Abstract
PTOV1 was recently identified as a novel gene and protein during a differential display screening for genes overexpressed in prostate cancer. The PTOV1 protein consists of two novel protein domains arranged in tandem, without significant similarities to known protein motifs. By immunohistochemical analysis, we have found that PTOV1 is overexpressed in 71% of 38 prostate carcinomas and in 80% of samples with prostate intraepithelial neoplasia. High levels of PTOV1 in tumors correlated significantly with proliferative index, as assessed by Ki67 immunoreactivity, and associated with a nuclear localization of the protein, suggesting a functional relationship between PTOV1 overexpression, proliferative status, and nuclear localization. In quiescent cultured prostate tumor cells, PTOV1 localized to the cytoplasm, being excluded from nuclei. After serum stimulation, PTOV1 partially translocated to the nucleus at the beginning of the S phase. At the end of mitosis, PTOV1 exited the nucleus. Transient transfection of chimeric green fluorescent protein-PTOV1 forced the entry of cells into the S phase of the cell cycle, as shown by double fluorescent imaging for green fluorescent protein and for Ki67, and also by flow cytometry. This was accompanied by greatly increased levels of cyclin D1 protein in the transfected cells. These observations suggest that overexpression of PTOV1 can contribute to the proliferative status of prostate tumor cells and thus to their biological behavior.
Prostate adenocarcinoma is the second most prevalent cancer among males in Western countries, with an incidence that increases in direct proportion with age. A large body of evidence indicates that activation of the pathways regulated by androgen receptor plays a central role in the development and malignant progression of prostate cancer. 1 Because this is a physiologically regulated pathway, the question remains as to the underlying mechanisms that can direct the activation of the androgen receptor-signaling routes toward undue responses, eventually leading to malignant transformation of the prostate epithelial cells. In a few instances of primary tumors, mutations in the androgen receptor have been found that permit its activation by steroids other than androgens, or even other stimuli, independent of steroids. 2 Most frequently, however, androgenreceptor mutations are a late event in the progression of prostate cancer, 3 and appear to be a major mechanism, along with androgen receptor gene amplification and overexpression, 4 by which these tumors become insensitive to androgens, and independent from these hormones for their growth. 5 This hormone-independent state is usually accompanied by an increase in the capacity of tumor cells to invade surrounding tissues and to form distant metastases. 6 In consequence, in the majority of prostate cancers other mechanisms must be invoked to explain the early events leading to the induction of the neoplastic phenotype.
Over the past few years, genetic and biochemical approaches have identified a number of molecular alterations that are associated with different stages of prostate tumorigenesis. 7-10 Systematic approaches usingmicroarray-based global transcriptome analysis have also attempted to identify markers and pathways altered in prostate cancer. 11-19 These studies have identified novel markers of early-stage prostate neoplasia, 14 as well as transcriptional signatures of advanced disease and poor prognosis. 18 In general, the possible biological significance of the altered expression of the genes identified in such analyses has not been addressed experimentally.
In our laboratory, we have recently identified a new protein, which we have designated PTOV1, that is overexpressed in a significant proportion of prostate adenocarcinomas. 20 Recent global transcriptional profile analyses by others 17 have found that PTOV1 is one of the genes most discriminant between the normal and carcinomatous prostate. Importantly, we have found that PTOV1 is also overexpressed in early neoplastic lesions, the prostate intraepithelial neoplasias. 20 At present, the function of this protein is not known, nor can it be deduced or suggested from its sequence, because it does not contain any known protein motifs. 20 In this report we present an extended immunohistochemical study on samples from 38 prostate adenocarcinomas showing a correlation between proliferating tumor cells and levels of PTOV1 expression in vivo. We also describe cell-cycle-associated changes in subcellular localization of PTOV1 in prostate cancer cell lines, and the consequences on the cell cycle of its overexpression by transient transfection. Our observations suggest that overexpression of PTOV1 can have a direct influence on the proliferative state of prostate cells.
Materials and Methods
Antibodies
Rabbit antibodies to human PTOV1 20 were affinity purified using Sulfolink columns from Pierce (Rockford, IL), on which the immunizing peptide was covalently immobilized. Monoclonal anti-Ki67 antibody was from Immunotek (Marseille, France), anti-p27Kip1 antibody was from Novocastra (Edinburgh, UK) and anti-cyclin D1 was from Santa Cruz Biotechnology (Santa Cruz, CA). Peroxidase-coupled anti-rabbit antibodies, tetramethyl-rhodamine isothiocyanate-coupled anti-rabbit antibodies were from Sigma (Alcobendas, Madrid, Spain) and fluorescein isothiocyanate-coupled anti-mouse antibodies were from DAKO (Glostrup, Denmark).
Immunohistochemistry
A total of 38 prostate adenocarcinomas from radical prostatectomies for T2 to T3 tumors were obtained from the files of the Department of Pathology of the Hospital Clinic of Barcelona. The mean patient age in this study was 63.5 ± 5.81 years (median, 64 years; range, 45 to 75 years). The mean preoperative serum prostate-specific antigen (PSA) was 11.50 ± 7.88 ng/ml (median, 9.6 ng/ml; range, 1.29 to 38 ng/ml). The Gleason sum score assigned to the radical prostatectomy specimens ranged from 4 to 9 and was evaluated as grade ≤6 (low, 17 patients) versus ≥7 (high, 21 patients). Normal or hyperplastic prostatic tissue could be evaluated in 29 of the cases as well as 10 high-grade prostatic intraepithelial neoplasia (PIN) lesions. Also, normal prostatic tissue from six patients who underwent cystoprostatectomy for bladder tumors were included in the study. Two-μm sections from multitissue blocks 21 harboring 6-mm-diameter selected fragments were deparaffinized in xylene, and rehydrated with graded alcohols, water, and phosphate-buffered saline (PBS). Antigen unmasking was performed with pressure cooker heating in ethylenediaminetetraacetic acid buffer, pH 7, for 2 minutes, slides were allowed to cool down for 5 minutes and were sequentially incubated in vertical humid chambers with PBS (3 minutes) background suppressor (DAKO S3022, 30 minutes at room-temperature) and the primary antibody (2 μg/ml overnight, humid chamber at 4°C). Slides were then rinsed twice in PBS and endogenous peroxidase-quenched with 2% hydrogen peroxide in methyl alcohol for 30 minutes. After rinsing twice in PBS, slides were incubated for 30 minutes with Envision anti-rabbit antibody (Envision, DAKO). The staining was developed with substrate chromogen solution (Envision, DAKO) and diaminobenzidine for 3 to 5 minutes. The slides were counterstained with Gill-I hematoxylin for 1 minute, dehydrated, and mounted with DPX solution. Negative controls consisted in the use of 100 μl of the primary antibody previously incubated at 37°C for 1 hour with 0.16 mg of the purified peptide, the latter previously resuspended in 0.1 N of acetic acid, and used as above in parallel with similar slides with nonadsorbed primary antibody. The use of such preadsorbed primary antibody gave clean negative results in all cases tested (not shown). A semiquantitative gradation of the intensity was performed with intensities ranging from weak (intensity level 1) to very strong (intensity level 4). A case was considered positive when >10% of the cells showed unequivocal staining (levels ≥1). Both cytoplasmic and nuclear staining were recorded.
A similar procedure was followed for detection of Ki67 except that incubation of the primary antibody was for 30 minutes at room temperature and was preceded by antigen unmasking in citrate buffer. Ki67 was evaluated by counting the total of positive cells in all tumor fragments of the same size. A correction factor was used when the fragments were only partially involved by tumor. Staining for p27Kip1 was performed with the streptavidin-alkaline phosphatase method (Biogenex, San Ramon, CA) as previously described 22 and the percentage of positive cells was recorded.
Cell Culture
All cell lines were obtained from the American Type Culture Collection (Rockville, MD). Cell lines PC-3, LNCaP, and COS7 were maintained in RPMI 1640 medium (Life Technologies, Inc., Grand Island, NY) supplemented with 10% heat-inactivated fetal bovine serum (Life Technologies, Inc.) at 37°C in an atmosphere of 5% CO2. The prostate cell line PZ-HPV-7 was maintained in Keratinocyte-Serum-Free Medium (Life Technologies, Inc.) supplemented with 5 ng/ml of human recombinant epidermal growth factor and 50 μg/ml of bovine pituitary extract.
Immunocytochemistry
Immunocytochemistry for confocal microscopy was performed as described. 20 Briefly, cells cultured on coverslips were washed with cold PBS, fixed with 4% paraformadehyde for 10 minutes, incubated with 50 mmol/L of NH4Cl for 30 minutes, and permeabilized with PBS/1% bovine serum albumin/0.1% saponin for 30 minutes. Samples were sequentially incubated with primary antibody diluted in permeabilization buffer for 2 hours, and secondary antibody for 1 hour, and mounted with Immuno-Fluore Mounting Medium (ICN, Costa Mesa, CA). Fluorescence was visualized by inverted fluorescence microscope DM IRBE (Leica, Wetzlar, Germany) and captured by a TCS-NT argon/krypton confocal laser (Leica).
Western Blotting
Cultured cells, washed with cold PBS, were lysed with Laemmli sample buffer [50 mmol/L Tris-HCl, pH 6.8, 2% (w/v) sodium dodecyl sulfate, 10% (v/v) glycerol, β-mercaptoethanol] heated at 95°C and centrifuged at 10,000 × g. Fifty to 100 μg of cell lysate were electrophoresed and transferred to nitrocellulose. Membranes were blocked for 1 hour with blocking buffer (5% nonfat dry milk in PBS/1% Tween-20) and incubated for 2 hours with anti-PTOV1 (5 μg/ml in blocking buffer), washed, and incubated with horseradish peroxidase-conjugated goat anti-rabbit Ig. Reactivity was detected with a chemiluminescent substrate (Amersham, Buckinghamshire, UK). For sequential Western blotting, membranes were stripped at 50°C according to the manufacturer’s instructions.
Transfections
Constructs for the expression of chimeric green fluorescent protein (GFP)-PTOV1 with GFP at the amino or carboxy termini of PTOV1 have been described previously. 20 Transient transfections were done with Lipofectamine (Life Technologies, Inc.). Twenty-four or 48 hours after transfection, cells were washed, fixed in 4% paraformaldehyde, processed for immunofluorescence with anti-PTOV1, mounted, and analyzed by confocal microscopy. Control transfections were performed with the vector pEGFP alone, for the expression of nonchimeric GFP, which consistently yielded a diffuse fluorescence distributed throughout the cells, without any subcellular specific localization.
Flow Cytometry
Cells were fixed in 70% ethanol in PBS at −20°C for at least 1 hour, washed several times with cold PBS, and treated with RNaseA (50 μg/ml) for 30 minutes at 37°C and incubated with propidium iodide (30 μg/ml). Cells were subsequently analyzed on a Coulter Epics XL flow cytometer (Coulter, Miami, FL) for GFP fluorescence (detection filter set at 525 nm) and DNA content (filter set at 675 nm). Cell aggregates were gated out, and 10,000 events were analyzed. For each cell type analyzed, untransfected cells were used to establish a threshold for green fluorescence (up to 102 arbitrary fluorescence units in a typical case), and which was taken as a threshold for positivity. GFP-positive and -negative populations were analyzed separately for DNA content and assigned to specific cell-cycle phases by applying the Multicycle cell-cycle analysis software (Phoenix Flow Systems, San Diego, CA).
Results
Immunohistochemical Analysis of Prostate Adenocarcinomas for the Expression of PTOV1 and Markers of Cell Proliferation
In our initial studies on tumor samples, most prostate adenocarcinomas analyzed by immunohistochemistry expressed high levels of PTOV1, without a correlation between these levels and clinicopathological parameters such as histological grade or clinical stage. 20 To test whether other parameters of in vivo biological activity, more directly associated with proliferative status, correlate with levels of PTOV1, we analyzed samples from 38 prostate adenocarcinomas and 6 normal prostates for the expression of PTOV1 and two other markers, the antigen Ki67, as an indicator of proliferating cells, 23 and the cyclin-dependent kinase inhibitor p27Kip1, a regulator of the G1 to S transition 24 that is frequently down-regulated in prostate cancer. 22,25,26 Immunohistochemistry for these markers was performed in serial or near-serial sections.
As previously reported, PTOV1 was mostly undetectable or showed a weak (level 1) cytoplasmic staining in normal prostate glandular epithelial cells (Figure 1) ▶ . Interestingly, a more intense (level 3) staining was seen in sporadic luminal cells in isolated glands, which was mostly cytoplasmic but in some cases also nuclear. Staining of serial sections showed that these PTOV1-positive cells most likely corresponded to chromogranin A-positive cells, a marker for neuroendocrine cells (data not shown). Also, a very strong PTOV1 expression (levels 3 to 4) was observed in endothelial cells, erythrocytes (Figure 1A) ▶ , and ganglion cells of vegetative ganglia. Therefore, most normal glandular epithelial cells usually do not express PTOV1, or do so at low levels, although several types of nonepithelial cells present in normal prostate structures can express PTOV1 at high levels.
Figure 1.
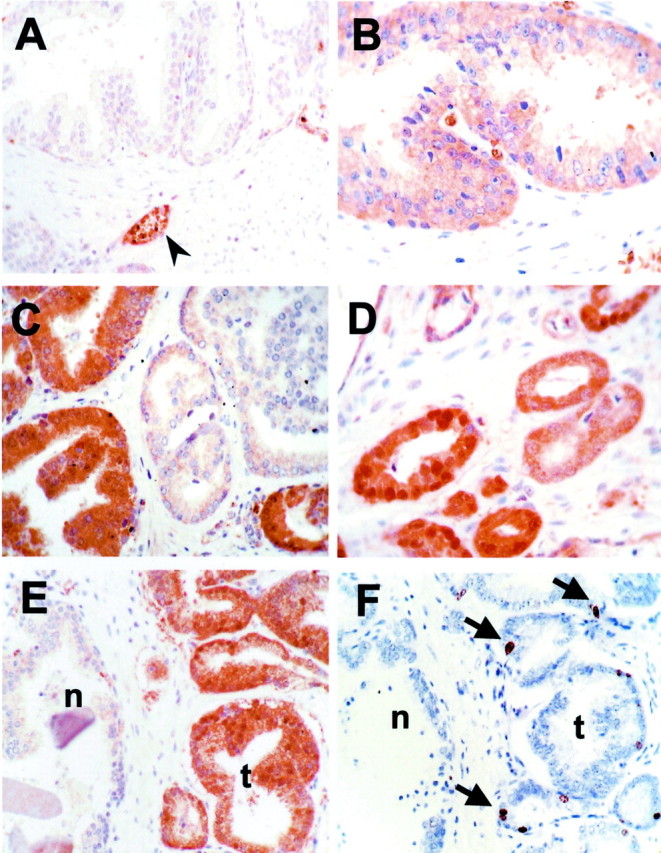
Immunohistochemical analysis of expression of PTOV1 and Ki67 in normal and tumor samples. A: Staining for PTOV1 of normal prostate epithelium, showing no detectable staining of glands, and a strong staining of blood vessels and red blood cells (arrowhead). B: High-grade PIN, showing a diffuse cytoplasmic staining for PTOV1 (level 2, see text and Materials and Methods). C: Staining for PTOV1 of a tumor sample, showing heterogeneous staining, with a strong positivity (level 3) in some glands, and faint to no staining in other tumoral glands. D: A tumor sample showing strong nuclear and cytoplasmic staining for PTOV1. E and F: Near-serial sections of a tumor sample containing glands with normal (n) and carcinomatous (t) morphology, stained for PTOV1 (E) and for Ki67 (F). Arrows indicate nuclei positive for Ki67 staining. Original magnifications, ×400.
Of the 38 adenocarcinoma samples analyzed, 27 (71%) overexpressed (ie, expression levels 2 to 3) PTOV1 in cells of carcinomatous appearance (Figure 1 ▶ and Table 1 ▶ ). The majority of the specimens positive for PTOV1 staining showed a homogeneous distribution of positive tumor cells throughout the carcinomatous areas, with >80% positive cells. Overexpression of PTOV1 was limited to the cytoplasm of carcinomatous cells in 16 cases, whereas it was strong (level 3) both in the nucleus and the cytoplasm in the remaining 11 cases (Table 2) ▶ . Two cases were an exception to this situation in that tumor cells had a strong nuclear staining in combination with a weak cytoplasmic staining for PTOV1. There was no obvious association between gland morphology (differentiation) or topographical area of the tumor (periphery versus central) with the levels or the pattern of PTOV1 expression. Ten tumors also contained abnormal glands with morphological characteristics of PIN. Of these, eight showed strong cytoplasmic or cytoplasmic and nuclear staining for PTOV1 (Figure 1 ▶ and Tables 1 and 2 ▶ ▶ ) without significant differences in intensity between basal or luminal cells, and also contained a higher number of Ki67-positive cells. In the tumor samples analyzed, the expression levels of PTOV1 in the epithelial cells of glands with a normal morphology (BPZ) were either very low (level 1) or undetectable (equal to or below background staining) (Figure 1 ▶ and Table 1 ▶ ).
Table 1.
Summary of Immunohistochemical Analyses of PTOV1 Expression in Prostate Cancer and Normal Prostate
| PTOV1 | P† | |||
|---|---|---|---|---|
| Level 0* (%) | Level 1 (%) | Level 2/3 (%) | ||
| Tumor | ||||
| Carcinoma | 0 | 11/38 (29) | 27/38 (71) | 0.0001 |
| PIN | 0 | 2/10 (20) | 8/10 (80) | 0.0001 |
| BPZ | 6/29 (20.7) | 22/29 (75.8) | 1/29 (3.4) | |
| Normal | 3/6 (50) | 3/6‡ (50) | 0 | |
*Level 0, no detectable staining; level 1, low; and level 2/3, high levels of expression (see Materials and Methods).
†P values obtained by comparisons with BPZ.
‡Focal expression in isolated glands (<10% of cells).
Table 2.
Expression of Ki67 and p27 in Relation to PTOV1 Levels and Subcellular Localization in Prostate Tumors
| PTOV1 level 1 | PTOV1 level 2/3 | P | |
|---|---|---|---|
| Ki67 index* | |||
| Total PTOV1 | 42.5 | 74.5 | 0.013 |
| PTOV1 C only | 42 | 77 | 0.015 |
| PTOV1 C and N | 0 | 83.6 | 0.010† |
| p27 (% positive cells) | 26 | 27 | 0.714 |
*Total PTOV1, average of Ki67 index for all samples; PTOV1 C only, average of Ki67 index for samples with PTOV1 localized to the cytoplasm only; PTOV1 C and N, average of Ki67 index for samples with PTOV1 localized both to the cytoplasm and the nucleus.
†P value obtained by comparison with Ki67 values of tumors expressing low levels of PTOV1, with PTOV1 localized to the cytoplasm only.
The same samples were analyzed by immunohistochemistry for the expression of Ki67 and p27Kip1. Expression of Ki67 was assessed in the form of an index, calculated as a function of positive nuclei per total nuclei in sections of comparable size (see Materials and Methods). Normal prostate epithelium did not express Ki67 at significant levels (index <1). In tumor samples, areas of normal morphology did not show Ki67 staining or did so in very few cells (index <1), whereas the carcinomatous areas from the same samples had indexes that ranged from 4 to more than 100 (Figure 1F) ▶ . When these indexes were compared with levels of expression of PTOV1, a significant correlation (P = 0.013) was found between Ki67 index and levels of PTOV1 (Table 2) ▶ , indicative of an association of high levels of PTOV1 with an active proliferative status. Of interest, all of the tumor samples with nuclear localization of PTOV1 showed high levels (levels 2 to 3) of expression of PTOV1 and had elevated Ki67 indexes (Table 2) ▶ . Expression of p27Kip1 protein was evaluated in 18 of the 38 tumors. As shown in Table 2 ▶ , no significant correlation was observed between levels of expression of PTOV1 and levels of p27Kip1. Patients with overexpression of PTOV1 (levels 2 or 3) showed higher PSA serum levels (12.62 ng/ml) compared to patients with low levels (level 1) (8.57 ng/ml), although this association did not reach statistically significant values (P = 0.056). When subcellular localization of PTOV1 was considered, independently of its levels of expression, patients whose samples showed nuclear expression of PTOV1 had significantly (P = 0.019) higher preoperative serum PSA levels (13.83 ng/ml) than did patients whose samples had an exclusively cytoplasmic localization of PTOV1 (8.74 ng/ml). Increased levels of expression of PTOV1 were not associated with a higher Gleason score (≥7, not shown), confirming our previous observations. 20 In conclusion, high levels of expression of PTOV1 in prostate cancer correlated with an active proliferative status, as determined by elevated Ki67 indexes. Also, overexpression of PTOV1 was frequently associated with its nuclear localization, both in carcinomatous and PIN glands. Furthermore, nuclear localization of PTOV1 in the tumor was significantly associated with higher serum PSA levels.
Changes in Subcellular Localization of PTOV1 as a Function of the Cell Cycle
The above observations suggested the existence of a direct relationship between the proliferative status of prostate epithelial cells and the levels and subcellular localization of PTOV1. In our initial description, PTOV1 was found mainly in the cytoplasm but also in the nucleus of prostate cancer cells. 20 To study if the subcellular localization of PTOV1 was dependent on particular stages of the cell cycle, PC-3 and PZ-HPV-7 prostate cancer cells were serum-starved for 48 hours, allowed to re-enter the cell cycle by re-addition of serum to the culture medium, and analyzed by immunocytochemistry for the expression of PTOV1. To monitor the proliferative status of the cells in these experiments, we used the marker Ki67, 23 an antigen that is not expressed in quiescent cells, that shows a nuclear pattern in the first part of S and a perinucleolar pattern toward the end of S, and is associated with condensed chromosomes throughout mitosis. 27 To further assess the cell-cycle status of the cells, double-fluorochrome immunocytochemistry was also performed for cyclin D1, which, after a mitogenic stimulus, exits the nucleus at the beginning of S. 28 After 48 hours of growth in serum-free medium, the majority of the cells were in proliferative quiescence as shown by lack of expression of Ki67, and PTOV1 was detected in a cytoplasmic localization, with nuclei clearly excluded from staining (Figure 2) ▶ . Nine hours after the addition of serum, Ki67 was detected in the nucleus of the majority (50 to 80%) of the cells, indicating entry into the S phase of the cell cycle, and PTOV1 was seen both in the cytoplasm and in the nucleus (Figure 2) ▶ . The nuclear localization of PTOV1 was maintained throughout the S phase, including the stage in which Ki67 concentrates around the nucleolus (Figure 2) ▶ . In mitotic cells, with Ki67 associated with condensed chromosomes, most of the signal for PTOV1 excluded the chromosomes (Figure 2) ▶ . Twenty-four hours after stimulation of quiescent cells with serum, PTOV1 was seen again mostly in the cytoplasm, although some nuclei retained positivity. This distribution of PTOV1 is equivalent to that originally observed in asynchronously growing cells. 20
Figure 2.
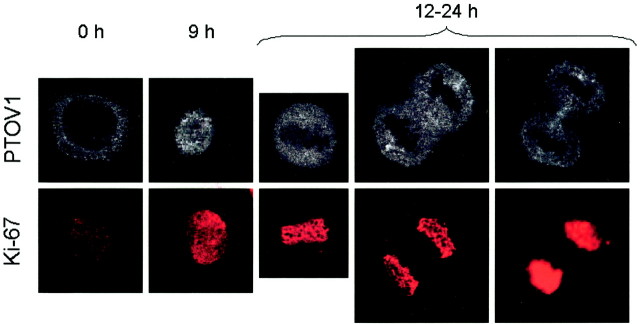
Nuclear localization of PTOV1 in proliferating cells. PZ-HPV-7 cells were growth factor-deprived for 48 hours, stimulated with 10% fetal bovine serum for the indicated times, and stained for PTOV1 or Ki67. Growth factor-deprived cells (time 0) that did not show staining for Ki67 (quiescence) showed a cytoplasmic staining for PTOV1 with nuclear exclusion. Nine hours after serum stimulation, coincident with a nuclear staining for Ki67, PTOV1 translocated into the nucleus. In dividing cells (12 hours after stimulation), PTOV1 is excluded from Ki67-stained condensed chromosomes.
Cyclin D1, which is necessary for the progression of the cell cycle from G1 to S phase, 28 is localized in the nucleus at late G1, and is exported from the nucleus or degraded at the beginning of the S phase. 29 We studied whether the nuclear shift of PTOV1 in serum-stimulated cells followed a similar pattern. In quiescent PC-3 cells, little or no specific staining for cyclin D1 was detected (Figure 3) ▶ . Nuclear translocation of the protein was observed at 3 hours after stimulation of the cells with serum, corresponding to approximately late G1 phase and up to 9 hours, corresponding to entry into S phase, in agreement with observations previously reported for other cell types. 29,30 Twelve hours after stimulation, a diffuse staining for cyclin D1 was observed in the cytoplasm, and weak or no staining was detectable at 24 hours (Figure 3) ▶ . Again, specific cytoplasmic staining for PTOV1 was observed in quiescent cells and 6 to 9 hours after serum stimulation PTOV1 entered the nucleus. In the majority of cells, the nuclear localization of PTOV1 was coincidental with the exit of cyclin D1 from the nucleus (Figure 3) ▶ . At 24 hours after serum stimulation, PTOV1 was observed mostly in the cytoplasm.
Figure 3.
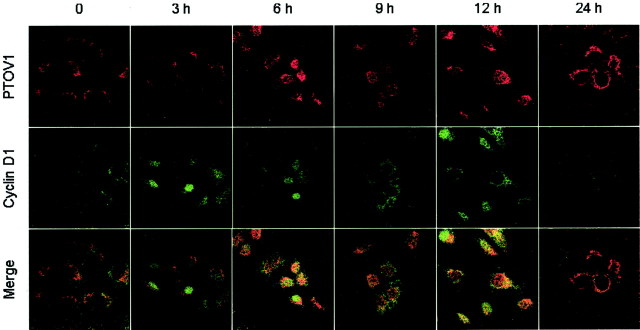
Nuclear entry of PTOV1 is concomitant with nuclear exit of cyclin D1. PC-3 cells were growth factor-deprived for 48 hours, stimulated with 10% fetal bovine serum for the indicated times, and stained by double fluorescence for PTOV1 and cyclin D1. In quiescent cells (time 0), PTOV1 is localized exclusively in the cytoplasm, whereas cyclin D1 is barely detectable. From 3 to 6 hours after stimulation with serum, cyclin D1 is detectable in the nucleus of most cells. PTOV1 is found in the nucleus at increasing proportion at 9 to 12 hours after serum stimulation, and its nuclear localization is most evident in cells in which cyclin D1 has been excluded from the nucleus (12 hours). At 24 hours after stimulation, cells show either a cytoplasmic or a nuclear localization of PTOV1, together with low levels of cyclin D1.
To determine whether these changes in subcellular localization of PTOV1 were accompanied by changes in protein levels, Western blotting was performed on serum-starved and serum-induced PC-3 and LNCaP cells. The levels of PTOV1 did not change significantly at any time after stimulation of these cells with serum (not shown). In conclusion, PTOV1 oscillates between the cytoplasm and the nucleus of prostate cancer cells in a cell-cycle-dependent manner, with nuclear concentration of the protein beginning in the S phase and peaking in mitosis. These changes in subcellular localization are not accompanied by changes in PTOV1 protein levels.
Effects of the Overexpression of PTOV1 on the Cell-Cycle Status of Prostate Cancer Cells
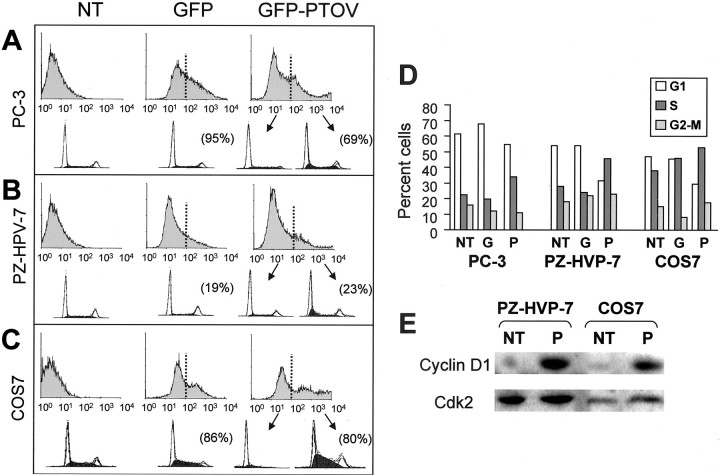
Because PTOV1 shows specific subcellular localizations that change according to the proliferative state, we reasoned that this protein could play a regulatory role in one or more of the cell-cycle transitions and that the artificial expression of high levels of PTOV1 in cultured cells could produce a significant accumulation of cells in that phase of the cell cycle at which this protein exerts a predominant regulatory role. For this purpose, we used a construct for the expression of PTOV1 with the green fluorescent protein (GFP) in frame at the amino terminus of PTOV1 (GFP-PTOV1). 20 This construct was introduced in transient transfection experiments with cationic liposomes into the prostate cells lines PC-3 and PZ-HPV-7, and the green monkey kidney fibroblast line COS7, and analyzed after 24 hours by counting the number of cells positive for both Ki67 and PTOV1 (Figure 4) ▶ . Controls for these experiments were untransfected cells and cells transfected with the vector pEGFP. As shown in Figure 4 ▶ , in PC-3, PZ-HPV-7, and COS7 cells the number of cells positive for Ki67 is significantly higher in cells transfected with the fusion protein GFP-PTOV1 compared to cells transfected with GFP only, suggesting that expression of PTOV1 induces entry of the cells into a proliferative state.
Figure 4.
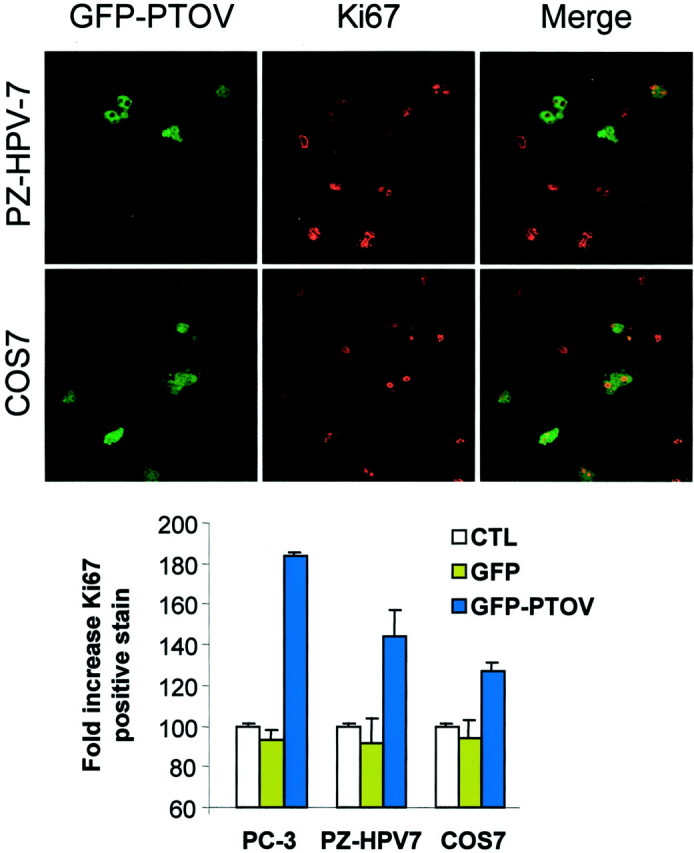
Overexpression of GFP-PTOV1 induces cell proliferation. PZ-HPV-7 and COS7 cells were transfected with GFP-PTOV1 (green fluorescence) and stained for Ki67 (red fluorescence). Cells positive for GFP-PTOV1, Ki67, or both were quantitated, and the relative numbers were compared to untransfected cells (histogram). As controls, cells were transfected with vector alone (GFP).
To study this effect more precisely, the cell-cycle status of cells overexpressing PTOV1 was assessed by flow cytometry 24 or 48 hours after the transfections. Three types of controls were used in these experiments. First, untransfected cells; second, transfections with GFP expression constructs that do not carry PTOV1; and, third, cells from the same transfections with the GFP-PTOV1 construct that show levels of GFP below background (GFP-negative population), and thus correspond to cells that either have not been transfected in those experiments or to cells that do not express the fusion protein above the established threshold. The transfection efficiencies ranged from 28 to >90%, measured as the percentage of cells with a GFP signal above background. For each transfection, the distribution of DNA and cell-cycle profile were analyzed separately for GFP-positive and GFP-negative populations, thus providing the cell-cycle profiles for cells expressing, or not expressing, the transfected proteins in the same experiment. Untransfected PC-3, PZ-HPV-7, and COS7 cells; cells transfected with GFP vector alone; and GFP-negative cells from the GFP-PTOV1 transfections had similar cell-cycle distributions (Figure 5) ▶ . For cells from transfection experiments, these distributions did not show significant differences between those harvested at 24 hours and those harvested at 48 hours after transfection. Therefore, the transfection procedure per se, or the expression of GFP alone, did not have significant effects on the cell-cycle profile of PC-3, PZ-HPV-7, or COS7 cells. In contrast, GFP-positive cells from transfections with GFP-PTOV1 showed a clear increase in the proportion of cells in the S phase when compared with any of the controls (Figure 5) ▶ . This increase in the proportion of cells in S was accompanied by a corresponding decrease of the population of cells in G1 (Figure 5) ▶ . These results confirm that the overexpression of PTOV1 promotes the entry into the S phase of the cell cycle of PC-3, PZ-HPV-7, and COS7 cells. In parallel experiments, we determined that cells forced to entry into S phase by the ectopic expression of PTOV1 contain significantly higher levels of the cyclin D1 protein, as shown by Western blotting of lysates from cells transfected with PTOV1 (Figure 5E) ▶ . These results show that PTOV1 induces progression through the cell cycle by enhancing entry into the S phase.
Figure 5.
Overexpression of GFP-PTOV1 forces entry into the S phase of the cell cycle. A—C Flow cytometry analysis of untransfected cells (NT), cells transfected with vector alone (GFP), or with GFP-PTOV1. The threshold for positivity for GFP was established as the levels corresponding to untransfected cells. Cells with green fluorescence below (not transfected) or above (transfected) this threshold were analyzed separately for DNA content. The histograms at the top of each panel correspond to GFP levels, and the histograms at the bottom of each panel correspond to analysis of DNA content and cell-cycle assignments. For each experiment, transfection efficiency is shown in brackets. D: Histograms of distribution of cell populations to the G1, S, or G2 to M phases of the cell cycle, corresponding to the flow cytometry analysis on the left panels. One representative experiment of four performed with each cell line is shown. E: Levels of cyclin D1 are elevated in cells transfected with GFP-PTOV1. PZ-HPV-7 or COS7 cells were either not transfected (NT) or transfected (P) with GFP-PTOV1, lysed 24 hours after transfection, and analyzed by Western blotting with antibodies to cyclin D1 or CDK2.
Discussion
We report here the cell-cycle-dependent shifts in subcellular localization of PTOV1, a novel protein of unknown function, and that its overexpression drives cells into a proliferative state. The distinguishing structural feature of PTOV1 is its duplicated domain, of which a single copy is present also in a second protein that we have called PTOV2. 20 Very recently deposited sequence entries (GenBank accession AAM20739) indicate that the latter protein could be identical to the transcriptional co-regulator ARC92. 31 Nevertheless, the PTOV domain does not resemble any other protein motif, 20 and thus examination of its sequence, per se, does not provide useful functional clues. Despite this lack of a priori functional knowledge, the fact that PTOV1 is overexpressed in a significant proportion of prostate cancers 17,20 led us to suggest that this gene and protein could have a role in the biological behavior of these tumors. 20 This notion is now reinforced by our observations, that successively associate 1) in vivo, high levels of PTOV1 with proliferative status and nuclear localization in carcinomatous cells; 2) in vitro, nuclear exclusion of PTOV1 with proliferative quiescence and nuclear localization with the S and M phases of the cell cycle; and 3) overexpression of transfected PTOV1 with induction of proliferation and entry into the S phase of the cell cycle.
The correlation found in tumor samples between high levels and nuclear localization of PTOV1 with elevated proliferative indexes, suggests a direct link of PTOV1 and the proliferative status of the tumor cells. This relationship was strongest in samples in which PTOV1 was localized in the nucleus. The statistically significant correlation found between tumors with PTOV1 expression in the nucleus and higher PSA serum levels might give additional support to a role for PTOV1 in the aggressive behavior of prostate tumors, because higher PSA levels are associated with more locally advanced disease. 32 On the other hand, the lack of correlation between PTOV1 expression and p27 levels, or the Gleason score, could be related to the fact that the cases analyzed did not include advanced stage tumors with highly undifferentiated morphologies. Loss of expression of p27 has been associated with advanced prostate tumors and high Gleason scores, 22,25,26 and therefore it would be interesting to study the levels of expression of PTOV1 in a set of samples corresponding to more advanced disease.
The correlation observed in tumor samples between Ki67 index and PTOV1 levels and subcellular localization prompted us to investigate in more detail the relationship between the cell-cycle status of the cells, the subcellular localization of PTOV1, and its overexpression. In our time-course experiments with PC-3 prostate cells, the nuclear translocation of PTOV1 occurred 9 hours after serum stimulation of quiescent cells, coincident with the exit of cyclin D1 from the nucleus, which marks the beginning of the S phase. 28 PTOV1 remained in the nucleus until the end of mitosis, suggesting a role for this protein not only in S, but also throughout G2 and M. The fact that entry of PTOV1 into the nucleus did not occur early after serum stimulation, but only after a 9-hour lag, suggests a functional link between nuclear translocation of PTOV1 and the cell-cycle regulatory machinery, and also that this translocation is not a direct or immediate consequence of the stimulus, as is the case for the nuclear import of certain proteins that are part of signaling cascades. 33,34 The enhanced entry of cells in S as a result of the forced expression of GFP-PTOV1 points to an active role of this protein in this stage of the cell cycle. A number of cell-cycle regulatory proteins undergo nuclear-cytoplasmic shifts in association with specific stages of the cell division cycle 28,30,35 and their localization to given subcellular compartments can determine the rate of progression through one or more cell-cycle transition points. 36 As discussed above, cyclin D1 localizes in the nucleus during G1, and is actively exported to the cytoplasm during the S phase, 28 in a process dependent on phosphorylation by GSK-3β 29 and on p21Cip1 and p27Kip1. 37 Mutant forms of cyclin D1 that fail to translocate in this way and do not form functional cyclin-CDK complexes cause the accumulation of cells in G1. 38 This could be relevant to our present findings, because the facts that PTOV1 drives the G1 to S transition and that the levels of cyclin D1 greatly increase after overexpression of GFP-PTOV1 point to the CDK4-cyclin D1 complex as a potential target for positive regulation by PTOV1, either by increasing the stability of the cyclin D1 subunit or by inducing its expression. Alternatively, PTOV1 could negatively regulate one or more inhibitors of the G1 cyclin-dependent kinases.
Taken together, our observations suggest that the overexpression of PTOV1 can play an active role in the proliferative status of neoplastic prostate epithelial cells, and thus in their biological behavior. The characterization of the molecular mechanisms by which PTOV1 exerts its mitogenic effects will require further exploration.
Acknowledgments
We thank J. Comas for expert advice with flow cytometry; M. Valeri for assistance with confocal microscopy; N. Agell for reagents and help with Western blotting; and M. Sanchez, supported in part by DAKO, for technical help.
Footnotes
Address reprint requests to Dr. Timothy Thomson, Instituto de Biología Molecular, Consejo Superior de Investigaciones Científicas, C. Jordi Girona 18-26, 08034 Barcelona, Spain. E-mail: ttobmc@cid.csic.es, or Dr. Rosanna Paciucci, Unitat de Recerca Biomèdica Vall d’Hebrón, P. Vall d’Hebrón 119-129, 08035 Barcelona, Spain. E-mail: rosan@hg.vhebron.es.
Supported by the Ministerio de Ciencia y Tecnología (SAF2001-1969 and SAF2000-0195 to T. M. T. and R. P.); the Fundación para la Investigación en Urología, Asociación Española de Urología (to J. M.); the Ministerio de Sanidad y Consumo (FIS 01/1519 to P. L. F.); Marató (TV3/1999 to R. P.); and the Fundación Científica, Asociación Española Contra el Cáncer (to P. L. F. and a predoctoral scholarship to A. S.).
R. P. and T. M. T. contributed equally to this work.
References
- 1.Jenster G: The role of the androgen receptor in the development and progression of prostate cancer. Semin Oncol 1999, 26:407-421 [PubMed] [Google Scholar]
- 2.Grossmann ME, Huang H, Tindall DJ: Androgen receptor signaling in androgen-refractory prostate cancer. J Natl Cancer Inst 2001, 93:1687-1697 [DOI] [PubMed] [Google Scholar]
- 3.Marcelli M, Ittmann M, Mariani S, Sutherland R, Nigam R, Murthy L, Zhao Y, DiConcini D, Puxeddu E, Esen A, Eastham J, Weigel NL, Lamb DJ: Androgen receptor mutations in prostate cancer. Cancer Res 2000, 60:944-949 [PubMed] [Google Scholar]
- 4.Linja MJ, Savinainen KJ, Saramaki OR, Tammela TL, Vessella RL, Visakorpi T: Amplification and overexpression of androgen receptor gene in hormone-refractory prostate cancer. Cancer Res 2001, 61:3550-3555 [PubMed] [Google Scholar]
- 5.Taplin ME, Bubley GJ, Shuster TD, Frantz ME, Spooner AE, Ogata GK, Keer HN, Balk SP: Mutation of the androgen-receptor gene in metastatic androgen-independent prostate cancer. N Engl J Med 1995, 332:1393-1398 [DOI] [PubMed] [Google Scholar]
- 6.Klein KA, Reiter RE, Redula J, Moradi H, Zhu XL, Brothman AR, Lamb DJ, Marcelli M, Belldegrun A, Witte ON, Sawyers CL: Progression of metastatic human prostate cancer to androgen independence in immunodeficient SCID mice. Nat Med 1997, 3:402-408 [DOI] [PubMed] [Google Scholar]
- 7.Blok LJ, Kumar MV, Tindall DJ: Isolation of cDNAs that are differentially expressed between androgen-dependent and androgen-independent prostate carcinoma cells using differential display PCR. Prostate 1995, 26:213-224 [DOI] [PubMed] [Google Scholar]
- 8.Chang GT, Blok LJ, Steenbeek M, Veldscholte J, van Weerden WM, van Steenbrugge GJ, Brinkmann AO: Differentially expressed genes in androgen-dependent and -independent prostate carcinomas. Cancer Res 1997, 57:3814-3822 [PubMed] [Google Scholar]
- 9.Lucas S, De Smet C, Arden KC, Viars CS, Lethe B, Lurquin C, Boon T: Identification of a new MAGE gene with tumor-specific expression by representational difference analysis. Cancer Res 1998, 58:743-752 [PubMed] [Google Scholar]
- 10.Stubbs AP, Abel PD, Golding M, Bhangal G, Wang Q, Waxman J, Stamp GW, Lalani EN: Differentially expressed genes in hormone refractory prostate cancer: association with chromosomal regions involved with genetic aberrations. Am J Pathol 1999, 154:1335-1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin B, Ferguson C, White JT, Wang S, Vessella R, True LD, Hood L, Nelson PS: Prostate-localized and androgen-regulated expression of the membrane-bound serine protease TMPRSS2. Cancer Res 1999, 59:4180-4184 [PubMed] [Google Scholar]
- 12.Xu J, Stolk JA, Zhang X, Silva SJ, Houghton RL, Matsumura M, Vedvick TS, Leslie KB, Badaro R, Reed SG: Identification of differentially expressed genes in human prostate cancer using subtraction and microarray. Cancer Res 2000, 60:1677-1682 [PubMed] [Google Scholar]
- 13.Luo JH, Yu YP, Cleply K, Lin F, Deflavia P, Dhir R, Finkelstein S, Michalopoulos G, Becich M: Gene expression analysis of prostate cancers. Mol Carcinog 2001, 33:25-35 [DOI] [PubMed] [Google Scholar]
- 14.Chaib H, Rubin MA, Mucci NR, Li L, Taylor JMG, Day ML, Rhim JS, MaCoska JA: Activated in prostate cancer: a PDZ domain-containing protein highly expressed in human primary prostate tumors. Cancer Res 2001, 61:2390-2394 [PubMed] [Google Scholar]
- 15.Chetcuti A, Margan S, Mann S, Russell P, Handeslman D, Rogers J, Dong Q: Identification of differentially expressed genes in organ-confined prostate cancer by gene expression array. Prostate 2001, 47:132-140 [DOI] [PubMed] [Google Scholar]
- 16.Dhanasekaran SM, Barrette TR, Ghosh D, Shah R, Varambally S, Kurachi K, Pienta KJ, Rubin MA, Chinnaiyan AM: Delineation of prognostic biomarkers in prostate cancer. Nature 2001, 412:822-826 [DOI] [PubMed] [Google Scholar]
- 17.Welsh JB, Sapinoso LM, Su AI, Kern SG, Wang-Rodriguez J, Moskaluk CA, Frierson HF, Jr, Hampton GM: Analysis of gene expression identifies candidate markers and pharmacological targets in prostate cancer. Cancer Res 2001, 61:5974-5978 [PubMed] [Google Scholar]
- 18.Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C, Tamayo P, Renshaw AA, D’Amico AV, Richie JP, Lander ES, Loda M, Kantoff PW, Golub TR, Sellers WR: Gene expression correlates of clinical prostate cancer behavior. Cancer Cell 2002, 1:203-209 [DOI] [PubMed] [Google Scholar]
- 19.Ernst T, Hargenhahn M, Kenzelmann M, Cohen CD, Bomrouhi M, Weninger A, Klären R, Gröne EF, Wiesel M, Güdemann C, Küstler J, Schott W, Staehler G, Kretzler M, Hollstein M, Gröne H-J: Decrease and gain of gene expression are equally discriminatory markers for prostate carcinoma. Am J Pathol 2002, 160:2169-2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benedit P, Paciucci R, Thomson TM, Valeri M, Nadal M, Càceres C, de Torres I, Estivill X, Lozano JJ, Morote J, Reventos J: PTOV1, a novel protein overexpressed in prostate cancer containing a new class of protein homology blocks. Oncogene 2001, 20:1455-1464 [DOI] [PubMed] [Google Scholar]
- 21.Fernández PL, Nayach I, Fernández E, Fresno L, Palacín A, Farré X, Campo E, Cardesa A: Tissue macroarrays (“microchops”) for gene expression analysis. Virchows Arch 2001, 438:591-594 [DOI] [PubMed] [Google Scholar]
- 22.Fernández PL, Arce Y, Farré X, Martínez A, Nadal A, Rey MJ, Peiró N, Campo E, Cardesa A: Expression of p27/kip1 is down-regulated in human prostate carcinoma progression. J Pathol 1999, 187:563-566 [DOI] [PubMed] [Google Scholar]
- 23.Gerdes J, Lemke H, Baisch H, Wacker H-H, Schwab U, Stein H: Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol 1984, 133:1710-1715 [PubMed] [Google Scholar]
- 24.Sherr C, Roberts JM: CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev 1999, 13:1501-1512 [DOI] [PubMed] [Google Scholar]
- 25.Guo Y, Lklar GN, Borkowski A, Kyprianou N: Loss of the cyclin-dependent kinase inhibitor p27(Kip1) protein in human prostate cancer correlates with tumor grade. Clin Cancer Res 1997, 3:2269-2274 [PubMed] [Google Scholar]
- 26.Cordon-Cardo C, Koff A, Drobnjak M, Capidieci P, Osman I, Millard SS, Gaudin PB, Fazzari M, Zhang ZF, Massague J, Scher HI: Distinct altered patterns of p27KIP1 gene expression in benign prostatic hyperplasia and prostatic carcinoma. J Natl Cancer Inst 1998, 90:1284-1291 [DOI] [PubMed] [Google Scholar]
- 27.Verheijen R, Kuijpers HJ, van Driel R, Beck JL, van Dierendonck JH, Brakenhoff GJ, Ramaekers FC: Ki-67 detects a nuclear matrix-associated proliferation-related antigen. II. Localization in mitotic cells and association with chromosomes. J Cell Sci 1989, 92:531-540 [DOI] [PubMed] [Google Scholar]
- 28.Baldin V, Lukas J, Marcote J, Pagano M, Draetta G: Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev 1993, 7:812-821 [DOI] [PubMed] [Google Scholar]
- 29.Diehl JA, Cheng M, Roussel MF, Sherr CJ: Glycogen synthase kinase-3b regulates cyclin D1 proteolysis and subcellular localization. Genes Dev 1998, 12:3499-3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang J, Kornbluth S: All aboard the cyclin train: subcellular trafficking of cyclins and their CDK partners. Trends Cell Biol 1999, 9:207-210 [DOI] [PubMed] [Google Scholar]
- 31.Naar AM, Beaurang PA, Zhou S, Abraham S, Solomon W, Tjian R: Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature 1999, 398:828-832 [DOI] [PubMed] [Google Scholar]
- 32.Douglas TH, Morgan TO, McLeod DG, Moul JW, Murphy GP, Barren R, III, Sesterhenn IA, Mostofi FK: Comparison of serum prostate specific membrane antigen, prostate specific antigen, and free prostate specific antigen levels in radical prostatectomy patients. Cancer 1997, 80:107-114 [DOI] [PubMed] [Google Scholar]
- 33.Chen RH, Sarnbecki C, Blenis J: Nuclear localization and regulation of ERK- and RSK-encoded protein kinases. Mol Cell Biol 1992, 12:915-927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brunet A, Roux D, Lenormand P, Dowd S, Kayse S, Pouysségur J: Nuclear translocation of p43/p44 mitogen-activated protein kinase is required for growth factor-induced gene expression and cell cycle entry. EMBO J 1999, 18:664-674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pines J, Hunter T: Human cyclins A and B1 are differentially located in the cell and undergo cell cycle-dependent nuclear transport. J Cell Biol 1991, 115:1-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takizawa CG, Morgan DO: Control of mitosis by changes in the subcellular location of cyclin-B1-Cdk1 and Cdc25C. Curr Opin Cell Biol 2000, 12:658-665 [DOI] [PubMed] [Google Scholar]
- 37.Alt JR, Gladden AB, Diehl JA: p21Cip1 promotes cyclin D1 nuclear accumulation via direct inhibition of nuclear export. J Biol Chem 2002, 277:8517-8523 [DOI] [PubMed] [Google Scholar]
- 38.Diehl JA, Sherr CJ: A dominant-negative cyclin D1 mutant prevents nuclear import of cyclin-dependent kinase 4 (CDK4) and its phosphorylation by CDK-activating kinase. Mol Cell Biol 1997, 17:7362-7374 [DOI] [PMC free article] [PubMed] [Google Scholar]