Abstract
High-level gains at 5p15, a chromosomal region including the human telomerase catalytic protein subunit (hTERT) gene, have been documented in several medulloblastomas. We therefore analyzed hTERT gene dosage in a group of medulloblastomas and other embryonal brain tumors using differential PCR. Amplification of the hTERT locus was detected in 15 of 36 (42%) tumors examined. To correlate gene amplification with message level, we used real-time quantitative PCR to measure hTERT mRNA in 50 embryonal brain tumors. hTERT mRNA was detected in all but one of these cases, and mRNA level correlated significantly with gene dosage (r = 0.82). Log-rank analysis of survival data revealed a trend toward poor clinical outcomes in patients with medulloblastomas containing high hTERT mRNA levels, but clinical follow-up was relatively short and the association was not statistically significant (P = 0.078). Comparative genomic hybridization was used to further analyze the tumor with the greatest hTERT gene dosage and mRNA level, a recurrent medulloepithelioma. hTERT was amplified in the recurrent tumor but not in the primary lesion, suggesting this locus can be involved in tumor progression. Our data indicate that hTERT gene amplification is relatively common in embryonal brain tumors, and that increased expression of hTERT mRNA may be associated with biologically aggressive tumor behavior.
Brain tumors are the most common solid neoplasms that occur in childhood. 1 Among them, embryonal tumors are the most frequently encountered malignant lesions. Included in the current World Health Organization classification are medulloblastoma, supratentorial primitive neuroectodermal tumor (sPNET), atypical teratoid/rhabdoid tumor (AT/RT), and medulloepithelioma. The major molecular changes in central nervous system (CNS) embryonal tumors are only partially understood. 2 One gene commonly involved in carcinogenesis that has not yet been analyzed in a significant number of embryonal brain tumors is hTERT, which encodes the telomerase catalytic protein subunit.
Telomerase is an enzyme synthesizing the repetitive nucleotide sequence TTAGGG in telomeres. Human telomerase consists of an RNA subunit, (hTR), which is widely expressed, and a protein subunit, telomerase reverse transcriptase (hTERT), whose expression is more tightly regulated. 3-5 The hTERT gene is located on chromosome 5 at 5p15.33; its expression is repressed in normal human somatic cells but is reactivated in most tumors (reviewed in 6 ). In many neoplasms, increased telomerase activity is associated with poor clinical outcomes. 6-11 While gene amplification has not generally been considered a common mechanism to increase telomerase activity in tumors, three recent reports have documented hTERT gene amplification in non-CNS primary tumors and tumor cell lines with concomitant increases in hTERT mRNA level. 12-14
Interestingly, high-level gains of chromosomal material in the 5p15 region have been detected in medulloblastomas, suggesting that the hTERT gene could be amplified in CNS embryonal tumors. 15,16 Data on telomerase in these tumors is sparse. To our knowledge, hTERT gene dosage and mRNA levels have never been analyzed in medulloblastoma or other CNS embryonal neoplasms. In a recent review of telomerase in brain tumors, Falchetti and colleagues 17 identified fewer than 10 CNS embryonal tumors from three studies in which telomerase enzymatic activity had been analyzed. We therefore used differential PCR and real-time RT-PCR to determine the relationship between hTERT gene copy number, hTERT mRNA expression, and clinical outcome in CNS embryonal tumors. We show that the hTERT gene is amplified in a significant number of cases, and that medulloblastoma patients with increased hTERT expression in their tumors have a trend toward worse clinical outcomes.
Materials and Methods
Clinical Samples
Tissue from 50 embryonal tumors resected between 1992 and 2002 at either the Johns Hopkins Hospital, Emory University Hospital, or L’hôpital Ste-Justine were used in these studies (Table 1) ▶ . The cases included 15 anaplastic medulloblastomas, 13 classic medulloblastomas, 10 nodular medulloblastomas, 8 supratentorial PNET, 2 medulloepitheliomas, 1 medullomyoblastoma, and 1 pineoblastoma. The median age of patients was 7 years (range, 6 months to 55 years) and 82% of the cases occurred in patients 18 years of age or less. The median follow-up for all patients was 19 months; the median follow-up in the medulloblastoma patients used for survival analysis was 20 months.
Table 1.
hTERT Molecular Analysis and Clinical Features in CNS Embryonal Tumors
| Samples | Sex | Age | Status | Follow-up (months) | Histology | hTERT RT-PCR | hTERT/5qSTS |
|---|---|---|---|---|---|---|---|
| 1 | M | 9 | P | A 18 | Classic MB | 0.0 | 2.0 |
| 2 | M | 21 | P | A 1 | Classic MB | 0.5 | 1.3 |
| 3 | M | 2 | P | D 10 | PNET | 0.7 | 0.8 |
| 4 | F | 13 | P | A 58 | Classic MB | 1.1 | 2.6 |
| 5 | F | 9 | P | A 4 | Nodular MB | 1.2 | NA |
| 6 | F | 9 | P | A 20 | Anaplastic MB-F | 1.3 | 0.6 |
| 7 | F | 2 | P | A 9 | Nodular MB | 1.3 | NA |
| 8 | F | 4 | P | A 8 | Classic MB | 1.8 | 6.0 |
| 9 | M | 5 | P | A 5M | PNET | 2.2 | 2.2 |
| 10 | M | 3 | P | A 15 | Nodular MB | 2.4 | NA |
| 11 | F | 16 | P | A 24 | Anaplastic MB-F | 2.7 | 1.2 |
| 12 | M | 23 | P | A 75 | Classic MB | 3.5 | 1.9 |
| 13 | F | <1 | P | D 0 | PNET | 4.9 | 0.4 |
| 14 | F | 18 | P | A 28 | Anaplastic MB | 5.4 | 0.4 |
| 15 | M | 11 | R | D 27 | Anaplastic MB | 5.5 | 1.0 |
| 16 | F | 4 | P | D 14 | PNET | 7.0 | 0.8 |
| 17 | M | 1 | P | A 14 | Nodular MB | 8.5 | NA |
| 18 | M | 4 | P | A 12 | Nodular MB | 10.0 | NA |
| 19 | F | 43 | P | A 23 | Anaplastic MB | 13.8 | 1.7 |
| 20 | M | 4 | P | A 9 | Nodular MB | 14.1 | 3.4 |
| 21 | M | 6 | P | A 49 | Anaplastic MB-F | 15.5 | 0.3 |
| 22 | M | 11 | R | A 73 | Anaplastic MB | 22.1 | 0.8 |
| 23 | M | 5 | R | A 21 | Medulloepithelioma | 22.6 | 2.3 |
| 24 | F | 7 | P | A 19 | Anaplastic MB-F | 26.9 | NA |
| 25 | F | 5 | P | A 3 | Anaplastic MB-F | 28.6 | 0.5 |
| 26 | M | 1 | P | D 0 | PNET | 30.3 | 2.1 |
| 27 | M | 2 | P | D 17 | Classic MB | 31.0 | NA |
| 28 | M | 5 | R | D 7 | Anaplastic MB | 31.8 | 3.1 |
| 29 | M | 6 | P | A 22 | Classic MB | 36.5 | NA |
| 30 | M | 12 | P | A 14 | Classic MB | 37.4 | 0.1 |
| 31 | F | 3 | P | D 2 | PNET | 41.0 | 3.6 |
| 32 | F | 2 | P | A 32 | Anaplastic MB-F | 48.1 | 0.4 |
| 33 | F | 16 | P | A 28 | Nodular MB | 50.7 | NA |
| 34 | M | 6 | P | A 25 | Nodular MB | 52.0 | NA |
| 35 | M | 9 | P | D 9 | Anaplastic MB | 52.4 | 3.2 |
| 36 | F | 3 | R | A 122 | PNET | 52.5 | 5.1 |
| 37 | F | 10 | P | A 82 | Classic MB | 59.7 | 3.7 |
| 38 | F | 3 | P | A 5 | Classic MB | 64.5 | 0.4 |
| 39 | M | 9 | P | A 21 | Nodular MB | 64.8 | NA |
| 40 | F | 12 | P | A 21 | Anaplastic MB-F | 65.2 | NA |
| 41 | F | 1 | P | A 14 | Anaplastic MB-F | 88.9 | NA |
| 42 | M | 12 | P | A 32 | Medullomyoblastoma | 125.1 | 1.2 |
| 43 | M | 39 | R | A 58 | Classic MB | 125.2 | 6.0 |
| 44 | F | 4 | P | D 3 | PNET | 129.5 | 2.5 |
| 45 | M | 26 | P | A 21 | Nodular MB | 132.0 | 1.0 |
| 46 | M | 5 | P | D 11 | Anaplastic MB | 133.0 | 1.6 |
| 47 | F | 55 | P | A 9 | Classic MB | 144.1 | NA |
| 48 | F | 22 | P | A 70 | Pineoblastoma | 152.3 | 3.2 |
| 49 | M | 15 | R | D 116 | Classic MB | 154.0 | 3.7 |
| 50 | F | 1 | R | D 31 | Medulloepithelioma | 603.6 | 17.6 |
* A, alive; D, deceased; F, focal; MB, medulloblastoma; P, primary; PNET, supratentorial primitive neuroectodermal tumor; R, recurrent.
Molecular Analyses
DNA and total RNA were extracted from snap-frozen tumor tissues using TRIZOL Reagent (Invitrogen, Carlsbad, CA) per the manufacturer’s instructions. RNA was then treated with DNase and further purified using the RNeasy Protocol (Qiagen, Valencia, CA). Quantitative RT-PCR was performed using the ABI Prism 7700 Sequence Detector (Applied Biosystems, Weiterstadt, Germany) with TaqMan One-Step RT-PCR Master Mix reagents (Applied Biosystems) according to the manufacturer’s instructions. PCR primers used for the analysis of hTERT expression were hTERT-1912F (forward primer: 5′-TACGTCGTGGGAGCCAGAAC-3′) and hTERT-1978R (reverse primer: 5′-CCTTCACCCTCGAGGTGAGA-3′). The TaqMan probe hTERT-1933T (5′-TTCCGCAGAGAAAAGAGGGCCGA-3′) was labeled with 6-FAM and TAMRA. Amplicon length was 67 bp. Final concentration of primers was 0.5 μmol/L; final concentration of the TaqMan probe was 0.2 μmol/L. cDNA amount was standardized in each reaction using expression of β-actin according to the manufacturer’s instructions (Applied Biosystems, Foster City, CA). All samples were analyzed in triplicate. Serial dilutions of total RNA from the D425 medulloblastoma cell line (ATCC) were used to generate standard curves and the expression of hTERT in all samples was calculated in relation to this. Gene copy number of hTERT was determined using differential PCR of sequences from hTERT on 5p and control sequences on 5q (5qSTS). The hTERT product was 260 bp long. The forward and reverse primers were 5′-GTGACCGTGGTTTCTGTGTG-3′ and 5′-GGGCCTCAGAGAGCTGAGTA-3′ respectively. The 5qSTS fragment was 199 bp in length; forward and reverse primers were 5′-TTGGTCCCCAGTAACTTGATG-3′ and 5′-TGTCCCTAGGATCTTGCTCAG-3′. PCR conditions are available on request. Bands were visualized and quantitated using a Fluor-S Multiimager (Bio-Rad, Hercules, CA). CGH was performed as previously described. 18 The SYSTAT 9 program was used for Kaplan-Meier survival curves and Breslow-Gehan log-rank analysis.
Results
The hTERT Gene is Amplified in 42% of CNS Embryonal Tumors
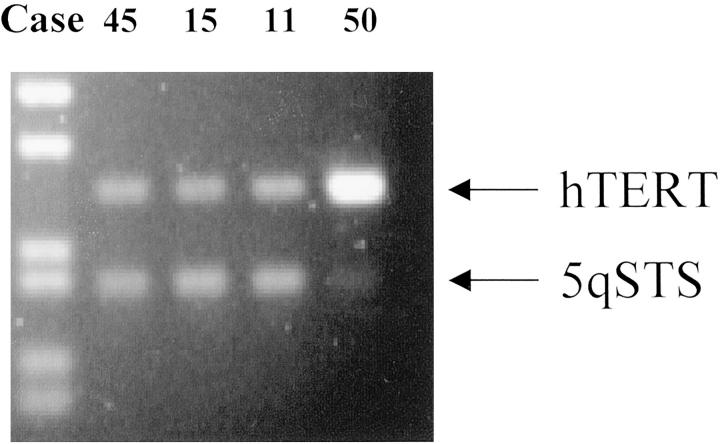
Amplification of the hTERT gene at 5p15.33 was detected using differential PCR following the methods of Waha and colleagues. 19 The hTERT/5qSTS DNA ratios in the 36 tumors analyzed ranged from 0.1 to 17.6 (Table 1 ▶ and Figure 1 ▶ ). Experimental replicates were performed in selected cases and yielded similar results. Analysis of hTERT/5qSTS DNA ratios in 8 normal subjects produced values ranging from 0.99 to 1.12, with a mean of 1.02. The hTERT/5qSTS DNA ratio threshold for amplification was therefore set at 2.17, which represents twice the mean ratio of the normal subjects plus three standard deviations, a standard established in earlier reports on this method. 19,20 Using this cut-off, hTERT gene amplification was detected in 15 of 36 (42%) CNS embryonal tumors (Table 1) ▶ . The hTERT/5qSTS ratios in amplified cases ranged from 2.2 to 17.6.
Figure 1.
hTERT gene amplification detected by differential PCR. In cases 45, 15, and 11, hTERT and 5qSTS sequences are amplified in a roughly 1:1 ratio. In case 50, the hTERT product is 17-fold more abundant, consistent with gene amplification at the locus.
hTERT mRNA Expression Level and Clinical Factors
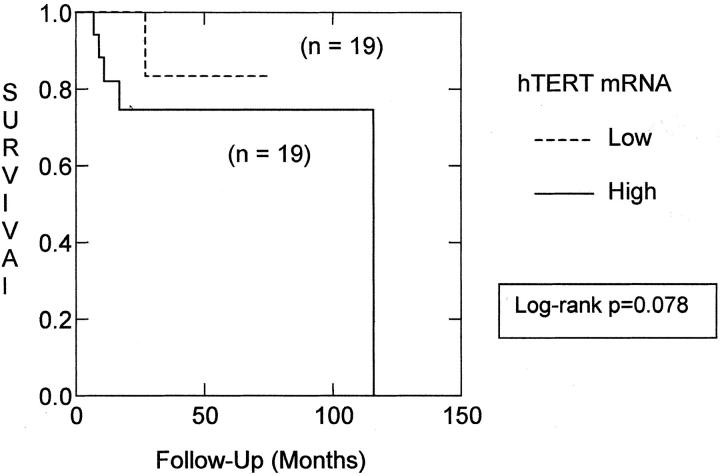
We used real-time RT-PCR to measure the level of hTERT in mRNA extracted from snap-frozen tumor tissue. hTERT mRNA was detected in 49 of 50 CNS embryonal tumors examined using this method. We divided these tumors into those with above-median hTERT expression, high hTERT, and below-median hTERT level, low hTERT (Table 1) ▶ . hTERT mRNA expression levels did not distinguish between different types of embryonal tumors, eg, PNET, medulloepithelioma, medulloblastoma, etc. The nodular, classic, and anaplastic medulloblastoma subtypes were also distributed roughly evenly with respect to hTERT level. However, as has been observed in other tumor types, high levels of hTERT were more common in patients who died from their disease. In the combined group of embryonal tumor patients, 9 of 25 (36%) with above-median hTERT died, compared to 4 of 25 (17%) with below-median hTERT. The survival difference was more pronounced within the diagnostically uniform group of medulloblastoma patients. Only one medulloblastoma patient with low hTERT died from his disease, compared to five deaths in the high hTERT medulloblastoma group. However, perhaps because of the relatively short follow-up time, log-rank analysis of Kaplan-Meier survival curves did not confirm significantly worse clinical outcomes in the high hTERT group of medulloblastoma patients (P = 0.078; Figure 2 ▶ ).
Figure 2.
High hTERT expression is associated with poor clinical outcome. Log-rank analysis of Kaplan-Meier survival curves for patients with above or below median hTERT mRNA level reveals a trend toward better clinical outcomes for patients in the low hTERT group (P = 0.078).
Gene Amplification is Associated with Increased hTERT mRNA Expression
Gene amplification often results in increased expression from the amplified locus. We therefore correlated hTERT copy number with hTERT mRNA expression. Linear regression analysis showed a strong association between increasing hTERT DNA copy number and increasing hTERT mRNA level. When comparing hTERT/5qSTS ratio to hTERT mRNA, the correlation coefficient was 0.82, a statistically significant value for this number of samples (P < 0.01). However, the copy number of hTERT at the DNA level did not appear to correlate with survival, and log-rank analysis of survival in patients with tumors amplified or nonamplified at the hTERT locus revealed no trend toward worse outcomes in cases with hTERT amplification.
CGH Analysis of a Medulloepithelioma with hTERT Gene Amplification
We chose to further investigate the molecular alterations in case 50, which represented the most extreme example of hTERT gene amplification and had a very high hTERT message level as well. The hTERT/5qSTS DNA ratio and hTERT mRNA level were several-fold higher in this recurrent medulloepithelioma than in any other CNS embryonal tumor. The patient was a one-year-old white female diagnosed with a medulloepithelioma located in the left cerebellar hemisphere when she was 16 months old. Initial therapy consisted of cisplatin, oral VP-16, and craniospinal radiation followed by peripheral blood autologous stem cell transplant. Seventeen months after the first operation, the tumor recurred in the posterior fossa and was again resected. Despite additional therapy, the tumor continued to progress locally and the patient died three months later.
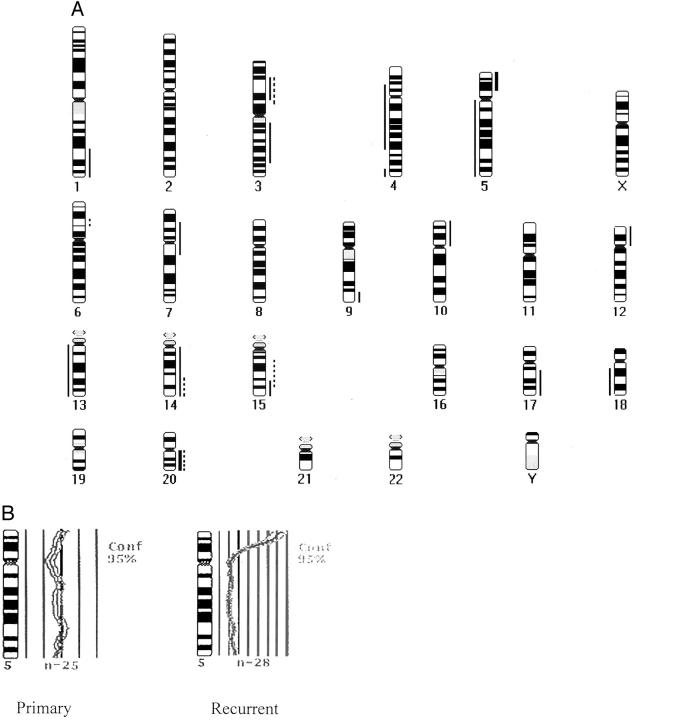
To confirm hTERT gene amplification, and to identify chromosomal changes occurring in medulloepithelioma, we analyzed DNA from both the original and the recurrent tumor. The chromosomal regions with DNA copy number alterations in the primary and recurrent medulloepithelioma are illustrated in Figure 3A ▶ . There were 5 chromosomal alterations in the primary tumor and 17 in the recurrent lesion. The gains in the primary medulloepithelioma were localized to 3p13–22, 6p21.2–21.3, 14q24-qter, 15q15–25, and 20q; gains in the recurrent medulloepithelioma were at 1q32-qter, 3p14–22, 3q13–25, 5p14-pter, 7p15-q11, 9q34-qter, 10p, 12p, 14q, 15q24-qter, 17q21-qter, and 20q (Figure 3) ▶ . There were two high-level gains in the recurrent tumor on chromosome 5p14-pter and 20q. Chromosome 5 profiles from the primary and recurrent tumor are shown in Figure 3B ▶ , where each line to the right or left of the central, dark band represents an incremental difference ration of 0.25. Thus the difference ratio at the hTERT locus in the amplified recurrent tumor is over 2.0, a change equal to or higher than what we have previously observed in medulloblastomas highly amplified at the myc loci. 15 There was no loss found in the primary medulloepithelioma. The losses in the recurrent medulloepithelioma involved 4p14-q28, 4q34-qter, 5q, 13q, and 18q12-qter.
Figure 3.
Chromosomal alterations in a medulloepithelioma detected by CGH. A: Thin vertical lines on the right side of the chromosome ideograms indicate gains; thin lines on the left represent losses. High-level gains are depicted as thick lines. Gains and losses in the primary and recurrent tumors from patient 50 are shown, with alterations in the primary lesion represented using dotted lines, and in the recurrent one using solid lines. B: While the primary tumor shows no gains or losses, the recurrent medulloepithelioma has a high level gain at 5p14-pter and loss of 5q. Each line represents a difference ratio of 0.25.
Discussion
The hTERT gene is located on chromosome 5 at 5p15.33, a region that is sometimes gained in CNS embryonal tumors such as medulloblastoma. 15,16 Recently, two groups demonstrated that the hTERT gene is amplified in several types of malignancy 13,14 ; however, to date, no information on hTERT amplification in solid tumors of the CNS has been published. We therefore analyzed hTERT gene dosage in a series of CNS embryonal tumors using comparative PCR, and identified hTERT gene amplification in 42% of the 36 samples examined. To our knowledge, this is the first analysis of hTERT gene copy number in CNS embryonal tumors, and one of only a few reports on hTERT gene amplification in any tumor type.
We identify a somewhat higher percentage of hTERT amplification in CNS embryonal tumors than previously reported for other tumor types. Using FISH, Zhang and colleagues 13 found amplification of the hTERT gene in 31% of 26 tumor cell lines and 29% of 58 primary tumors examined, including lung tumors, cervical tumors, breast carcinoma, and neuroblastomas. The same group also described hTERT gene amplification in 24% of 88 cervical carcinomas, and an association between elevated hTERT protein expression and gene amplification. 12 A second group recently analyzed hTERT in lung cancer cell lines using several methods, documenting increased hTERT gene dosage in 6 of 20 samples. They also found a correlation between increased hTERT gene dosage, hTERT mRNA expression and telomerase activity. 14
High hTERT expression or increased telomerase activity has correlated with clinical factors in a number of tumors, including Wilms’ tumor, lymphoma, breast cancer, non-small-cell lung cancer, and urothelial carcinoma. 11-14 We therefore evaluated hTERT message levels in CNS embryonal tumors using quantitative RT-PCR, detecting mRNA in 49 of 50 tumors. In both the full group of CNS embryonal tumors and the clinically more homogeneous subset of individuals with medulloblastomas, more than twice as many deaths were seen when tumors contained high levels of hTERT mRNA. However, log-rank analysis of survival curves did not confirm the significance of this difference (P = 0.078). Most of the patients involved in our study had their tumors removed within the last few years, and the follow-up available is therefore relatively short (median 20 months). This short follow-up period may have contributed to the lack of significance in our study, and additional investigations will be needed to confirm the prognostic power of hTERT expression in these CNS tumors.
It will also be important to correlate hTERT mRNA expression with telomerase enzymatic activity in CNS embryonal tumors, as the link between these remains uncertain. While all of the telomerase positive brain tumors studied by Cabuy and de Ridder 21 contained hTERT message, they failed to detect telomerase activity in some hTERT-expressing tumors. It has been suggested that the expression of non-functional hTERT spice variants can result in hTERT positive tumors that lack telomerase enzymatic acitivity. 22 However, many authors report good correlations between hTERT message level and telomerase enzymatic activity. 23-25
Intratumoral heterogeneity of hTERT expression and telomerase activity could have contributed to some of the differences reported in these earlier studies. Regional variations in hTERT expression and telomerase activity have been detected in some CNS gliomas. 21,26 Similar heterogeneity at the level of hTERT DNA amplification or hTERT expression could be present in the embryonal tumors we analyzed. We attempted to extend our studies of hTERT expression to the protein level using immunohistochemistry, which would also enable localization of expression to specific tumor regions. However, while the highly amplified medulloepithelioma was diffusely immunopositive for hTERT, in the other 13 cases we found no correlation between DNA, RNA and protein levels (data not shown).
Gene amplification in tumors often results in increased mRNA expression from the amplified locus. In CNS embryonal tumors, we found a significant positive correlation between increasing gene dosage and hTERT message level. In some individual cases gene amplification was not associated with elevated message level, a finding similar to that observed for other oncogenes. For example, Grotzer and colleagues 27 recently found that c-myc gene amplification was only associated with high mRNA levels in a subset of medulloblastomas. Similarly, we have identified medulloblastomas amplified at the N-myc locus, but with low levels of N-myc mRNA (unpublished results). In 20 CNS embryonal tumors we detected hTERT mRNA, but no amplification was found at the locus by differential PCR. Presumably, other transcriptional regulators direct hTERT expression in these tumors. c-Myc, 28-30 Sp1, 31 and estrogen 32 have all been implicated as hTERT transcriptional regulators (reviewed in 33 ), and could play a role in the diverse hTERT message levels we detected. The large number of embryonal tumors we found with unamplified hTERT and low-level hTERT mRNA expression also suggests that only modest telomerase expression levels may be required for formation of these neoplasms. Indeed, it is possible that telomerase-independent mechanisms of transformation are operative in PNET, as has been suggested for CNS gliomas. 34
We examined chromosomal alterations in a recurrent medulloepithelioma with very high hTERT levels using CGH. High-level DNA gains were detected at 5p15, confirming the hTERT amplification identified using differential PCR. Interestingly, CGH analysis of the primary lesion from this patient detected no gains of 5p, indicating that hTERT amplification can first occur during tumor progression. However, most of the embryonal brain tumors with hTERT amplification in our study were not recurrent lesions, suggesting that as is the case in gliomas these alterations can occur early in tumorigenesis as well. 34 The overall number of chromosomal abnormalities was much higher in the recurrent medulloepithelioma than in the primary one, suggesting molecular progression can occur in these rare embryonal neoplasms. A similar accumulation of chromosomal changes has been observed in recurrence or metastasis of medulloblastomas. 15,35 Thus our CGH analysis of this medulloepithelioma adds to the growing body of evidence suggesting CNS embryonal tumors can progress both microscopically and molecularly.
In conclusion, we have shown that the hTERT oncogene is amplified in a significant proportion of medulloblastomas and other CNS embryonal neoplasms. This gene amplification correlates with increased expression of hTERT mRNA, and implicates, for the first time, changes at the hTERT locus in the evolution of primitive neuroepithelial tumors of the CNS. Finally, our correlation of hTERT expression with survival suggests hTERT message level could be a useful molecular prognostic marker.
Acknowledgments
We thank Krista Summers for help with specimen preparation and Amy Schuster for CGH analysis. Pat Goldthwaite and Linda Hershon assisted with clinical data management.
Footnotes
Address reprint requests to Charles G. Eberhart, M.D., Ph.D. Department of Pathology, The Johns Hopkins University School of Medicine, Ross Research Building, Room 558, 720 Rutland Avenue, Baltimore, MD 21205-2196. E-mail: ceberha@jhmi.edu.
Supported in part by grants from the Children’s Cancer Foundation of Baltimore (to P.C.B.) and The Brain Tumor Society (to C.G.E.). C.G.E. is a recipient of a Burroughs Wellcome Fund Career Award in the Biomedical Sciences.
References
- 1.Kleihues P, Cavenee WK: Tumors of the Nervous System 2000. IARC Press, Lyon, France
- 2.Ellison D: Classifying the medulloblastoma: insights from morphology and molecular genetics. Neuropathol Appl Neurobiol 2002, 28:257-282 [DOI] [PubMed] [Google Scholar]
- 3.Feng J, Funk WD, Wang SS, Weinrich SL, Avilion AA, Chiu CP, Adams RR, Chang E, Allsopp RC, Yu J, Le S, West MD, Harley CB, Andrews WH, Greider CW, Villeponteau B: The RNA component of human telomerase. Science 1995, 269:1236-1241 [DOI] [PubMed] [Google Scholar]
- 4.Harrington L, McPhail T, Mar V, Zhou W, Oulton R, Bass MB, Arruda I, Robinson MO: A mammalian telomerase-associated protein. Science 1997, 275:973-977 [DOI] [PubMed] [Google Scholar]
- 5.Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR: Telomerase catalytic subunit homologs from fission yeast and human. Science 1997, 277:955-959 [DOI] [PubMed] [Google Scholar]
- 6.Hiyama E, Hiyama K: Clinical utility of telomerase in cancer. Oncogene 2002, 21:643-649 [DOI] [PubMed] [Google Scholar]
- 7.Komiya T, Kawase I, Nitta T, Yasumitsu T, Kikui M, Fukuoka M, Nakagawa K, Hirashima T: Prognostic significance of hTERT expression in non-small cell lung cancer. Int J Oncol 2000, 16:1173-1177 [DOI] [PubMed] [Google Scholar]
- 8.Wisman GB, Knol AJ, Helder MN, Krans M, de Vries EG, Hollema H, de Jong S, van der Zee AG: Telomerase in relation to clinicopathologic prognostic factors and survival in cervical cancer. Int J Cancer 2001, 91:658-664 [DOI] [PubMed] [Google Scholar]
- 9.Lee BK, Diebel E, Neukam FW, Wiltfang J, Ries J: Diagnostic and prognostic relevance of expression of human telomerase subunits in oral cancer. Int J Oncol 2001, 19:1063-1068 [DOI] [PubMed] [Google Scholar]
- 10.Lehner R, Enomoto T, McGregor JA, Shroyer AL, Haugen BR, Pugazhenthi U, Shroyer KR: Quantitative analysis of telomerase hTERT mRNA and telomerase activity in endometrioid adenocarcinoma and in normal endometrium. Gynecol Oncol 2002, 84:120-125 [DOI] [PubMed] [Google Scholar]
- 11.Marchetti A, Pellegrini C, Buttitta F, Falleni M, Romagnoli S, Felicioni L, Barassi F, Salvatore S, Chella A, Angeletti CA, Roncalli M, Coggi G, Bosari S: Prediction of survival in stage I lung carcinoma patients by telomerase function evaluation. Lab Invest 2002, 82:729-736 [DOI] [PubMed] [Google Scholar]
- 12.Zhang A, Zheng C, Hou M, Lindvall C, Wallin KL, Angstrom T, Yang X, Hellstrom AC, Blennow E, Bjorkholm M, Zetterberg A, Gruber A, Xu D: Amplification of the telomerase reverse transcriptase (hTERT) gene in cervical carcinomas. Genes Chromosomes Cancer 2002, 34:269-275 [DOI] [PubMed] [Google Scholar]
- 13.Zhang A, Zheng C, Lindvall C, Hou M, Ekedahl J, Lewensohn R, Yan Z, Yang X, Henriksson M, Blennow E, Nordenskjold M, Zetterberg A, Bjorkholm M, Gruber A, Xu D: Frequent amplification of the telomerase reverse-transcriptase gene in human tumors. Cancer Res 2000, 60:6230-6235 [PubMed] [Google Scholar]
- 14.Saretzki G, Petersen S, Petersen I, Kolble K, von Zglinicki T: hTERT gene dosage correlates with telomerase activity in human lung cancer cell lines. Cancer Lett 2002, 176:81-91 [DOI] [PubMed] [Google Scholar]
- 15.Eberhart CG, Kratz JE, Schuster A, Goldthwaite P, Cohen KJ, Perlman EJ, Burger PC: Comparative genomic hybridization detects an increased number of chromosomal alterations in large cell/anaplastic medulloblastomas. Brain Pathol 2002, 12:36-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reardon DA, Michalkiewicz E, Boyett JM, Sublett JE, Entrekin RE, Ragsdale ST, Valentine MB, Behm FG, Li H, Heideman RL, Kun LE, Shapiro DN, Look AT: Extensive genomic abnormalities in childhood medulloblastoma by comparative genomic hybridization. Cancer Res 1997, 57:4042-4047 [PubMed] [Google Scholar]
- 17.Falchetti ML, Larocca LM, Pallini R: Telomerase in brain tumors. Childs Nerv Syst 2002, 18:112-117 [DOI] [PubMed] [Google Scholar]
- 18.Hu J, Wills M, Baker BA, Perlman EJ: Comparative genomic hybridization analysis of hepatoblastomas. Genes Chromosomes Cancer 2000, 27:196-201 [DOI] [PubMed] [Google Scholar]
- 19.Waha A, Rollbrocker B, Wiestler OD, von Deimling A: A polymerase chain reaction-based assay for the rapid detection of gene amplification in human tumors. Diagn Mol Pathol 1996, 5:147-150 [DOI] [PubMed] [Google Scholar]
- 20.Rollbrocker B, Waha A, Louis DN, Wiestler OD, von Deimling A: Amplification of the cyclin-dependent kinase 4 (CDK4) gene is associated with high cdk4 protein levels in glioblastoma multiforme. Acta Neuropathol (Berl) 1996, 92:70-74 [DOI] [PubMed] [Google Scholar]
- 21.Cabuy E, de Ridder L: Telomerase activity and expression of telomerase reverse transcriptase correlated with cell proliferation in meningiomas and malignant brain tumors in vivo. Virchows Arch 2001, 439:176-184 [DOI] [PubMed] [Google Scholar]
- 22.Ulaner GA, Hu JF, Vu TH, Oruganti H, Giudice LC, Hoffman AR: Regulation of telomerase by alternate splicing of human telomerase reverse transcriptase (hTERT) in normal and neoplastic ovary, endometrium and myometrium. Int J Cancer 2000, 85:330-335 [PubMed] [Google Scholar]
- 23.Hoang-Vu C, Boltze C, Gimm O, Poremba C, Dockhorn-Dworniczak B, Kohrle J, Rath FW, Dralle H: Expression of telomerase genes in thyroid carcinoma. Int J Oncol 2002, 21:265-272 [PubMed] [Google Scholar]
- 24.Chong EYY, Pang JCS, Ko CW, Poon WS, Ng HK: Telomere length and telomerase catalytic subunit expression in non-astrocytic gliomas. Pathol Res Pract 2000, 196:691-699 [DOI] [PubMed] [Google Scholar]
- 25.Takakura M, Kyo S, Kanaya T, Tanaka M, Inoue M: Expression of human telomerase subunits and correlation with telomerase activity in cervical cancer. Cancer Res 1998, 58:1558-1561 [PubMed] [Google Scholar]
- 26.Weil RJ, Wu YY, Vortmeyer AO, Moon YW, Delgado RM, Fuller BG, Lonser RR, Remaley AT, Zhuang Z: Telomerase activity in microdissected human gliomas. Mod Pathol 1999, 12:41-46 [PubMed] [Google Scholar]
- 27.Grotzer MA, Hogarty MD, Janss AJ, Liu X, Zhao H, Eggert A, Sutton LN, Rorke LB, Brodeur GM, Phillips PC: MYC messenger RNA expression predicts survival outcome in childhood primitive neuroectodermal tumor/medulloblastoma. Clin Cancer Res 2001, 7:2425-2433 [PubMed] [Google Scholar]
- 28.Wu KJ, Grandori C, Amacker M, Simon-Vermot N, Polack A, Lingner J, Dalla-Favera R: Direct activation of TERT transcription by c-MYC. Nat Genet 1999, 21:220-224 [DOI] [PubMed] [Google Scholar]
- 29.Wang J, Xie LY, Allan S, Beach D, Hannon GJ: Myc activates telomerase. Genes Dev 1998, 12:1769-1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greenberg RA, O’Hagan RC, Deng H, Xiao Q, Hann SR, Adams RR, Lichtsteiner S, Chin L, Morin GB, DePinho RA: Telomerase reverse-transcriptase gene is a direct target of c-Myc but is not functionally equivalent in cellular transformation. Oncogene 1999, 18:1219-1226 [DOI] [PubMed] [Google Scholar]
- 31.Takakura M, Kyo S, Kanaya T, Hirano H, Takeda J, Yutsudo M, Inoue M: Cloning of human telomerase catalytic subunit (hTERT) gene promoter and identification of proximal core promoter sequences essential for transcriptional activation in immortalized and cancer cells. Cancer Res 1999, 59:551-557 [PubMed] [Google Scholar]
- 32.Kyo S, Takakura M, Kanaya T, Zhuo W, Fujimoto K, Nishio Y, Orimo A, Inoue M: Estrogen activates telomerase. Cancer Res 1999, 59:5917-5921 [PubMed] [Google Scholar]
- 33.Kyo S, Inoue M: Complex regulatory mechanisms of telomerase activity in normal and cancer cells: how can we apply them for cancer therapy? Oncogene 2002, 21:688-697 [DOI] [PubMed] [Google Scholar]
- 34.Chong EY, Lam PY, Poon WS, Ng HK: Telomerase expression in gliomas including the nonastrocytic tumors. Hum Pathol 1998, 29:599-603 [DOI] [PubMed] [Google Scholar]
- 35.Leonard JR, Cai DX, Rivet DJ, Kaufman BA, Park TS, Levy BK, Perry A: Large cell/anaplastic medulloblastomas and medullomyoblastomas: clinicopathological and genetic features. J Neurosurg 2001, 95:82-88 [DOI] [PubMed] [Google Scholar]