Abstract
Alternative splicing of fibronectin-like type III (FNIII) repeats of tenascin-C (Tn-C) generates a number of splice variants. The distribution of large variants, typical components of provisional extracellular matrices that are up-regulated during tumor stroma remodeling, was here studied by immunoblotting and immunohistochemistry using a monoclonal antibody against the FNIII B domain (named 4C8MS) in a series of human breast cancers. Large Tn-C variants were found at only low levels in normal breast tissues, but were highly expressed at invading sites of intraductal cancers and in the stroma of invasive ductal cancers, especially at invasion fronts. There was a positive correlation between the expression of large Tn-C variants and the cell proliferation rate determined by immunolabeling of the Ki-67 antigen. Of the Tn-C recombinant fragments (all FNIII repeats or mFNIII FL, the conserved FNIII domain only, the epidermal growth factor-like domain, and the fibrinogen-like domain) which were expressed by CHO-K1 cells transfected with mouse Tn-C cDNAs, only the mFNIII FL enhanced in vitro migration and mitotic activity of mammary cancer cells derived from a Tn-C-null mouse. Addition of 4C8MS blocked the function of mFNIII FL. These findings provide strong evidence that the FNIII alternatively spliced region has important roles in tumor progression of breast cancer.
During tumor progression, the cancer stroma becomes remodeled by both tumor cells and stromal cells, and protein components of the extracellular matrix (ECM) are dynamically changed by degradation and neosynthesis. Cellular interaction with the ECM strongly influences the behavior of cancer and stromal cells, resulting in modulation of cell growth, migration, differentiation, and apoptosis. 1-3 Compositional change of the ECM in cancer stroma is thus a key determinant of tumor growth and cancer progression. A variety of ECM glycoproteins, such as tenascin-C (Tn-C) and fibronectin, are overexpressed in cancer stroma. In addition, splice variants of these proteins, which are generally absent in normal adult tissues, become predominant. 4-11
It has been reported that overexpression of Tn-C in breast cancer is related to a poor prognosis, and local and distant recurrence, 12-14 this being attributable to the ability to promote cell migration and proliferation demonstrated in vitro. 15,16 Tn-C is a hexameric glycoprotein, each subunit consisting of a TA (Tn assembly) domain; 14 + 1/2 epidermal growth factor (EGF)-like domains, a variable number of fibronectin type III (FN III) repeats, and a C-terminal fibrinogen-related domain (FBG). 17-21 The size of Tn-C monomers varies as a result of alternative splicing in the FN III repeats at the pre-mRNA level. There are eight conserved FN III repeats (designated as numbers 1–8) and, in the case of human Tn-C, up to nine alternatively spliced FN III repeats (designated as letters A-D) inserted between the conserved repeats 5 and 6. In adults, the smallest Tn-C variant in which the alternatively spliced domain is spliced out is present in static tissues such as cartilage, 22 whereas large variants containing the alternatively spliced FNIII domains in various combinations are found in developing tissues 23-25 and in pathological tissues which undergo remodeling, as with regeneration, inflammation, and tumorigenesis. 8,26-29 Recent studies have demonstrated that some splice variants are specific to diseased tissues. After balloon catheterization, smooth muscle cells express a Tn-C isoform containing only A1 and A2 repeats. 30 Transcripts for splice variants including the AD2 repeat are found in oral cancer tissues, but not in normal and pre-malignant tissues. 31 Immunohistochemistry using a recombinant antibody to the spliced repeat C has shown that the repeat is not detectable in normal adult tissues, not or barely detectable in malignant epithelial tumors, but abundant in glioblastomas. 32 From quantitative analysis using antibodies against alternative spliced domains, an increased fraction of Tn-C containing A1-A4 appears related to the tumor stage of colorectal cancer, while domain D is down-regulated in metastasizing cancers. 10 In breast tissues, expression of two Tn-C variants, one containing domain D and the other both B and D, was found to be associated with invasive carcinomas in one study. 11
Malignant properties of tumors highly expressing Tn-C may to some extent be dependent on functions of alternative spliced domains of the FNIII repeats, since it has been reported that the alternatively spliced region, FNIII A-D, promotes migration and proliferation of endothelial cells through interaction with annexin II. 33-35 Thus an involvement of Tn-C with the repeats in angiogenesis is possible. The spliced region is further known to support attachment of chick embryonic neurons, 36 to mediate neurite outgrowth and guidance, 37,38 and to promote proliferation of hemopoietic precursor cells in bone marrow. 39 In addition, the receptor protein tyrosine phosphatase β present on glial tumor cells binds to FNIII A1, 2, and 4, suggesting it to be an adhesion receptor to ECM. 40 However, it is unclear whether the alternatively spliced region of Tn-C directly enhances progressive behavior of tumor cells.
The aim of this study, therefore, was to examine the expression and distribution of large Tn-C splice variants in breast tissues using a novel monoclonal antibody (mAb) specific to FNIII B. An investigation of direct effects of recombinant Tn-C fragments containing the alternatively spliced region on migration and proliferation of mammary cancer cells was also included.
Materials and Methods
Purification of Tn-C and Tn-C Recombinant Fragments
To produce a monoclonal antibody specific for large Tn-C splice variants, we first prepared a recombinant protein containing FNIII A4-D domains of human Tn-C (Figure 1A) ▶ . Complementary DNA encoding the region was generated by PCR using human fetal brain Marathon-Ready cDNA (BD Biosciences Clontech, Palo Alto, CA) as the template and the primers shown in Table 1 ▶ . PCR products were cloned into the BamHI and HindIII sites of the pQE30 expression vector (Qiagen, Hilden, Germany) and the sequence confirmed. For protein expression, single colonies of E. coli JM109 cells transformed with the construct were grown in LB medium. Expression of the recombinant protein was induced by addition of 1 mmol/L IPTG. The bacteria were collected by centrifugation at 7,000 × g for 15 minutes, and resuspended in lysis buffer (phosphate buffer pH 7.4, 20 mmol/L imidazole, 1% Tween 20, 6 mol/L urea) with protease inhibitor cocktail tablets (Roche Diagnostic, Basel, Switzerland). The mixture was stirred for 30 minutes at room temperature and then centrifuged at 15,000 × g for 30 minutes at 4°C, the supernatant being collected and applied to a HisTrap column (Amersham Pharmacia, Buckinghamshire, UK). Recombinant proteins were eluted with elution buffer (PB pH 7.4, 0.5 mol/L imidazole, 1% Tween-20, 6 mol/L urea). Recombinant hFNIII A4-C, A4-B, and A4 fragments were also prepared by the same procedures using the reverse primers shown in Table 1 ▶ .
Figure 1.
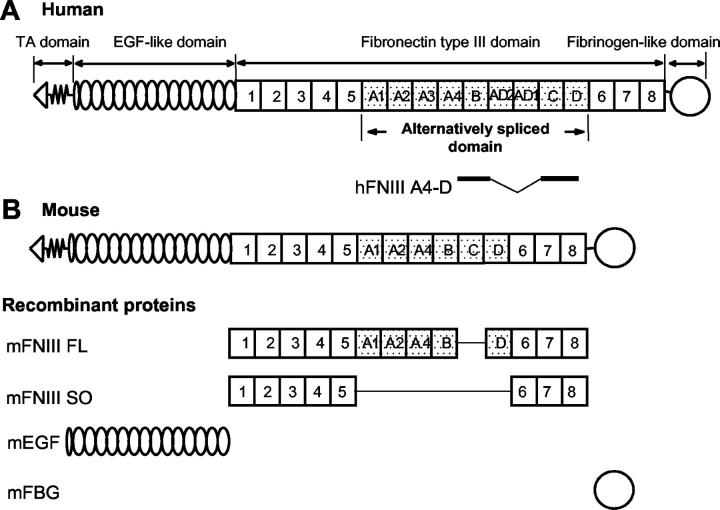
Multidomain structure of human (A) and mouse (B) Tn-Cs. The amino-termini of six arms are joined to form a hexamer. Each arm consists of 14 + 1/2 EGF domains, 8–15 FN III domains, depending on alternative RNA splicing, and a single fibrinogen-like domain. The universal FNIII domains (FNIII repeats 1–5 and FNIII repeats 6–8) are present in all Tn-C variants. The largest Tn-C variant also contains nine alternatively spliced FNIII (FNIII repeats A1-D), which are missing in the shortest splice variant. Mouse Tn-C lacks FNIII A3, AD2, and AD1 repeats. A scheme of recombinant proteins of mouse Tn-C domains is shown. mFNIII FL does not contain the C domain known as a glioma-associated repeat.
Table 1.
PCR Primers Used for the Generation of Recombinant Proteins Issued from the Alternatively Spliced Domain of Human and Mouse Tn-C
| Recombinant protein | Primers (5′-3′) |
|---|---|
| hFNIII A4-D | F:CTGGGATCCCAGGTGCAGGAGGTCAACAAAGTG |
| R:CCCAAGCTTGGGGGCAAGTAGGGTTATTTCCAG | |
| hFNIII A4-C | R:TACAAGCTTGTAACAATCTCAGCCCTCAAG |
| hFNIII A4-B | R:GACAAGCTTGTCGTGGCTGTGGCACTGATG |
| hFNIII A4 | R:CACAAGCTTGTGGAGGCCTCAGCAGAGTACTG |
| mFNIII FL | F:AAGGAATTCCTTCTGAGGTGTCCCCTCCCAAA |
| R:AACTCGAGGGAATGGGTACAGGAGTCCAAT | |
| mFNIII SO | F(1):AAGGAATTCCTTCTGAGGTGTCCCCTCCCAAA |
| R(5):AACCTCGAGGCCTTCACACGTGCAGGCTTGC | |
| F(6):ACGAATTCTGTCGACGGCCATGGGTTCTCCGAAGG | |
| R(8):AACTCGAGGGAATGGGTACAGGAGTCCAAT | |
| mEGF | F:AACGAATTCGCTGTGTCTGTGAACCAGGCTGG |
| R:TCCTCGAGAGCAGTCTATCCCTGTGTAACC | |
| mFBG | F:TTGGAATTCCCAGGGACTGCTCTCAAGCAATG |
| R:CGGGCAGCGGCCGCTGCCCGCTTACGCCTGCCT |
To analyze functions of Tn-C domains, we also prepared recombinant fragments of mouse Tn-C using a cell expression system. Complementary DNAs encoding EGF-like repeats (mEGF), FNIII repeats (mFNIIII FL and SO), and the FBG-like domain (mFBG) of mouse Tn-C (Figure 1B) ▶ were generated by PCR using mouse Tn-C cDNA as a template and the primers shown in Table 1 ▶ . The complete cDNA of mouse Tn-C was cloned from a cDNA library of a mammary cancer cell line. 21 Restriction sites were included in the primers to facilitate directional cloning and PCR products were cloned into the pSecTag2A expression vector having 6xHis and myc tags (Invitrogen, Carlsbad, CA), and the sequences confirmed. Since the template Tn-C cDNA includes five alternatively spliced repeats of the FNIII domain, repeats of FNIII 1–5, A1, A2, A4, B, D, and 6–8 comprised cDNA for mFNIII FL. The plasmids containing FNIII 1–5 were then restricted with XhoI, and those containing FNIII 6–8 were restricted with SalI and XhoI. These constructs were ligated again, and then the plasmids for mFNIII SO were completed. For transfection, CHO-K1 cells were used and grown in Iscove’s modified Dulbecco’s medium (IMDM, Sigma, St. Louis, MO) supplemented with 10% fetal calf serum (FCS, Gibco-BRL, Grand Island, NY) under 95% air/5% CO2 atmosphere at 37°C. The constructs were transfected into CHO-K1 cells with Lipofectamin plus reagent (Invitrogen). Forty-eight hours later, the cells were cultured in medium containing Zeocin (Invitrogen) to select stable transfectants. The clones producing the recombinant proteins were chosen by Western blotting using anti-myc-tag antibody (MBL, Nagoya, Japan), and subcloned three times. Conditioned media containing the proteins were collected. The recombinant proteins were precipitated with 50% ammonium sulfate, and purified using a HisTrap column (Amersham Pharmacia). Purified proteins of Tn-C domains from conditioned media of stable transfectants are shown in Figure 2 ▶ . Human Tn-C was purified from conditioned media of a glioma cell line, U251MG, by a previously described method. 15 It is referred to as intact Tn-C in this paper.
Figure 2.
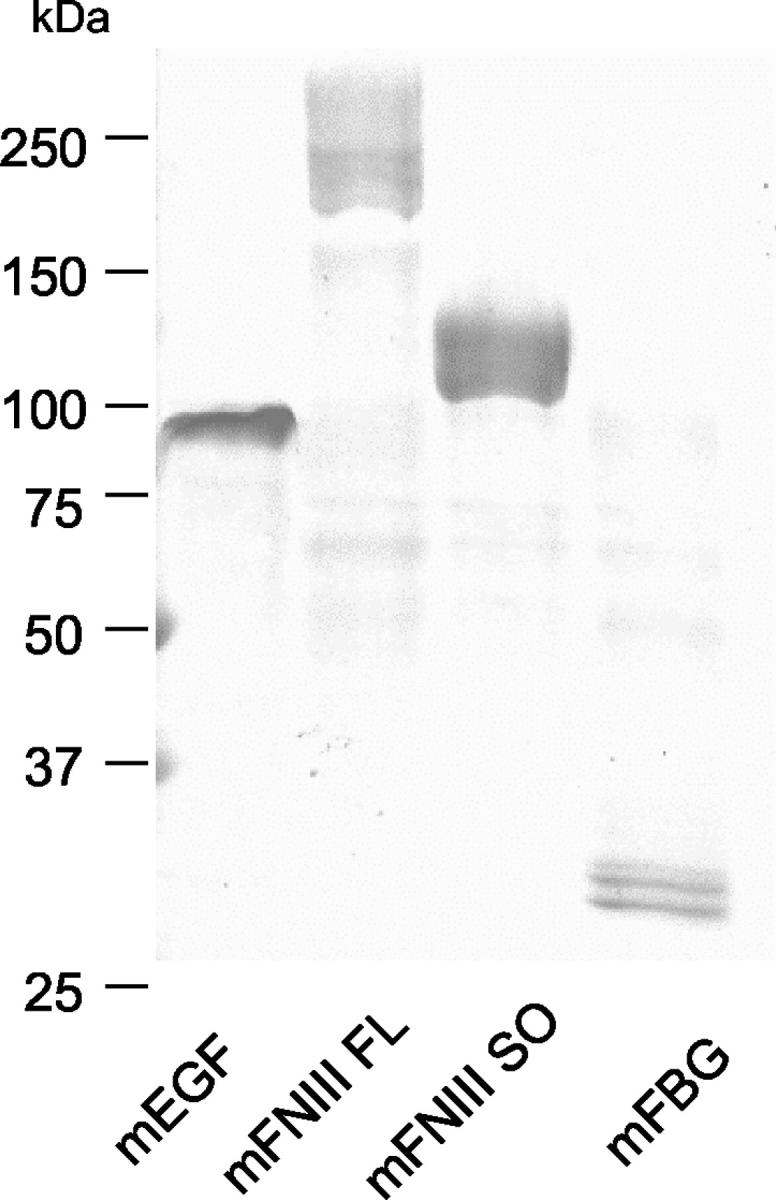
Purified recombinant proteins of mouse Tn-C domains. Recombinant proteins were purified from the conditioned media using His-trap column, electrophoresed in sodium dodecyl sulfate polyacrylamide gel (2 to 15%), and stained with Coomassie blue.
Monoclonal Antibodies Against Tn-C
To obtain higher efficiencies of antibody production, we immunized congenic Tn-C-null mice obtained by back-crossing Tn-C-null mice originally generated by Saga et al 41 with BALB/c mice. They received 100 μg of the recombinant FNIII A4-D protein mixed with Freund’s complete adjuvant, and after 2 weeks, the antigen mixture with Freund’s incomplete adjuvant. Three days before cell fusion, the mice were hyperimmunized with the antigen by i.v. injection. Hybridomas were prepared by fusion of immunized mouse spleen cells with myeloma cells (Sp2/0) in polyethylene glycol. Positive hybridoma clones were detected by enzyme immunoassay using plates coated with native human Tn-C and peroxidase-conjugated anti-mouse IgG antibodies (Biorad, Hercules, CA). After being cloned three times, they were transferred to serum-free medium (Wako, Osaka, Japan) with 20 units/ml IL-6 (Roche Diagnostic) and grown. The conditioned media were collected by centrifugation, and then the antibodies were purified by 50% ammonium sulfate fractionation, followed by affinity-purification using a HiTrap protein A column (Amersham Pharmacia). The isotype of the monoclonal antibodies was determined with the mouse monoclonal antibody isotype kit (Amersham Lifescience, Piscataway, NJ) and was IgG1 κ.
Immunoblotting
Four breast tissue samples were obtained from specimens taken at therapeutic surgeries for breast cancers. Small pieces of normal parts and cancer tissues were isolated by gross examination and stored at −80°C. Aliquots (100 μg) were then homogenized in 1 ml of 10 mmol/L Tris-buffered saline (TBS, pH 7.6) containing 0.2% Nonidet P-40 and protease inhibitor cocktail tablets (Complete mini, Roche, Manheim, Germany), and centrifuged at 15,000 × g for 10 minutes. The pellets were resuspended in TBS, and washed twice with centrifugation, then resuspended in 1 ml of TBS containing 2 mol/L urea, and the slurries were gently shaken for 1 hour on ice. They were centrifuged at 15,000 × g for 15 minutes, and the supernatants were collected. The samples were applied to polyacrylamide gels and electrophoresed by the method of Laemmli. The electrophoresed proteins were blotted onto Immobilon membranes (Millipore Japan, Tokyo, Japan), blocked with a blocking buffer (TBS, pH 7.6 and 0.5% skimmed milk), and incubated with monoclonal anti-Tn-C antibodies (4F10TT or 4C8MS, 1 μg/ml) at 4°C overnight. The monoclonal antibody 4F10TT against the EGF-like repeats of Tn-C was as previously reported. 42 The membranes were then washed in the blocking buffer and successively treated with peroxidase-labeled goat anti-mouse IgG Fab’ (1:400, MBL, Nagoya, Japan) for 1 hour at room temperature. The reactive bands were developed with diaminobenzidine (DAB)/H2O2 solution.
Immunohistochemistry
Immunohistochemical analysis was performed on 10 fiboroadenomas and 22 invasive ductal carcinomas using archival samples which were fixed with formalin and routinely processed for embedding in paraffin. All sections were cut at 4 μm and placed on silane-coated glass slides (DAKO Japan, Kyoto, Japan). For 4F10TT, the sections were incubated in 0.4% pepsin (1:60,000, Sigma) in 0.01N HCl for 20 minutes at 37°C to retrieve the antigens. The sections were incubated in 0.3% H2O2 in methanol for 15 minutes to block endogenous peroxidase activity. This proteinase treatment conversely abolished the positive reaction with the 4C8MS antibody. Treatment with 0.1% sapponin in distilled water for 20 minutes or heating in an autoclave was effective for the retrieval of antigens. Sections were treated with superblock solution (Scytek Laboratories, Logan, UT) or 10% normal goat serum before incubation with anti-Tn-C antibodies (1 to 10 μg/ml) overnight at 4°C. After being washed, sections were treated with a commercially available LSAB kit (Scytek) or peroxidase-conjugated anti-mouse IgG Fab’ (MBL), followed by color development with DAB/H2O2 solution. Light counterstaining with hematoxylin was performed to aid orientation. Various tissue samples from autopsy cases were also examined.
For double immunohistochemistry with 4C8MS and Ki-67 antibodies, the sections were treated with 4C8MS, followed by the incubation with peroxidase-conjugated anti-mouse IgG Fab’, and color development by a DAB/H2O2 solution. The immune complexes were then removed by treatment with 0.01N HCl solution. Next the sections were exposed to the Ki-67 antibody (clone MIB-1, Dako Japan), and again treated with peroxidase-conjugated anti-mouse IgG Fab’. The second coloring reaction was achieved with a Ni/DAB/H2O2 solution. Therefore, the 4C8MS immunoreactivity was brown in color, whereas the Ki-67 immunoreactivity was blue. Finally the sections were lightly counterstained with methylgreen solution.
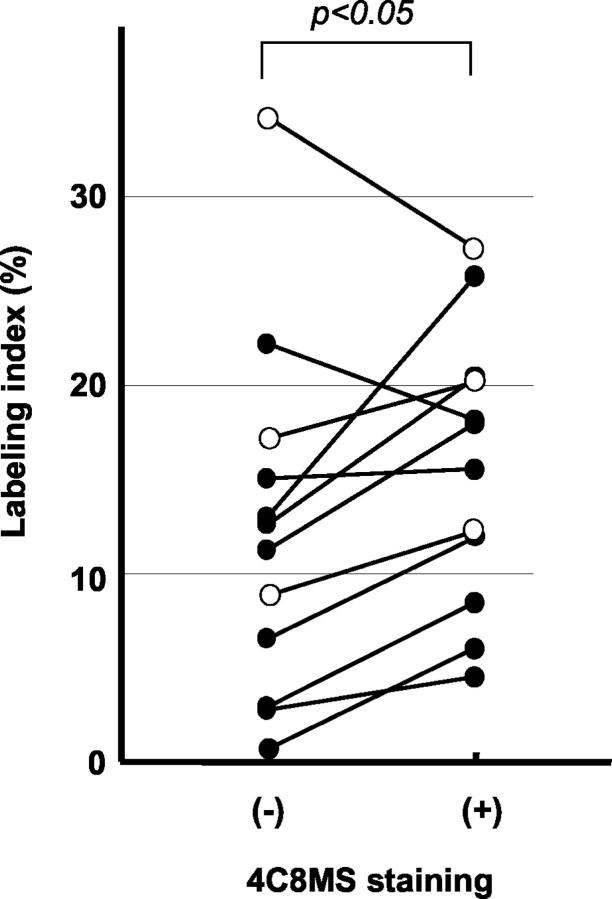
To examine whether expression of large Tn-C variants was related to cell proliferation in human cancer tissues, 12 cases having sufficient areas for analysis were chosen from fully invasive carcinomas. When the 4C8MS-staining of cancer stroma was equivalent to or stronger than that of the muscular arteries in the same tissue, the immunolabeling was considered as positive. Labeling indices for Ki-67-positive nuclei in three fields (1 mm2 each) positive for 4C8MS was compared with those for three negative fields close to the positive fields.
Migration Assay
The mouse mammalian cancer cell line, GHOM5E, was established from a spontaneously developing tumor in a congenic Tn-C-null mouse of the GRS/A strain. By transwell migration assay using cell culture inserts (8-μm pore size, Becton Dickinson Labware, Franklin Lakes, NJ), we examined effects of intact Tn-C or its recombinant fragments on cancer cell migration. Into the inner chamber, 5 × 104 cells were plated in 0.5% bovine serum albumin (BSA)/serum-free IMDM, and the medium containing 0.5% BSA and 5% FCS as a chemoattractant was poured in the outer chamber (Falcon 24-well plate, Becton Dickinson Labware). After addition of the proteins tested into the medium of the upper chamber, the cells were allowed to migrate to the lower membrane surface for 18 hours. After the cells on the upper surface were wiped off, the inserts were fixed and stained with 0.1% crystal violet (Sigma) in 10% ethanol. Stained cells on the lower surface in three fields of 1 mm2 were counted under a 10× objective lens.
BrdU Incorporation Assay
The cells were grown on coverslips in IMDM/10% FCS for 16 hours, washed with serum-free medium, and then incubated in the medium with 0.1% FCS. Twenty-four hours thereafter, Tn-C or the fragments were added into the medium at final concentration of 1 to 10 μg/ml. After incubation for 12 hours, the cells were labeled with 8-bromodeoxyuridine (BrdU 10 μg/ml) for 2 hours, fixed with 70% ethanol at −20°C for 30 minutes, and treated with 1N HCl solution at room temperature for 20 minutes. Labeled nuclei were detected with monoclonal anti-BrdU antibody (DAKO Japan) and anti-mouse IgG conjugated with peroxidase (MBL). The percentage of the labeled nuclei was determined.
Antibody Blocking
Three μg of mouse Tn-C fragments were incubated with 15 μg of the 4C8MS Fab’ fragment for 30 minutes at room temperature and then added into the medium for BrdU incorporation and migration assays. As a control antibody, an isotype-matched monoclonal antibody against V protein of parainfluenza virus type II (clone 53–1) (kindly provided by the Department of Microbiology, Mie University School of Medicine, Mie, Japan) was used. The experiment was performed in triplicate.
Statistical Analysis
For quantitative analysis the arithmetical mean values and standard deviations were calculated and compared by independent Student’s or Welch’s t-tests. The statistical significance of differences of the rates for Ki-67 positive cells between the large Tn-C variant distributions was tested with the paired t-test.
Results
Monoclonal Antibody Characterization
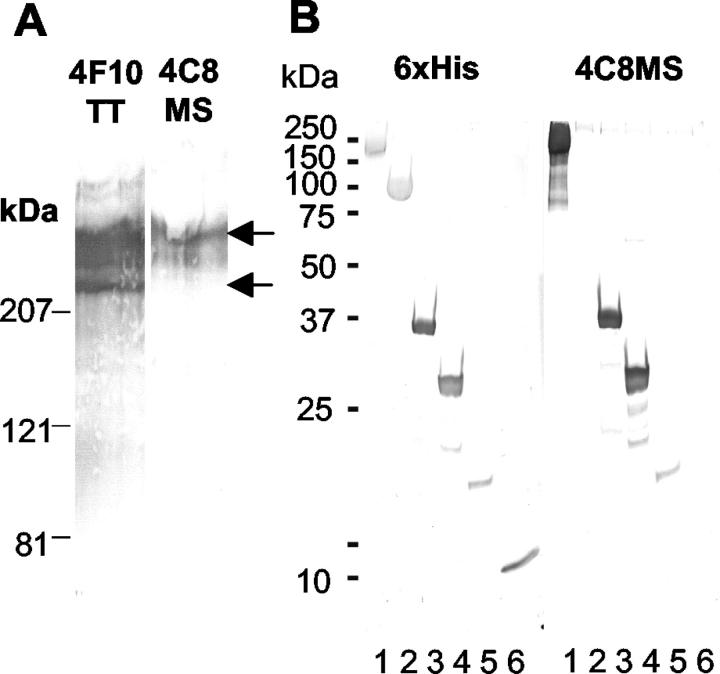
For the characterization of the antibodies used in this study, immunoblots of intact Tn-C from human glioma cells were analyzed with the mAbs 4F10TT and 4C8MS. Antibody 4F10TT labeled larger Tn-C variants and the smallest variant of human glioma Tn-C, whereas 4C8MS labeled larger variants but not the smallest one (Figure 3A) ▶ . Immunoblotting using recombinant proteins with human FNIII A4-D regions showed 4C8MS to react with FNIII repeats A4-D, A4-C and A4-B, but not A4 (Figure 3B) ▶ . Thus, the epitope recognized by 4C8MS is located on FNIII repeat B. When the binding of the antibody to mouse Tn-C recombinant proteins was examined, 4C8MS reacted with mFNIII FL but not mFNIII SO, showing a cross-reactivity with mouse FNIII repeat B (Figure 3B) ▶ .
Figure 3.
Characterization of monoclonal antibodies by immunoblotting. With intact Tn-C purified from human glioma supernatant, 4F10TT showed bands around 350 kd and at 210 kd (arrows), while 4C8MS did so only at 350 kd (A). Recombinant proteins mFNIII FL (lane 1), mFNIII SO (lane 2), hFNIIIA4-C (lane 3), hFNIIIA4-C (lane 4), hFNIIIA4-B (lane 5), and hFNIIIA4 (lane 6) which have 6xHis tags were visualized with an anti-6xHis antibody (left in B). mFNIII FL, hFNIIIA4-D, hFNIIIA4-C, and hFNIIIA4-B were labeled by 4C8MS, but mFNIII SO and hFNIIIA4 were not (right in B), indicating that 4C8MS can react with FNIII B repeats of human and mouse Tn-C.
Expression and Distribution of Large Tn-C Variants
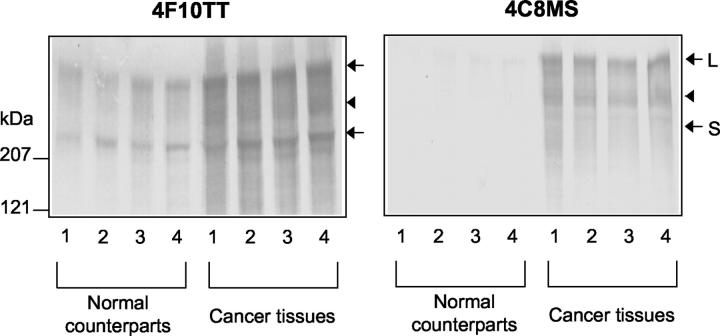
In immunoblotting of normal breast tissue extracts, 4F10TT labeled two main bands at 210 kd (the smallest variant, S) and 350 kd (the largest variant, L), whereas 4C8MS labeled faintly the 350 kd band (Figure 4) ▶ . In breast cancer tissue extracts, 4F10TT strongly labeled bands at 210 kd and 350 kd. In addition, positive labeling was also visible at a position around 250 kd (arrowhead). The 4C8MS labeled the two bands at 250 kd and 350 kd but not the 210 kd band. Thus, expression of the large Tn-C variants was found to be higher in breast cancers than in normal tissues.
Figure 4.
Immunoblot analysis of tissue extracts from breast cancers and normal tissues using 4C8MS and 4F10TT. Four samples of cancer tissues and normal counterparts collected from four cases of mastectomies were examined. When the extracts were analyzed with 4F10TT, bands at 350 (L), 250 (arrowhead) and 210 kd (S) were intensely labeled in breast cancers, but only weakly in normal tissues (left). With 4C8MS, 350 and 250 kd bands are apparently limited to breast cancers (right).
Next we analyzed breast cancer tissues with the same antibodies by immunohistochemistry. Different staining patterns of 4F10TT and 4C8MS were observed (Figure 5,A and B) ▶ . In normal tissues, 4F10TT labeled a thin layer beneath the basement membrane zone of the ducts and acini. Muscular layers of vessels and the basement membrane of capillaries were also intensely positive. Immunostaining by 4C8MS was usually absent around normal ducts and acini, but those associated with inflammation were sometimes positive. Muscular arteries were positive but not capillary vessels. In cases of fibroadenomas, 4F10TT showed apparent labeling of the basement membrane zone of the epithelial components, and the fibrous structures of the stroma, whereas 4C8MS labeling was weak in the stroma (Figure 5, C and D) ▶ . We examined 22 cases of invasive ductal carcinoma including 4 predominantly intraductal, 12 scirrhous, and 6 non-scirrhous lesions. In the intraductal cases, the sub-basement membrane zone of the duct containing intraductal cancer components and the stroma surrounding ducts were strongly labeled by 4F10TT (Figure 5E) ▶ . Conversely, little labeling by 4C8MS was observed in the stroma around ducts (Figure 5F) ▶ , but positive labeling appeared in sites where cancer cells were invading into the surrounding tissues (Figure 5G) ▶ . In scirrhous carcinomas, the stroma among the small cancer nests was densely positive for 4F10TT staining (Figure 5H) ▶ . 4C8MS also labeled the cancer stroma, stronger staining being frequently present at invasion fronts than in the central regions of tumors (Figure 5, I and J) ▶ . In non-scirrhous cancers, 4C8MS-positive areas were also found at the tumor borders (Figure 5L) ▶ . Foci of cancer cells having cytoplasmic staining and intercellular 4C8MS-positive deposits were observed in 9 cases (Figure 5, G and M) ▶ . With double immunostaining of Ki-67 antigen and large Tn-C variants, the areas positive for the latter had an elevated index, compared with those negative for 4C8MS (paired t-test, P < 0.05, Figure 6 ▶ ).
Figure 5.
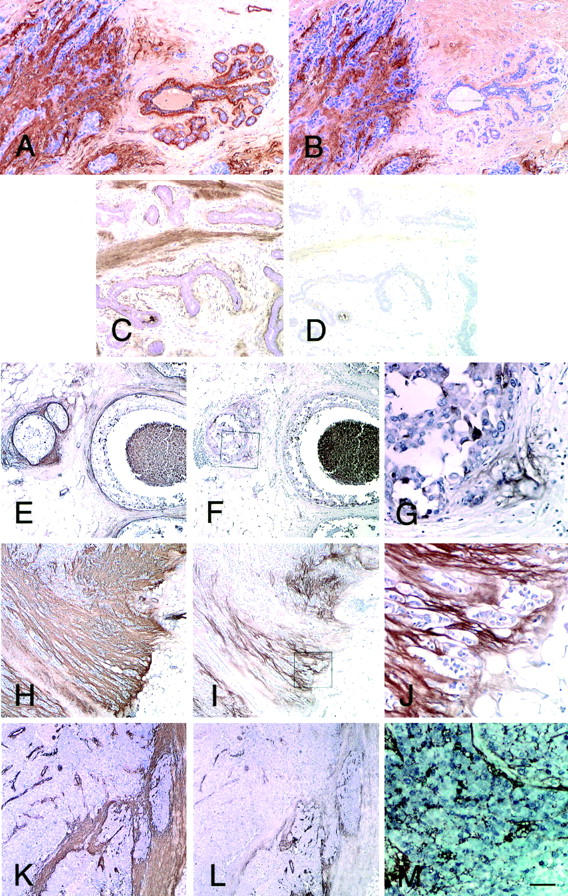
Expression of Tn-C in normal mammary glands, benign lesions, and breast cancers determined by immunohistochemistry using 4F10TT and 4C8MS. In normal mammary glands, all Tn-C variants labeled by 4F10TT are positive in the thin layer beneath the basement membrane zone of the ducts and acini (right in A), but the large Tn-C variants labeled by 4C8MS are absent (right in B). In contrast, both the antibodies demonstrate strong binding in the cancer stroma (left in A and B). In a fibroadenoma, the basement membrane zone of the epithelial components and the fibrous structures of the stroma are positive for 4F10TT (C), but only weak labeling of the stroma is evident with 4C8MS (D). In an intraductal lesion, the basement membrane zone containing cancer cells is strongly positive for 4F10TT (E), but not 4C8MS (F). The site of periductal invasion of cancer cells is, however, positive for 4C8MS (F). Panel G is a higher magnification in a square of panel F. In this case, cytoplasmic staining of cancer cells is apparent. In a scirrhous lesion, 4F10TT diffusely labels the cancer stroma (H), while 4C8MS demonstrate binding particularly near the invasion front (I, J). In a non-scirrhous invasive cancer also, 4F10TT labeling is extensive in cancer stroma (K), but 4C8MS labeling is limited to the tumor border (L). Double staining of Ki-67 (blue) and large Tn-C with 4C8MS (brown) is shown in M. Note the intercellular deposition of Tn-C (M). Nuclei were counterstained with hematoxylin in A–L and with methylgreen in M. Bar in M, 100 μm for A–D, 200 μm for E, F, H, I, K and L, 50 μm for G, J, and M.
Figure 6.
Relationship of cell proliferation with expression of large Tn-C variants in human breast cancers. The labeling index with the Ki-67 antigen was compared between areas with strong staining for 4C8MS and areas without the staining in 12 invasive ductal carcinomas. In the positive area for 4C8MS, the labeling index was significantly higher than in the negative area (paired t-test P < 0.05). Closed circles denote scirrhous cancers, and open circles denote non-scirrhous examples.
Data for immunoreactivity of 4C8MS with various normal tissues obtained from autopsy cases are summarized in Table 2 ▶ . Immunoreactivity was focal in the renal medulla, and weak in epithelial basement membrane zones and muscular layers of the alimentary tract (data not shown).
Table 2.
Reactivity of the 4F10TT and 4C8MS mAbs with Human Normal Adult Tissues
| Organ | Tissue | mAb | |
|---|---|---|---|
| 4F10 | 4C8 | ||
| Esophagus | Basement membrane | + | − |
| Muscular layer of mucosa | + | − | |
| Muscular layer | + | − | |
| Stomach | Basement membrane | + | ± |
| Muscular layer of mucosa | + | − | |
| Muscular layer | + | − | |
| Intestine | Basement membrane | + | ± |
| Muscular layer of mucosa | + | ± | |
| Muscular layer | + | ± | |
| Liver | Sinusoidal capillaries | + | − |
| Central veins | + | − | |
| Kidney | Medulla | + | + |
| Cortex | + | − | |
| Adrenal gland | Medulla | + | − |
| Cortex | + | − | |
| Pancreas | + | − | |
| Lung | Alveolar wall | + | − |
| Cartilage | Perichondrium | + | − |
| (Trachea) | Chondrocytes | + | − |
| Blood vessel | Muscular artery | + | + |
| Capillary vessel | + | − | |
| Cardiac muscle | − | − | |
Effects of the Tn-C FNIII Domain Containing Alternatively Spliced Sites on Migration of Tn-C-Null Mammary Cancer Cells
We have reported that intact Tn-C promotes cell migration in a wound closure assay in HEp 2 cells. 15 In the present study, we also confirmed that intact Tn-C also promotes migration and proliferation in a transwell migration assay with the mammary cancer cell line GHOM5E, derived from Tn-C-null mouse to minimize the effect of intrinsic Tn-C production (Figure 7) ▶ . In our immunohistochemical study, the large variants were expressed in invasion sites and fronts of cancer cells, suggesting involvement in cancer invasion. Next, we examined the effects of Tn-C variants on migration of mammary cancer cells, paying special attention to functional sites between the alternatively spliced variants. Purified proteins of Tn-C domains from conditioned media of stable transfectants are shown in Figure 2 ▶ . In a transwell assay, addition of mFNIII FL (containing alternatively spliced sites) significantly promoted cell migration, although the effect seemed lower than that of intact Tn-C. In contrast, mFNIII SO, mEGF, and mFBG showed no effects on cell migration.
Figure 7.
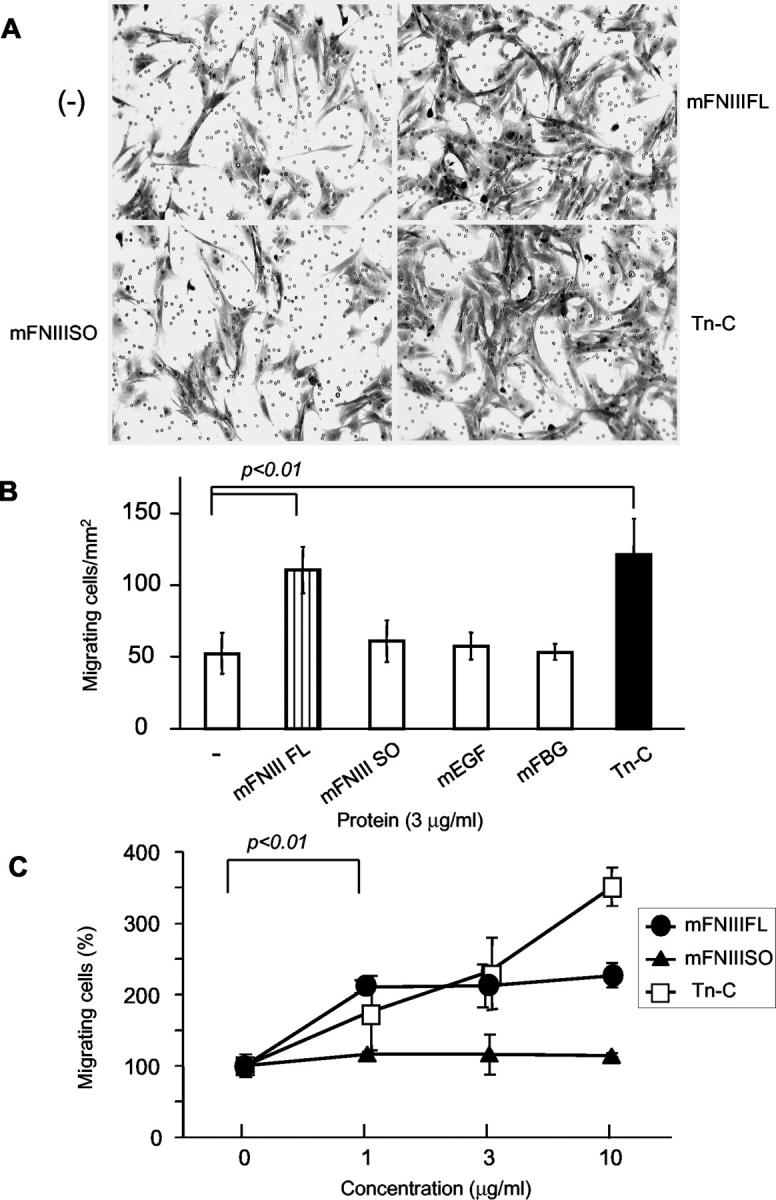
Effects of Tn-C domains and intact Tn-C on migration of mouse mammary tumor cells in a transmigration assay. Cells were plated on the cell culture insert, treated with the proteins, and allowed to migrate for 18 hours. Cells migrating through the membrane were stained with 0.1% crystal violet (A). Intact Tn-C and mFNIII FL significantly promoted tumor cell migration, but mFNIII SO, mEGF, and mFBG did not (B). In C, results for promotive effects of various protein concentrations are shown. The cells were counted in three fields of 1 mm2 in each insert, and the data are expressed as averages and standard deviations from three independent experiments. Data in C are relative to the control without Tn-C.
Effects of Tn-C FNIII Domain Containing Alternatively Spliced Sites on Proliferation of Tn-C-Null Mammary Cancer Cells
We previously demonstrated that intact Tn-C also promotes cell proliferation in a BrdU incorporation assay in HEp 2 cells. 15 In the present study, we showed that expression of the large variants was associated with high labeling index of Ki-67 in breast cancers (Figure 6) ▶ . Therefore we investigated effects of Tn-C domains on cell proliferation using a BrdU incorporation assay. The labeling index of Tn-C-null cancer cells, GHOM5E, was increased by addition of mFNIII FL as well as intact Tn-C at various concentrations (Figure 8) ▶ . Addition of mFNIII SO, mEGF, and mFBG was without effect.
Figure 8.
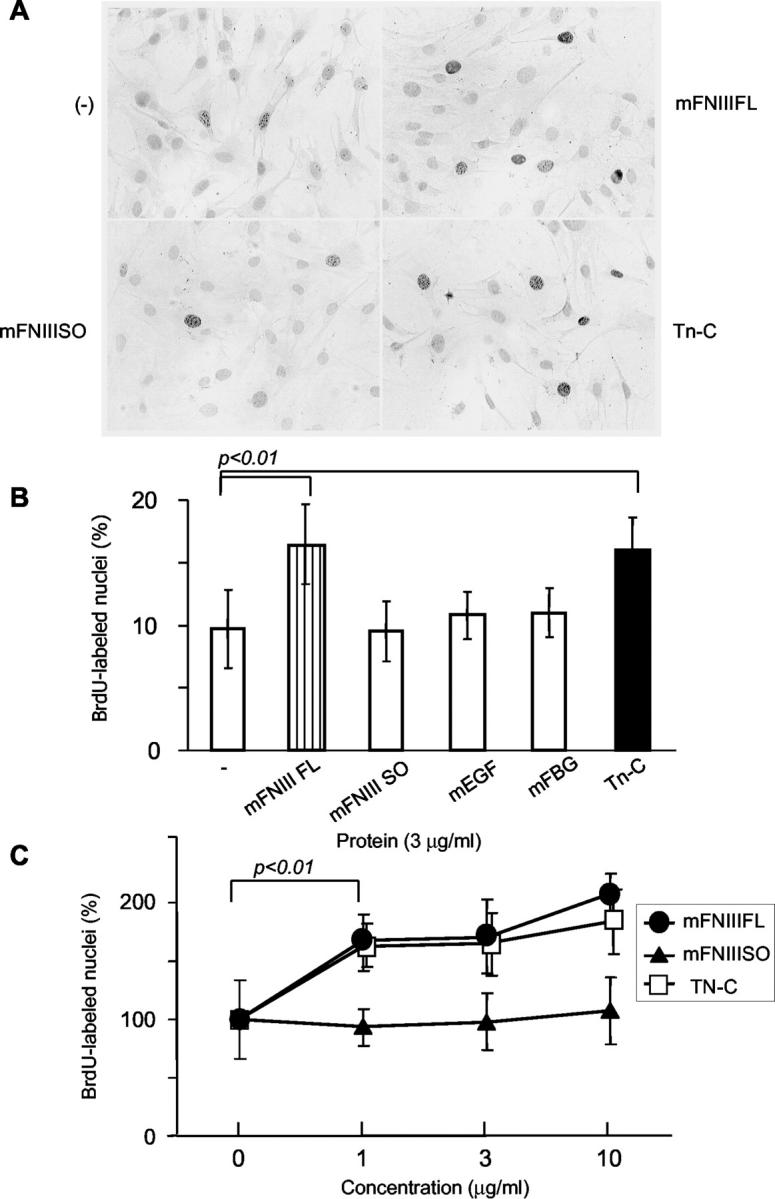
Effects of Tn-C domains and intact Tn-C on proliferation of mouse mammary tumor cells in a BrdU incorporation assay. After addition of the proteins tested, cells were incubated for 12 hours, and treated with BrdU for 2 hours. The cells labeled with BrdU were visualized by immunocytochemistry (A). Intact Tn-C and mFNIII FL significantly increased the BrdU labeling index, but mFNIII SO, mEGF, and mFBG did not (B). In C, the effects in various concentrations of intact Tn-C and mFNIII FL are shown. Data in C are relative to the control without Tn-C. All data are average and SD values from at least three independent experiments.
Effects of mAb 4C8MS to the Alternatively Spliced Domain B on Cell Migration and Proliferation
Having shown that the alternatively spliced FNIII repeats of Tn-C promote tumor cells proliferation and migration, we tried next to confirm this function by blocking with 4C8MS which binds to FNIII repeat B. When mFNIII FL were incubated with 4C8MS before the addition into the medium, the effects on migration and proliferation were abolished (Figure 9) ▶ . The same amount of control antibody did not exert any influence. Pretreatment of intact Tn-C with mAbs, either 4C8MS or 4F10TT, did not affect cell migration or the mitotic labeling index (data not shown).
Figure 9.
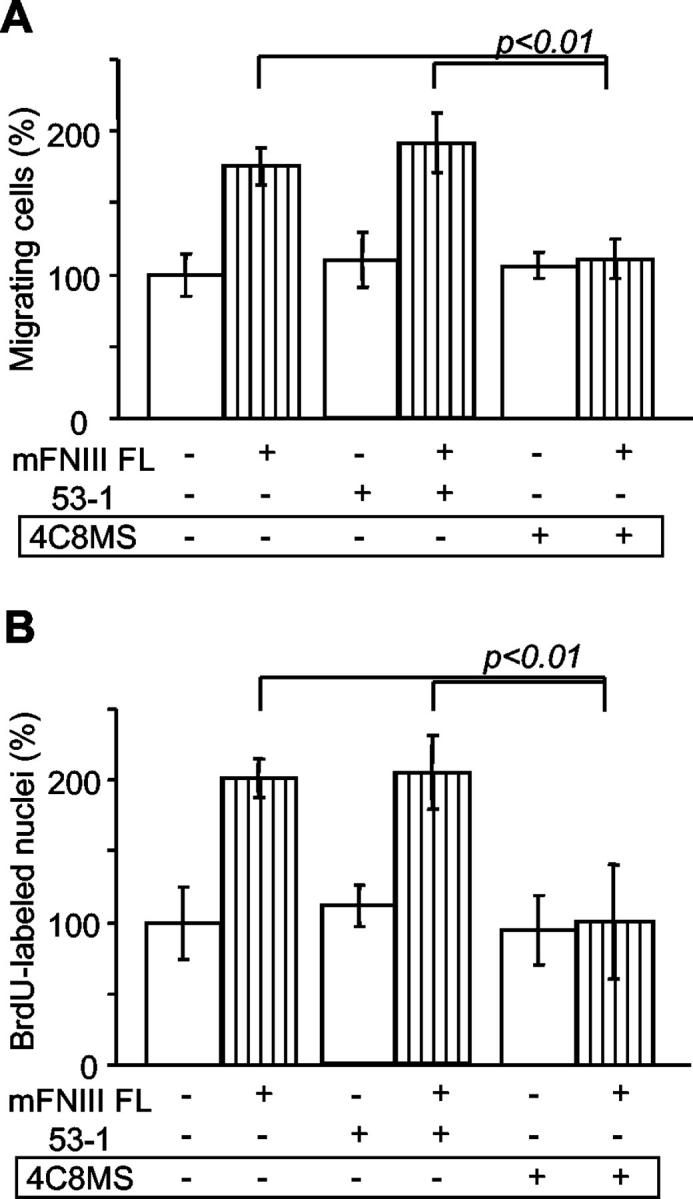
Blockage of promotive effects of mFNIII FL on migration and proliferation of cancer cells by antibody 4C8MS. Before addition to the medium, mFNIII FL (3 μg/ml) was incubated with the antibodies (15 μg/ml) for 30 minutes. When mFNIII FL was pre-incubated with 4C8MS, the promotive effect on cell migration (A) and BrdU incorporation (B) was completely abolished. Control antibody 53–1 has no effect.
Discussion
Detection of Tn-C expression in breast tissues has dramatically changed since the first discovery of this glycoprotein, reflecting improvement in the quality of materials and methods applied. Originally only reported as a stromal marker of epithelial malignancy in breast tissues, 43 it is now clear that Tn-C is also expressed around normal ducts and benign lesions of the breast. 44-46 Since comparisons of Tn-C expression and outcome of patients yielded conflicting results, the distribution and site of synthesis in breast cancer have been considered to have greater prognostic significance than its presence or absence. 12-14,47 Furthermore, although it has been evident for a long time that Tn-C has multiple variants produced by alternative splicing of its RNA, data on the distribution and biological functions of Tn-C variants in normal and diseased breast tissues has remained scarce. The lack of good monoclonal antibodies against Tn-C different variants, reactive in archived breast tissues, is partly to blame.
In this study, we therefore first examined expression of Tn-C in human breast cancers by means of immunoblotting and immunohistochemistry using a novel monoclonal antibody, 4C8MS, against FNIII B in the alternatively spliced region. Results of our immunoblotting generally paralleled those in a previous report comparing two other monoclonal antibodies. 8 Analysis by immunoblotting with BC-7 showed two bands of Tn-C at 330 kd and 190 kd in extract from breast cancer tissues, but only the lower band in the normal and benign tissues. When BC-2 against FNIII A was used, only the higher band was labeled in cancer tissue extracts. Our immunoblotting using 4C8MS antibody also showed that Tn-C including the alternative spliced FNIII repeat B was rare in normal tissues, but abundant in the cancer stroma. Despite some differences in Tn-C bands in normal tissue with the two studies, possibly due to the differences in the method of tissue extraction, monoclonal antibodies, or the blotting procedures, both indicate that the large variants are a major fraction of Tn-C deposited in the breast cancer stroma.
This was here confirmed by our immunohistochemical analysis of breast tissues. With the 4F10TT, Tn-C expression was detected in the sub-basement membrane zones around ducts and acini as well as vessels in normal breast tissues, and in the cancer stroma. In contrast, using the 4C8MS, large Tn-C variants were almost exclusively found in the cancer stroma. A similar finding has been recently reported using mAb αIIIB with frozen sections of breast cancers. 11 In addition, differences in patterns of Tn-C were found depending on expression site, with more intense staining by 4C8MS at invasion borders, contrasting with weaker staining in the central area of cancers. Large Tn-C variants may be preferentially expressed at the border, and the weaker staining in the central areas possibly illustrates their degradation, since they are known to be more susceptible to matrix metalloproteinase than the small variant. 48 Furthermore, while strong staining of basement membrane zones of the ducts containing intraductal cancer was observed with 4F10TT, the staining of 4C8MS was absent. The smallest variant (with complete omission of the alternatively spliced region) binds to fibronectin with higher affinity than the large variants and is incorporated into the matrix. 49,50 This difference in binding capability between the Tn-C variants may account for the variation in staining patterns between the two antibodies. In chick tissues, the intermediate sized and largest isoforms of Tn-C (containing variable numbers or all alternatively spliced domains, respectively) are known to be distributed in smooth muscle and underneath the epithelial lining of the villi, paralleling our immunohistochemical analysis of human tissues. 49
In a previous study on breast cancers using a conventional Tn-C antibody, it was shown that expression at invasion sites correlates with a higher proliferation rate of cancer cells and is a predictor of local recurrence and metastasis. 13,14 We have showed that Tn-C molecules expressed at the invasion fronts of tumors may chiefly be of the large variant type. We also found a positive correlation between their expression and proliferation of cancer cells. Since motility of cancer cells might be activated during invasion, it is plausible that the enhanced expression at the invasion front is involved in cell migration. Promotive effects of Tn-C on proliferation and migration of malignant epithelial cells in vitro has been reported. 15,16,51 In the present study, we prepared recombinant proteins of Tn-C domains and investigated their functions using the mouse mammary tumor cell line established from Tn-C knockout mouse, and only the recombinant protein of FNIII repeats including alternative spliced region promoted both proliferation and migration of the cells. Furthermore, the 4C8MS, specific to FNIII B, neutralized this promoting effect, providing compelling evidence that the alternative spliced FNIII region is a functional domain for proliferation and migration. The 4C8MS could not neutralize the effects of purified intact Tn-C. Since intact Tn-C is a mixture of the large variants containing various combinations of the alternatively spliced repeats, the large variants lacking FNIII B repeat may be also effective.
An earlier study demonstrated that intact Tn-C and recombinant proteins of FNIII A-D repeats cause down-regulation of focal adhesion integrity in endothelial cells. 33 Adhesion of endometrial epithelial cells to Matrigel is also disrupted by intact Tn-C and the recombinant fragments of the repeats. 52 Addition of the fragments also increased endothelial cell motility in a wound closure assay. 35 Tn-C and these functional fragments are considered to induce an intermediate state of adherence, characterized by a restructuring of focal adhesions and stress fibers, and to facilitate cell migration. 53 In the present study, addition of Tn-C and recombinant proteins containing the repeats A-D into the upper chamber also promoted active locomotion of the cancer cells on the membrane, followed by increased transmigration to the lower surface. Furthermore, we found that addition of the repeats caused increase in the proliferating fraction of the cancer cells, in line with results for endothelial cells treated with Tn-C fragment repeats A-D. 35 A recent study using a recombinant large Tn-C variant demonstrated an anti-adhesive mechanism and interference with cell binding to the HepII/syndecan-4 site in fibronectin through direct binding of the protein to the 13th FNIII repeat. 51 When cells adhere to fibronectin matrix, the interaction of integrin α5β1 and syndecan-4 attenuates cell proliferation. By binding to fibronectin, Tn-C inhibits syndecan-4 activation and impairs the signaling, resulting in enhanced tumor cell proliferation. Although a domain for Tn-C binding to fibronectin has not yet been identified, the alternative spliced region may be thus a candidate.
In conclusion, the present study provided evidence that large Tn-C variants are preferentially expressed in breast cancer tissues. Immunohistochemistry and in vitro experiments using recombinant fragments indicate that they are also closely associated with migration and proliferation of breast cancer cells. In addition, our novel monoclonal antibody specific to FNIII B, 4C8MS, reactive with paraffin-embedded tissues, is useful for exploration of large Tn-C variant expression in pathological tissues.
Footnotes
Address reprint requests to Toshimichi Yoshida, Department of Pathology, Mie University School of Medicine, 2–174 Edobashi, Tsu, Mie, 514-8507 Japan. E-mail: t-yosida@doc.medic.mie-u.ac.jp.
Supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology, Japan.
References
- 1.Donjacour A, Cunha G: Stromal regulation of epithelial function. Cancer Treat Res 1991, 53:335-364 [DOI] [PubMed] [Google Scholar]
- 2.Liotta L, Kohn E: The microenvironment of the tumour-host interface. Nature 2001, 411:375-379 [DOI] [PubMed] [Google Scholar]
- 3.Wernert N: The multiple roles of tumor stroma. Virchows Arch 1997, 430:433-443 [DOI] [PubMed] [Google Scholar]
- 4.Carnemolla B, Balza E, Siri A, Zardi L, Nicotra M, Bigotti A, Natali P: A tumor-associated fibronectin isoform generated by alternative splicing of messenger RNA precursors. J Cell Biol 1989, 108:1139-1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oyama F, Hirohashi S, Shimosato Y, Titani K, Sekiguchi K: Deregulation of alternative splicing of fibronectin pre-mRNA in malignant human liver tumors. J Biol Chem 1989, 264:10331-10334 [PubMed] [Google Scholar]
- 6.Pujuguet P, Hammann A, Moutet M, Samuel JL, Martin F, Martin M: Expression of fibronectin ED-A+ and ED-B+ isoforms by human and experimental colorectal cancer: contribution of cancer cells and tumor-associated myofibroblasts. Am J Pathol 1996, 148:579-592 [PMC free article] [PubMed] [Google Scholar]
- 7.Matsumoto E, Yoshida T, Kawarada Y, Sakakura T: Expression of fibronectin isoforms in human breast tissue: production of extra domain A+/extra domain B+ by cancer cells and extra domain A+ by stromal cells. Jpn J Cancer Res 1999, 90:320-325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borsi L, Carnemolla B, Nicolo G, Spina B, Tanara G, Zardi L: Expression of different tenascin isoforms in normal, hyperplastic, and neoplastic human breast tissues. Int J Cancer 1992, 52:688-692 [DOI] [PubMed] [Google Scholar]
- 9.Kalembeyi I, Yoshida T, Iriyama K, Sakakura T: Analysis of tenascin mRNA expression in the murine mammary gland from embryogenesis to carcinogenesis: an in situ hybridization study. Int J Dev Biol 1997, 41:569-573 [PubMed] [Google Scholar]
- 10.Dueck M, Riedl S, Hinz U, Tandara A, Möller P, Herfarth C, Faissner A: Detection of tenascin-C isoforms in colorectal mucosa, ulcerative colitis, carcinomas, and liver metastases. Int J Cancer 1999, 82:477-483 [DOI] [PubMed] [Google Scholar]
- 11.Adams M, Jones JL, Walker RA, Pringle JH, Bell SC: Changes in tenascin-C isoform expression in invasive and preinvasive breast disease. Cancer Res 2002, 62:3289-3297 [PubMed] [Google Scholar]
- 12.Ishihara A, Yoshida T, Tamaki H, Sakakura T: Tenascin expression in cancer cells and stroma of human breast cancer and its prognostic significance. Clin Cancer Res 1995, 1:1035-1041 [PubMed] [Google Scholar]
- 13.Jahkola T, Toivonen T, von Smitten K, Blomqvist C, Virtanen I: Expression of tenascin in invasion border of early breast cancer correlates with higher risk of distant metastasis. Int J Cancer 1996, 69:445-447 [DOI] [PubMed] [Google Scholar]
- 14.Jahkola T, Toivonen T, Virtanen I, von Smitten K, Nordling S, von Boguslawski K, Haglund C, Nevanlinna H, Blomqvist C: Tenascin-C expression in invasion border of early breast cancer: a predictor of local and distant recurrence. Br J Cancer 1998, 78:1507-1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshida T, Yoshimura H, Numata H, Sakakura Y, Sakakura T: Involvement of tenascin-C in proliferation and migration of laryngeal carcinoma cells. Virchows Arch 1999, 435:496-500 [DOI] [PubMed] [Google Scholar]
- 16.Orend G, Chiquet-Ehrismann R: Adhesion modulation by anti-adhesive molecules of the extracellular matrix. Exp Cell Res 2000, 261:104-110 [DOI] [PubMed] [Google Scholar]
- 17.Jones FS, Jones PL: The tenascin family of ECM glycoproteins: structure, function, and regulation during embryonic development and tissue remodeling. Dev Dyn 2000, 218:235-259 [DOI] [PubMed] [Google Scholar]
- 18.Aukhil I, Slemp C, Lighter V, Nishimura K, Briscoe G, Erickson H: Purification of hexabrachion (tenascin) from cell culture conditioned medium, and separation from a cell adhesion factor. Matrix 1990, 10:98-111 [DOI] [PubMed] [Google Scholar]
- 19.Nies DE, Hemesath TJ, Kim JH, Gulcher JR, Stefansson K: The complete cDNA sequence of human hexabrachion (tenascin). J Biol Chem 1991, 266:2818-2823 [PubMed] [Google Scholar]
- 20.Siri A, Carnemolla B, Saginati M, Leprini A, Casari G, Baralle F, Zardi L: Human tenascin: primary structure, pre-mRNA splicing patterns and localization of the epitopes recognized by two monoclonal antibodies. Nucleic Acids Res 1991, 19:525-531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saga Y, Tsukamoto T, Jing N, Kusakabe M, Sakakura T: Murine tenascin: cDNA cloning, structure, and temporal expression of isoforms. Gene 1991, 104:177-185 [DOI] [PubMed] [Google Scholar]
- 22.Mackie EJ, Tucker RP: Tenascin in bone morphogenesis: expression by osteoblasts and cell type-specific expression of splice variants. J Cell Sci 1992, 103:765-771 [DOI] [PubMed] [Google Scholar]
- 23.Joester A, Faissner A: Evidence for combinatorial variability of tenascin-C isoforms and developmental regulation in the mouse central nervous system. J Biol Chem 1999, 274:17144-17151 [DOI] [PubMed] [Google Scholar]
- 24.Maseruka H, Ridgway A, Tullo A, Bonshek R: Developmental changes in patterns of expression of tenascin-C variants in the human cornea. Invest Ophthalmol Vis Sci 2000, 41:4101-4107 [PubMed] [Google Scholar]
- 25.Sahlberg C, Aukhil I, Thesleff I: Tenascin-C in developing mouse teeth: expression of splice variants and stimulation by TGF β and FGF. Eur J Oral Sci 2001, 109:114-124 [DOI] [PubMed] [Google Scholar]
- 26.Hindermann W, Berndt A, Borsi L, Luo X, Hyckel P, Katenkamp D, Kosmehl H: Synthesis and protein distribution of the unspliced large tenascin-C isoform in oral squamous cell carcinoma. J Pathol 1999, 189:475-480 [DOI] [PubMed] [Google Scholar]
- 27.Latijnhouwers MA, de Jongh GJ, Bergers M, de Rooij M, Schalkwijk J: Expressions of tenascin-C splice variants by human skin cells. Arch Dermatol Res 2000, 292:446-454 [DOI] [PubMed] [Google Scholar]
- 28.Ghert MA, Jung ST, Qi W, Harrelson JM, Erickson HP, Block JA, Scully SP: The clinical significance of tenascin-C splice variant expression in chondrosarcoma. Oncology 2001, 61:306-314 [DOI] [PubMed] [Google Scholar]
- 29.Matsumoto K, Hiraiwa N, Yoshiki A, Ohnishi M, Kusakabe M: Tenascin-C expression and splice variant in habu snake venom-induced glomerulonephritis. Exp Mol Pathol 2002, 72:186-195 [DOI] [PubMed] [Google Scholar]
- 30.Wallner K, Shah PK, Sharifi BG: Balloon catheterization induces arterial expression of new tenascin-C isoform. Atherosclerosis 2002, 161:75-83 [DOI] [PubMed] [Google Scholar]
- 31.Mighell AJ, Thompson J, Hume WJ, Markham AF, Robinson PA: Human tenascin-C: identification of a novel type III repeat in oral cancer and of novel splice variants in normal, malignant, and reactive oral mucosae. Int J Cancer 1997, 72:236-240 [DOI] [PubMed] [Google Scholar]
- 32.Carnemolla B, Castellani P, Ponassi M, Borsi L, Urbini S, Nicolo G, Dorcaratto A, Viale G, Winter G, Neri D, Zardi L: Identification of a glioblastoma-associated tenascin-C isoform by a high affinity recombinant antibody. Am J Pathol 1999, 154:1345-1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphy-Ullrich JE, Lightner VA, Aukhil I, Yan YZ, Erickson HP, Höök M: Focal adhesion integrity is down-regulated by the alternatively spliced domain of human tenascin. J Cell Biol 1991, 115:1127-1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung CY, Erickson HP: Cell surface annexin II is a high affinity receptor for the alternatively spliced segment of tenascin-C. J Cell Biol 1994, 126:539-548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chung CY, Murphy-Ullrich JE, Erickson HP: Mitogenesis, cell migration, and loss of focal adhesions induced by tenascin-C interacting with its cell surface receptor, annexin II. Mol Biol Cell 1996, 7:883-892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fischer D, Brown-Ludi M, Schulthess T, Chiquet-Ehrismann R: Concerted action of tenascin-C domains in cell adhesion, anti-adhesion, and promotion of neurite outgrowth. J Cell Sci 1997, 110:1513-1522 [DOI] [PubMed] [Google Scholar]
- 37.Meiners S, Mercado ML, Nur-e-Kamal MS, Geller HM: Tenascin-C contains domains that independently regulate neurite outgrowth and neurite guidance. J Neurosci 1999, 19:8443-8453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meiners S, Nur-e-Kamal MS, Mercado ML: Identification of a neurite outgrowth-promoting motif within the alternatively spliced region of human tenascin-C. J Neurosci 2001, 15:7215-7225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seiffert M, Beck SC, Schermutzki F, Müller CA, Erickson HP, Klein G: Mitogenic and adhesive effects of tenascin-C on human hematopoietic cells are mediated by various functional domains. Matrix Biol 1998, 17:47-63 [DOI] [PubMed] [Google Scholar]
- 40.Adamsky K, Schilling J, Garwood J, Faissner A, Peles E: Glial tumor cell adhesion is mediated by binding of the FNIII domain of receptor protein tyrosine phosphatase β(RPTPβ) to tenascin C. Oncogene 2001, 20:609-618 [DOI] [PubMed] [Google Scholar]
- 41.Saga Y, Yagi T, Ikawa Y, Sakakura T, Aizawa S: Mice develop normally without tenascin. Genes Dev 1992, 6:1821-1831 [DOI] [PubMed] [Google Scholar]
- 42.Imanaka-Yoshida K, Hiroe M, Yasutomi Y, Toyozaki T, Tsuchiya T, Noda N, Maki T, Nishikawa T, Sakakura T, Yoshida T: Tenascin-C is a useful marker for disease activity in myocarditis. J Pathol 2002, 197:388-394 [DOI] [PubMed] [Google Scholar]
- 43.Mackie EJ, Chiquet-Ehrismann R, Pearson CA, Inaguma Y, Taya K, Kawarada Y, Sakakura T: Tenascin is a stromal marker for epithelial malignancy in the mammary gland. Proc Natl Acad Sci USA 1987, 84:4621-4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Howeedy AA, Virtanen I, Laitinen L, Gould NS, Koukoulis GK, Gould VE: Differential distribution of tenascin in the normal, hyperplastic, and neoplastic breast. Lab Invest 1990, 63:798-806 [PubMed] [Google Scholar]
- 45.Lightner VA, Marks JR, McCachren SS: Epithelial cells are an important source of tenascin in normal and malignant human breast tissue. Exp Cell Res 1994, 210:177-184 [DOI] [PubMed] [Google Scholar]
- 46.Yoshida T, Matsumoto E, Hanamura N, Kalembeyi I, Katsuta K, Ishihara A, Sakakura T: Co-expression of tenascin and fibronectin in epithelial and stromal cells of benign lesions and ductal carcinomas in the human breast. J Pathol 1997, 182:421-428 [DOI] [PubMed] [Google Scholar]
- 47.Jahkola T, Toivonen T, Nordling S, von Smitten K, Virtanen I: Expression of tenascin-C in intraductal carcinoma of human breast: relationship to invasion. Eur J Cancer 1998, 34:1687-1692 [DOI] [PubMed] [Google Scholar]
- 48.Siri A, Knauper V, Veirana N, Caocci F, Murphy G, Zardi L: Different susceptibility of small and large human tenascin-C isoforms to degradation by matrix metalloproteinases. J Biol Chem 1995, 270:8650-8654 [DOI] [PubMed] [Google Scholar]
- 49.Chiquet-Ehrismann R, Matsuoka Y, Hofer U, Spring J, Bernasconi C, Chiquet M: Tenascin variants: differential binding to fibronectin and distinct distribution in cell cultures and tissues. Cell Regul 1991, 2:927-938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chung CY, Erickson HP: Glycosaminoglycans modulate fibronectin matrix assembly and are essential for matrix incorporation of tenascin-C. J Cell Sci 1997, 110:1413-1419 [DOI] [PubMed] [Google Scholar]
- 51.Huang W, Chiquet-Ehrismann R, Moyano JV, Garcia-Pardo A, Orend G: Interference of tenascin-C with syndecan-4 binding to fibronectin blocks cell adhesion and stimulates tumor cell proliferation. Cancer Res 2001, 61:8586-8594 [PubMed] [Google Scholar]
- 52.Julian J, Chiquet-Ehrismann R, Erickson HP, Carson DD: Tenascin is induced at implantation sites in the mouse uterus and interferes with epithelial cell adhesion. Development 1994, 120:661-671 [DOI] [PubMed] [Google Scholar]
- 53.Murphy-Ullrich JE: The de-adhesive activity of matricellular proteins: is intermediate cell adhesion an adaptive state? J Clin Invest 2001, 107:785-790 [DOI] [PMC free article] [PubMed] [Google Scholar]