Abstract
Expression-profiling studies have helped define genetic changes associated with carcinogenesis. Determining which alterations in gene expression are causally associated with cancer and which result from the general dysregulation in gene expression that is characteristic of malignancies remains a problem. Transcriptional profiling of early lesions (small cancers or precancers) holds promise for identifying biologically important changes in gene expression. There are, however, technical barriers to the study of small tumors. The total number of cells available for analysis is limiting. It is also often difficult to distinguish cancer cells from normal proliferating cells in frozen sections that are typically used as a source of RNA. Here we describe an ethanol fixation and paraffin-embedding protocol that preserves tissue architecture and cellular morphology of the mouse endometrium, and allows for the recovery of high-quality RNA from microdissected cells. We performed GeneChip expression profiling using RNA from 800 to 4400 cells microdissected from ethanol-fixed, paraffin-embedded uteri. Endometrial adenocarcinomas exhibited changes in the levels of a number of messages known to be abnormally expressed in cancer, and differential expression of additional transcripts not previously implicated in carcinogenesis. We confirmed increased Amd1 expression in RNAs from mouse endometrial carcinomas that were hybridized to GeneChips and validated overexpression of this transcript in additional tumors.
Gene expression profiling is a powerful method for defining genetic changes that may contribute to the development of cancer. Microarray technology has been used in identifying genes differentially expressed in a variety of bulk tumors that consist of cancerous and noncancerous, normal cells. 1-5 The advent of laser capture microdissection (LCM) and RNA amplification methodologies has made it possible to examine the gene expression signatures of specific cell populations in tissues of interest. 6,7 LCM can be used to isolate malignant epithelium from the heterogeneous cellular milieu of a bulk tumor. Starting with small amounts of total RNA, T7-based RNA amplification allows for generation of sufficient amounts of labeled anti-sense RNA (aRNA) for hybridization to nucleotide arrays. 8,9 LCM and RNA amplification have been used in profiling the expression patterns of laser-captured neuronal subtypes and microdissected ductal carcinoma cells. 9,10 Frozen sections are often used as the source of tissue in transcription profiling experiments because RNA extracted from microdissected frozen cells is generally of high quality. 9,11,12 Good preservation of histology and cellular RNA have been achieved using frozen sections of tissues fixed in sucrose. 13 The cellular morphology in frozen sections of many tissues, including the endometrium, is suboptimal, and as such it can be difficult to distinguish precancerous and cancerous lesions from normal proliferating cells. Although existing fixation protocols and tissue processing methods provide good histology, 11,12 preservation of cellular RNA for gene expression-profiling applications can remain problematic. There is a need for alternative tissue fixation and processing protocols that yield good histology and preserve cellular RNA for microarray studies.
Expression profiling of cancer cells microdissected from large tumors has brought forth valuable insights into the molecular signatures of malignancies. 9,14-16 It is widely accepted that cancer cells accumulate numerous genetic defects as they divide. 17,18 Consequently, when profiling large tumors that have undergone many cell divisions it can be difficult to distinguish early changes in gene expression, some of which are likely to represent causes of cancer, from secondary genetic alterations that accumulate late in tumorigenesis. Profiling gene expression in precancers or in cancers that have undergone fewer cell divisions may help identify genes differentially expressed early in tumorigenesis. Genes dysregulated early in tumor development could then be evaluated in functional studies in an attempt to determine their roles in cancer formation.
In this study we describe an ethanol fixation and paraffin-embedding protocol that preserves tissue architecture and cellular morphology of the endometrium. Our fixation and processing methods also allow for recovery of high-quality cellular RNA from microdissected cells. Gene expression profiling was performed using RNA from 800 to 4400 cancer and normal cells microdissected from ethanol-fixed, paraffin-embedded tissues. Analysis of GeneChip data revealed differential expression of a number of transcripts abnormally expressed in other types of cancer, and changes in the levels of other messages not previously implicated in carcinogenesis. Overrepresentation of Amd1 in mouse endometrial cancers was confirmed in RNA specimens that were used in GeneChip hybridizations, and increased levels of this transcript was validated in additional cancers.
Materials and Methods
Tissue Processing and Staining
Approval was obtained from the Washington University Institutional Animal Care and Use Committee for all experiments involving animals. Uteri were removed from 9-month-old C57BL/6J X FVB/N F1 hybrid mice (Jackson Laboratories, Bar Harbor, ME; Taconic, Germantown, NY) that had been given subcutaneous injections (2 μg/day) of diethylstilbestrol (Sigma, St. Louis, MO) in corn oil on neonatal days 1 to 5. Diethylstilbestrol treatment of newborn female mice has been shown previously to result in a variety of uterine abnormalities, including endometrial hyperplasia and carcinoma. 19,20 Mouse uteri were cut into three pieces (two uterine horns and a junction that included the upper vagina) to facilitate penetration of tissue by the fixative, and sandwiched between two pieces of absorptive paper to prevent the uterine horns from twisting during fixation. Tissues were then placed into cassettes, immersed in ice-cold (4°C) 70% or 80% ethanol, and fixed for 16 or 24 hours at 4°C. After fixation, the uteri were transferred to room temperature, 70% ethanol for approximately 1 hour, and then processed and embedded in paraffin blocks at the Tissue Procurement Core at the Washington University School of Medicine/Barnes-Jewish Hospital Siteman Cancer Center. Specimens were processed using the Tissue-Tek V.I.P. processor (Sakura Finetek, Inc., Torrance, CA), and embedded in conventional or low-melting temperature paraffin (Sherwood Medical, St. Louis, MO) using the Leica EG1160 apparatus (Leica Microsystems, Wetzlar, Germany).
Five- to 6-μm sections were mounted on slides and stained with hematoxylin and eosin as follows: xylenes for 5 minutes (twice), 100% ethanol for 1 minute (three times), 95% ethanol for 1 minute (twice), 70% ethanol for 1 minute (once), deionized water for 1 minute (once), Mayer’s hematoxylin (Sigma, St. Louis, MO) for 2.5 minutes (once), deionized water for 30 seconds (once), Scott’s tap water substitute bluing solution (Fisher Scientific, Pittsburgh, PA) for 1 minute (once), deionized water for 30 seconds (once), 70% ethanol for 1 minute (once), 95% ethanol for 1 minute (once), alcoholic eosin (95%) (Sigma, St. Louis, MO) for 20 seconds (once), 100% ethanol for 1 minute (twice), and xylenes for 3 minutes (twice). Tissue slides for microdissection were stained as follows: xylene for 1.5 minutes (once), 10 dips in 100% ethanol (once), 10 dips in 95% ethanol (once), 10 dips in 70% ethanol (once), 10 dips in deionized water (once), 10 dips in Mayer’s hematoxylin (once), five dips in deionized water (once), five dips in Scott’s tap water substitute (once), 10 dips in 70% ethanol (once), 10 dips in 95% ethanol (once), five dips in alcoholic eosin (once), 20 dips in 100% ethanol (twice), and xylenes for 1.5 minutes (once).
LCM and RNA Purification
LCM of well-differentiated mouse endometrial cancers and uninvolved endometrial epithelium was performed using the PixCell II apparatus (Arcturus Engineering, Mountain View, CA). One thousand laser pulses (15-μm beam setting; 40 to 45 mW power setting) were sufficient to capture approximately 1000 cells. For gene expression validation studies, three cancers were microdissected: Ca D (850 cells), Ca E (300 cells), and Ca F (675 cells). Histologically normal endometrial epithelium (∼1000 cells/case) was microdissected from the same uterine tissues. RNA was purified from microdissected tissues using the Arcturus PicoPure RNA Isolation Kit (Arcturus).
RNA Analysis
RNA specimens were analyzed by microcapillary electrophoresis on Agilent LabChips (Agilent, Wilmington, DE) or by conventional denaturing gel electrophoresis on mini formaldehyde gels. Because of the limited quantities of RNA recovered from the small, microdissected cancers and the requirement for ≥5 ng of total RNA for LabChip analysis, a mini gel system was devised in which less than 1 ng of total RNA could be evaluated. One μl of RNA prepared from the LCM tissues was loaded on 5 × 6- cm 1% agarose denaturing gels and electrophoresed for 10 to 15 minutes at 90 V. The gels were then stained with SYBR Gold dye (Molecular Probes, Eugene, OR), and RNA was visualized using UV light (250 nm wavelength). The quantity of RNA was estimated by comparison to standards of known concentration, and the RNA quality was assessed by the ratio of 28S and 18S rRNA bands.
Generation of Biotinylated aRNA Targets for GeneChip Hybridizations
Targets for hybridization to Affymetrix U74Av2 GeneChips were generated using two rounds of T7-based RNA linear amplification. 8 Bacillus subtillis transcripts were added to all RNA samples before amplification. 9 The starting copy numbers of these control transcripts were as follows: 1000 copies of trp, 5000 copies of thr, 20,000 copies of phe, and 100,000 copies of lys. Based on an average of 10 ng of total starting cellular RNA (1000 cells worth of RNA), the preamplification copy number of each B. subtillis transcript was equivalent to an estimated 1 copy per cell of trp, 5 copies per cell of thr, 20 copies per cell of phe, and 100 copies per cell of lys. RNA samples were subjected to one round of linear amplification using the Arcturus RiboAmp RNA Amplification Kit (Arcturus). Biotin-labeled ribonucleotides were incorporated into the aRNA during the second round of amplification using Enzo’s BioArray HighYield RNA transcript Labeling Kit (T7) (Enzo Diagnostics Inc., Farmingdale, NY). The biotinylated aRNA was treated with DNase, purified on Qiagen RNeasy columns (Qiagen, Valencia, CA), and the yield of labeled aRNA determined using ultraviolet light absorbance. Biotinylated aRNA was then fragmented and delivered to the Multiplexed Gene Analysis Core Facility at the Siteman Cancer Center, where it was hybridized to mouse U74Av2 GeneChips (up to 12,488 mouse genes represented) according to Affymetrix protocols (Affymetrix, Santa Clara, CA).
Microarray Data Analysis
Hybridization signals were made comparable between arrays by individually scaling the signals on each chip to an average intensity of 1500 signal units using Microarray Analysis Suite 5.0 (Affymetrix). Scaled array data were imported into DNA-Chip analyzer (dChip) version 1.2 for filtering and analysis. 21 Three filtering parameters were applied to the scaled GeneChip data: 1) transcripts that had hybridization signals of >100,000 units were eliminated because of possible skewing in the representation of these transcripts during RNA amplification or GeneChip signal scaling; 2) transcripts that did not give hybridization signals of ≥300 units on two or more of the seven GeneChips were considered noise for the purposes of this experiment and excluded; and 3) transcripts that did not meet a minimum threshold for variability in hybridization signals of ≥0.4, as determined by the ratio of the SD of hybridization signals for a transcript on all GeneChips/the mean hybridization signal of that transcript, were also disregarded. Filtered transcripts that were differentially expressed by more than or equal to twofold in all cancers compared to both normal samples were identified using dChip. 21
Validation Studies
First-strand cDNA templates for quantitative, real-time polymerase chain reaction (QPCR) experiments were synthesized using the ThermoScript RT-PCR system (Invitrogen Corp., Carlsbad, California). Random hexamer-primed reverse transcription was performed on 1 μg of each linearly amplified and biotinylated aRNA for cancer specimens Ca pool 1, Ca pool 2, Ca A, Ca B, and Ca C and for each of their corresponding normal aRNA samples N pool 1, N pool 2, N A, N B, and N C, respectively. Biotinylated normal aRNAs N A, N B, and N C were only used in validation studies and were not hybridized to GeneChips. For cancer and normal RNAs D, E, and F, first-strand cDNA was generated using random hexamer-primed reverse transcription of 1 μg of linearly amplified aRNA (two rounds) that was not biotinylated. Primers were designed using Primer Express software (Applied Biosystems, Foster City, CA). Amd1-specific primers were as follows: forward, 5′-CCAGAAGATTGACGGCTTTAAAC-3′; reverse, 5′-TGAACATAGCACTCTGGCAATCA-3′. Gapdh was selected as a reference for the amount of input RNA in each reaction tube because GeneChip data showed minimal variability in the hybridization signals of this transcript across all arrays. Gapdh-specific primers were as follows: forward, 5′-ACAATGAATACGGCTACAGCAACA-3′; reverse 5′-TCTTACTCCTTGGAGGCCATGTAG-3′. QPCR was performed using SYBR Green PCR reagents (Applied Biosystems), and product accumulation was detected by real-time on the Sequence Detector 5700 system (Applied Biosystems). The efficiencies of the Amd1 and Gapdh assays were similar (not shown). Expression levels of Amd1 were compared between cancer and normal endometrium in each animal using the ΔCt approach (Applied Biosystems, User Bulletin no. 2). All QPCR experiments were performed in quadruplicate and included no reverse transcriptase controls and no template controls (not shown).
Results and Discussion
Ethanol-Fixed, Paraffin-Embedded Tissues Give Good Histology and Yield High-Quality RNA
Ethanol-based fixation and paraffin-embedding methods have been used to preserve cellular morphology, DNA, and protein in a variety of tissues such as kidney, breast, and prostate. 11,12,22,23 However, cellular RNA isolated from ethanol-fixed tissues can show substantial degradation. We examined the endometrial histology in murine uterine tissues that were fixed in 70% or 80% ethanol at 4°C for 16 or 24 hours, and processed and embedded in paraffin using standard protocols. Mouse uteri fixed in 70% ethanol gave endometrial histology superior to that of specimens fixed in 80% ethanol (not shown). Furthermore, the endometrial histology of mouse uteri fixed in 70% ethanol for 16 hours was comparable to that of tissues fixed in 70% ethanol for 24 hours. Fixation for longer than 16 hours increased the likelihood of RNA degradation (data not shown).
We modified tissue-processing and paraffin-embedding protocols for recovery of high-quality RNA from our ethanol-fixed tissues. We reduced the number of steps, the duration of each step, and the temperature at which each tissue-processing step is performed. Modifications also included the use of low-melting temperature wax for paraffin embedding of mouse uteri (Table 1) ▶ . These modifications in tissue processing did not compromise endometrial histology. Uteri that were fixed in 70% ethanol for 16 hours at 4°C, processed according to our modified protocol, and embedded in low-melting temperature paraffin yielded endometrial histology comparable to that obtained using conventional formalin fixation and tissue-processing methods (Figure 1) ▶ .
Table 1.
Modified Tissue Processing Protocol for Preservation of RNA in Ethanol-Fixed Tissues
| Tissue processing step | Duration, minutes | Temperature, °C |
|---|---|---|
| 70% Ethanol | 10 | 35† |
| 80% Ethanol | 10 | 35† |
| 95% Ethanol* | 15 | 35† |
| 100% Ethanol* | 15 | 35† |
| Xylene | 10 | 35† |
| Xylene | 15 | 35† |
| Low-melt paraffin | 15 | 56‡ |
| Low-melt paraffin | 15 | 56‡ |
| Low-melt paraffin | 15 | 56‡ |
| Low-melt paraffin | 15 | 56‡ |
| Total minutes | 135 |
*Single 95% and 100% ethanol steps rather than conventional duplicate steps for each.
†Temperature reduced from 40°C to 35°C.
‡Low-melting temperature paraffin steps at 56°C for 60 minutes instead of standard 58°C incubations for 120 minutes. Processing time reduced from the conventional 450 total minutes to 135 minutes.
Figure 1.
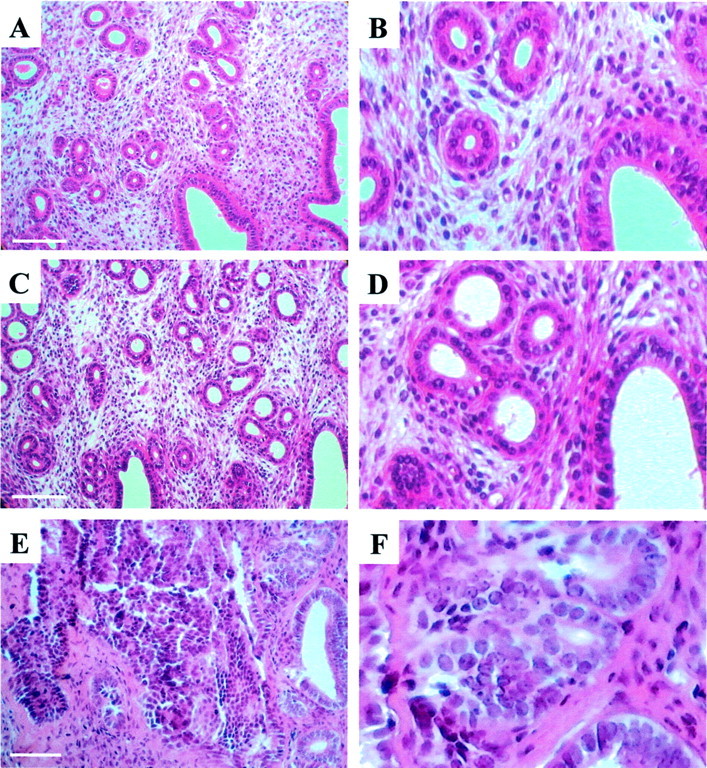
Representative examples of mouse uterine sections stained with H&E. A: Formalin-fixed normal mouse endometrium; C: ethanol-fixed normal mouse endometrium. E: Section through an ethanol-fixed uterus containing endometrial adenocarcinoma. A, C, and E: Low magnification; B, D, and F: high magnification. Note the good preservation of tissue architecture and cellular morphology in ethanol-fixed tissues. Scale bars, 100 μm.
To test the quality of cellular RNA in our ethanol-fixed, paraffin-embedded tissues, we evaluated RNA specimens prepared from microdissected endometrial epithelial cells by microcapillary or mini formaldehyde gel electrophoresis. Representative examples of high-quality, semidegraded, and degraded RNA specimens are shown in Figure 2 ▶ . RNA samples analyzed by microcapillary electrophoresis that showed a 28S/18S rRNA ratio of approximately two were classified as high quality (Figure 2 ▶ , lanes 3 and 5). Fifteen of 27 RNA specimens (55%) were of high quality based on microcapillary analysis. We were able to assess the quality and quantity of as little as 0.6 ng of total RNA using mini gels, allowing us to conserve as much RNA as possible for transcription profiling experiments (data not shown). In some cases, it was difficult to estimate the ratio of 28S/18S rRNA on the mini formaldehyde gels. Therefore, RNA samples that showed intact 28S and 18S bands on the formaldehyde gels were considered of high quality (Figure 2 ▶ , lane 8). The 28S and 18S rRNA bands were both evident in 35 of 57 RNA samples (61%) that were analyzed on the formaldehyde gels. The average amount of RNA recovered from 1000 microdissected cells was approximately 10 ng of total RNA.
Figure 2.
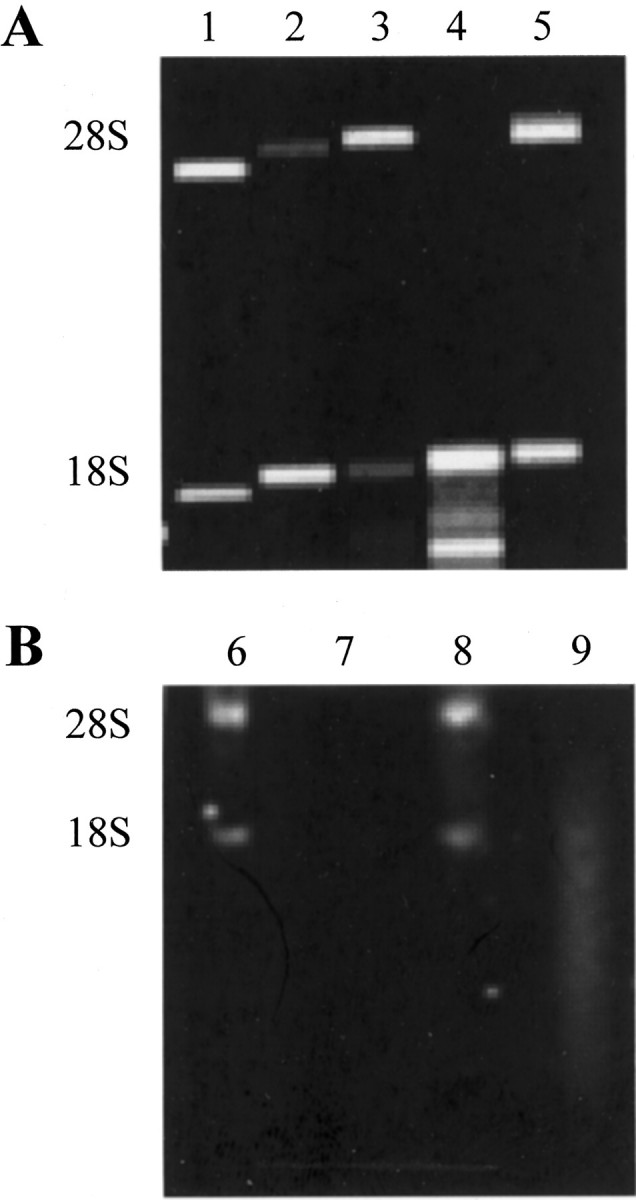
Qualitative and quantitative assessment of RNA from microdissected tissues. All tissues were fixed in ethanol and embedded in paraffin. Representative examples of Agilent LabChip and mini formaldehyde gel analysis are shown in A and B, respectively. Lanes 1 and 6: High-quality RNA standards (lane 1, 45 ng total RNA; lane 6, 1 ng total RNA). Lanes 3, 5, and 8: High-quality RNA with a 28S/18S rRNA ratio of ∼2 (lane 3, 5.16 ng total RNA; lane 5, 5.46 ng total RNA; lane 8, 1 ng total RNA). Lanes 2, 4, and 9: Semidegraded RNA. Note the 28S/18S rRNA ratio of <2 in lane 2, and the absence of a 28S rRNA band in lanes 4 and 9 (lane 2, 19.26 ng total RNA; lane 4, 14.4 ng total RNA; lane 9, 2 ng total RNA). Lane 7: Completely degraded RNA.
The fixation and tissue processing methods that we optimized for these studies may prove useful in preserving the cellular morphology and RNA in other tissues, particularly those that do not give good frozen histology.
Gene Expression Profiling of Cancer and Normal Cell Populations Microdissected from Ethanol-Fixed, Paraffin-Embedded Tissues
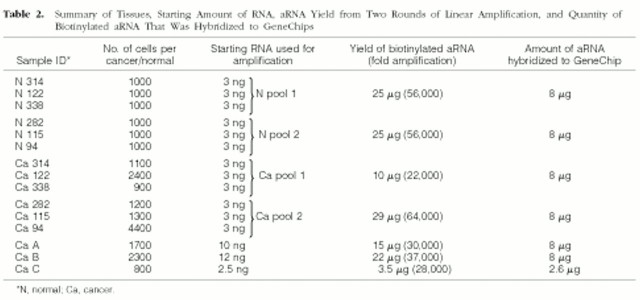
Gene expression profiling of large tumors has provided valuable information on molecular changes that are observed late in tumorigenesis. 1-4,9,14-16 Transcriptional profiling of precancers and early/small cancers will help identify early genetic events that may represent causes of carcinogenesis. Tumor cells (well-differentiated endometrial adenocarcinoma) and uninvolved (histologically normal) epithelium were microdissected from ethanol-fixed, paraffin-embedded tissues. For expression-profiling studies, we used RNA specimens that were analyzed on mini formaldehyde gels and showed both 18S and 28S rRNA bands. Four RNA pools were prepared. Two cancer RNA pools were generated, each of which was made up of equal amounts of cancer cell RNA from three different animals (Table 2 ▶ , Ca pools 1 and 2). Two normal RNA pools were prepared, each of which contained equal amounts of RNA from uninvolved endometrial epithelium from the same tissues used to generate the cancer RNA pools (Table 2 ▶ , N pools 1 and 2). RNA from three additional cancers (RNA not pooled) was also included in this study (Table 2 ▶ , Ca A, Ca B, and Ca C). Biotinylated aRNA for GeneChip hybridizations was generated using two rounds of T7-based linear amplification. 8 Two rounds of linear amplification yielded an average 42,000-fold increase in RNA quantity, consistent with results reported previously. 9 The percentage of transcripts called “present” on our GeneChips ranged from 23 to 27%, similar to earlier findings. 9 Affymetrix software gives “present” calls to transcripts for which the specific hybridization signals are higher than background noise signals. Approximately 3000 transcripts were called “present” on each GeneChip. The number of different transcripts detected on at least one of the seven arrays was 5241, or 42% of the 12,488 transcripts that are represented on the U74Av2 GeneChip.
Table 2.
Summary of Tissues, Starting Amount of RNA, aRNA Yield from Two Rounds of Linear Amplification, and Quantity of Biotinylated aRNA That Was Hybridized to GeneChips
Set numbers of copies of B. subtillis control transcripts were added to all RNA specimens before amplification. 9 The hybridization signals of these transcripts provided a measure of the linearity of the amplification process, and a rough indication of the sensitivity of the experiment. The scaled signals of each of the bacterial transcripts showed ≤2.3-fold variability between any two samples (Figure 3A ▶ , hybridization signals). The uniformity in amplification of B. subtillis transcripts is an indication that cellular RNA was amplified uniformly in all specimens. Scaled signal ratios of thr to trp transcripts (added to starting RNA at ∼5 copies per cell and ∼1 copy per cell, respectively) were close to five in each sample after amplification, as would be expected from the preamplification copy numbers of these two transcripts (Figure 3A) ▶ . Based on the linearity in amplification of trp and thr transcripts, we believe that after amplification there should have been appropriate representation of cellular transcripts that were expressed at one to five copies per cell in each of the starting RNA samples (Figure 3A ▶ , overlap of red dashed curve and the black dotted line). The hybridization signals of the trp transcripts were detectable on all seven GeneChips, with an average scaled signal of 10,000 units (Figure 3A) ▶ . Thus, we expect that the sensitivity of this experiment was sufficient to allow us to monitor transcripts that were present at one copy per cell in the preamplified RNA samples. The signal ratios of phe to thr transcripts (phe added to starting RNA at ∼20 copies per cell) were approximately three for most specimens rather than the anticipated ratios of four. Scaled signal ratios of lys to phe (lys added to starting RNA at ∼100 copies per cell) were close to two, well below the expected ratios of five. This indicates that there may have been skewing in the representation of the phe and lys transcripts either as a consequence of nonlinear RNA amplification or signal scaling (Figure 3A ▶ , divergence of red dashed curve from the black dotted line). The hybridization signals for cellular transcripts that are expressed at ≥20 copies per cell, such as messages of ribosomal protein genes Rpl 7, Rpl 30, Rps 11, and Rps 28, were therefore considered unreliable indicators of actual expression levels of these transcripts. Consequently, we filtered out transcript with hybridization signals >100,000 signal units before analyzing GeneChip data.
Figure 3.
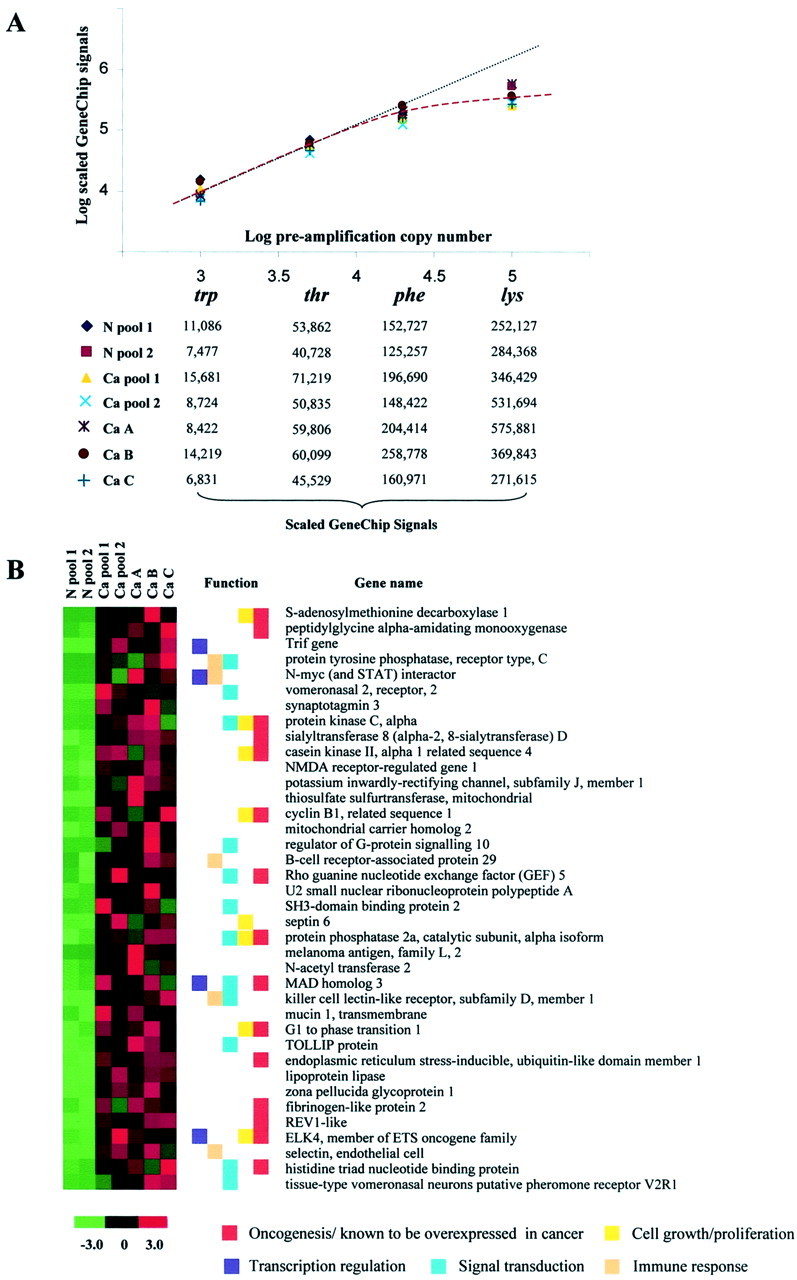
Hybridization signals of amplification controls and visual representation of the GeneChip signals for 38 transcripts overrepresented in mouse endometrial cancer. A: Logarithmic (log10) plot of the preamplification copy number of B. subtillis transcripts versus the scaled GeneChip signals of these control transcripts in each sample after two rounds of linear amplification, GeneChip hybridization, and signal scaling. The signals for the trp and thr transcripts show a linear correlation (black dotted line), while the scaled signals for the phe and lys messages appear to diverge from the linear plot (red dashed curve). B: A heat map of the 38 annotated transcripts that were overexpressed in all five cancers compared to both normal aRNA specimens that were hybridized to GeneChips. Red indicates an increase in signal intensity and green signifies a decrease in signal levels, both compared to the overall mean signal intensity for each transcript across all GeneChips (black). Colored boxes denote biological function(s) and/or involvement in oncogenesis.
We applied three filtering criteria to hybridization data, which reduced the number of transcripts to ∼6600 linearly amplified cellular transcripts. Thirty-one filtered transcripts were underexpressed by more than or equal to twofold in all five cancers compared to both normal pools (data not shown). Filtered transcripts that had a signal ratio of more than or equal to two in all cancers compared to both normal pools were considered overrepresented in cancer cells. Fifty-eight transcripts, 38 of which were messages of annotated genes and 20 were unknown expressed sequence tags, satisfied this criterion. The 38 annotated transcripts that were overexpressed in cancer cells are shown in Figure 3 ▶ . The Gene Ontology database (http://godatabase.org) was used to assign functions to the annotated transcripts, and Monochromatic SAGE/cDNA Virtual Northerns on the Cancer Genome Anatomy Project website (http://cgap.nci.nih.gov) were used in determining if any of the 38 transcripts are known to be overexpressed in cancer. The expression levels of 15 transcripts (40%) were statistically higher (P < 0.05) in at least one tumor type compared to the corresponding normal tissue (Figure 3B ▶ , red boxes). 24 Twenty-two of the 38 transcripts (60%) are involved in cell proliferation, cell signaling, transcription regulation, and/or the immune response, cellular processes shown previously to be abnormally regulated in cancer (Figure 3B ▶ , colored boxes). 1-4 There was limited functional annotation for the 16 remaining annotated transcripts that were overrepresented in the endometrial cancers that were investigated. The genes from which these transcripts are derived may be of interest for more in-depth molecular investigations.
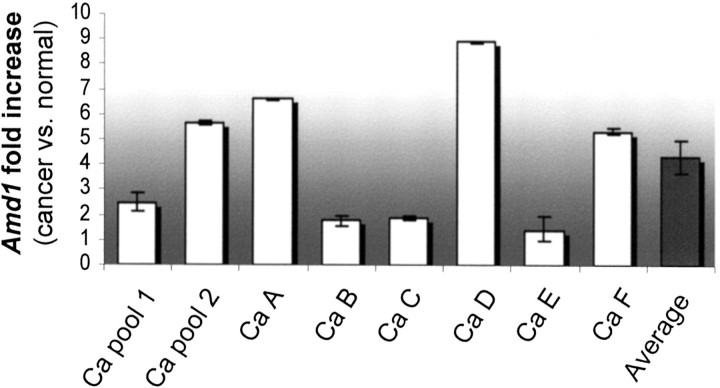
The results of transcription profiling studies must be confirmed and validated in additional cancer specimens. GeneChip data indicated that S-adenosylmethionine decarboxylase1 (Amd1) was overexpressed in the five cancer aRNA samples compared to both normal pools. The average Amd1 hybridization signal was 16,000 units for the cancer specimens and 5400 units for the normal pools, approximately threefold higher in cancer compared to normal RNA. Amd1 catalyzes the decarboxylation of S-adenosylmethionine, which in turn serves as an aminopropyl donor for the biosynthesis of the polyamines spermidine and spermine. 25 Amd1 is involved in cell growth, cell cycle progression, and development. 26-28 Tumor progression and metastasis are associated with increases in Amd1 levels. 29,30 Quantitative, real-time PCR (QPCR) confirmed overrepresentation of Amd1 in Ca pool 1 ×2.5-fold and in Ca pool 2 ×5.7-fold relative to transcript levels in their corresponding N pools 1 and 2, respectively (Figure 4) ▶ . These fold increases in Amd1 levels were comparable to the expected fold increases in message levels based on GeneChip data (2.3-fold increase in Ca pool 1 compared to N pool 1, and a 2.7-fold increase in Ca pool 2 compared to N pool 2). Amd1 levels were also higher in RNA from individual cancers Ca A, Ca B, and Ca C, compared to transcript levels in their respective normal epithelial RNA samples that were not hybridized to GeneChips (Figure 4) ▶ . Amd1 was overexpressed in RNA from two of three additional small, microdissected cancer specimens (300 to 850 cells) compared to message levels in normal epithelial RNA from each animal (Figure 3 ▶ , B, Ca D, Ca E and Ca F). Comparing Amd1 levels in individual cancers (Ca A to Ca F) to message levels in the corresponding uninvolved endometrial epithelium from those animals, we saw an average increase of 4.7-fold in Amd1 levels in cancer (Figure 4 ▶ , average). This average fold increase in Amd1 expression is within the range of fold increases observed in Ca pools 1 and 2 (Figure 4) ▶ . Confirmation of the increase in Amd1 levels in Ca pools 1 and 2 and in individual tumors Ca A, Ca B, and Ca C, and observing a similar increase in two of three additional cancers (Ca D and Ca F) validates the GeneChip expression data and suggests that Amd1 is overexpressed in diethylstilbestrol-promoted murine endometrial cancers.
Figure 4.
Validation of increased expression of Amd1 in mouse endometrial cancer. Quantitative, real-time PCR verifying overexpression of Amd1 in the five cancer RNA samples hybridized to GeneChips (Ca pools 1 and 2, and Ca A, Ca B, and Ca C), and validating increased expression of Amd1 in additional cancers (Ca D and Ca F). The average fold increase in Amd1 levels in specimens Ca A to Ca F (electronic pool represented by the average column) is within the range of fold increases seen for Ca pools 1 and 2 (biological pools). The fold increases were obtained by comparing Amd1 levels in each cancer to transcript levels in uninvolved normal endometrium from the same animal (see Materials and Methods).
Starting with ethanol-fixed, paraffin-embedded tissues, we were able to profile gene expression in small, microdissected endometrial cancers and uninvolved uterine endometrium. Of the 58 transcripts that were overrepresented in our series of cancers, many are involved in cell proliferation, signal transduction, and regulation of transcription, or have been shown to be up-regulated in other types of cancer. The transcripts with no known roles in tumorigenesis are candidates for larger scale expression analysis and functional studies. The methodologies used in these experiments should prove useful in gene expression-profiling studies of other types of cancers, precancerous lesions or tissues for which limited numbers of cells are available
Note Added in Proof
Analysis of RNA prepared from paraffin-embedded tissues stored for more than six months revealed a reduction in quality compared to specimens processed immediately.
Acknowledgments
We thank Mrs. Mary Ann Mallon for her help with animal care, tissue recovery, and processing; Dr. Mark A. Watson for his advice regarding experimental design and preparation of this manuscript; Drs. Elise C. Kohn, John W. Gillespie, and Lu Charboneau for sharing with us their expertise in ethanol fixation of tissues and LCM; the Alvin J. Siteman Cancer Center at Washington University School of Medicine and Barnes-Jewish Hospital in St. Louis, Missouri, for the use of the Multiplexed Gene Analysis Core and the Tissue Procurement Core; and Dr. Barak Cohen for his input on GeneChip data analysis.
Footnotes
Address reprint requests to Dr. Paul J. Goodfellow, Washington University School of Medicine, Department of Surgery, Campus Box 8109, 660 South Euclid Ave., St. Louis, MO 63106-1029. E-mail: goodfellowp@msnotes.wustl.edu.
Supported in part by the National Institutes of Health [R01 CA71754 to P. J. G., P30 CA91842 (P.I. Timothy J. Eberlein) and Genomic Analysis training grant T32 HG00045 (P.I. Sean Eddy)].
References
- 1.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D: Molecular portraits of human breast tumours. Nature 2000, 406:747-752 [DOI] [PubMed] [Google Scholar]
- 2.Prakash K, Pirozzi G, Elashoff M, Munger W, Waga I, Dhir R, Kakehi Y, Getzenberg RH: Symptomatic and asymptomatic benign prostatic hyperplasia: molecular differentiation by using microarrays. Proc Natl Acad Sci USA 2002, 28:7598-7603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hippo Y, Taniguchi H, Tsutsumi S, Machida N, Chong JM, Fukayama M, Kodama T, Aburatani H: Global gene expression analysis of gastric cancer by oligonucleotide microarrays. Cancer Res 2002, 62:233-240 [PubMed] [Google Scholar]
- 4.Mutter GL, Baak JP, Fitzgerald JT, Gray R, Neuberg D, Kust GA, Gentleman R, Gullans SR, Wei LJ, Wilcox M: Global expression changes of constitutive and hormonally regulated genes during endometrial neoplastic transformation. Gynecol Oncol 2001, 83:177-185 [DOI] [PubMed] [Google Scholar]
- 5.Giordano TJ, Shedden KA, Schwartz DR, Kuick R, Taylor JM, Lee N, Misek DE, Greenson JK, Kardia SL, Beer DG, Rennert G, Cho KR, Gruber SB, Fearon ER, Hanash S: Organ specific molecular classification of primary lung, colon, and ovarian adenocarcinoma using gene expression profiles. Am J Pathol 2001, 159:1231-1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA, Liotta LA: Laser capture microdissection. Science 1996, 274:998-1001 [DOI] [PubMed] [Google Scholar]
- 7.Van Gelder RN, Von Zastrow ME, Yool A, Dement WC, Barchas JD, Eberwine JH: Amplified RNA synthesized from limited quantities of heterogenous cDNA. Proc Natl Acad Sci USA 1990, 87:1663-1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang E, Miller LD, Ohnmacht GA, Liu ET, Marincola FM: High-fidelity mRNA amplification for gene profiling. Nature Biotechnol 2000, 18:457-459 [DOI] [PubMed] [Google Scholar]
- 9.Luzzi V, Holtschlag V, Watson MA: Expression profiling of ductal carcinoma in situ by laser capture microdissection and high-density oligonucleotide arrays. Am J Pathol 2001, 158:2005-2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo L, Salunga RC, Guo H, Bittner A, Joy KC, Galindo JE, Xiao H, Rogers KE, Wan JS, Jackson MR, Erlander MG: Gene expression profiles of laser-captured adjacent neuronal subtypes. Nat Med 1999, 5:117-122 [DOI] [PubMed] [Google Scholar]
- 11.Gillespie JW, Best CJM, Bichsel VE, Cole KA, Greenhut SF, Hewitt SM, Ahram M, Gathright YB, Merino MJ, Strausberg RL, Epstein JI, Hamilton SR, Gannot G, Baibakova GV, Calvert VS, Flaig MJ, Chuaqui RF, Herring JC, Pfeifer J, Petricoin EF, Linehan M, Duray PH, Bova S, Emmert-Buck MR: Evaluation of non-formalin tissue fixation for molecular profiling studies. Am J Pathol 2002, 160:449-457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldsworthy SM, Stockton PS, Trempus CS, Foley JF, Maronpot RR: Effects of fixation on RNA extraction and amplification from laser capture microdissected tissue. Mol Carcinog 1999, 25:86-91 [PubMed] [Google Scholar]
- 13.Parlato R, Rosica A, Cuccurullo V, Mansi L, Macchia P, Owens JD, Mushinski JF, De Felice M, Bonner RF, Di Lauro R: A preservation method that allows recovery of intact RNA from tissues dissected by laser capture microdissection. Anal Biochem 2002, 300:139-145 [DOI] [PubMed] [Google Scholar]
- 14.Leethanakul C, Patel V, Gillespie J, Pallente M, Ensley JF, Koontongkaew S, Liotta LA, Emmert-Buck M, Gutkind JS: Distinct pattern of expression of differentiation and growth-related genes in squamous cell carcinomas of the head and neck revealed by the use of laser capture microdissection and cDNA arrays. Oncogene 2000, 19:530-536 [DOI] [PubMed] [Google Scholar]
- 15.Crnogorac-Jurcevic T, Efthimiou E, Nielsen T, Loader J, Terris B, Stamp G, Baron A, Scarpa A, Lemoine NR: Expression profiling of microdissected pancreatic adenocarcinomas. Oncogene 2002, 21:4587-4594 [DOI] [PubMed] [Google Scholar]
- 16.Zou TT, Selaru FM, Xu Y, Shustova V, Yin J, Mori Y, Shibata D, Sato F, Wang S, Olaru A, Deacu E, Liu TC, Abraham JM, Meltzer SJ: Application of cDNA microarrays to generate a molecular taxonomy capable of distinguishing between colon cancer and normal colon. Oncogene 2002, 21:4855-4862 [DOI] [PubMed] [Google Scholar]
- 17.Hanahan D, Weinberg RA: The hallmarks of cancer. Cell 2000, 100:57-70 [DOI] [PubMed] [Google Scholar]
- 18.Hahn WC, Weinberg RA: Modeling the molecular circuitry of cancer. Nat Rev Cancer 2002, 2:331-341 [DOI] [PubMed] [Google Scholar]
- 19.Newbold RR, Bullock BC, McLachlan JA: Uterine adenocarcinoma in mice following developmental treatment with estrogens: a model for hormonal carcinogenesis. Cancer Res 1990, 50:7677-7681 [PubMed] [Google Scholar]
- 20.Couse JF, Davis VL, Hanson RB, Jefferson WN, McLachlan JA, Bullock BC, Newbold RR, Korach KS: Accelerated onset of uterine tumors in transgenic mice with aberrant expression of the estrogen receptor after neonatal exposure to diethylstilbestrol. Mol Carcinog 1997, 19:236-242 [DOI] [PubMed] [Google Scholar]
- 21.Schadt EE, Li C, Ellis B, Wong WH: Feature extraction and normalization algorithms for high-density oligonucleotide gene expression array data. J Cell Biochem 2001, 37:S120-S125 [DOI] [PubMed] [Google Scholar]
- 22.Giannella C, Zito FA, Colonna F, Paradiso A, Marzullo F, Alaibac M, Schittuli F: Comparison of formalin, ethanol, and histochoice fixation on the PCR amplification from paraffin-embedded breast cancer tissue. Eur J Clin Chem Clin Biochem 1997, 35:633-635 [PubMed] [Google Scholar]
- 23.Ben-Ezra J, Johnson DA, Rossi J, Cook N, Wu A: Effect of fixation on the amplification of nucleic acids from paraffin-embedded material by the polymerase chain reaction. J Histochem Cytochem 1991, 39:351-354 [DOI] [PubMed] [Google Scholar]
- 24.Lal A, Lash AE, Altschul SF, Velculescu V, Zhang L, McLendon RE, Marra MA, Prange C, Morin PJ, Polyak K, Papadopoulos N, Vogelstein B, Kinzler KW, Strausberg RL, Riggins GJ: A public database for gene expression in human cancers. Cancer Res 1999, 59:5403-5407 [PubMed] [Google Scholar]
- 25.Heby O, Persson L: Molecular genetics of polyamine biosynthesis in eukaryotic cells. Trends Biochem Sci 1990, 143:424-430 [DOI] [PubMed] [Google Scholar]
- 26.Chattopadhyay MK, Tabor CW, Tabor H: Absolute requirement of spermidine for growth and cell cycle progression of fission yeast (Schizosaccharomycespombe). Proc Natl Acad Sci USA 2002, 99:10330-10334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kramer DL, Chang BD, Chen Y, Diegelman P, Alm K, Black AR, Roninson IB, Porter CW: Polyamine depletion in human melanoma cells leads to G1 arrest associated with induction of p21WAF1/CIP1/SDI1, changes in the expression of p21-regulated genes, and a senescence-like phenotype. Cancer Res 2001, 61:7754-7762 [PubMed] [Google Scholar]
- 28.Nishimura K, Nakatsu F, Kashiwagi K, Ohno H, Saito T, Igarashi K: Essential role of S-adenosylmethionine decarboxylase in mouse embryonic development. Genes Cells 2002, 7:41-47 [DOI] [PubMed] [Google Scholar]
- 29.Bettuzzi S, Davalli P, Astancolle S, Carani C, Madeo B, Tampieri A, Corti A, Saverio B, Pierpaola D, Serenella A, Cesare C, Bruno M, Auro T, Arnaldo C: Tumor progression is accompanied by significant changes in the levels of expression of polyamine metabolism regulatory genes and clusterin (sulfated glycoprotein 2) in human prostate cancer specimens. Cancer Res 2000, 60:28-34 [PubMed] [Google Scholar]
- 30.Hardin MS, Mader R, Hurta RA: K-FGF mediated transformation and induction of metastatic potential involves altered ornithine decarboxylase and S-adenosylmethionine decarboxylase expression—role in cellular invasion. Mol Cell Biochem 2002, 233:49-56 [DOI] [PubMed] [Google Scholar]