Abstract
All blood capillaries consist of endothelial tubes surrounded by mural cells referred to as pericytes. The origin, recruitment, and function of the pericytes is poorly understood, but the importance of these cells is underscored by the severe cardiovascular defects in mice genetically devoid of factors regulating pericyte recruitment to embryonic vessels, and by the association between pericyte loss and microangiopathy in diabetes mellitus. A general problem in the study of pericytes is the shortage of markers for these cells. To identify new markers for pericytes, we have taken advantage of the platelet-derived growth factor (PDGF)-B knockout mouse model, in which developing blood vessels in the central nervous system are almost completely devoid of pericytes. Using cDNA microarrays, we analyzed the gene expression in PDGF-B null embryos in comparison with corresponding wild-type embryos and searched for down-regulated genes. The most down-regulated gene present on our microarray was RGS5, a member of the RGS family of GTPase-activating proteins for G proteins. In situ hybridization identified RGS5 expression in brain pericytes, and in pericytes and vascular smooth muscle cells in certain other, but not all, locations. Absence of RGS5 expression in PDGF-B and PDGFRβ-null embryos correlated with pericyte loss in these mice. Residual RGS5 expression in rare pericytes suggested that RGS5 is a pericyte marker expressed independently of PDGF-B/Rβ signaling. With RGS5 as a proof-of-principle, our data demonstrate the usefulness of microarray analysis of mouse models for abnormal pericyte development in the identification of new pericyte-specific markers.
Pericytes are associated with the abluminal wall of microvessels, ie, arterioles, venules, and capillaries. They can be distinguished from other perivascular cell types by their location within the microvascular basement membrane. Pericytes, such as vascular smooth muscle cells (VSMCs; the mural cells of larger vessel), are mesenchymal cells of presumed mesodermal or neuroectodermal origin. 1,2 The absence, reduced numbers, or altered migration of pericytes in certain knockout mouse mutants have shed light on mechanisms involved in their recruitment to endothelial tubes, and on the function of mural cells during embryonic development. Null mutants for transforming growth factor-β, endoglin, activin receptor-like kinase-1 (Alk-1), and SMAD5 all appear to fail to recruit mural coats to their vessels and die around mid-gestation from cardiovascular defects, together implicating transforming growth factor-β signaling in early VSMC/pericyte formation. 3-6 Null mutants for platelet-derived growth factor (PDGF)-B/PDGF receptor-β (PDGFRβ) fail to recruit proper numbers of pericytes in conjunction with angiogenic sprouting in many organs, leading to perinatal death from rupturing microaneurysms and edema. 7-9 Because PDGF-B is expressed by sprouting capillary endothelial cells and PDGFRβ by developing pericytes, paracrine PDGF-B/Rβ signaling appears to control pericyte proliferation and migration during developmental angiogenesis. 10
Pericytes express markers in common with VSMCs, such as α-smooth muscle actin (α-SMA), desmin, and PDGFRβ, which together with the ultrastructural appearance of pericytes suggest that they belong to the VSMC lineage. However, the structural and biochemical diversity of pericytes 2 and the suggested existence of other perivascular pericyte-like mesenchymal cells 11 and of perivascular cells with macrophage-like properties 12 make pericytes hard to define. Pericytes have also been proposed to have the capacity to trans-differentiate into fibroblast-like cells, 13 osteoblasts, chondrocytes, and adipocytes, 11 adding to the confusion regarding the origin, identity, and potency of these cells.
Based on their distribution and expression of contractile proteins, pericytes have been assumed to participate in hemodynamic regulation. Pericytes may also regulate transport across the capillary wall and capillary permeability. 1 During angiogenesis, contact between pericytes and endothelial cells has been suggested to stabilize vessels, promote endothelial survival, and inhibit endothelial cell proliferation. 14-17 These ideas are primarily based on the view that pericyte recruitment lags behind endothelial sprouting, and that a window of pericyte absence allows vascular plasticity resulting in growth, survival, remodeling, or regression of the endothelium depending on the presence or absence of growth factors such as vascular endothelial growth factor (VEGF). However, it has also been proposed that pericytes may have a direct role in the sprouting process by migrating ahead of the endothelium and expressing VEGF. This function may be of particular importance in corpus luteum angiogenesis. 18 Despite these numerous suggested roles and potencies of pericytes, our understanding of these cells remains limited.
Little is known about the gene expression profile of pericytes, and clearly, the lack of specific markers for pericytes constitutes an obstacle in studies of these cells. The small number and scattered distribution of pericytes, and their propensity to change properties on in vitro culture, calls for new approaches to map their gene expression in vivo and search for novel specific markers of these cells. In the present study, we have taken advantage of the fact that the brain microvessels of PDGF-B- and PDGFRβ-deficient mice have severely reduced numbers of pericytes. We used cDNA microarrays to compare the gene expression in the heads of PDGF-B- or PDGFRβ-deficient mouse embryos with littermate controls and looked for genes with down-regulated expression in the mutants. The most down-regulated gene present on our arrays was RGS5 (regulator of G-protein signaling 5), a protein implicated in heterotrimeric G-protein signaling. In situ hybridization confirmed that RGS5 is a marker for brain pericytes in mouse embryos, but also revealed its expression in other but not all populations of embryonic pericytes and SMCs.
Materials and Methods
Animals
Mice carrying a targeted disruption of the PDGF-B or PDGFRβ genes were bred as hybrids of C57BL6 and 129SV. 7,8 The morning of the vaginal plug was counted as E0.5.
Microarray
Chip Design
Microarray chips were printed with 1350 randomly chosen mouse sequence-verified expressed sequence tags from the IMAGE consortium (purchased from Invitrogen Ltd., Renfrewshire, Scotland), 5 yeast clones, and 100 selected control genes. After amplification with polymerase chain reaction, the clones were purified and resuspended in 3× standard saline citrate (SSC) and 0.2% sarcosyl and printed with GMS 417 spotter (Affymetrix, Santa Clara, CA) on γ-amino propyl silane-coated CMT-Gap slides (Corning Int., London, UK). Postprocessing of the glasses was performed as described. 19
RNA Recovery and Labeling
Total RNA from head was extracted from PDGFB−/−, PDGFB+/−, and PDGFB+/+ mice using Qiagen RNeasy kits (VWR International AB, Stockholm, Sweden) and quantified with UV-spectrophotometric analysis. Total RNA (100 μg) was primed with 2 μg of oligo-dT primer and labeled in a reverse-transcription reaction with Cy3-dUTP or Cy5-dUTP (Amersham AB, Uppsala, Sweden) according to standard protocols (http://cmgm.stanford.edu/pbrown). The differentially labeled targets were combined, mixed with 1 μl of 10 μg/μl yeast tRNA and 1 μl of 10 μg/μl polyA RNA, vacuum-dried, and resuspended in 20 μl of DIGeasy hybridization buffer (Roche Diagnostics GmbH, Mannheim, Germany). The hybridization mix was placed at 100°C for 2 minutes and 37°C for 30 minutes before being added to the chip.
Microarray Hybridization and Scanning
Hybridization was performed in a 40°C waterbath for 12 to 18 hours under lifter coverslips (Histolab, Göteborg, Sweden) in ArrayIt hybridization cassettes (TeleChem International Inc., Sunnyvale, CA). After hybridization, the slides were washed in 2× SSC and 0.1% sodium dodecyl sulfate at room temperature for 5 minutes, 1× SSC for 5 minutes and finally 0.1× SSC for 5 minutes. The slides were scanned with ScanArray 3000 confocal microarray scanner (Packard Bioscience, Meriden, CT) at laser intensity and photomultiplier tube settings, giving the best dynamic range for each chip in respective channel.
Data Preparation
Image segmentation and spot quantification was performed with the ImaGene software (Biodiscovery, Marina Del Rey, CA). After median local background subtraction, the log-transformed ratios were normalized for signal intensity variation in a nonlinear intensity-dependent way using the loess function in the S-Plus software (MathSoft Inc., Surrey, UK) as described elsewhere. 19,20 The hybridizations were repeated five times (two of them with dye swap). Genes were classified as down-regulated using two criteria: the normalized ratio was more than twofold in at least four of the five repetitions and the gene showed down-regulation in all five of five repetitions. After this filtering, mean expression values were calculated using the data from all five repetitions.
Immunohistochemistry
The following antibodies were used: biotin-conjugated isolectin B4 from Bandeiraea simplicifolia (L-2140, Sigma, St. Louis, MO) for staining of endothelial cells, anti-NG2 chondroitin sulfate proteoglycan (AB5320, Chemicon Int., Harrow, U.K.), both after incubation of sections in PBLEC (phosphate-buffered saline, pH 6.8, 1% Triton X-100, 0.1 mmol/L CaCl2, 0.1 mmol/L MgCl2, 0.1 mmol/L MnCl2) and anti-PDGFRβ (SC432, Santa Cruz Biotechnology, Santa Cruz, CA) after standard tissue treatment and biotin-conjugated goat anti-rabbit Ig (E0432, DAKO, A/S, Copenhagen, Denmark). Antibody binding was detected with streptavidin-conjugated horseradish peroxidase (D0397, DAKO) using standard protocols.
In Situ Hybridization
Nonradioactive in situ hybridization was performed on 14-μm-thick sections with digoxigenin-labeled RNA probes (Boehringer-Mannheim) visualized by alkaline phosphatase-conjugated antibodies, as described by Boström and colleagues 21 with modifications outlined by Lindahl and colleagues. 9 An RGS5 expressed sequence tag was obtained from Incyte Genomics (St. Louis, MO), subcloned in a pBluescript vector and used to generate sense and anti-sense digoxigenin-labeled probes according to standard protocols. PDGF-B and -Rβ sense and anti-sense probes were generated as described. 9 No hybridization signal was obtained with any of the sense probes, which were used as negative controls. Whole-mount in situ hybridization was performed as described 22 with the following changes; hindbrains were treated with proteinase K (10 μg/ml) for 10 minutes. Hybridization buffer contained 50% formamide, 1.3× SSC, 5 mmol/L ethylenediaminetetraacetic acid, 50 μg/ml yeast RNA, 0.2% Tween-20, 0.8% sodium dodecyl sulfate, and 100 μg/ml heparin. Posthybridization wash was performed in SSC-based buffers. Post-anti-digoxigenin antibody wash was done in maleic acid buffered saline containing Tween-20.
Results
cDNA Microarray Hybridizations Identify RGS5 as a Down-Regulated Gene in PDGF-B-Null Mutant Mice
PDGF-B knockout embryos die perinatally from widespread microvascular hemorrhage and leakage. Rupturing microaneurysms become abundant at E17 to E19, so to limit secondary influences of the hemorrhage on the gene expression profile, we decided to look for differentially expressed genes at E15.5, before the onset of severe hemorrhaging, but at a time when central nervous system (CNS) vascularization, including pericyte recruitment, is already quite advanced in wild-type mice. Whole heads from PDGF-B−/− embryos and littermate +/− and +/+ controls were used to prepare labeled targets for cDNA array hybridization. Although the pericyte-deficient state is most severe in the CNS, and somewhat less dramatic in other organs, 9,10 we chose to analyze whole heads to minimize variation in the tissue sampling for target labeling. We also wanted to minimize the time required for tissue preparation to preserve the in vivo transcription profile. The results of these experiments showed that out of the 1350 unigene set of expressed sequence tags (ESTs) present on the chips, 54 ESTs (P value <0.05) were more than twofold down-regulated (Table 1) ▶ and three ESTs were found to be more than twofold up-regulated (data not shown) in PDGF-B−/− heads compared with wild-type or PDGF-B+/− littermates. Several of these ESTs represent markers for erythrocytes or leukocytes (denoted in italics in Table 1 ▶ ). This is not unexpected because PDGF-B knockout mutants develop anemia, 7 most likely secondary to bleeding caused by the vascular defects. 23 Most of the genes on the list were, however, unexpected. The most down-regulated gene, encoding a member of the RGS family of regulators of G protein signaling, RGS5, caught our attention, because this gene has previously been reported to have a vascular expression pattern. 24,25
Table 1.
Genes Down-Regulated More Than Two-Fold in PDGFB−/− Mouse Embryos in Comparison with Control Littermates
| Gene | Hit in Celera database | e-Value with Celera hit | Image ID | Unigene ID | -fold (log2) | P value |
|---|---|---|---|---|---|---|
| Regulator of G-protein signaling, RGS5 | 439412 | Mm.20954 | 6.87 | 0.000 | ||
| Hemoglobin beta, pseudogene bh3 | 313981 | Mm.2329 | 5.91 | 0.044 | ||
| Cytochrome C oxidase, subunit VIIIa | 481408 | Mm.14022 | 5.21 | 0.026 | ||
| EST | Hemoglobin τ 1-chain | 1 × 10−110 | 419003 | 4.02 | 0.041 | |
| Hemoglobin alpha, adult chain 1 | 316082 | Mm.16820 | 3.97 | 0.030 | ||
| Tumor-associated mucin 1 | 482312 | Mm.16193 | 3.74 | 0.019 | ||
| Antigen identified by monoclonal antibody MRC OX-2 | 391081 | Mm.8649 | 3.37 | 0.006 | ||
| Lymphocyte antigen 6 complex (LY-6) | 534886 | Mm.15889 | 2.63 | 0.002 | ||
| Proliferating cell nuclear antigen | 390311 | Mm.7141 | 2.12 | 0.018 | ||
| EST | αCrystallin B-chain | 2 × 10−98 | 424655 | 2.05 | 0.003 | |
| EST | 571955 | 2.02 | 0.002 | |||
| EST | Syntaxin 3A | 4 × 10−94 | 445430 | 1.92 | 0.005 | |
| EST | Chymotrypsin B | 6 × 10−85 | 407693 | 1.91 | 0.021 | |
| Ribosomal protein L30 | 616239 | Mm.3487 | 1.89 | 0.021 | ||
| Myeloblastosis oncogene (c-myb) | 618475 | Mm.1202 | 1.85 | 0.008 | ||
| B-cell leukemia/lymphoma 3 (bcl-3) | 597868 | Mm.1068 | 1.84 | 0.000 | ||
| EST | GTPase-activating protein RAS | 8 × 10−7 | 354360 | 1.79 | 0.001 | |
| EST | 60S Ribosomal protein L38 | 1 × 10−133 | 483411 | 1.67 | 0.038 | |
| Erythroid α-spectrin, Spna1 | 538325 | Mm.2528 | 1.61 | 0.002 | ||
| EST | Cyclin-dependent kinases regulatory subunit | 1 × 10−101 | 424470 | 1.57 | 0.023 | |
| Milk fat globule-EGF factor 8 protein, Mfge8 | 478473 | Mm.1451 | 1.56 | 0.002 | ||
| EST | Similar to Flightless I (Drosophila) | 1–10−72 | 390479 | 1.53 | 0.005 | |
| Tight junction protein 1 (ZO1), Tip1 | 579645 | Mm.4342 | 1.52 | 0.008 | ||
| EST | Similar to Kallikrein (Human) | 9 × 10−18 | 329597 | 1.49 | 0.000 | |
| Nuclear, factor, erythroid derived 2, like 2 (NF-E2) | 635541 | Mm.1025 | 1.46 | 0.001 | ||
| β2 microglobulin | 596438 | Mm.163 | 1.44 | 0.029 | ||
| Transcription factor GIF (TIEG) | 597716 | Mm.4292 | 1.44 | 0.030 | ||
| EST | Tryptophanyl-tRNA synthetase | 0.0 | 478820 | 1.43 | 0.001 | |
| EST | Serine/threonine kinase 11 | 2 × 10−61 | 437421 | 1.42 | 0.018 | |
| Mini chromosome maintenance deficient 4 homolog (S. cerevisiae) | 586181 | Mm.1500 | 1.41 | 0.022 | ||
| Hemoglobin X, α-like embryonic chain in Hba complex | 604555 | Mm.545 | 1.41 | 0.049 | ||
| Creatine kinase, muscle | 608246 | Mm.2375 | 1.34 | 0.008 | ||
| OPA-containing protein 1 mRNA | 368355 | Mm.20873 | 1.32 | 0.013 | ||
| Immunoglobulin J chain precursor | 619073 | Mm.1192 | 1.31 | 0.001 | ||
| TA-9 ATP synthase β chain homolog mRNA | 573942 | Mm.30112 | 1.29 | 0.003 | ||
| Heparan sulfate proteoglycan 1, cell surface-ass. (fibroglycan, syndecan-2) | 535688 | Mm.29350 | 1.29 | 0.011 | ||
| EST | Polyadenylate-binding protein | 1 × 10−135 | 476649 | Mm.12267 | 1.28 | 0.004 |
| Gas5 growth arrest specific protein | 605306 | Mm.884 | 1.27 | 0.022 | ||
| EST | NADH dehydrogenase (ubiquinone) 1 α subcomplex 2 | 7 × 10−90 | 608660 | Mm.29867 | 1.27 | 0.022 |
| EST | Similar to filamin C γ (actin binding protein-280-human) | 1 × 10−64 | 426427 | Mm.34355 | 1.25 | 0.029 |
| EST | Endothelial differentiation, lysophosphatidic acid G-protein-coupled receptor 2, Edg2 | 2 × 10−50 | 574735 | Mm.4772 | 1.22 | 0.011 |
| Type VI collagen α 3 subunit | 574605 | Mm.7562 | 1.17 | 0.038 | ||
| EST | Malonyl-CoA decarboxylase, Mlycd | 1 × 10−138 | 318951 | Mm.20260 | 1.17 | 0.021 |
| Rho-associated, coiled-coil forming protein kinase p160 ROCK-1 | 598406 | Mm.6710 | 1.16 | 0.011 | ||
| EST | Signal recognition particle 19kD protein | 0.0 | 493659 | Mm.29452 | 1.15 | 0.036 |
| ELAV-like 1 (Hu antigen R) | 634761 | Mm.21766 | 1.09 | 0.013 | ||
| Nuclear factor of kappa light chain gene enhancer in B-cells 1, p105 (NF-κB) | 575105 | Mm.3420 | 1.09 | 0.019 | ||
| ATP-binding cassette 1 | 579422 | Mm.369 | 1.07 | 0.039 | ||
| EST | Peroxiredoxin 5, Prdx5 | 5 × 10−88 | 514837 | 1.05 | 0.022 | |
| EST | β2 adrenergic receptor | 1 × 10−137 | 573046 | Mm.5598 | 1.04 | 0.011 |
| EST | AMP deaminase 2 AMPD2 related | 1 × 10−106 | 332415 | 1.04 | 0.022 | |
| EST | Microtubule-associated protein 4, Mtap4 | 1 × 10−106 | 538121 | 1.03 | 0.024 | |
| ASMA | 2491 | 1.01 | 0.028 | |||
| EST | 26S protease regulatory subunit 7 | 3 × 10−29 | 389926 | Mm.2462 | 1.01 | 0.002 |
Clones that encode previously known endothelial, smooth muscle, and basement membrane markers are in bold. Markers for erythrocytes or leukocytes are denoted in italics.
RGS5 Is Expressed by Developing Pericytes and VSMCs
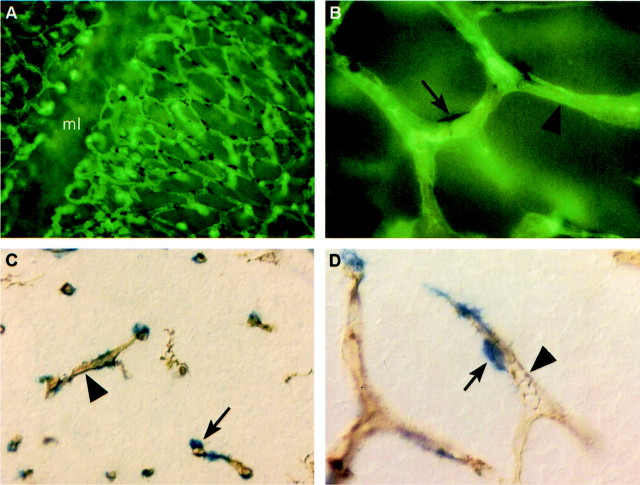
Combined RGS5 in situ hybridization and staining for brain capillary endothelium by isolectin B4 on E12.5 mouse hindbrain whole-mounts (Figure 1, A and B) ▶ and E14.5 forebrain sections (Figure 1, C and D) ▶ revealed that RGS5 was expressed by cells in association with brain microvessels. At high magnification it was evident that RGS5 was not expressed in the endothelial cells. Instead it was expressed in cells associated with the abluminal side of the endothelium. The distribution of these cells is typical for pericytes, which are expected to cover part of the abluminal endothelial surface (Figure 1, B and D ▶ , arrows) while leaving part of it free (Figure 1, B and D ▶ , arrowheads). We next compared the RGS5-expression with expression patterns of other known markers for pericytes. The most common used markers, α-SMA and desmin, are not appreciably expressed in mouse embryonic brain before E16.5. 10 We therefore compared the RGS5 expression pattern with those of PDGFRβ and the NG2 chondroitin sulfate proteoglycan. In the brain and around larger blood vessels at most locations in the embryo, the expression patterns of RGS5, PDGFRβ, and NG2 were primarily overlapping (Figure 2; A to F) ▶ . RGS5 and PDGFRβ transcripts and NG2 immunoreactivity were detected in cells corresponding to pericytes surrounding capillaries (Figure 2; A to C ▶ , arrows), VSMCs around arteries and arterioles (Figure 2; D to F ▶ , and data not shown), and in kidney mesangial cells (Figure 2; G to I) ▶ , whereas veins showed limited or no detectable levels of expression (Figure 2; D to F) ▶ . This confirms and extends previous analyses of PDGFRβ 9,10 and NG2 26 expression.
Figure 1.
RGS5 expression in brain pericytes. Whole mount in situ hybridization of flat-mounted wild-type hindbrain (ventricular view of vascular plexus in subventricular zone; ml, midline) at E12.5 (A and B) and in situ hybridization on sections from wild-type embryos at E14.5 (C and D) shows RGS5 expression (arrows) in tight association with microvessels in the brain. Double staining with isolectin B4 (arrowheads), which labels endothelial cells, indicates that RGS5 is expressed by pericytes and not in endothelial cells.
Figure 2.

Comparison of RGS5, PDGFRβ, and NG2 expression in E14.5 mouse embryos. In the brain, RGS5 (A) is expressed in cells in the capillary wall and this mimics the pattern of PDGFRβ (B) and NG2 (C) expression. Similar expression of the three markers was also seen in larger arteries (D–F) and in mesangial cells in the kidney (G–I). In the lung and gut, the expression patterns of RGS5 and PDGFRβ and NG2 are not completely overlapping. In the lung, PDGFRβ- and NG2-positive cells can be found scattered in the mesenchyme at areas corresponding to ongoing angiogenesis (K and L, asterisk), whereas RGS5-positive cells are mainly found around developing airways, in cells that are also positive for NG2 expression (J and L, arrowheads). All three markers are expressed in VSMCs surrounding the developing lung arterial branches (J–L, arrows). In the gut, PDGFRβ- and NG2-positive cells are found in mesenchymal cells immediately subjacent to the epithelium (N and O, arrows), whereas RGS5 is expressed by cells located more peripherally in the gut wall at the expected site of the developing SMC layers (M, arrowhead). NG2 is also expressed by these developing SMC layers (O, arrowheads). a, artery; v, vein; p, podocytes; e, endothelial layer; M, mesangial core.
RGS5 Expression in Lung and Gut Is Associated with Visceral Rather than Vascular SMCs
Whereas the expression patterns of RGS5, PDGFRβ, and NG2 overlapped at many sites, they differed in the developing lung and gut. Lung pericytes have previously been demonstrated to be PDGFRβ-positive 10 and we confirmed that a scattered population of PDGFRβ-positive cells was present in the lung mesenchyme in regions harboring ongoing angiogenesis (Figure 2K ▶ , asterisk). PDGFRβ was also expressed in VSMCs around developing lung arteries (Figure 2K ▶ , arrow) and a similar pattern was seen by staining with NG2 (Figure 2L) ▶ . In addition, NG2 expression was seen surrounding developing bronchial branches (Figure 2L ▶ , arrowhead). The lung RGS5 expression pattern was different from that of PDGFRβ, but overlapping with that of NG2; no scattered RGS5-positive cells were seen at the presumed location of developing microvessels (Figure 2J ▶ , asterisk). Some weakly RGS5-positive cells were found in association with lung arterial branches (Figure 2J ▶ , arrow), but the dominant RGS5 expression pattern corresponded to mesenchymal cells surrounding developing proximal airways, probably corresponding to developing bronchial SMCs (Figure 2J ▶ , arrowhead). In the gut, PDGFRβ and NG2 both seemed to be expressed by mesenchymal cells immediately subjacent to the epithelium, at a location where angiogenesis is known to be active (Figure 2, N and O ▶ , arrows). The location of these cells also suggests that they may be progenitors of the later developing muscularis mucosae and villus SMCs. NG2, but not PDGFRβ-expression, was also seen further out in the intestinal wall in the developing external muscle layers (Figure 2O ▶ , arrowheads) and in the SMCs of larger intestinal blood vessels. RGS5, on the other hand, was not expressed at detectable levels in the inner mesenchymal layer (Figure 2M ▶ , arrow). Instead, its expression was more peripheral, and seemed to occur in cells located within the developing external muscle layers (Figure 2M ▶ , arrowhead). Thus, in endodermally derived organs such as lung and gut, RGS5 expression appears to locate to visceral rather than vascular smooth muscle, and overlaps with the expression of NG2, but not PDGFRβ, at this site.
RGS5 Expression in PDGF-B and PDGFRβ Knockout Tissues
Although the expression of RGS5, PDGFRβ, and NG2 was highly similar in brain pericytes it was difficult, using currently available protocols, to provide convincing evidence of their co-expression in the same cells using double labeling. As an alternative approach, we therefore compared RGS5, PDGFRβ, and NG2 expression in mouse embryos lacking PDGF-B. Genetic deficiency of PDGF-B or PDGFRβ leads to loss of most pericytes in the developing CNS. 10,27 In the case of PDGF-B−/− mice, this is reflected by a dramatic reduction in the density of PDGFRβ-positive cells 9 (Figure 3, C and D) ▶ . Similarly, both RGS5- and NG2-positive cells were dramatically reduced in number in PDGF-B−/− brain tissue in comparison with PDGF+/− brain (Figure 3, A and B and E and F) ▶ . This does not reflect a major change in vascular density, as shown by isolectin staining (Figure 3, G and H) ▶ . The reduction of RGS5-positive cells in PDGF-B−/− mutants was not complete; a small proportion of residual cells expressing the marker remained in brain (example shown in Figure 3H ▶ , arrow), as well as in larger arteries (Figure 3J ▶ , arrow). This is in agreement with a small number (<5%) of remaining pericytes in PDGF-B−/− mutants detected by electron microscopy 9 and by the XlacZ4 marker 28 and NG2 immunohistochemistry (A Abramsson et al, unpublished observations). Thus, RGS5 reproduced the pattern of expression of PDGFRβ and NG2 in both wild-type and PDGF-B−/− mutant CNS, arguing strongly that the same population of CNS pericytes expresses all three markers.
Figure 3.

RGS5, PDGFRβ, and NG2 expression in PDGF-B−/− embryos. The developing CNS of E14.5 PDGFB−/− embryos exhibits loss of RGS5-positive cells in comparison with PDGF-B+/− embryos (B). This loss correlates with the expression pattern of PDGFRβ and NG2 seen in PDGFB−/− (D and F) mice, confirming the pericyte-specific expression of RGS5 in the brain. A small number of RGS5-positive pericytes appear to persist in PDGF-B−/− embryonic brain (a single blue spot is seen in B), which would be in agreement with previous studies demonstrating some residual pericytes in PDGF-B−/− brain. 9,10 To confirm that the rare spots of RGS5 staining corresponded to pericytes, double labeling for RGS5 and endothelial cells was performed. These stainings showed that the residual RGS5-positive cells had pericytic location in the capillary wall (H, arrow). Using this method, the reduction in number, but not complete absence, of RGS5-positive VSMC was also observed in developing arteries (compare I and J; a residual RGS5-positive VSMC in the PDGF-B−/− artery is indicated by an arrow).
Recently, a novel PDGFRβ agonist was identified and named PDGF-D. 29,30 Because the developmental pattern of expression of this factor has not yet been determined, we wanted to exclude the possibility that the residual RGS5- and NG2-expressing cells seen in PDGF-B-null embryos reflected PDGF-D-dependent signaling via PDGFRβ. We therefore determined the RGS5 and NG2 expression patterns in PDGFRβ−/− embryos. Also these mutants showed a major loss of RGS5- and NG2-positive cells, however as in PDGF-B−/− embryos, a small number of residual pericytes positive for both markers were found (data not shown).
Discussion
We demonstrate here that cDNA microarray hybridization analysis of PDGF-B mutant mouse embryos provides a method to identify genes with their expression restricted to pericytes and VSMCs. Given the small number of genes present on our array (≈3% of the total number of mouse genes) and the fact that a complex tissue was used as RNA source in which pericytes/VSMCs represent less than 5% of the cells, this result is encouraging. The use of purified capillary fragments from PDGF-B mutants as an RNA source should significantly increase the sensitivity of the method, and hence allow for the identification of pericyte-specific RNA species that are less abundantly expressed than RGS5. Arrays containing a broader coverage of the mouse genome are now also available, allowing for the screening of genes representing ∼50% of the mouse genome. We should also be able to screen for pericyte markers at a genome-wide level by using dedicated chips generated from microvascular libraries.
We establish that RGS5 is a novel marker for pericytes and VSMCs in most organs, although in endodermally derived organs (lung and gut), visceral rather than vascular SMCs are labeled. RGS5 is an interesting example of such a marker, because it is involved in the regulation of intracellular signaling. RGS5 is a member of a family of more than 25 proteins that negatively regulate G-protein coupled receptor signaling by acting as GTPase-activating proteins for Gα subunits, thereby limiting the time during which the Gα subunit remains GTP-bound and active. 31 RGS5 acts as a GTPase-activating protein for Giα and Gqα and attenuates signaling induced by angiotensin II, endothelin-1, sphingosin-1-phosphate, and PDGF in cultured cells. 32,33 Whether these or other pathways are regulated by RGS5 in pericytes in vivo remains to be established.
Mouse RGS5 was originally identified in pituitary, heart, and skeletal muscle 34 and expression of the human counterpart has been demonstrated at high abundance in heart, lung, skeletal muscle, and small intestine, and at lower levels in brain, placenta, liver colon, and leukocytes. 35 RGS5 has also been identified by microarray analysis of genes expressed in the media of human aorta but not vena cava, suggesting specific arterial expression. 24 In a search for genes differentially expressed in a genetic rat model for stroke, Kirsch and colleagues 25 found RGS5 to be down-regulated, and they localized RGS5 expression in the rat CNS to capillary endothelium and the plexus choroideus.
Our results, and also an expression analysis by Cho and co-workers, 33 demonstrate that RGS5 is selectively expressed in pericytes and in VSMCs surrounding developing arteries. It is likely that the brain endothelial RGS5 expression reported by Kirsch and colleagues 25 also represents pericytic expression, as pericytes are abundant on brain capillaries, and tightly associated with the endothelial cells. Our double-labeling experiments exclude RGS5 expression at detectable levels in mouse embryonic brain endothelial cells. Pericytes are more difficult to define than endothelial cells because of fewer available markers. This is particularly problematic in embryos, in which most pericytes lack detectable expression of the regular pericyte/VSMC markers α-SMA and desmin. However, both NG2 and PDGFRβ label embryonic brain pericytes and we found that their expression was similar to that of RGS5. Additional support for a pericytic expression of RGS5 came from the analysis of mouse mutants that are specifically depleted in pericytes. In both PDGF-B and PDGFRβ null mice, the changed RGS5 expression pattern perfectly matched the known sites of pericyte deficiency, whereas RGS5 expression remained at sites where pericytes or VSMCs persist. These results also argue that RGS5 expression is not directly controlled by PDGF-B/PDGFRβ signaling. This conclusion is also consistent with in vitro data, showing that RGS5 expression is only marginally influenced by exposure of cultured VSMCs to PDGF. 33
In summary, using microarray analysis of a pericyte-deficient mouse mutant, a knockout of PDGF-B, we have identified a new marker for developing pericytes and VSMCs. As such, RGS5 will provide a valuable complement to existing markers, in particular because it is expressed in pericytes in early embryos, in which the classical pericyte and SMC markers, such as α-SMA and desmin, are not expressed at detectable levels. We anticipate a functional role of RGS5 in pericytes and VSMCs but this remains a speculation until appropriate functional analyses, eg, gene knockouts, are available. Our experiment demonstrates the power of applying microarray analysis to well-characterized mouse mutant tissues as an approach to identify novel markers for cells and signaling pathways. We expect that further refinement of the approach used here, eg, by using enriched microvascular preparations and gene arrays with broader coverage will prove useful in the search for additional molecules with pericyte-specific expression.
Acknowledgments
We thank Helen Hjelm and Monica Elmestam for excellent technical assistance and Phil Soriano for the PDGFRβ mutant mice.
Footnotes
Address reprint requests to Christer Betsholtz, Department of Medical Biochemistry, The Sahlgrenska Academy at Göteborg University, P.O. Box 440, SE 405 30 Göteborg, Sweden. E-mail: christer.betsholtz@medkem.gu.se.
Supported by the Novo Nordisk Foundation, the Swedish Cancer Foundation, IngaBritt and Arne Lundberg Foundation, the European Community, and Göteborg University.
References
- 1.Allt G, Lawrenson JG: Pericytes: cell biology and pathology. Cells Tissues Organs 2001, 169:1-11 [DOI] [PubMed] [Google Scholar]
- 2.Sims DE: Diversity within pericytes. Clin Exp Pharmacol Physiol 2000, 27:842-846 [DOI] [PubMed] [Google Scholar]
- 3.Dickson MC, Martin JS, Cousins FM, Kulkarni AB, Karlsson S, Akhurst RJ: Defective haematopoiesis and vasculogenesis in transforming growth factor beta-1 knock-out mice. Development 1995, 121:1845-1854 [DOI] [PubMed] [Google Scholar]
- 4.Li DY, Sorensen LK, Brooke BS, Urness LD, Davis EC, Taylor DG, Boak BB, Wendel DP: Defective angiogenesis in mice lacking endoglin. Science 1999, 284:1534-1537 [DOI] [PubMed] [Google Scholar]
- 5.Yang X, Castilla LH, Xu X, Li C, Gotay J, Weinstein M, Liu PP, Deng CX: Angiogenesis defects and mesenchymal apoptosis in mice lacking SMAD5. Development 1999, 126:1571-1580 [DOI] [PubMed] [Google Scholar]
- 6.Oh SP, Seki T, Goss KA, Imamura T, Yi Y, Donahoe PK, Li L, Miyazono K, ten Dijke P, Kim S, Li E: Activin receptor-like kinase 1 modulates transforming growth factor-beta 1 signaling in the regulation of angiogenesis. Proc Natl Acad Sci USA 2000, 97:2626-2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levéen P, Pekny M, Gebre-Medhin S, Swolin B, Larsson E, Betsholtz C: Mice deficient for PDGF B show renal, cardiovascular, and hematological abnormalities. Genes Dev 1994, 8:1875-1887 [DOI] [PubMed] [Google Scholar]
- 8.Soriano P: Abnormal kidney development and hematological disorders in PDGF beta-receptor mutant mice. Genes Dev 1994, 8:1888-1896 [DOI] [PubMed] [Google Scholar]
- 9.Lindahl P, Johansson BR, Levéen P, Betsholtz C: Pericyte loss and microaneurysm formation in PDGF-B-deficient mice. Science 1997, 277:242-245 [DOI] [PubMed] [Google Scholar]
- 10.Hellström M, Kalén M, Lindahl P, Abramsson A, Betsholtz C: Role of PDGF-B and PDGFR-beta in recruitment of vascular smooth muscle cells and pericytes during embryonic blood vessel formation in the mouse. Development 1999, 126:3047-3055 [DOI] [PubMed] [Google Scholar]
- 11.Canfield AE, Doherty MJ, Wood AC, Farrington C, Ashton B, Begum N, Harvey B, Poole A, Grant ME, Boot-Handford RP: Role of pericytes in vascular calcification: a review. Z Kardiol 2000, 89:20-27 [DOI] [PubMed] [Google Scholar]
- 12.Thomas WE: Brain macrophages: on the role of pericytes and perivascular cells. Brain Res Brain Res Rev 1999, 31:42-57 [DOI] [PubMed] [Google Scholar]
- 13.Sundberg C, Ivarsson M, Gerdin B, Rubin K: Pericytes as collagen-producing cells in excessive dermal scarring. Lab Invest 1996, 74:452-466 [PubMed] [Google Scholar]
- 14.Orlidge A, D’Amore PA: Inhibition of capillary endothelial cell growth by pericytes and smooth muscle cells. J Cell Biol 1987, 105:1455-1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirschi KK, Rohovsky SA, Beck LH, Smith SR, D’Amore PA: Endothelial cells modulate the proliferation of mural cell precursors via platelet-derived growth factor-BB and heterotypic cell contact. Circ Res 1999, 84:298-305 [DOI] [PubMed] [Google Scholar]
- 16.Benjamin LE, Hemo I, Keshet E: A plasticity window for blood vessel remodelling is defined by pericyte coverage of the preformed endothelial network and is regulated by PDGF-B and VEGF. Development 1998, 125:1591-1598 [DOI] [PubMed] [Google Scholar]
- 17.Benjamin LE, Golijanin D, Itin A, Pode D, Keshet E: Selective ablation of immature blood vessels in established human tumors follows vascular endothelial growth factor withdrawal. J Clin Invest 1999, 103:159-165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reynolds LP, Grazul-Bilska AT, Redmer DA: Angiogenesis in the corpus luteum. Endocrine 2000, 12:1-9 [DOI] [PubMed] [Google Scholar]
- 19.Scheidl S, Nilsson S, Kalen M, Hellstrom M, Takemoto M, Håkansson J, Lindahl P: mRNA expression profiling of laser microbeam microdissected cells from slender embryonic structures. Am J Pathol 2002, 160:801-813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dudoit S, Yang YH, Callow MJ, Speed TP: Technical report no. 578. Department of Biochemistry, Stanford University School of Medicine, 2000
- 21.Boström H, Willetts K, Pekny M, Levéen P, Lindahl P, Hedstrand H, Pekna M, Hellström M, Gebre-Medhin S, Schalling M, Nilsson M, Kurland S, Törnell J, Heath JK, Betsholtz C: PDGF-A signaling is a critical event in lung alveolar myofibroblast development and alveogenesis. Cell 1996, 85:863-873 [DOI] [PubMed] [Google Scholar]
- 22.Hogan B, Beddington R, Costantini F, Lacy E: Manipulating the Mouse Embryo: A Laboratory Manual. 1994. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
- 23.Kaminski W, Lindahl P, Lin NL, Broudy VC, Crosby JR, Swolin B, Bowen-Pope DF, Martin P, Ross R, Betsholtz C, Raines EW: The basis of hematopoietic defects in PDGF-B and PDGFRbeta deficient mice. Blood 2001, 97:1990-1998 [DOI] [PubMed] [Google Scholar]
- 24.Adams LD, Geary RL, McManus B, Schwartz SM: A comparison of aorta and vena cava medial message expression by cDNA array analysis identifies a set of 68 consistently differentially expressed genes, all in aortic media. Circ Res 2000, 87:623-631 [DOI] [PubMed] [Google Scholar]
- 25.Kirsch T, Wellner M, Luft FC, Haller H, Lippoldt A: Altered gene expression in cerebral capillaries of stroke-prone spontaneously hypertensive rats. Brain Res 2001, 910:106-115 [DOI] [PubMed] [Google Scholar]
- 26.Ozerdem U, Grako KA, Dahlin-Huppe K, Monosov E, Stallcup WB: NG2 proteoglykan is expressed exclusively by mural cells during vascular morphogenesis. Dev Dyn 2001, 222:218-227 [DOI] [PubMed] [Google Scholar]
- 27.Lindahl P, Karlsson L, Hellstrom M, Gebre-Medhin S, Willetts K, Heath JK, Betsholtz C: Alveogenesis failure in PDGF-A-deficient mice is coupled to lack distal spreading of alveolar smooth muscle cell progenitors during lung development. Development 1997, 124:3943-3953 [DOI] [PubMed] [Google Scholar]
- 28.Enge M, Bjarnegård M, Gerhardt H, Gustafsson E, Kalén M, Asker N, Hammes H-P, Shani M, Fässler R, Betsholtz C: Endothelium-specific platelet-derived growth factor-B ablation mimics diabetic retinopathy. EMBO J 2002, 21:4307-4316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bergsten E, Uutela M, Li X, Pietras K, Ostman A, Heldin C-H, Alitalo K, Eriksson U: PDGF-D is a specific, protease activated ligand for the PDGF beta-receptor. Nat Cell Biol 2001, 3:512-516 [DOI] [PubMed] [Google Scholar]
- 30.LaRochelle WJ, Jeffers M, McDonald WF, Chillakuru RA, Giese NA, Lokker NA, Sullivan C, Boldog FL, Yang M, Vernet C, Burgess CE, Fernandes E, Deegler LL, Rittman B, Shimkets J, Shimkets RA, Rothberg JM, Lichtenstein HS: PDGF-D, a new protease-activated growth factor. Nat Cell Biol 2001, 3:517-521 [DOI] [PubMed] [Google Scholar]
- 31.Cho H, Kehrl JH: RGS proteins, modulators of heterotrimeric G-protein signaling: their physiological regulation and emerging in vivo functions. Curr Top Biochem Res 2000, 2:216 [Google Scholar]
- 32.Zhou J, Moroi K, Nishiyama M, Usui H, Seki N, Ishida J, Fukamizu A, Kimura S: Characterization of RGS5 in regulation of G protein-coupled receptor signaling. Life Sci 2001, 68:1457-1469 [DOI] [PubMed] [Google Scholar]
- 33.Cho H, Kozaka T, Bondjers C, Betsholtz C, Kehrl J: Pericyte-specific expression of RGS5: implications for PDGF and EDG receptor signaling during vascular maturation. EMBO J (in press) [DOI] [PubMed]
- 34.Chen C, Zheng B, Han J, Lin SC: Characterization of a novel mammalian RGS protein that binds to Galpha proteins and inhibit pheromone signaling in yeast. J Biol Chem 1997, 272:8679-8685 [DOI] [PubMed] [Google Scholar]
- 35.Seki N, Sugano S, Suzuki Y, Nakagawara A, Ohira M, Muramatsu M, Saito T, Hori T: Isolation, tissue expression, and chromosomal assignment of human RGS5, a novel G-protein signal regulator gene. J Hum Genet 1998, 43:202-205 [DOI] [PubMed] [Google Scholar]