Abstract
Collagen degradation by matrix metalloproteinases is the limiting step in reversing liver fibrosis. Although collagen production in cirrhotic livers is increased, the expression and/or activity of matrix metalloproteinases could be normal, increased in early fibrosis, or decreased during advanced liver cirrhosis. Hepatic stellate cells are the main producers of collagens and matrix metalloproteinases in the liver. Therefore, we sought to investigate whether they simultaneously produce α1(I) collagen and matrix metalloproteinase-13 mRNAs. In this communication we show that expression of matrix metalloproteinase-13 mRNA is reciprocally modulated by tumor necrosis factor-α and transforming growth factor-β1. When hepatic stellate cells are co-cultured with hepatocytes, matrix metalloproteinase-13 mRNA is up-regulated and α1(I) collagen is down-regulated. Injuring hepatocytes with galactosamine further increased matrix metalloproteinase-13 mRNA production. Confocal microscopy and differential centrifugation of co-cultured cells revealed that matrix metalloproteinase-13 is localized mainly within hepatic stellate cells. Studies performed with various hepatic stellate cell lines revealed that they are heterogeneous regarding expression of matrix metalloproteinase-13. Those with myofibroblastic phenotypes produce more type I collagen whereas those resembling freshly isolated hepatic stellate cells express matrix metalloproteinase-13. Overall, these findings strongly support the notion that α1(I) collagen and matrix metalloproteinase-13 mRNAs are reciprocally modulated.
Liver fibrosis results from excess deposition of extracellular matrix components, mainly type I collagen that is produced by hepatic stellate cells (HSCs). 1-4 These cells can undergo a phenotypic change named activation. During activation HSCs acquire a different morphology and function, express higher levels of fibrillar collagens, and develop a contractile apparatus that includes the up-regulation of nonskeletal myosin and α-actin. 1-5 Although the activity of matrix metalloproteinases (MMPs) may be normal or increased 6,7 and the half-life of collagen I and III is decreased by 50%, 8 the capacity of HSCs to degrade fibrillar collagens may be hampered by two key factors: firstly, lack of access to digest collagen fibrils within thick, highly cross-linked collagen bundles; 8,9 and secondly, the up-regulation of tissue inhibitors of metalloproteinases (TIMPs) that blocks MMP activity. 10-16 Indeed, under conditions in which TIMP-1 is overexpressed 17 or its expression up-regulated, 18,19 there is increased accumulation of collagen in the liver. Conversely, during resolution of liver fibrosis in a reversible rat model, interstitial collagenase expression (MMP-1/MMP-13) remains unaltered, whereas TIMP-1 and TIMP-2 expression is down-regulated. 20
Another important event taking place during resolution of liver fibrosis is apoptosis of myofibroblasts with concomitant decrease in α-smooth muscle-positive cells. 20 Indeed, data in the literature have documented a higher susceptibility of activated cells to undergo apoptosis than quiescent HSCs. 21 Based on these findings, it is difficult to reconcile the sustained expression of MMPs observed during resolution of liver fibrosis, 20,22 unless MMPs are produced preferentially by nonactivated HSCs. Indeed, this possibility is supported by the observation that collagen and MMP-13 production are reciprocally modulated in cultured HSCs. 8 Accordingly, cells that produce an excess of type I collagen, such as myofibroblast within fibrous septa, should express less MMPs than quiescent HSCs expressing low levels of type I collagen. To test this hypothesis we took advantage of several HSC clones developed in our laboratory that have distinct phenotypes. 23,24 Although two of these clones resemble quiescent HSCs, two others are more myofibroblastic. In this communication we show that those clones with a myofibroblastic phenotype expressing high levels of α1(I) procollagen and elastin mRNAs, do not express MMP-2 or MMP-13, and that these MMPs cannot be induced by tumor necrosis factor (TNF)-α. Conversely, those HSC clones expressing lower levels of α1(I) collagen and elastin mRNA express these MMPs that are further induced by TNF-α or by co-culture with hepatocytes. We also showed that when co-cultured with HSCs, hepatocytes induce MMP-13 mRNA expression while down-regulating α1(I) collagen gene expression. Moreover, similar to events occurring during wound healing in skin, 25 hepatocyte injury induced with galactosamine further up-regulates MMP-13 mRNA expression.
Materials and Methods
Chemicals
Minimum essential medium was purchased from Mediatech, Inc. (Herndon, VA). Nonessential amino acids, penicillin, and streptomycin were obtained from Life Technologies, Inc. (Rockville, MD). Bovine serum albumin and acetone were purchased from Sigma Chemical Co. (St. Louis, MO). TNF-α, transforming growth factor (TGF)-β1, and the protease inhibitor cocktail Complete were obtained from Roche Molecular Biochemicals (Indianapolis, IN). Affi-gel and horseradish peroxidase-conjugated goat anti-mouse IgG were obtained from Bio-Rad Life Science Research Products (Hercules, CA). [α-32P]-dCTP was obtained from Amersham Life Science (Arlington Heights, IL).
Cell Culture
NFSC and CFSC cell lines derived from normal and CCl4-fibrotic rat livers have been previously characterized. 23,24 Clones CFSC-8B, CFSC-3H, CFSC-2G, and CFSC-5H were derived from the CFSC line as described. 24 All clones expressed known markers of HSCs, including α-smooth muscle actin, glial fibrillary acidic protein, and nestin. Cells (1 × 106) were cultured in minimum essential medium with l-glutamine supplemented with 10% fetal bovine serum, nonessential amino acids, 100 IU/ml penicillin, and 100 μg/ml streptomycin at 37°C in a humidified atmosphere with 5% CO2 until they reached confluence (48 hours). After this time, the medium was replaced by a serum-free medium containing 0.2% bovine serum albumin and cells were further incubated for 24 hours. For time-course experiments, HSC clones were plated and maintained in culture for various time periods using the same culture conditions described above (see specific experiments). In some experiments, CFSC-8Bs were cultured as described above and the serum-containing minimum essential medium replaced with a serum-free medium that contained 0.2% bovine serum albumin. These cells were cultured for an additional period of 12 hours followed by treatment with TNF-α (20 ng/ml) for 12 hours. Controls consisted of HSCs maintained in culture under identical conditions but without the addition of cytokine.
Primary cultures of mouse HSCs prepared as previously described 26 were used for some experiments. These cells were cultured in serum-free medium as described for rat HSC clones and treated with either 20 ng/ml of TNF-α or 8 ng/ml of TGF-β1. Twenty-four hours after incubation with each cytokine, cells were harvested for extraction of total RNA and Northern analysis of α1(I) collagen, MMP-13, and S14 mRNAs.
Total RNA Extraction and Northern Analysis
Total RNA was extracted from HSC cultures and co-cultures by the method of Chomczynski and Sacchi. 27 Aliquots of 10 μg of RNA were subjected to electrophoresis on 1% agarose-formaldehyde gels and transferred to nylon membranes (GeneScreen; Dupont, Boston, MA). Prehybridizations and hybridizations were performed as previously described. 23,24 The following 32P-labeled cDNA probes were used: rat α1(I) procollagen 28 fibronectin, 29 MMP-13, 30 MMP-2, 31 TIMP-1, 32 MMP-9, 33 colony-stimulating factor-1 (CSF-1), ribosomal protein S14, and human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (ATCC, Rockville, MD). The relative intensities of the generated [α-32P]-cDNA/mRNA signals were determined either by phosphor imaging analysis (Molecular Dynamics, Sunnyvale, CA) or by densitometric analysis of autoradiographies. Values are reported as means of triplicate experiments ± SD after correcting for loading differences using the signals generated by GAPDH or S14 ribosomal protein mRNAs.
Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR) Amplification of MMP-13, α1(I) Collagen, and Albumin mRNAs
In some experiments we measured the expression of albumin, α1(I) collagen and MMP-13 mRNA by RT-PCR following standard procedures. 34 For these experiments, the following primers were used: albumin, 5′AAGGCACCCCGATTACTCCG3′ and 5′TGCGAAGTCACCCATCACCG3′; MMP-13, 5′AAAGAACATGGTGACTTCTACC3′ and 5′ACTGGATTCCTTGAACGTC3′; α1(I) collagen, 5′GACATCCCTGAAGTCAGCTGC3′ and 5′TCCCTTGGGTCCCTCGAC3′.
The concentrations of primers and the number of amplification cycles required to obtain linear rates was determined experimentally. For albumin, amplification primer concentration was 0.05 μmol/L and the number of cycles 16. For α1(I) collagen, primer concentration was 0.2 μmol/L and the number of cycles 23. For MMP-13 primer concentration was 0.5 μmol/L and the number of cycles 29.
Co-Cultures of CFSC-8B with CFSC-3H
CFSC-8B and CFSC-3H were co-cultured at different ratios as indicated in the text. Cells were cultured in minimum essential medium supplemented with 10% fetal bovine serum, nonessential amino acids, and antibiotics until they reached confluence (48 hours). After this time, the medium was replaced with a serum-free medium containing 0.2% bovine serum albumin. Approximately 24 hours after replacing the culture medium, total RNA was extracted as described above. 27
Co-Cultures of Hepatocytes and HSCs
Co-cultures were prepared with freshly isolated hepatocytes and the HSC clone CFSC-8B as previously described. 35,36 Hepatocytes were isolated from Sprague-Dawley rats by a two-step perfusion method using collagenase, as previously described. 37 Viability of hepatocytes was determined by the trypan blue exclusion test and only cells with viability greater than 90% were used. Frozen stocks of CFSC-8B were thawed and maintained in culture as described above. Confluent dishes were trypsinized and 1 × 106 cells seeded as described previously. Forty-eight hours after plating, triplicate dishes were used for counting the number of HSCs with a hemocytometer. Co-cultures with freshly isolated hepatocytes were then established by plating the cells at ratios ranging from 1:1 to 5:1 (hepatocytes:HSCs). Co-cultures were maintained for 24 to 48 hours in a serum-free hormonally defined medium 35,36 and total RNA was extracted as described above. 27 In duplicate co-cultures maintained for 24 hours, 5 mmol/L of galactosamine was added and 24 hours later culture medium and cells harvested separately. Lactic dehydrogenase was measured in the culture medium using a commercially available kit (Sigma Diagnostics, St. Louis, MO) and total RNA was extracted from the cells. As controls for these experiments we used monocultures of CFSC-8B, and hepatocytes and co-cultures without any treatment.
To determine the expression of α1(I) collagen and MMP-13 mRNA in individual cell populations in co-cultures maintained for 48 hours, the culture medium was removed, the cells washed twice with Lefferts medium with EGTA, 37 followed by incubation with bacterial collagenase for 5 minutes at 37°C. Detached cells were separated by differential centrifugation as described. 37 Total RNA was extracted from the cells and the expression of α1(I) collagen, MMP-13, and albumin mRNAs determined by RT-PCR as described above.
Western Blot Analysis
HSCs were cultured to confluence, washed with phosphate-buffered saline (PBS), and incubated in serum-free medium for 12 hours. Culture media conditioned by HSCs were removed and proteins precipitated with 50% acetone at 4°C for 1 hour. Pellets were resuspended in 1 ml of 20 mmol/L phosphate buffer, pH 7.1, containing 40 μl of the protease inhibitor cocktail Complete. Before precipitation, excess albumin was eliminated by Affi-gel chromatography following the manufacturer’s recommendations. Pellets were resuspended in 2× Laemmli sample buffer 38 and protein concentration was determined with the BCA protein assay kit (Pierce, Rockford, IL). HSCs remaining in the culture dishes were lysed by the direct addition of sodium dodecyl sulfate lysis buffer and protein concentration was determined as described above. Aliquots containing 30 μg of protein were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 8% polyacrylamide gels and transferred to Immobilon-P membranes (Millipore Co., Bedford, MA). Membranes were blocked for 1 hour in 3% low-fat milk in 50 mmol/L Tris, pH 7.5, 0.5 mol/L NaCl, 0.01% Tween 20, before incubating for 2 hours with a 1:2000 dilution of the MMP-13 monoclonal antibody (Neomarkers, Fremont, CA). After extensive washing of the membranes with Tween 20/Tris-buffered saline, bound antibodies were detected with horseradish peroxidase-conjugated goat anti-mouse IgGs diluted 1:3000 in Tris-buffered saline containing 3% low-fat milk. Membranes were washed and developed with a horseradish peroxidase chemiluminescence detection reagent (ECL Renaissance System; NEN Life Sci Products, Boston, MA).
Confocal Immunomicroscopy
Co-cultures of hepatocytes and CFSC-8B were prepared on coverslips and maintained in culture for 48 hours. Coverslips were rinsed with PBS, fixed with 4% paraformaldehyde for 10 minutes, permeabilized with cold acetone for 15 minutes, and then rinsed with PBS and blocked with 1% bovine serum albumin-2% goat serum for 1 hour. Coverslips were incubated overnight in a humidified chamber with either a monoclonal antibody against MMP-13 (1:200) (see Western Blot Analysis) and polyclonal antibodies against albumin (1:200) (ICN Cappel, Costa Mesa, CA) or an antibody to MMP-13 (1:200) and an anti-desmin antibody (1:10) (ICN Cappel). After several washes with PBS, coverslips were incubated for 1 hour with either one of the following fluorescent-tagged antibodies diluted 1:300: CY3-labeled goat anti-mouse (Chemicon International, Temecula, CA) or fluorescein-labeled goat anti-rabbit (Molecular Probes, Eugene, OR). Coverslips were washed with 0.1%Tween-20/PBS and were mounted on glass slides with Gel/Mount from Biomeda Corp. (Foster City, CA). Images were obtained using AX70 Olympus photomicroscope and a Meridian Ultra confocal microscope and processed with Adobe Photoshop software v5.0.2.
Results
HSC Clone CFSC-8B Expresses Constitutively MMP-2 mRNA
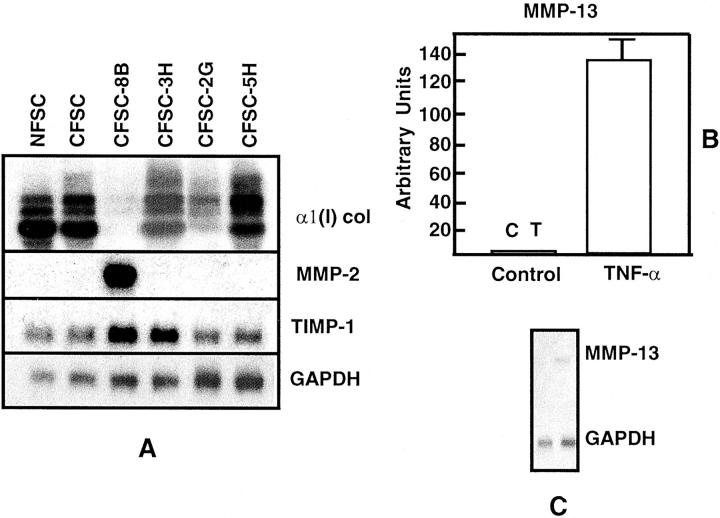
We have previously shown that HSCs are heterogeneous regarding expression of α1(I) procollagen mRNA. 23,24 To determine whether HSCs are also heterogeneous regarding expression of MMPs, and whether MMP-2, MMP-9, and/or MMP-13 mRNA levels reciprocally correlate with that of α1(I) procollagen mRNA, the following experiments were performed: Steady-state levels of MMP-2, MMP-9, MMP-13, and TIMP-1 mRNAs in the maternal HSC lines NFSC and CFSC, which are derived from normal and CCl4-cirrhotic rat liver, respectively, were measured. In addition, we determined the levels of expression of these mRNAs in four clones derived from CFSC, namely CFSC-8B, CFSC-2G, CFSC-3H, and CFSC-5H. As shown in Figure 1A ▶ , steady-state levels of α1(I) procollagen, MMP-2, and TIMP-1 mRNAs measured in confluent cultures varied significantly among the various HSC clones investigated. Interestingly, only clone CFSC-8B expressed MMP-2 mRNA. Moreover, although all clones expressed TIMP-1 (Figure 1A) ▶ , neither the maternal CFSC line nor the four clones investigated, expressed MMP-9 (data not shown). In contrast to these findings, none but CFSC-8B expressed measurable levels of MMP-13 mRNA that was up-regulated by TNF-α administration (Figure 1, B and C) ▶ .
Figure 1.
HSCs are heterogeneous with regard to expression of MMP-2 and MMP-13 mRNAs. Representative Northern blots performed with 10 μg of total RNA extracted from the maternal HSC lines NFSC and CFSC, from four clones derived from CFSC (A), and from CFSC-8B treated with 20 ng/ml of TNF-α (B). The figure shows that while the cell lines and clones expressed variable levels of α1(I) procollagen and TIMP-1 mRNAs, only clone CFSC-8B expressed MMP-2 (A). None of the HSCs tested expressed detectable levels of MMP-9 (data not shown) and only CFSC-8B expressed MMP-13 mRNA whose expression was up-regulated by TNF-α. The histogram in B corresponds to triplicate experiments performed with total RNA extracted 12 hours after culturing cells with TNF-α (see Materials and Methods). Values were corrected for loading differences using GAPDH mRNA as an internal control, and are expressed as means ± SD. C corresponds to a representative Northern blot.
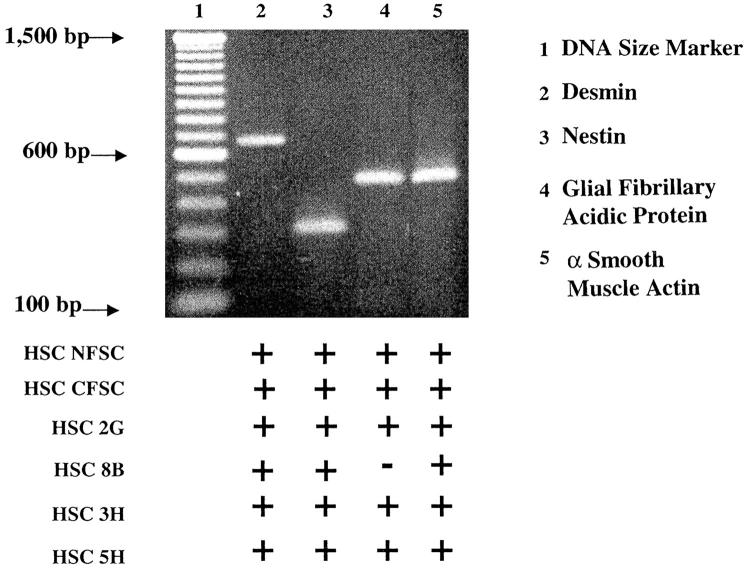
Because of the marked heterogeneity of the HSCs we used primers described by others to determine by RT-PCR the expression of various HSC markers, namely α-smooth muscle actin, desmin, glial fibrillary acidic protein, and nestin in the various HSC lines and clones (Figure 2) ▶ . Except for CFSC-8B whose amplicon for glial fibrillary acidic protein was barely detectable, all of the other cells expressed strong signals for all of the selected markers. However, their expression was quite heterogeneous and signal intensities for each marker varied from clone to clone. These results confirmed the heterogeneity of these cells and clearly indicated that HSC lines and clones isolated in our laboratory are bona fide HSCs with an activated phenotype.
Figure 2.
PCR analysis of desmin, nestin, glial fibrillary acidic protein, and α-smooth muscle actin in maternal HSC lines (NFSC and CFSC) and in the four clones derived from CFSC (2G, 8B, 3H, and 5H). This figure shows a representative gel of the transcripts detected using total RNA extracted from each of the HSC lines and clones that summarizes the findings obtained. Although the signal intensities varied from cell to cell, all expressed strong signals for most HSC markers. However, levels of expression of glial fibrillary acidic protein were barely detectable in CFSC-8B.
Expression of MMP-2 mRNA in CFSC-8B Increases with Time in Culture
To further investigate expression of MMPs and TIMP-1 in CFSC-8B cells, time-course experiments were performed. As shown in Figure 3 ▶ , expression of MMP-2, but not that of TIMP-1 mRNA, was dependent on cell density. Highest levels of MMP-2 mRNA expression were observed 96 hours after plating when the cells were confluent, with values close to eightfold higher than those obtained 24 hours after trypsinization and plating (Figure 3) ▶ . To further determine whether the time-dependent increase of MMP-2 mRNA expression was specific we investigated the pattern of expression of various other mRNAs known to be expressed by HSCs. As shown in Figure 3 ▶ , expression of CSF-1 mRNA, a chemoattractant chemokine, was highest in freshly plated HSCs and decreased with time in culture, whereas neither fibronectin nor α1(I) procollagen mRNA followed the same pattern of expression. Although changes in α1(I) procollagen mRNA were unremarkable, expression of fibronectin mRNA increased significantly with confluence of the cells (Figure 3) ▶ .
Figure 3.
Time-dependent expression of MMP-2, TIMP-1, α1(I) procollagen, fibronectin, and CSF-1 mRNAs by cultured CFSC-8B. Northern blot analysis was performed with 10 μg of total RNA. The first time point was obtained with RNA extracted 24 hours after plating HSCs. The zero time point was obtained after trypsinization of confluent cultures before plating. Left: Histograms obtained with data from triplicate experiments. Results are expressed as arbitrary units ± SD and were corrected for loading differences after hybridization with a GAPDH cDNA. Right: A representative Northern blot.
Co-Culturing CFSC-8B with Hepatocytes, but Not with Other HSC Clones, Up-Regulates Expression of MMP-13 mRNA
The aim of these experiments was to investigate whether CFSC-8B could influence expression of MMP-13 mRNA in HSC clones that do not produce it and whether hepatocytes could enhance the expression of this mRNA in CFSC-8B. To this end, firstly, we prepared co-cultures in which the ratio of CFSC-8B to CFSC-3H was decreased from 100 to 0%. As shown in Figure 4, A and B ▶ , expression of MMP-2 mRNA was highest in cultures containing 100% CFSC-8B and it decreased steadily as the proportion of CFSC-3H was increased. As also shown in the Figure 4 ▶ , these co-cultures did not contain detectable levels of MMP-13 mRNA irrespectively of which cell was the predominant type in culture. On the other hand, steady state levels of α1(I) procollagen mRNA by co-cultured cells reflected the abundance of CFSC-3H cells in the co-culture (Figure 4, A and B) ▶ . These data suggested that CFSC-8B did not induce CFSC-3H to express MMP-2 and that, in general, expression of MMP-2 and α1(I) procollagen mRNAs reflected the abundance of CFSC-8B and CFSC-3H, respectively. To further determine whether TNF-α could up-regulate expression of MMP-2 and MMP-13 mRNA in these co-cultures, cells cultured as described above were treated with 20 ng/ml of TNF-α. As clearly shown in Figure 4B ▶ , MMP-13 mRNA was up-regulated but again, the response was a reflection of the abundance of CFSC-8B in the co-culture. Surprisingly, the expression of MMP-2 in this co-culture system was not affected by TNF-α.
Figure 4.
Expression of α1(I) procollagen, MMP-2, and MMP-13 mRNAs by co-cultures of CFSC-8B and CFSC-3H. Cells were plated to contain 100%, 75%, 50%, 25%, and 0% CFSC-8B and the expression of the aforementioned mRNAs determined after 12 hours of co-culture. Northern analysis was performed with 10 μg of total RNA as described above. Right: Histograms of triplicate experiments. Results are expressed as fold change compared to controls and are means ± SD after correction for loading differences using GAPDH mRNA as an internal control. Left: A representative blot. Note that MMP-2 mRNA expression reflects the proportion of CFSC-8B in the co-culture, while that of α1(I) procollagen mRNA reflects the number of CFSC-3H. Although TNF-α had no effect on MMP-2 mRNA expression it up-regulated the expression of MMP-13 mRNA.
Expression of MMP-13, but Not MMP-2 mRNAs, Is Increased in Co-Cultures Containing CFSC-8B and Freshly Isolated Hepatocytes
In vivo, HSCs are in close association with other cell types including hepatocytes, and when hepatic injury occurs, their quiescent phenotype is transformed into that of active myofibroblasts. Because cross-talk among the various cell types present in the liver plays a key role in maintaining homeostasis of the liver ecosystem, 39 we considered it important to investigate whether hepatocytes could modulate the expression of MMPs by HSCs. To this end, we used a co-culture system of hepatocytes and HSCs that has been previously characterized in our laboratory. 35,36 However, we used HSC clone CFSC-8B instead of CFSC-2G, because as shown in this communication, the former and not the latter, expresses MMP-2 and MMP-13 mRNAs. As shown in Figure 5 ▶ , freshly isolated hepatocytes maintained in culture for either 24 or 48 hours did not express MMP-2 or MMP-13 mRNAs. As expected, CFSC-8B expressed very low levels of MMP-13 and contained high basal levels of MMP-2 mRNAs (Figures 1 and 5) ▶ . Remarkably, on co-culturing CFSC-8B with freshly isolated hepatocytes, expression of MMP-13 mRNA was up-regulated, whereas that of α1(I) collagen and MMP-2 mRNAs was down-regulated (Figure 5, A and B ▶ , respectively). Interestingly, levels of expression of MMP-13 mRNA were dependent on the ratio of hepatocytes to HSCs plated in culture. Thus, as shown in Figure 6A ▶ , only co-cultures containing ratios of five hepatocytes per one HSC expressed significant levels of MMP-13 mRNA and this level of expression was sustained in co-cultures containing ratios of up to 8 to 10 hepatocytes/HSCs. Moreover, up-regulation of MMP-13 was dependent on cell-cell contact and co-cultures in which the cells were separated by a Millipore filter did up-regulate express MMP-13 mRNA (data not shown).
Figure 5.
Expression of α1(I) procollagen, MMP-13 (A) and MMP-2 (B) mRNAs by cultured hepatocytes, CFSC-8B, and co-cultures of hepatocytes plus CFSC-8B. Northern blot analysis was performed with 10 μg of total RNA as described in Materials and Methods. Note that while neither hepatocytes nor CFSC-8B cultured alone expressed detectable levels of MMP-13, co-cultures containing both cell types expressed this mRNA and values remained elevated for 24 and 48 hours of co-culture (A). In contrast to these findings, while CFSC-8B expressed high levels of MMP-2 mRNA, this mRNA was not expressed by hepatocytes and its expression level was significantly lower in co-cultures (B). A shows that α1(I) procollagen mRNA expression was down-regulated by 24 and 48 hours of co-culture. Detection of this mRNA was deliberately overexposed to detect the very low levels expressed by co-cultures. Even under these conditions, α1(I) procollagen mRNA was not detected in cultured hepatocytes.
Figure 6.
Northern (A) and Western blot (B) analyses of MMP-13 expression by co-cultures prepared with various ratios of CFSC-8B cells and hepatocytes. Note that expression of MMP-13 mRNA increased as the ratio of hepatocytes/CFSC-8B was increased in co-culture. Values were maximal above a ratio of 5:1 and remained elevated even at ratios of 10:1. Similarly, the presence of MMP-13 protein in culture media was detectable only with co-cultures containing five hepatocytes/CFSC-8B. Note that while MMP-13 present in the culture media of CFSC-8B cultured alone corresponds to procollagenase, that collected from the co-cultures is the active form. The intensity of the signals do not reflect the actual concentration of MMPs. Western blots correspond to two distinct experiments, different volumes of culture medium were concentrated (10-fold versus 25-fold) and different development times were used to detect the protein.
Expression of MMP-13 mRNA in this co-culture system was accompanied by production and secretion of the corresponding protein to the culture medium as determined by Western blot analysis (Figure 6B) ▶ . Because, basal levels of expression of MMP-13 mRNA in CFSC-8B monocultures were very low, 15 ml of conditioned culture medium obtained from ∼25 × 106 cells were precipitated to detect the protein (see Materials and Methods), whereas only 5 ml of medium were needed to detect the protein in co-cultures. Figure 6B ▶ shows that whereas the culture media of CFSC-8B monocultures contained pro-MMP-13 with an apparent molecular mass of 64.5 kd, that present in the culture medium of the co-cultures contained the processed enzyme with an apparent molecular mass of 49 kd.
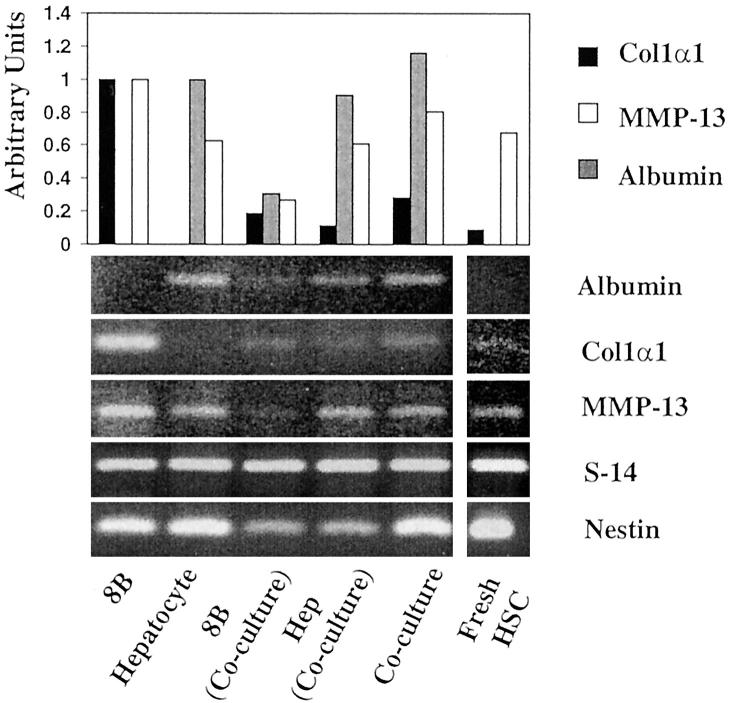
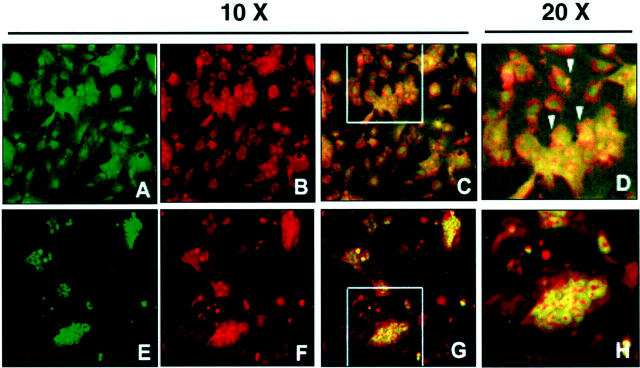
Because the apparent low levels of expression of α1(I) collagen mRNA by co-cultures could result from the large contribution of hepatocyte mRNA to total RNA extracted from the cells, we extracted RNA from cells isolated after collagenase digestion and differential centrifugation. As illustrated in Figure 7 ▶ , HSC fraction was relatively free of hepatocytes as demonstrated by the very low levels of expression of albumin mRNA by RT-PCR. Nonetheless, this cell fraction expressed nestin mRNA, a marker for HSCs. In contrast to this result, the hepatocyte fraction contained some residual HSCs, and therefore expressed α1(I) collagen and nestin mRNAs. It is noteworthy to mention, however, that expression of α1(I) collagen mRNA in the purified HSC fraction was very low as compared to that expressed by CFSC-8B before co-culturing with hepatocytes and these cells did not express significant levels of MMP-13. Conversely, MMP-13 mRNA expression was significantly increased in the population of HSCs that remained attached to hepatocytes (Figure 8) ▶ . Because this fraction contained both cell types it was important to determine whether hepatocytes were also contributing to the production of MMP-13 mRNA. Therefore, we grew co-cultures in coverslips and incubated them with antibodies to MMP-13 and albumin for the detection of MMP-13 within the hepatocytes and desmin and MMP-13 antibodies for detecting MMP-13 within HSCs. As illustrated in Figure 8 ▶ , hepatocytes, as determined by the presence of immunoreactive albumin do not appear to contain significant amounts of MMP-13. However, CFSC-8Bs, as characterized by desmin staining express the immunoreactive enzyme. As expected, there is staining outside the CFSC-8B that likely corresponds to secreted enzyme that remains bound to extracellular matrix components produced by the co-cultures.
Figure 7.
RT-PCR analysis of transcripts of albumin, α1(I) collagen, MMP-13, S14, and nestin expressed by monocultures of CFSC-8B(8B), hepatocytes, freshly isolated HSCs, co-cultures of hepatocytes and CFSC-8B, and CFSC-8B (8B, co-culture), and hepatocytes (Hep Co-culture) isolated after co-culturing the cells for 48 hours. All of the reactions were performed as described using forward and reverse primers of the control gene (S14) and that under investigation. Although signal intensities for the same gene are comparable, signal intensities of the various genes investigated are not. The number of cycles required for optimal amplification of each gene was different (see Materials and Methods). The bars correspond to duplicate experiments. Although the cells isolated after co-culture are contaminated, nonetheless the data show that collagen gene expression by HSCs obtained from co-cultures is very low as compared to monocultured cells. It is noteworthy to indicate that HSCs contaminating the hepatocyte fraction express the highest levels of MMP-13 mRNA.
Figure 8.
Confocal immunomicroscopy of co-cultures of hepatocytes with CFSC-8B. The cells were co-cultured for 48 hours and fixed as described. The upper row (A–D) corresponds to double-antibody staining for desmin (HSC marker, green) and MMP-13 (red). The lower row (E–H) represents double staining for albumin (hepatocyte marker, green) and MMP-13 (red). A–C and E–G were taken with ×10 magnification. D and H are a ×2 enlargement of the areas indicated in C and G, respectively. Please note that orange-yellow color (arrows) is only observed in HSCs and not in hepatocytes.
To determine whether injuring hepatocytes in this co-culture system would result in changes in MMP-13 mRNA expression, we added 5 mmol/L of galactosamine to the co-cultures and measured 24 hours later the expression of MMP-13 mRNA. As illustrated in Figure 9 ▶ , co-cultures treated with 5 mmol/L of galactosamine contained more lactic dehydrogenase in the culture medium, thus suggesting that hepatocytes were indeed injured by the amino sugar. Concomitantly, the expression of MMP-13 mRNA was increased.
Figure 9.
This figure corresponds to duplicate experiments in which co-cultures of hepatocytes and CFSC-8B were treated with 5 mmol/L of galactosamine. The table shows that treatment with galactosamine induced hepatocyte injury as determined by the increase in LDH activity in the culture medium. The Northern blot is a representative blot showing the increased expression of MMP-13 in galactosamine-treated as compared to control co-cultures.
Expression of α1(I) Collagen and MMP-13 mRNAs Is Reciprocally Modulated in Early Passaged Mouse HSCs
Cell lines are excellent models to study cell physiology. However, they may differ from freshly isolated and/or early passaged cells in some functions. Therefore, we considered it important to determine whether the reciprocal modulation of MMP-13 and α1(I) collagen mRNA observed in HSC lines also occurred in early passaged mouse cultured with either TNF-α or TGF-β for 24 hours. As shown in Figure 10 ▶ , expression of MMP-13 mRNA was increased in cells treated with TNF-α whereas that of α1(I) collagen mRNA was decreased. Conversely, in cells treated with TGF-β1, expression of α1(I) collagen mRNA was elevated and that of MMP-13 mRNA was decreased.
Figure 10.
Data presented in this figure support the notion that there is a reciprocal modulation of α1(I) collagen and MMP-13 mRNAs in early passaged (five to six times) mouse HSCs treated with 20 ng/ml of TNF-α or 8 ng/ml of TGF-β1 for 12 hours. The figure illustrates that while TNF-α induced the expression of MMP-13 mRNA it inhibited that of α1(I) collagen, TGF-β1 induced the expression of α1(I) collagen mRNA and down-regulated that of MMP-13.
Discussion
Resolution of liver fibrosis is a complex process requiring discontinuation of the injurious agent and restoration of the proper balance of factors necessary to decrease deposition and enhance degradation of collagen. 7,9,40,41 This balance could be restored either by decreasing production of inhibitors of MMPs, namely TIMPs, or by increasing production and/or activity of MMPs. Of the many MMPs described thus far, only interstitial MMPs, such at types 1, 8, and 13 have the potential of degrading native type I collagen, and therefore, in playing a role in resolution of liver fibrosis. Of these MMPs, type 8 is expressed by neutrophils, 42 whereas fibroblasts and HSCs secrete MMP-13. 43 However, although HSCs can produce MMP-13 and secrete the proenzyme, as shown in this communication, they are unable to convert it to active MMP-13. Thus, an additional component necessary to activate collagen degradation is the contribution of other cells in the production of the proteolytic activities needed to activate MMPs. 44 Therefore, the process is complex and involves different cell types, various proteolytic activities and their inhibitors. As shown in this communication, the culture medium of co-cultures of HSCs and hepatocytes contain the processed enzyme, as determined by its apparent molecular weight. Although hepatocytes produce factors necessary for activation of proMMP-13, we cannot rule out from our experiments whether hepatocytes induce in HSCs the proteolytic cascade needed for activation of pro-MMP-13.
We have previously shown that HSC clones derived from a cell line obtained from a single CCl4-cirrhotic rat liver are heterogeneous regarding expression of type I collagen genes. 24 In this communication we extended our observations and demonstrated that although all of the cells express TIMP-1, only one of them, CFSC-8B expresses MMP-2 and MMP-13 and responds to TNF-α with increased expression of MMP-13. The phenotype of the HSCs that expressed MMPs was similar to freshly isolated HSCs. Conversely, those HSCs that expressed high levels of α1(I) procollagen also produced high levels of elastin mRNA (data not shown), a marker of active fibrogenesis in vivo 45 and their phenotype resembled that of myofibroblasts. This HSC heterogeneity observed in our cell lines has been also shown to occur in vivo. 4 HSCs are quite heterogeneous regarding expression of desmin and α-smooth muscle actin in vivo 4,46 and significant differences in susceptibility to programmed cell death in myofibroblasts as compared to HSCs have been reported. 21 Unfortunately, we do not have specific markers to determine whether extreme phenotypes such as those reported in this communication exist. Thus, the results obtained need to be interpreted with caution because experiments were performed with HSC lines and clones, and these could represent extreme situations occurring in immortalized cells. Nonetheless, the HSCs express bona fide markers of activated HSCs, and therefore, this possibility, namely the presence of diverse HSC phenotypes needs to be further investigated. Moreover, they also appear to reflect events occurring in vivo and in vitro in different experimental conditions. 8,25
As also shown in this communication, expression of MMP-13 by CFSC-8B was accompanied by a decline in α1(I) procollagen gene expression. This and other findings 8,47 strongly suggest that α1(I) procollagen and MMP-13 mRNAs are reciprocally modulated and therefore, transcription factors required for the activation of collagen may down-regulate MMPs and vice versa. A similar reciprocal modulation was observed in early passaged HSCs treated with TNF-α or TGF-β1. Indeed, recent findings suggest that Smad3, a transcription factor required for TGF-β1-mediated up-regulation of type I collagen genes, is involved in down-regulation of MMP-1 expression by an indirect mechanisms involving the co-transcriptional regulator p300. 48 This reciprocal modulation of collagen and MMPs has been also demonstrated in vivo. Studies performed during healing of wounds have revealed that expression of MMP-13 mRNA precedes α1(I) procollagen gene up-regulation and that MMP-13 expression decreases substantially when collagen deposition reaches maximal levels. 25 Altogether, these findings suggest that in healing responses, there is activation of several concerted mechanisms that can result in excess deposition of scar tissue. These include the simultaneous up-regulation of TIMP-1 and type I collagen mRNAs and down-regulation of MMPs involved in degradation of interstitial collagens.
The data presented in this communication demonstrated that only those HSCs with phenotypes resembling early passaged HSCs (CFSC-8B) expressed MMP-2 and MMP-13 mRNAs, and that levels of expression of these MMPs were up regulated by TNF-α. Moreover, co-culturing HSCs with hepatocytes resulted in further up-regulation of MMP-13 mRNA. Because this co-culture system is only functional when cells establish cell-cell contact with formation of gap-junctional complexes, 35 the data suggest that the close relation between hepatocytes and HSCs in the space of Disse could contribute to MMP-13 expression and activation in the liver. However, because of excess collagen deposition in the space of Disse this close association between the two cell types is lost in fibrotic livers, a factor that could further contribute to decreased MMP production and/or activation.
In summary, our data indicate that irrespectively of the HSC markers expressed by these cell lines, they have two distinct phenotypes. Those with a myofibroblastic phenotype, based on their morphology and expression of type I collagen and elastin mRNAs (CFSC-3H and CFSC-5H), are unable to produce MMPs to degrade extracellular matrix. On the other hand, HSCs resembling activated HSCs, but without a full myofibroblastic phenotype (CFSC-8B), express type I collagen and MMPs. However, when these cells are engaged in type I collagen production, they significantly down-regulate MMP-13 mRNA expression and vice versa. Thus, these two mRNAs appear to be reciprocally modulated. A better understanding of the molecular events involved in this reciprocal modulation could lead to the development of novel targets for anti-fibrogenic therapy.
Acknowledgments
We thank Dr. József Czégé from the Biomedical Instrumentation Center at the Uniformed Services University of the Health Sciences for expert technical assistance in the use of the confocal microscope.
Footnotes
Address reprint requests to Marcos Rojkind, M.D., Ph.D., Chief, Experimental Pathology Section (T2-209), Department of Clinical Investigation, Walter Reed Army Medical Center, 6900 Georgia Ave., NW, Washington, DC 20307. E-mail: marcos.rojkind@na.amedd.army.mil.
Supported by the National Institute of Alcohol Abuse and Alcoholism (grants RO1AA09231 and RO1AA10541 to M. R., and RO1AA12196 to P. G.).
B. S. was on leave from the Department of Medicine I, Klinikum B. Franklin, Free University of Berlin, Hindenburgdamm 30, 12200 Berlin, Germany, and his stay in NY was supported in part with fellowships from the German Academic Exchange Service (DAAD), Bonn, Germany, and the Boehringer Ingelheim Foundation, Stuttgart, Germany.
“The opinions or assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting the views of the Department of the Army or the Department of Defense.”
The present address of A. M. R. E. is Lady Davis Institute for Medical Research, McGill University, Montreal Canada.
The present address of D. S. is Department of Medicine I Friedrich-Alexander-University Erlangen-Nürnberg Krankenhausstrasse 12 91054 Erlangen Germany.
The present address of P. G. is Center for Scientific Review, National Institutes of Health, Bethesda MD.
The present address of N.M.-C. is Dirección de Investigación, Instituto Nacional de Perinatología, Lomas Virreyes, Mexico DF.
The present address of M.A. is Universidad Juárez de Durango, Facultad de Medicina Durango, Mexico.
References
- 1.Eng FJ, Friedman SL: Fibrogenesis I: new insights into hepatic stellate cell activation: the simple becomes complex. Am J Physiol 2000, 279:G7-G11 [DOI] [PubMed] [Google Scholar]
- 2.Pinzani M, Marra F: Cytokine receptors and signaling in hepatic stellate cells. Semin Liver Dis 2001, 21:397-416 [DOI] [PubMed] [Google Scholar]
- 3.Rockey DC: Hepatic blood flow regulation by stellate cells in normal and injured liver. Semin Liver Dis 2001, 21:337-349 [DOI] [PubMed] [Google Scholar]
- 4.Geerts A: History, heterogeneity, developmental biology, and functions of quiescent hepatic stellate cells. Semin Liver Dis 2001, 21:311-335 [DOI] [PubMed] [Google Scholar]
- 5.Ogata I, Saez CG, Greenwel P, Ponce ML, Geerts A, Leinwand L, Rojkind M: Rat liver fat-storing cell lines express sarcomeric myosin heavy chain mRNA and protein. Cell Motil Cytoskel 1993, 26:125-132 [DOI] [PubMed] [Google Scholar]
- 6.Maruyama K, Feinman L, Okazaki I, Lieber CS: Direct measurement of neutral collagenase activity in homogenates from baboon and human liver. Biochim Biophys Acta 1981, 658:124-131 [DOI] [PubMed] [Google Scholar]
- 7.Maruyama K, Feinman L, Fainsilber Z, Nakano M, Okazaki I, Lieber CS: Mammalian collagenase increases in early alcoholic liver disease and decreases with cirrhosis. Life Sci 1982, 30:1379-1384 [DOI] [PubMed] [Google Scholar]
- 8.Rojkind M: Role of metalloproteinases in liver fibrosis. Alcohol Clin Exp Res 1999, 23:934-939 [PubMed] [Google Scholar]
- 9.Vater CA, Harris ED, Jr, Siegel RC: Native cross-links in collagen fibrils induce resistance to human synovial collagenase. Biochem J 1979, 181:639-649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knittel T, Mehde M, Kobold D, Saile B, Dinter C, Ramadori G: Expression patterns of matrix metalloproteinases and their inhibitors in parenchymal and non-parenchymal cells of rat liver: regulation by TNF-alpha and TGF-beta1. J Hepatol 1999, 30:48-60 [DOI] [PubMed] [Google Scholar]
- 11.Knittel T, Mehde M, Grundmann A, Saile B, Scharf JG, Ramadori G: Expression of matrix metalloproteinases and their inhibitors during hepatic tissue repair in the rat. Histochem Cell Biol 2000, 113:443-453 [DOI] [PubMed] [Google Scholar]
- 12.Arthur MJ, Iredale JP, Mann DA: Tissue inhibitors of metalloproteinases: role in liver fibrosis and alcoholic liver disease. Alcohol Clin Exp Res 1999, 23:940-943 [DOI] [PubMed] [Google Scholar]
- 13.McCrudden R, Iredale JP: Liver fibrosis, the hepatic stellate cell and tissue inhibitors of metalloproteinases. Histol Histopathol 2000, 15:1159-1168 [DOI] [PubMed] [Google Scholar]
- 14.Arthur MJ: Fibrogenesis II: metalloproteinases and their inhibitors in liver fibrosis. Am J Physiol 2000, 279:G245-G249 [DOI] [PubMed] [Google Scholar]
- 15.Benyon RC, Arthur MJ: Extracellular matrix degradation and the role of hepatic stellate cells. Semin Liver Dis 2001, 21:373-384 [DOI] [PubMed] [Google Scholar]
- 16.Arthur MJ, Mann DA, Iredale JP: Tissue inhibitors of metalloproteinases, hepatic stellate cells and liver fibrosis. J Gastroenterol Hepatol 1998, 13:S33-S38 [DOI] [PubMed] [Google Scholar]
- 17.Yoshiji H, Kuriyama S, Miyamoto Y, Thorgeirsson UP, Gomez DE, Kawata M, Yoshii J, Ikenaka Y, Noguchi R, Tsujinoue H, Nakatani T, Thorgeirsson SS, Fukui H: Tissue inhibitor of metalloproteinases-1 promotes liver fibrosis development in a transgenic mouse model. Hepatology 2000, 32:1248-1254 [DOI] [PubMed] [Google Scholar]
- 18.Iredale JP, Benyon RC, Arthur MJ, Ferris WF, Alcolado R, Winwood PJ, Clark N, Murphy G: Tissue inhibitor of metalloproteinase-1 messenger RNA expression is enhanced relative to interstitial collagenase messenger RNA in experimental liver injury and fibrosis. Hepatology 1996, 24:176-184 [DOI] [PubMed] [Google Scholar]
- 19.Greenwel P, Rojkind M: Accelerated development of liver fibrosis in CCl4-treated rats by the weekly induction of acute phase response episodes: upregulation of alpha1(I) procollagen and tissue inhibitor of metalloproteinase-1 mRNAs. Biochim Biophys Acta 1997, 1361:177-184 [DOI] [PubMed] [Google Scholar]
- 20.Iredale JP, Benyon RC, Pickering J, McCullen M, Northrop M, Pawley S, Hovell C, Arthur MJ: Mechanisms of spontaneous resolution of rat liver fibrosis. Hepatic stellate cell apoptosis and reduced hepatic expression of metalloproteinase inhibitors. J Clin Invest 1998, 102:538-549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saile B, Knittel T, Matthes N, Schott P, Ramadori G: CD95/CD95L-mediated apoptosis of the hepatic stellate cell. A mechanism terminating uncontrolled hepatic stellate cell proliferation during hepatic tissue repair. Am J Pathol 1997, 151:1265-1272 [PMC free article] [PubMed] [Google Scholar]
- 22.Iredale JP: Hepatic stellate cell behavior during resolution of liver injury. Semin Liver Dis 2001, 21:427-436 [DOI] [PubMed] [Google Scholar]
- 23.Greenwel P, Schwartz M, Rosas M, Peyrol S, Grimaud JA, Rojkind M: Characterization of fat-storing cell lines derived from normal and CCl4-cirrhotic livers: differences in the production of interleukin-6. Lab Invest 1991, 65:644-653 [PubMed] [Google Scholar]
- 24.Greenwel P, Rubin J, Schwartz M, Hertzberg E, Rojkind M: Liver fat-storing cell clones obtained from a CCl4-cirrhotic rat are heterogeneous with regard to proliferation, expression of extracellular matrix components, interleukin-6 and connexin 43. Lab Invest 1993, 69:210-216 [PubMed] [Google Scholar]
- 25.Liu X, Levenson SM, Chang TH, Steinberg JJ, Imegwu O, Rojkind M: Molecular mechanisms underlying wound healing acceleration by Staphylococcus aureus peptidoglycan. Wound Repair Regen 1996, 4:470-476 [DOI] [PubMed] [Google Scholar]
- 26.Inagaki Y, Truter S, Garret LA, De Crombrugghe B, Nemoto T, Kobayashi K, Greenwel P: Activation of proα2(I) collagen promoter during hepatic fibrogenesis in transgenic mice. Biochem Biophys Res Commun 1998, 250:606-611 [DOI] [PubMed] [Google Scholar]
- 27.Chomczynski P, Sacchi N: Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 1987, 162:156-159 [DOI] [PubMed] [Google Scholar]
- 28.Genovese C, Rowe D, Kream B: Construction of DNA sequences complementary to rat α(1) and α(2) collagens mRNA and their use in studying the regulation of type I collagen synthesis by 1,25-dihydroxyvitamin D. Biochemistry 1984, 23:6210-6216 [DOI] [PubMed] [Google Scholar]
- 29.Schwarzbauer JE, Tamkun JW, Lemischka IR, Hynes RO: Three different fibronectin mRNAs arise by alternative splicing within the coding region. Cell 1983, 35:421-431 [DOI] [PubMed] [Google Scholar]
- 30.Henriet P, Rousseau GG, Eeckhout Y: Cloning and sequencing of mouse collagenase cDNA. Divergence of mouse and rat collagenases from the other mammalian collagenases. FEBS Lett 1992, 310:175-178 [DOI] [PubMed] [Google Scholar]
- 31.Reponen P, Sahlberg C, Huhtala P, Hurskainen T, Thesleff I, Tryggvason K: Molecular cloning of murine 72-kDa type IV collagenase and its expression during mouse development. J Biol Chem 1992, 267:7856-7862 [PubMed] [Google Scholar]
- 32.Coulombe B, Skup D: In vitro synthesis of the active tissue inhibitor of metalloproteinases encoded by a complementary DNA from virus-infected murine fibroblasts. J Biol Chem 1988, 263:1439-1443 [PubMed] [Google Scholar]
- 33.Thesleff I, Tryggvason K: High expression of 92-kDa type IV collagenase (gelatinase B) in the osteoclasts lineage during mouse development. J Cell Biol 1994, 124:1091-1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beverley SM: Amplification of RNA by PCR. Ausubel FM Brent R Kingston RE Moore DD Seidman JG Smith JA Struhl K eds. Current Protocols in Molecular Biology, 1992, vol 2.:pp 15.4.1-15.4.6 Wiley Interscience, New York [Google Scholar]
- 35.Rojkind M, Novikoff PM, Greenwel P, Rubin J, Rojas-Valencia L, Campos de Carvalho A, Stockert R, Spray D, Hertzberg EL, Wolkoff A: Characterization and functional studies on rat liver fat-storing cell line and freshly isolated hepatocyte co-culture system. Am J Pathol 1995, 276:1508-1520 [PMC free article] [PubMed] [Google Scholar]
- 36.Fontana L, Jerez D, Rojas-Valenica L, Solis-Herruzo A, Greenwel P, Rojkind M: Ethanol induces the expression of α1(I) procollagen mRNA in a co-culture system containing a liver stellate cell-line and freshly isolated hepatocytes. Biochem Biophys Acta 1997, 1362:135-144 [DOI] [PubMed] [Google Scholar]
- 37.Neufeld DS: Isolation of rat hepatocytes. Methods Mol Biol 1997, 75:145-151 [DOI] [PubMed] [Google Scholar]
- 38.Laemmli UK: Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227:680-685 [DOI] [PubMed] [Google Scholar]
- 39.Rojkind M, Greenwel P: The liver as a bioecological system. Arias IM Jakoby WB Popper H Schachter D Shafritz DA eds. The Liver Biology and Pathobiology, ed 2 1988:pp 1269-1285 Raven Press Ltd., New York
- 40.Perez-Tamayo R, Montfort I, Gonzalez E: Collagenolytic activity in experimental cirrhosis of the liver. Exp Mol Pathol 1987, 47:300-308 [DOI] [PubMed] [Google Scholar]
- 41.Montfort I, Perez-Tamayo R, Alvizouri AM, Tello E: Collagenase of hepatocytes and sinusoidal liver cells in the reversibility of experimental cirrhosis of the liver. Virchows Arch B Cell Pathol Incl Mol Pathol 1990, 59:281-289 [DOI] [PubMed] [Google Scholar]
- 42.Baramova E, Foidart JM: Matrix metalloproteinase family. Cell Biol Int 1995, 19:239-242 [PubMed] [Google Scholar]
- 43.Murphy G, Knauper V, Cowell S, Hembry R, Stanton H, Butler G, Freije J, Pendas AM, Lopez-Otin C: Evaluation of some newer matrix metalloproteinases. Ann NY Acad Sci 1999, 878:25-39 [DOI] [PubMed] [Google Scholar]
- 44.Maruyama K, Okazaki I, Kobayashi T, Suzuki H, Kashiwazaki K, Tsuchiya M: Collagenase production by rabbit liver cells in monolayer culture. J Lab Clin Med 1983, 102:543-550 [PubMed] [Google Scholar]
- 45.Scheuer PJ, Maggi G: Hepatic fibrosis and collapse: histological distinction by orecin staining. Histopathology 1980, 4:487-490 [DOI] [PubMed] [Google Scholar]
- 46.Schmitt-Graff A, Desmouliere A, Gabbiani G: Heterogeneity of myofibroblast phenotypic features: an example of fibroblastic cell plasticity. Virchows Arch 1994, 425:3-24 [DOI] [PubMed] [Google Scholar]
- 47.Liu X, Chang TH, Levenson SM, Rojkind M: Wound fluids from saline- and S. aureus peptidoglycan inoculated sponges induce expression of metalloproteinase 13 (MMP-13) mRNA by cultured rat fibroblasts. Wound Repair Regen 1997, 5:348-354 [DOI] [PubMed] [Google Scholar]
- 48.Yuan W, Varga J: Transforming growth factor-beta repression of matrix metalloproteinase-1 in dermal fibroblasts involves Smad3. J Biol Chem 2001, 276:38502-38510 [DOI] [PubMed] [Google Scholar]