, below detection.
c +++, high level; ++, intermediate level; +, low level.
d lasR and rhlR genes sequenced
Phenotypic characterization of TA and ID P. aeruginosa isolates
(i) In vitro autoinducer production
We extracted and quantified the two autoinducers from culture supernatants of TA and ID isolates. All isolates produced both autoinducers in vitro, though levels varied according to the site of isolation (Table 2). Figure 1A and 1B show the mean production of both AIs for the isolates obtained from TA and ID for each individual patient. For the three patients ID isolates consistently produced higher amounts of 3-oxo-C12-HSL than TA isolates (Fig. 1A). When all the ID isolates were compared to the TA isolates the mean production of 3-oxo-C12-HSL was increased in the ID group (1.6 × 10-6M; Standard Error [SE], 4.8 × 10-7M), compared to the TA group (4.0 × 10-7M; SE, 7.4 × 10-8M, p = 0.12) without reaching statistical significance, possibly due to the small sample size. In contrast the TA isolates of all patients produced higher amounts of C4-HSL than the ID isolates (Fig. 1B), and the mean C4-HSL concentration was significantly higher for the TA isolates group (1.2 × 10-5 M; SE, 1.7 × 10-6M) than for the ID isolates group (3.1 × 10-6M; SE, 5.4 × 10-7M; p = 0.04). Interestingly all isolates (including genotypes C and P), whatever their site of collection, produced higher levels of C4-HSL than of 3-oxo-C12-HSL. The mean of the C4-HSL/3-oxo-C12-HSL ratios was 38.5 for the TA group compared to 5.1 for the ID group (p = 0.002; Fig. 1C). Excluding the TA isolates with genotype C and P did not affect these results significantly. Therefore the relative amounts of AIs produced in vitro were significantly different between TA and ID isolates. The fact that all P. aeruginosa isolates tested were QS-proficient, i.e. able to produce both AIs in vitro, supports a role for QS during late stages of colonization of intubated patients both in the biofilm formed on intubation devices and in the lung.
Figure 1.
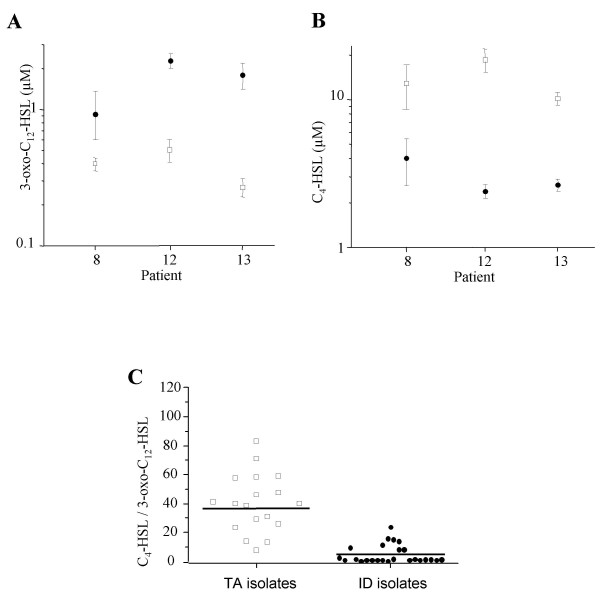
Autoinducer production of P. aeruginosa isolates from tracheal aspirates and intubation devices. 3-oxo-C12-HSL (A) and C4-HSL (B) production by P. aeruginosa isolates recovered from tracheal aspirates (open squares, TA isolates) and intubation devices (plain circles, ID isolates) of 3 colonized patients (8, 12 and 13). The production of the two autoinducers was determined in culture supernatants by specific bioassays. Error bars correspond to the standard errors of the means. (C) Ratios of AI concentrations (C4-HSL/3-oxo-C12-HSL) produced by 18 isolates recovered from tracheal aspirates and 25 isolates recovered from intubation devices. The horizontal line indicates the mean of the ratios.
(ii) Production of QS-dependent virulence factors
To further characterize the P. aeruginosa isolates, we determined their in vitro production of three virulence factors that depend upon an active QS-circuitry, namely, elastase, protease, and rhamnolipids [4]. We assayed the production of proteases and rhamnolipids using semi-quantitative plate assays. Important fluctuations in the production of rhamnolipids were observed (Table 2). This finding was not surprising as we had previously found fluctuations among QS-associated phenotypes occurring during the colonization of intubated patients [18]. LasB elastase is the most potent elastase produced by P. aeruginosa and is one of the major virulence factors controlled by QS [4]. We quantified the in vitro elastase production of the clinical isolates by means of the Elastin Congo Red assay at the same point in the growth curve (early stationary phase) as for determination of AIs production. For each patient the isolates obtained from the TA displayed higher elastolytic activities than those obtained from the ID (Fig. 2A). When the isolates were grouped by site of isolation, the mean elastase production was higher in the TA group (0.57; SE, 0.02) than in the ID group (0.23; SE, 0.02; p < 0.001; Fig. 2B). These results did not differ when all the genotypes were included in the TA isolates group. These results suggest that P. aeruginosa strains originating from the lungs of intubated patients produce higher amounts of elastase than isolates that were resident inside the biofilm on the intubation device.
Figure 2.
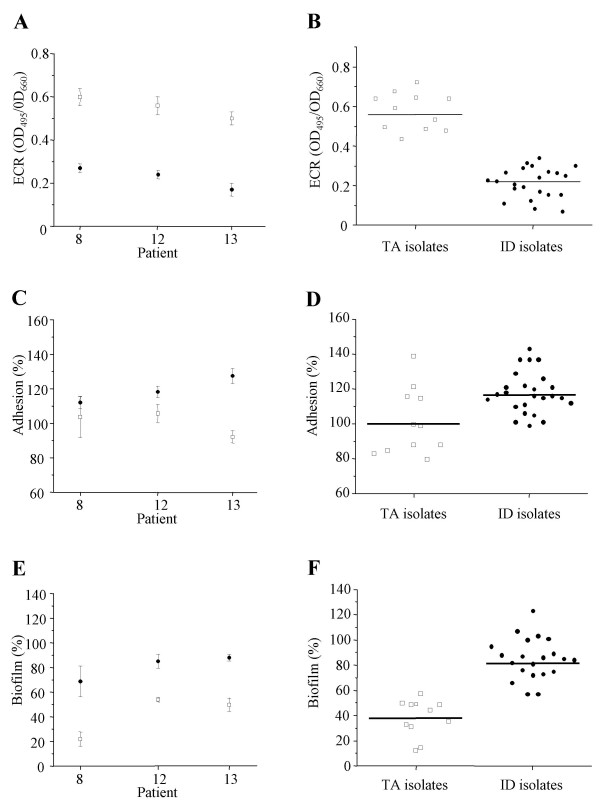
Quorum-sensing dependent phenotypes of P. aeruginosa isolates from tracheal aspirates and intubation devices. Elastase production (A and B), adhesion capacity (C and D) and biofilm formation (E and F) by P. aeruginosa isolates recovered from tracheal aspirates (open squares, TA isolates) and intubation devices (plain circles, ID isolates). Elastolytic activities were determined by ECR assays and expressed as the OD measured at 495 nm per unit of OD 660 nm. The adhesion capacities and biofilm formation were expressed as the percentage relative to the level obtained with the wild-type P. aeruginosa strain PT5. (A, C and E) Results individualized for each patient (8, 12 and 13), error bars correspond to the standard errors of the means. (B, D and F) Mean values and variances for 11 tracheal aspirate isolates and 25 isolates recovered from the intubation devices. For each individual isolate the determinations were performed twice in triplicates.
(iii) Adhesion and biofilm formation capacity
QS has been suggested to be involved in the formation and differentiation of biofilms [7,8,13]. The impact of the type of surface and medium used in biofilm formation assays appears to influence the relationship between QS and biofilm formation [20]. We therefore decided to measure biofilm formation with our well-established static system using sterile PVC coupons obtained from intubation devices [8]. In our opinion this model simulates the best the conditions found on intubation devices in patients. Both adhesion and biofilm formation capacities were compared to the laboratory strain P. aeruginosa PT5 (100%). For each individual patient P. aeruginosa isolates recovered from the surface of the ID adhered more efficiently than their counterparts collected from the TA (Fig. 2C). When all the isolates were grouped by site of isolation the mean adhesion capacity was slightly increased in the ID group without reaching statistical significance (118.0%; SE, 4.0%) as compared to the TA group (101.3%; SE, 5.2%; p = 0.19; Fig. 2D). The ID isolates for each patient demonstrated a greater ability to form biofilm than their TA counterparts (Fig. 2E). This difference was statistically significant when the strains were grouped according to their site of isolation (ID isolates: 79.3%; SE, 6.3%, TA isolates: 39.1%; SE, 9.8% p= 0.01, Fig. 2F). Again these results were not affected when genotypes C and P were included in the TA group. These observations indicate that P. aeruginosa isolates originating from the biofilm of intubation devices tend to adhere more efficiently and have higher biofilm production capacities than their counterparts isolated from the lungs.
Correlations between autoinducer production and quorum-sensing dependent phenotypes
(I) Correlations between autoinducer and elastase production
In vitro, the production of elastase is under the joint control of both the las and rhl QS-systems [4,9]. In sputum of CF patients the expression of lasR has been correlated with the accumulation of lasB transcripts [21]. Other investigators have described a weak correlation between the in vitro transcription levels of lasR and lasB in 50% of environmental and clinical isolates [22]. We wondered whether the production of elastase would be correlated with the production of autoinducers. For these analyses we included only genotypes B and D, which were present in both TA and ID isolates. We observed a trend for a positive correlation between 3-oxo-C12-HSL and elastase production for the TA group (r = 0.58; p = 0.06). This positive correlation became statistically significant when genotypes C and P were included (r = 0.63, p = 0.006; Fig. 3A). In contrast for ID isolates we observed a statistically significant negative correlation between elastase and 3-oxo-C12-HSL production (r = -0.41, p = 0.04). Furthermore, C4-HSL production was not correlated with elastase activity, whatever the isolate group studied (TA isolates: r =-0.38, p = 0.25 and ID isolates: r = 0.02, p = 0.70). These results suggest that the level of 3-oxo-C12-HSL is positively correlated to elastase production for isolates from tracheal aspirates of colonized patients, but not for P. aeruginosa isolates from biofilms covering the intubation devices.
Figure 3.
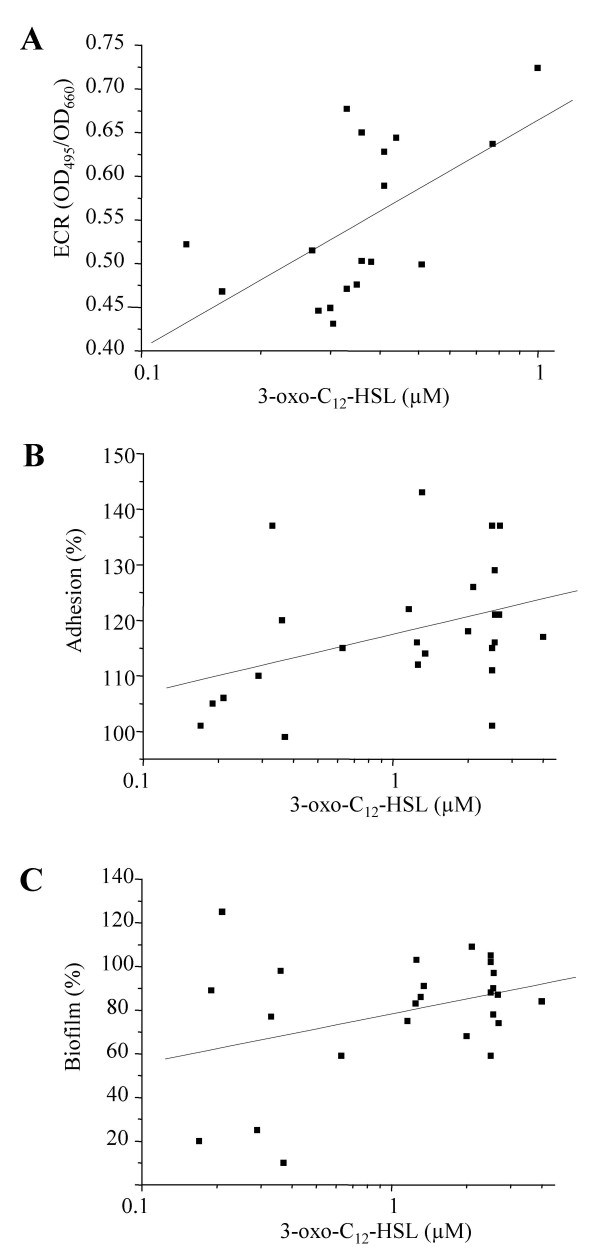
Correlations between autoinducer production and quorum-sensing dependent phenotypes. Relations between levels of 3-oxo-C12-HSL and elastase production by P. aeruginosa isolates recovered from tracheal aspirations (A), and adhesion capacity (B) as well as biofilm formation (C) by P. aeruginosa isolates recovered from intubation devices. (A) For each isolate the production of 3-oxo-C12-HSL and elastase was measured in aliquots taken at the same time during the growth curve, and plotted against each other. (B and C) For each isolate aliquots for 3-oxo-C12-HSL determinations were taken at an identical time during the growth curve. Adhesion capacities and biofilm formation are expressed compared to wild type strain PT5 (100%). Each determination is the mean of two independent experiments performed in triplicate.
(II) Correlation between autoinducer production, adhesion and biofilm formation capacities
We then examined correlations between autoinducer production and adhesion capacity. There was a weak, statistically not significant, correlation between 3-oxo-C12-HSL production and adhesion capacity for isolates from the ID group (r = 0.30, p = 0.14) (Fig. 3B). No such correlation was found for isolates from the TA group (r = 0.33, p = 0.32), even when genotypes C and P were included in the analysis (r = 0.30, p = 0.22). We found no correlation between C4-HSL production and adhesion for both ID isolates (r = 0.0; p = 0.99) and TA isolates (n = 11: r = 0.34; p = 0.30, and n = 18: r = 0.16; p = 0.53).
Similarly we observed a weak, statistically not significant, correlation between 3-oxo-C12-HSL and biofilm production for isolates of the ID group (r = 0.33, p = 0.11, Fig. 3C), but not for those recovered from TA (r = 0.25, p = 0.46), even after inclusion of genotypes C and P in the analysis (r = 0.17, P = 0.49). There was no correlation between C4-HSL and biofilm production for the ID (r = -0.18, p = 0.31), and TA isolates (r = -0.18, p = 0.60), despite the inclusion of genotypes C and P (r = 0.16, p = 0.52). Together these observations suggest that the production of 3-oxo-C12-HSL but not of C4-HSL, might be associated with in vitro adherence and biofilm formation capacities among isolates recovered from the biofilm on intubation devices.
Discussion
Respiratory tract colonization of intubated patients by P. aeruginosa is initiated by the colonization of the intubation device by bacteria either originating from the digestive flora of the patient (endogenous acquisition) or transmitted via handling of the device by health care workers (exogenous acquisition) [23]. This colonization requires efficient bacterial adherence to the inert surface and the formation of a biofilm [3]. P. aeruginosa then reach the respiratory tract either by leakage around the cuff of the intubation device, by shearing of secretions containing embedded bacteria from the device during the inspiratory air flow, or by contamination and direct inoculation of the respiratory tract by suction tubes introduced through the intubation device by health care workers [23-25].
Both in vitro studies and animal models have suggested that QS plays a major role in the virulence of P. aeruginosa [11]. However in situ and in patient data concerning the potential role of QS during colonization and infection by P. aeruginosa in humans remain scarce. QS-activity has been linked to the detection of P. aeruginosa AIs in the lungs of cystic fibrosis patients [13-16], as well as in lung transplanted patients [17]. So far studies have only shown that most of the isolates originating from colonized intubated patients are QS-proficient [18,22].
In the present study we show that the two P. aeruginosa autoinducers, 3-oxo-C12-HSL and C4-HSL, can be detected in situ in biofilms covering intubation devices retrieved from patients colonized by P. aeruginosa. Moreover all P. aeruginosa isolates collected either from these biofilms or from tracheal aspirates produced these AIs in vitro. Whereas 3-oxo-C12-HSL has been previously shown to play a role in the differentiation of P. aeruginosa biofilms [7,26], C4-HSL seems to be important during the maturation stage of biofilm development [27], for the total amount of biofilm formed [8], and for the maintenance of biofilm architecture [28]. In our study, isolates recovered from the biofilm of intubation devices produced higher levels of C4-HSL than 3-oxo-C12-HSL in vitro. Previous observations have suggested that C4-HSL is produced in higher quantities than 3-oxo-C12-HSL in P. aeruginosa biofilms in the lungs of CF patients [13,14]. Singh et al. have even suggested that this AI ratio might serve as a biomarker for the biofilm mode of growth, as the mucoid isolates from the sputum of CF patients inverted their AI ratio when sub-cultured in liquid medium [13]. In contrast, in our study a higher level of C4-HSL as compared to 3-oxo-C12-HSL was observed when isolates were grown in liquid cultures, suggesting that this ratio is not useful as a biomarker for biofilm growth for these isolates. None of the isolates collected from our patients had a mucoid phenotype. This discrepancy could therefore be explained by inherent differences in biofilm development patterns between mucoid and non-mucoid strains. So far there is no evidence that non-mucoid P. aeruginosa colonizing the lung of intubated patients grows in biofilms. Such a hypothesis deserves further investigations.
Autoinducers have been previously recovered from biofilms formed on urinary tract catheters [19] and our results show for the first time that QS-signaling molecules are also produced in biofilms covering intubation devices. Whether the local concentrations of AIs on the intubation devices that we measured are physiologically relevant and are sufficient to activate the expression of QS-dependent genes remains speculative. However microarray analyses have shown that QS is a continuum and that the expression of some genes is already activated even at low AI concentrations [9]. It is therefore likely that even low concentrations of AIs on the intubation devices are physiologically relevant.
Intriguingly, the in vitro production of both AIs varied with the site of collection. Isolates from tracheal aspirates produced higher levels of C4-HSL but lower levels of 3-oxo-C12-HSL than their genotypically identical counter-parts isolated from intubation devices, even after several in vitro passages. This observation suggests that different microenvironments select P. aeruginosa isolates with various AI production capacities. Moreover the production of elastase, and the capacity to adhere to an inert surface and to produce a biofilm also varied with the site of isolation. Tracheal aspirate isolates produced higher levels of elastase, but were less able to adhere and produce biofilm than their counter-parts recovered from intubation devices. LasB elastase is one of the major virulence factors controlled by the P. aeruginosa QS-circuit [4]. In vitro its production is regulated by the QS-circuit and depends mainly on the production of 3-oxo-C12-HSL [9,10]. LasB elastase is believed to allow P. aeruginosa to invade surrounding tissues by its broad range enzymatic activity and to facilitate blood stream invasion by degradation of elastin fibers in the lamina propria of blood vessels [4].
The observed differences in phenotypes between isolates obtained from tracheal aspirates, compared to those from the intubation devices, make sense. Isolates growing in the biofilm on intubation devices might not invest energy in producing elastase, but might be primed to adhere and form biofilms, whereas isolates growing in the lungs might find an advantage in producing elastase. For tracheal aspirate isolates, levels of 3-oxo-C12-HSL production correlated positively with the capacity to produce elastase. In contrast for isolates originating from the intubation device, 3-oxo-C12-HSL levels did not correlate with elastase production, but correlated weakly with the capacity to adhere and to form biofilms on inert surfaces. These observations suggest that the previously described control of LasR on elastase production might not apply to all isolates depending on the site they were collected from. It seems that the microenvironment on the intubation devices selects for phenotypes that produce high levels of 3-oxo-C12-HSL but without concomitant increased elastase production. In contrast the lower lung environment seems to select for isolates that produce less 3-oxo-C12-HSL but with a positive control of this AI on elastase production. Two of our patients (12 and 13) received antimicrobial therapies during their mechanical ventilation. We can therefore not exclude that different concentrations of these drugs at the two sites might have selected different phenotypes. However this seems unlikely as isolates of patient 8 presented the same differences in phenotype in the absence of any exposure to antimicrobial therapies.
We detected no correlations between the levels of C4-HSL production and elastase activity, adhesion or biofilm formation. Strikingly, these phenotypes remained stable for each isolate independently of its origin even after several in vitro passages. This suggests that mutations in specific regulators, that affect the production of AIs and the expression of QS-dependent phenotypes might be selected in particular microenvironments. Indeed lasR and rhlR mutants of P. aeruginosa strains isolated from intubated patients have been previously described [18]. Such mutations influence the bacteria capacity to produce AIs, and are expected to affect QS-dependent phenotypes. We therefore sequenced both lasR and rhR in 6 TA and 6 ID isolates (Table 2). None of them harbored mutations compared to the wild type strain PAO1 (data not shown). This explains why all the isolates analyzed in the present study were QS-proficient. Phenotypic variations not linked to QS but also influencing biofilm formation have been described in P. aeruginosa isolates from CF patients [29]. The nature of the mutational events possibly selected either during biofilm growth on intubation devices, or inside the lung are presently under investigation. The identification of these events would have major implications for our understanding of bacterial selection in specific environments and the dynamics of bacterial populations.
QS-inhibition has been suggested as a new therapeutical approach against P. aeruginosa infections. For instance macrolides interfere with the production of AIs [8,30] and were shown to retard biofilm formation [31]. Our result might be important if QS-inhibitors that interfere with the production and/or activity of one, or both, of the autoinducers are used in the prevention or treatment of P. aeruginosa VAP. Indeed, our data suggest that inhibition of QS might have different effects on P. aeruginosa isolates depending on their microenvironnement.
Conclusion
Our study was limited to the analysis of three patients; therefore it is difficult to draw general conclusions from our observations. However the results were consistent between these patients and reached statistical significance despite the small number. Our results showed that during colonization of intubated patients P. aeruginosa autoinducers are produced in the biofilms covering the intubation devices. The capacity to produce autoinducers and the expression of QS-dependent phenotypes vary with the site of isolation. Further studies will be required to investigate the mechanism that led to selection of specific phenotypes in a particular microenvironment directly in vivo and to determine how this impacts the use of anti-quorum-sensing strategies.
Methods
Patient population and clinical sample collection
We collected the intubation devices of eight patients known to be colonized by P. aeruginosa and hospitalized in the surgical intensive care service of the University Hospital Geneva. All patients, except patient 14, have been described previously (Table 1) [18]. Following extubation the endotracheal tubes were wrapped in a sterile cloth and transported without delay to the laboratory were they were stored at -70°C until use. Sterile cotton swabs were used to detach bacteria embedded in biofilm formed on the cuff, on the exterior surface of the intubation device (ID). Pseudomonas isolates were identified on selective Cetrimide agar plates. In order to avoid compiling identical P. aeruginosa isolates, we collected individual clones based on morphologic differences of their respective colony forming units (CFU). A total of 25 ID isolates were collected from patients 8, 12 and 13, and stored at -70°C (Table 2).
We obtained tracheal aspirates by suctioning secretions with a sterile tube introduced deeply inside the lower respiratory tract through the intubation device from three patients (patients 8, 12 and 13) either on the day before, or on the day of extubation. The tracheal aspirates (TA) were plated on selective agar plates (Cetrimide 0.03%) in order to identify Pseudomonas isolates [14]. Similarly to ID isolates, special care was taken to select isolates with different morphologic phenotypes according to their CFUs. A total of 18 different TA isolates were identified and stored at -70°C (Table 2).
For further assays both ID and TA isolates were taken from the frozen stocks and grown in liquid cultures.
Media and culture conditions
Cultures for extraction of cell-to-cell signaling molecules and production of elastase were grown in Luria-Bertani (LB) broth. M63 medium [32] was used for bacterial adhesion assays and AP medium [33] was used for biofilm formation assays. No differences in growth rates were observed between the strains in these media.
Extraction and quantification of acylated homoserine lactones (AIs) from intubation devices and liquid cultures
Previous studies have suggested that secretions colonized by bacteria accumulate above the cuff of intubation devices and form biofilms [34-36]. Leakage of bacteria around the cuff might play a major role in the development of ventilator-associated pneumonia [25,37]. Consequently, we selected the cuff of the intubation devices for the quantification of AIs. To extract AIs; cuffs were aseptically cut off the intubation devices, incubated for 30 minutes at 37°C in 5 ml sterile saline containing 0.025% dithiothreitol (DTT) to resuspend the biofilms formed on the cuff, and extracted twice by the addition of 1 volume of acidified ethyl acetate for 5 minutes at -20°C. To extract AIs from liquid cultures of both ID and TA isolates, the strains were incubated in LB broth at 37°C with shaking. Aliquots of filtered culture supernatants, taken after 8 hours of growth (early stationary phase), were subjected to 2 extractions in 1 volume of acidified ethyl acetate and stored at -70°C. Aliquots of the extracts obtained from the cuffs and liquid cultures of ID and TA isolates were evaporated under nitrogen gas and the concentrations of 3-oxo-C12-HSL and C4-HSL determined using the reporter strains E. coli MGλ1,4 (pPCS1) [38] and P.aeruginosa PAO-JP2 (pECP61.5) [39] respectively. For each determination standard curves were obtained with the wild type strain PAO1. The amount of AIs recovered from the intubation devices was expressed as nmoles/cuff.
Determination of bacterial genotype
Bacterial strains were genotyped by Random Amplified Polymorphic DNA (RAPD) analysis as previously described using the Ready-to-Go Kit (Amersham) [18] using primer 208 (ACGGCCGACC) [40].
Production of protease, rhamnolipids and elastase
Total extracellular protease [41] and rhamnolipid [42] production of both ID and TA isolates were determined semi-quantitatively by agar plate assays. Elastase production was quantified using the Elastin Congo Red elastolysis assay [38,41]. Culture supernatants were collected at the same point in the bacterial growth curve as for AI extraction.
Assessment of bacterial adhesion and biofilm formation assay
Adhesion capacity was measured for both ID and TA isolates as the ability of bacteria to adhere to the wells of 96-well microtiter plates. The assay was performed as previously described [18]. Results were expressed as percent adhesion with respect to the PT5 wild type strain. The ability of bacteria to form biofilm in vitro was determined in a static model as previously described [8]. Briefly, overnight LB cultures of ID and TA isolates were diluted to an OD660 of 0.5 and incubated for a further 6 hours at 37°C with shaking until cells had entered early stationary phase. Sterile calibrated coupons of PVC tracheal tubing were immersed in the suspension and incubated statically at 37°C for 1 h, then the PVC coupons were transferred to 6-well cell culture plates and incubated statically at 37°C for 72 h in AP medium. The extent of biofilm formation was measured using crystal violet staining [43]. Results were expressed as percent biofilm formation with respect to the PAO1 wild type strain.
Statistical analysis
We used the Student's t test to compare mean production of AIs and virulence factors by isolates from tracheal aspirates (TA) and the cuff of the intubation device (ID). Correlations between AIs production and QS-dependent phenotypes were calculated using a robust estimator of the Pearson's correlation coefficient. To account for the correlation among multiple observations in the same patient, standard errors and p values were computed using a first-order Taylor-series linearization method. All analyses were conducted using Stata 6.0 statistical software (StataCorp, 1999). p < 0.05 was considered statistically significant.
Authors' contributions
SFB carried out the experiments and drafted the manuscript. CE conducted the statistical analysis. TK participated in the design of the study and participated in the analysis of the results. JAR organized the sample collection. CVD conceived the study, supervised the experiments and their analysis, and wrote the final manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We would like to thank J. Pugin, and B. Ricou for supplying us with the intubation devices necessary for the study, P. Wood for helping during the preparation of the manuscript and R. Comte for technical assistance. This work was supported by grants from the Programme Commun de Recherche en Génie Biomédical, Genève, Lausanne 1999–2002 and from the Swiss National Science Foundation (3231–51940.97 and 3200–052189.97) to CVD.
Contributor Information
Sabine Favre-Bonté, Email: favre@biomserv.univ-lyon1.fr.
Eric Chamot, Email: echamot@uab.edu.
Thilo Köhler, Email: Thilo.Kohler@medecine.unige.ch.
Jacques-A Romand, Email: Jacques-Andre.Romand@hcuge.ch.
Christian van Delden, Email: christian.vandelden@medecine.unige.ch.
References
- Dunn M, Wunderink RG. Ventilator-associated pneumonia caused by Pseudomonas infection. Clin Chest Med. 1995;16:95. [PubMed] [Google Scholar]
- Levine SA, Niederman MS. The impact of tracheal intubation on host defenses and risks for nosocomial pneumonia. Clin Chest Med. 1991;12:523. [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Van Delden C, Iglewski BH. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg Infect Dis. 1998;4:551. doi: 10.3201/eid0404.980405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latifi A, Winson MK, Foglino M, Bycroft BW, Stewart GS, Lazdunski A, Williams P. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol Microbiol. 1995;17:333. doi: 10.1111/j.1365-2958.1995.mmi_17020333.x. [DOI] [PubMed] [Google Scholar]
- Passador L, Cook JM, Gambello MJ, Rust L, Iglewski BH. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science. 1993;260:1127. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. The involvement of cell-to cell signals in the development of bacterial biofilm. Science. 1998;280:295. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- Favre-Bonté S, Köhler T, Van Delden C. Biofilm formation by Pseudomonas aeruginosa : role of the C4-HSL cell-to-cell signal and inhibition by azithromycin . J Antimicrob Chemother. 2003;52:598–604. doi: 10.1093/jac/dkg397. [DOI] [PubMed] [Google Scholar]
- Schuster M, Lostroh CP, Ogi T, Greenberg EP. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol. 2003;185:2066. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J Bacteriol. 2003;185:2080. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumbaugh KP, Griswold JA, Hamood AN. The role of quorum sensing in the in vivo virulence of Pseudomonas aeruginosa. Microbes Infect. 2000;2:1721. doi: 10.1016/S1286-4579(00)01327-7. [DOI] [PubMed] [Google Scholar]
- Geisenberger O, Givskov M, Riedel K, Hoiby N, Tümmler B, Eberl L. Production of N-acyl-L-homoserine lactones by P. aeruginosa isolates from chronic lung infections associated with cystic fibrosis. FEMS Microbiol Lett. 2000;184:273. doi: 10.1111/j.1574-6968.2000.tb09026.x. [DOI] [PubMed] [Google Scholar]
- Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;407:762. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- Favre-Bonté S, Pache JC, Robert J, Blanc D, Pechère JC, Van Delden C. Detection of Pseudomonas aeruginosa cell-to-cell signals in lung tissue of cystic fibrosis patients. Microb Pathog. 2002;32:143. doi: 10.1006/mpat.2001.0487. [DOI] [PubMed] [Google Scholar]
- Erickson DL, Endersby R, Kirkham A, Stuber K, Vollman DD, Rabin HR, Mitchell I, Storey DG. Pseudomonas aeruginosa quorum-sensing systems may control virulence factor expression in the lungs of patients with cystic fibrosis. Infect Immun. 2002;70:1783. doi: 10.1128/IAI.70.4.1783-1790.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton B, Rodgers HC, Camara M, Knox AJ, Williams P, Hardman A. Direct detection of N-acylhomoserine lactones in cystic fibrosis sputum. FEMS Microbiol Lett. 2002;207:1. doi: 10.1111/j.1574-6968.2002.tb11019.x. [DOI] [PubMed] [Google Scholar]
- Ward C, Camara M, Forrest I, Rutherford R, Pritchard G, Daykin M, Hardman A, de Soyza A, Fisher AJ, Williams P, Corris PA. Preliminary findings of quorum signal molecules in clinically stable lung allograft recipients. Thorax. 2003;58:444. doi: 10.1136/thorax.58.5.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denervaud V, TuQuoc P, Blanc D, Favre-Bonté S, Krishnapillai V, Reimmann C, Haas D, Van Delden C. Characterization of cell-to-cell signaling-deficient Pseudomonas aeruginosa strains colonizing intubated patients. J Clin Microbiol. 2004;42:554. doi: 10.1128/JCM.42.2.554-562.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stickler DJ, Morris NS, McLean RJ, Fuqua C. Biofilms on indwelling urethral catheters produce quorum-sensing signal molecules in situ and in vitro . Appl Environ Microbiol. 1998;64:3486. doi: 10.1128/aem.64.9.3486-3490.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrout JD, Chopp DL, Just CL, Hentzer M, Givskov M, Parsek MR. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol Microbiol. 2006 doi: 10.1111/j.1365-2958.2006.05421.x. [DOI] [PubMed] [Google Scholar]
- Storey DG, Ujack EE, Rabin HR, Mitchell I. Pseudomonas aeruginosa lasR transcription correlates with the transcription of lasA, lasB, and toxA in chronic lung infections associated with cystic fibrosis. Infect Immun. 1998;66:2521. doi: 10.1128/iai.66.6.2521-2528.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrol S, Olliver A, Pier GB, Andremont A, Ruimy R. Transcription of quorum-sensing system genes in clinical and environmental isolates of Pseudomonas aeruginosa. J Bacteriol. 2003;185:7222. doi: 10.1128/JB.185.24.7222-7230.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastre J, Fagon JY. Ventilator-associated pneumonia. Am J Respir Crit Care Med. 2002;165:867. doi: 10.1164/ajrccm.165.7.2105078. [DOI] [PubMed] [Google Scholar]
- Craven DE, Steger KA. Epidemiology of nosocomial pneumonia. New perspectives on an old disease. Chest. 1995;108:1S. doi: 10.1378/chest.108.2_supplement.1s. [DOI] [PubMed] [Google Scholar]
- Rello J, Sonora R, Jubert P, Artigas A, Rue M, Valles J. Pneumonia in intubated patients: role of respiratory airway care. Am J Respir Crit Care Med. 1996;154:111. doi: 10.1164/ajrccm.154.1.8680665. [DOI] [PubMed] [Google Scholar]
- De Kievit TR, Gillis R, Marx S, Brown C, Iglewski BH. Quorum-sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl Environ Microbiol. 2001;67:1865. doi: 10.1128/AEM.67.4.1865-1873.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol. 2002;184:1140. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey ME, Caiazza NC, O'Toole GA. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J Bacteriol. 2003;185:1027. doi: 10.1128/JB.185.3.1027-1036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenkard E, Ausubel FM. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature. 2002;416:740. doi: 10.1038/416740a. [DOI] [PubMed] [Google Scholar]
- Tateda K, Comte R, Pechère JC, Köhler T, Yamaguchi K, Van Delden C. Azithromycin inhibits quorum sensing in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2001;45:1930–1933. doi: 10.1128/AAC.45.6.1930-1933.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis RJ, Iglewski BH. Azithromycin retards Pseudomonas aeruginosa biofilm formation. J Clin Microbiol. 2004;42:5842. doi: 10.1128/JCM.42.12.5842-5845.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SE, Kaye SA, Hamood AN, Iglewski BH. Characterization of Pseudomonas aeruginosa mutants that are deficient in exotoxin A synthesis and are altered in expression of regA, a positive regulator of exotoxin A. Infect Immun. 1994;62:897. doi: 10.1128/iai.62.3.897-903.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry JM, Pina SE, Mattingly SJ. Role of energy metabolism in conversion of nonmucoid Pseudomonas aeruginosa to the mucoid phenotype. Infect Immun. 1992;60:1329. doi: 10.1128/iai.60.4.1329-1335.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahul P, Auboyer C, Jospe R, Ros A, Guerin C, el Khouri Z, Galliez M, Dumont A, Gaudin O. Prevention of nosocomial pneumonia in intubated patients: respective role of mechanical subglottic secretions drainage and stress ulcer prophylaxis. Intensive Care Med. 1992;18:20. doi: 10.1007/BF01706421. [DOI] [PubMed] [Google Scholar]
- Inglis TJ, Millar MR, Jones JG, Robinson DA. Tracheal tube biofilm as a source of bacterial colonization of the lung. J Clin Microbiol. 1989;27:2014. doi: 10.1128/jcm.27.9.2014-2018.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis TJ, Lim TM, Ng ML, Tang EK, Hui KP. Structural features of tracheal tube biofilm formed during prolonged mechanical ventilation. Chest. 1995;108:1049. doi: 10.1378/chest.108.4.1049. [DOI] [PubMed] [Google Scholar]
- Valles J, Artigas A, Rello J, Bonsoms N, Fontanals D, Blanch L, Fernandez R, Baigorri F, Mestre J. Continuous aspiration of subglottic secretions in preventing ventilator- associated pneumonia. Ann Intern Med. 1995;122:179. doi: 10.7326/0003-4819-122-3-199502010-00004. [DOI] [PubMed] [Google Scholar]
- Pearson JP, Pesci EC, Iglewski BH. Role of Pseudomonas aeruginosa las and rhl quorum-sensing systems in the control of elastase and rhamnolipid biosynthesis genes. J Bacteriol. 1997;179:5756. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Delden C, Comte R, Bally M. Stringent response activates quorum sensing and modulates cell density-dependent gene expression in Pseudomonas aeruginosa. J Bacteriol. 2001;183:5376. doi: 10.1128/JB.183.18.5376-5384.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahenthiralingam E, Campbell ME, Foster J, Lam JS, Speert DP. Random amplified polymorphic DNA typing of Pseudomonas aeruginosa isolates recovered from patients with cystic fibrosis. J Clin Microbiol. 1996;34:1129. doi: 10.1128/jcm.34.5.1129-1135.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Delden C, Pesci EC, Pearson JP, Iglewski BH. Starvation selection restores elastase and rhamnolipid production in a Pseudomonas aeruginosa quorum-sensing mutant. Infect Immun. 1998;66:4499. doi: 10.1128/iai.66.9.4499-4502.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler T, Van Delden C, Curty LK, Hamzehpour MM, Pechère JC. Overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa. J Bacteriol. 2001;183:5213–5222. doi: 10.1128/JB.183.18.5213-5222.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]


