Abstract
We screened expressed sequence tag databases for genes with up-regulated expression in inflammatory bowel diseases. A gene encoding a regenerating protein (REG)-like protein called RELP was identified and characterized. The relp gene encodes a major transcript of 1518 nucleotides, and two truncated splice variants. Unlike the reg genes, which form a cluster in chromosome 2, relp maps to chromosome 1p12–13.1. The predicted translation product is a 158-amino acid preprotein, showing 43% to 47% similarity to the REG proteins. It contains a 22-amino acid signal peptide, and a conserved calcium-dependent carbohydrate-recognition domain. Complementary DNA for the orthologous mouse gene was also cloned. The RELP protein is constitutively expressed in epithelial neuroendocrine cells of the small intestine and in parietal cells of the gastric mucosa. An up-regulated expression of RELP was seen in epithelial cells of inflammatory mucosa in ulcerative colitis and Crohn’s disease, in regenerating epithelial borders of gastric ulcers, and in metaplastic epithelium in the antrum and the esophagus. Our findings suggest that RELP might be involved in inflammatory and metaplastic responses of the gastrointestinal epithelium.
Inflammatory bowel diseases (IBDs) are thought to result from an inappropriate activation of the mucosal immune system. 1-3 Several known susceptibility genes, such as NOD2, an activator of the nuclear factor-κB4, CD11 integrins, IFN-γ, NRAMP2, T-cell receptor α/δ complex, and MHC 2 are functional parts of the immune surveillance machinery that might be implicated in the development of IBDs. The molecular pathways associated with the tissue injury and regeneration in the mucosal epithelium, although equally important to the disease process, are less well-known. The risk of colorectal cancer is elevated both in patients with ulcerative colitis 5 and with Crohn’s disease. 6 This indicates that the epithelial cells are not inactive bystanders in the ongoing inflammation, but are engaged in the cross-talk between external stimuli and mucosal responses. Oxygen radicals produced by the neutrophils in the inflamed mucosa are genotoxic and contribute to the increased risk of oncogenic transformation in IBDs. 7,8
We screened an Incyte LifeSeq database (Incyte Pharmaceuticals, Palo Alto, CA) for transcripts especially abundant in the gastrointestinal tract of ulcerative colitis- and Crohn’s disease-derived libraries. The aim was to find proteins associated with the IBD-related regenerative responses. Regenerating protein-like protein (RELP) was selected for detailed characterization because of its abundant expression in the libraries of interest, and of its homology with cell-growth- and migration-associated regenerating (REG) proteins. 9-21 The structure of the gene and the predicted protein product, as well as the expression of the RELP protein in normal tissues and IBDs were investigated.
Materials and Methods
cDNA Cloning and Sequencing
RELP mRNA was detected as a cluster of expressed sequence tags (EST) in small-intestine- and colon-derived libraries in the Life Seq database (Incyte Pharmaceuticals). A full-length cDNA insert, encoding for the predicted RELP preprotein, was acquired and verified by sequencing. The human RELP nucleotide sequence data reported in this paper have been submitted to the GenBank/EMBL/DNA databases under accession numbers AY126670-AY126672.
A blast search in the NCBI EST database using human RELP cDNA yielded three highly homologous mouse sequences. The corresponding clones (IMAGE clone IDs 717371, 1079498, and 1096767) were acquired and sequenced. The cDNA for the mouse orthologue of RELP was subcloned into a pCR 2.1-TOPO vector (Invitrogen, Carlsbad, CA).
Antibodies
A C-terminal (CAEMSSNNNFLTWSSNE) RELP-derived peptide was synthesized, coupled to keyhole limpet hemocyanin, and used to produce polyclonal antibodies in rabbits. The sera were tested for reactivity against the peptide with enzyme-linked immunosorbent assay (ELISA), and the positive batches were affinity-purified. Anti-FLAG epitope antibodies were obtained from Sigma (ANTI-FLAG M2 Monoclonal Antibody; St. Louis, MO), and monoclonal mouse antibodies against chromogranin A from Chemicon (Temecula, CA). Secondary antibodies, HRP-conjugated swine anti-rabbit and rabbit anti-mouse immunoglobulins (IgGs), TRITC-conjugated swine anti-rabbit IgGs, and fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse Ig were obtained from Dako (Glostrup, Denmark).
In Situ Hybridization
Human tissue samples were obtained from the Department of Pathology, University of Helsinki, in accordance with the Ethical Committee of the University Hospital. Formalin-fixed, paraffin-embedded tissue samples were cut into 5- to 7-μm sections, mounted on silane-coated glass slides, deparaffinized in xylene, and rehydrated through a graded series of ethanol solutions (100% to 70%). Thereafter, the samples were permeabilized, treated with proteinase K (Finnzymes, Helsinki, Finland), and postfixed in 3% paraformaldehyde. Positive charges were blocked by soaking the slides in 0.25% (v/v) acetic anhydride in 100 mmol/L triethanolamine. Finally, the slides were equilibrated in a solution of 4X standard saline citrate (SSC; 1X SSC is 150 mmol/L NaCl, 15 mmol/L sodium citrate, pH 7.0)/50% (v/v) deionized formamide at 37°C for 10 minutes. Prehybridization and hybridization were carried out in a solution composed of 40% formamide, 10% dextran sulfate, 4X SSC, 10 mmol/L dithiothreitol (DTT), 1X Denhardt’s solution (0.2 mg/ml each, bovine serum albumin, polyvinylpyrrolidone, and Ficoll), 1 mg/ml herring sperm DNA, and 1 mg/ml yeast tRNA. Relp-specific probes were prepared using the TA cloning kit (Invitrogen, San Diego, CA). A polymerase chain reaction (PCR)-amplified 0.4-kb RELP cDNA insert was cloned into the pCR-II vector (Invitrogen), and used as a template for antisense and sense digoxigenin-labeled RNA probes (RNA Labeling Kit; Boehringer-Mannheim, Mannheim, Germany). After hybridization the slides were washed twice in 2X SSC at 37°C for 5 minutes and then in 1X SSC at 37°C for 10 minutes followed by incubation with 5 μg/ml RnaseA (Boehringer-Mannheim) at 37°C for 30 minutes. After washing twice in 0.5X SSC at room temperature for 10 minutes the sections were incubated with alkaline phosphatase-conjugated anti-digoxigenin-AP Fab Fragments (dilution 1:250; Boehringer-Mannheim). The signals were visualized by incubating the sections in NBT/BCIP Stock Solution (Boehringer-Mannheim) for 1.5 hours.
Immunohistochemistry
Four-μm sections from formalin-fixed, paraffin-embedded tissues were mounted on 3-aminopropyl-triethoxy-silane-coated slides (Sigma), deparaffinized, and heated twice for 5 minutes in a microwave oven (650W) before exposure to the first antibody (polyclonal rabbit anti-RELP peptide antibody, dilution 1:2000). Elite ABC Kit (Vectastain; Vector Laboratories, Burlingame, CA) was used for immunoperoxidase staining, visualized with 3-amino-9-ethylcarbazole (Sigma).
Nucleotide Sequencing of the Human Relp Gene
A human genomic plasmid artificial chromosome (PAC) clone containing the genomic sequence of relp was obtained from GenomeSystems, Inc. (St.Louis, MO). The 120-kb insert was amplified by PCR with RELP-specific primers, subcloned into a TA vector (Invitrogen), and sequenced with vector-derived and RELP-specific primers. The sequencing data were used to complement and verify the genomic sequence derived from the public database.
Fluorescence in Situ Hybridization (FISH)
A human genomic PAC clone containing the relp gene was used as a probe to localize the relp gene in the human chromosomes. The PAC plasmid was labeled with biotin-16-dUTP by nick translation. Slides with human interphase and metaphase nuclei were hybridized with the probes, and the signal was detected with avidin-conjugated FITC and amplified with biotinylated anti-avidin antibodies. After washing, the slides were counterstained with 4,6-diamino-2-phenyl-indole (DAPI; ICN Biomedicals, Costa Mesa, CA), and mounted in an antifade solution.
Dot Blot and Northern Blot Analyses
Human Multiple-Tissue Expression (MTE) Array and Multiple Tissue Northern (MTN) blots Human II and Human III products (Clontech Laboratories, Inc., Palo Alto, CA) were used for dot blot and Northern blot analyses, with 32P-labeled (Multiprime DNA Labeling System kit; Amersham Pharmacia Biosciences, Inc., Helsinki, Finland), full-length RELP cDNA as a probe. For autoradiography, filters were exposed to Kodak Biomax MS film for 1 to 3 days.
In Vitro Translation
A cDNA fragment containing the full-length sequence of RELP cDNA was subcloned into the eukaryotic expression vector pCDNA 3 (Invitrogen) for expression regulated by the T7 RNA polymerase promoter. Rabbit reticulocyte lysate, with or without canine pancreatic microsomal membranes (Promega, Madison, WI), in the presence of 35S-methionine (Amersham International’s Redivue L-35S-methionine, Amersham Pharmacia Biosciences, Inc.), was used for the induction of expression. Proteins obtained by in vitro translation were analyzed by sodium dodecyl sulfate polyacrylamide (12%) gel electrophoresis (SDS-PAGE), and visualized by autoradiography.
RELP Protein Expression in Cell Lines, Western Blot, and Immunofluorescence Staining
A cDNA-encoding RELP-C-terminal FLAG epitope fusion protein was amplified using 5′ AGCATGGCTTCCAGAAGCATG 3′ and 5′ CTACTTGTCATCGTCATCCTTGTAATCTGGTCGGTACTTGCA 3′ as forward and reverse primers, respectively, and cloned into a pcDNA 3.1/V5-HisTOPO vector (Invitrogen). Construct was transfected into COS 7 cells using Transfectam reagent (Promega, Madison, WI) according to the manufacturer’s instructions. The cells were lysed in Pawson’s buffer (50 mmol/L HEPES pH 7.0, 150 mmol/L NaCl, 10% glycerol, 1% Triton X-100, 1.5 mmol/L MgCl2, 1 mmol/L ethylene glycol-bis-N′N′N′N′-tetraacetic acid (EGTA), 100 mmol/L NaF, 10 mmol/L Na4P2O7). The proteins were separated by 12% SDS-PAGE and analyzed by Western blotting using the affinity-purified rabbit anti-RELP C-terminal peptide antibodies and monoclonal anti-FLAG antibodies. Horseradish peroxidase (HRP)-conjugated swine anti-rabbit and rabbit anti-mouse immunoglobulins were used as secondary antibodies and detected by enhanced chemiluminescence.
MKN45, a gastric cancer cell line cell, was a gift from Dr. A. Ristimäki (Molecular/Cancer Laboratory, Biomedicum, University of Helsinki). The MKN45 cells were transiently transfected to express the RELP-C-terminal FLAG epitope fusion protein, fixed with 3.5% paraformaldehyde, and permeabilized with 0.05% NP-40 in PBS. Anti-FLAG antibodies (20 μg/ml) and FITC-conjugated goat anti-mouse antibodies (1:30) were used to visualize the fusion protein in the transfected cells. The images were obtained using a confocal microscope and associated software (Leica SP2 Laser scanning microscope; Leica Microsystems, Jena, Germany).
Double Immunofluorescence Staining
Tissue sections of formalin-fixed and paraffin-embedded normal duodenal mucosa were double-stained with the polyclonal rabbit peptide antibody against RELP (1:30; 25 μg/ml), and a monoclonal mouse antibody against chromogranin A (1:5000; 0.2 μg/ml), detected with tetramethylrhodamine isothiocyanate-conjugated swine anti-rabbit immunoglobulins and FITC-conjugated goat anti-mouse immunoglobulins. For controls, primary antibodies were replaced with the IgG fractions of non-immune rabbit and mouse sera.
Results
Chromosomal Localization and Structure of the Relp Gene
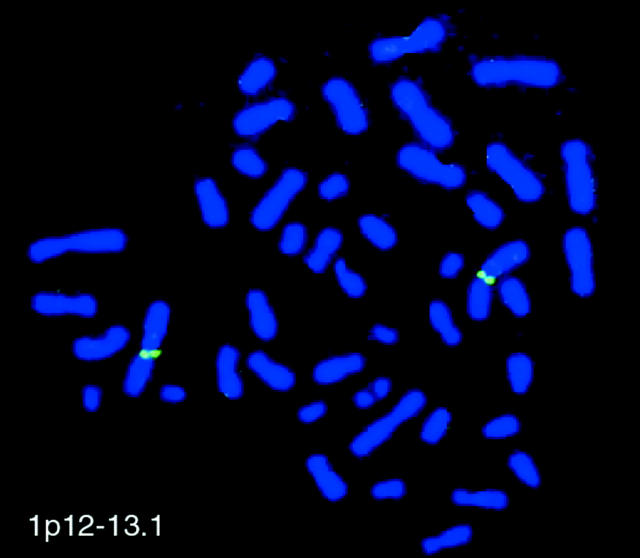
Fluorescent in situ hybridization showed an exclusive signal for the relp-specific probe on chromosome 1 band p12 to 13.1 (Figure 1) ▶ . The result agrees with the electronic localization of the relp gene to human chromosome 1 (Homo sapiens, chromosome 1 clone RP5–10425I8 map p11 to 13.2, GenBank Accession AL359752).
Figure 1.
Chromosomal localization of RELP gene by fluorescence in situ hybridization. Metaphase chromosome spread from human lymphocytes was hybridized with fluorescently-labeled PAC clone containing the RELP gene. The signals locate the gene in chromosome 1p12–13.1.
Partial sequencing data of the human PAC clone containing the relp gene was combined with the GenBank-derived sequencing data, to define the gapless sequence of the gene. The size of the relp gene was determined to comprise 17557 bp. It consists of 7 exons separated by 6 introns (Figure 2A) ▶ . All of the exon-intron junctions follow the GT-AG rule, except for the truncated splice variant 1, where the sixth intron anomalously ends with ml (Table 1) ▶ . The lengths of exons I, II, III, IV, V, VI, and VII are 172, 174, 161, 98 (845 in the truncated splice variant 2), 137, 106, and 669 (183 in the truncated splice variant 1) bp, respectively. Due to differential splicing, exon II is not represented in all transcripts. The first truncated splice variant lacks exon VI and most of exon VII, and the second has an extended exon IV, with an in-frame stop codon. A 2000-kb fragment of the 5′-flanking region of the transcription initiation site of the RELP gene was analyzed by Genomatix (http:genomatix.gsf.de/mat fam) and the Hamming-Clustering method for TATA Signal Prediction in Eukaryotic Genes (http://125.itba.mi.cnr.it/∼webgene/wwwHC TATA.html). No apparent functional promoter was predictable in the region. Putative binding sites for Cdx-2 mammalian caudal-related intestinal transcription factor, interferon regulatory factor-1, steroidogenic factor 1 (SF1), serum response factor (SFR1), GATA binding factor 3 (GATA3), c-Ets-1 and hepatic nuclear factor 4 (HNF4) were found at 250, 232, 213, 156, 151, 144, and 66 nucleotides upstream from the presumed transcription initiation site. The analysis showed no TATA box, suggesting that the transcription might be driven from multiple initiation sites.
Figure 2.

A: Structure of the RELP gene. Exons (boxes), protein coding regions (filled areas), and untranslated regions (open areas) are shown. The second exon, present in a subset of messages only, is shown gray. B: The complete RELP cDNA and the deduced amino acid sequence of the predicted preprotein. The cDNA sequence shows the starting ATG (bold) with Kozaks’ consensus translational start site (underlined), the stop codon (bold), and the polyadenylation signal AATAAA (underlined). The protein sequence shows the putative signal sequence (italics), the predicted glycosylation site (bold), and the carbohydrate-recognizing domain (underlined). Nucleotide sequence and corresponding amino acid sequence of two RELP splice variants are shown.
Table 1.
Exon-Intron Junctions of the reg IV Gene
| Exon no. | mRNA location | Gene location | Exon-intron junctions* |
|---|---|---|---|
| 1 | 1–172 | 1–172 | AAACAGgtaatcctcctcgcggttga |
| 2 | 173–346 | 2379–2552 | tggattttcttgacaacagAATGCC...TTGCAGgtaagaacaaaatctcttaa |
| 3 | 347–507 | 2818–2878 | acctgtgcaaatgtgttcagATTTGC...TGGGTGgtgagtatccatcacgtag |
| 4 | 508–605 | 8414–8511 | tgccttttgtttatgtgcagATATCA...GCCGAGgtaagaaacttgctgcctgc |
| 5 | 606–743 | 11717–11854 | ggacctctccttgtacatagCTCGAG...CAGAAGgtaacctcagacttggccac |
| 6 | 744–849 | 12969–13074 | tccatctcctgttgttgcagAGGCAG...ATAACAgtaagttataatgcaccacc |
| 7 | 850–1452 | 16886–17488 | atctgtttattttcttacagACTTTT |
*, Exon sequences in upper case, introns in lower case.
RELP cDNA and the Primary Structure of the RELP Protein
RELP cDNA comprises 1518 nucleotides. The protein coding region is made up of 474 bp of nucleotides encoding a preprotein of 158 amino acids. The 5′-untranslated and 3′-untranslated regions contain 440 and 601 nucleotides, respectively. A polyadenylation signal (AATAAA) is located 510 bp downstream of the termination codon (Figure 2B) ▶ . The calculated molecular weight of RELP is 18.229 kd, and the isoelectric point is 9.128. The aminoterminus of RELP is highly hydrophobic and contains a putative cleavable signal sequence of 22 amino acids, predicted by the SignalP program (SignalP V1.1 server at http://genome.cbs.dtu.dk/services/SignalP).
In the protein sequence of RELP, residues 37 through 155 represent a conserved calcium-dependent carbohydrate recognition domain (CRD) (Figure 2B) ▶ . The four conserved cysteines and the two optional cysteines are CRD associated, involved in intramolecular disulphide bonds, and are conserved in RELP. Residues 50 through 53 represent a putative N-glycosylation site (NWS; Figure 2B ▶ ). The secondary structure of RELP is predicted to be similar to that of human REG1α, and the global folds of these proteins appear to be closely related, as shown by successful modeling (MSI Insight II) of RELP 3-D structure with the published fold of REG1α as a template (data not shown).
By its primary structure, human RELP is 37% identical and 47% similar to human REG1α, and 34% identical and 43% similar to human pancreatitis-associated protein (pap1). Human RELP is also similar to other single CRD domain proteins, such as chicken ovocleidin (36% identical, 44% similar) and snake venom galactose-specific lectin (36% identical, 43% similar).
Mouse Orthologue of RELP
The mouse orthologue of RELP was identified in a rodent EST database. The cDNA of the putative mouse RELP is 70% identical to that of human RELP (Figure 3A) ▶ and the predicted mouse and human preproteins are 66% identical and 72% similar to each other (Figure 3B) ▶ .
Figure 3.

A: Alignment of the human RELP cDNA coding region with that of the mouse orthologue. Starting ATG and stop codons are underlined. B: Alignment of the human RELP preprotein with the mouse orthologue.
In Vitro Translation of RELP
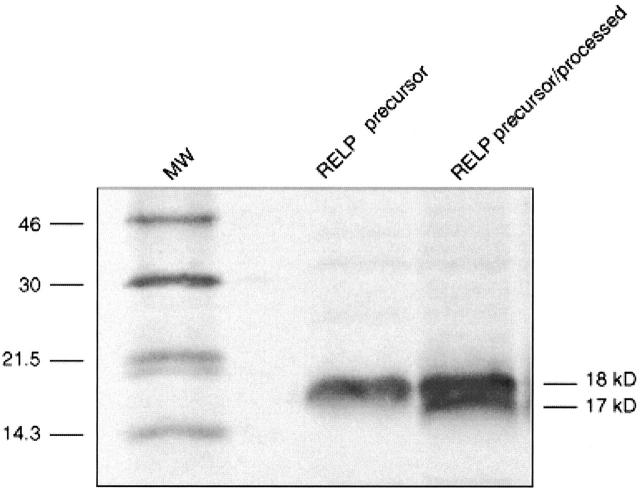
In vitro translation of RELP resulted in a protein product with an apparent molecular weight of 18 kd as analyzed by SDS-PAGE, in agreement with the calculated molecular weight of RELP (18.2 kd). Since RELP was predicted to contain a cleavable signal peptide, the translation reaction was repeated in the presence of canine microsomal membranes to verify the expected processing of the preprotein. SDS-PAGE analysis of the resulting protein products confirmed the cleavage of a signal peptide by establishing the presence of a 17 kd protein, representing the mature form of RELP (Figure 4) ▶ .
Figure 4.
In vitro expression and processing of the RELP protein. The translation product of RELP cDNA and the canine microsomal membrane-processed version were labeled with 35S-methionine and analyzed by SDS-PAGE.
Detection of RELP mRNA by Dot Blot and Northern Blot Analysis in Human Tissues
Dot blot analysis verified the electronic Northern-based finding revealed expression of RELP message mainly in the gastrointestinal tract including the duodenum, jejunum, ileum, ileocecum, appendix, descending colon, pancreas, and a lower amount in the stomach and other parts of the colon (Figure 5A) ▶ . After a lengthened exposure time, the autoradiographs showed hybridization signals in samples from the prostate and testes. Northern blot hybridization with the RELP cDNA probe detected a single 1.5-kb message in the RNA extracted from samples from the stomach, small intestine, colon, prostate, and testes. In the same assay, RELP mRNA was not detected in samples from the thyroid, spinal cord, adrenal gland, bone marrow, spleen, thymus, ovaries, or blood leukocytes (Figure 5, B and C) ▶ .
Figure 5.
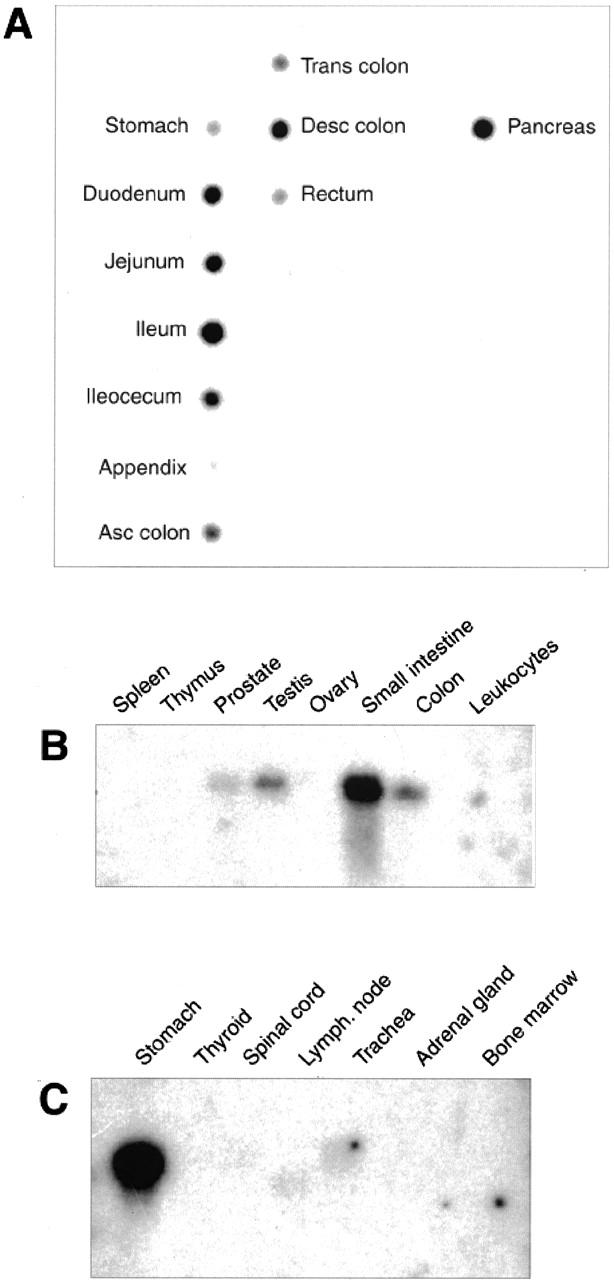
Expression of RELP mRNA in human tissues. Dot blot analysis (A) and Northern blot analysis (B and C).
To confirm that the hybridization signals were not cross-reactions with transcripts of other REG genes, the putative RELP messages from the small intestine, prostate, and fetal kidney were RT-PCR amplified using coding-region-specific primers, and sequenced. The sequencing data verified the identity of the amplicons with the original full-length RELP clone acquired from the Incyte Genomics.
Expression of the RELP Protein in Human Tissues
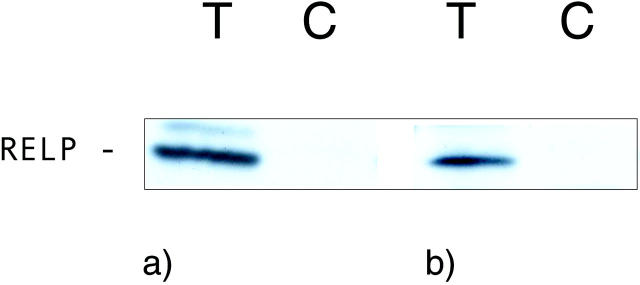
The polyclonal anti-RELP C-terminal peptide antibodies and the monoclonal anti-FLAG antibodies detected the same major 18-kd band in Western blotting of lysates from COS 7 cells transiently expressing the RELP C-terminal FLAG construct (Figure 6) ▶ . Immunohistochemical staining with the anti-RELP antibodies revealed the presence of RELP in the cytoplasm of selected epithelial cells in the duodenal (Figure 7a) ▶ and ileal mucosa (Figure 8a) ▶ . No specific immunostaining of duodenal mucosa was seen with anti-RELP antibodies pre-absorbed with the relevant RELP peptide (Figure 7b) ▶ . Double immunofluorescence staining demonstrated a colocalization of RELP and chromogranin A in the neuroendocrine cells of the duodenal epithelium. In situ hybridization verified the presence of RELP mRNA in cells with the distribution in the duodenal mucosa identical to that of neuroendocrine cells (Figure 7, c–g) ▶ . This finding demonstrates a constitutive transcription and translation of the relp gene in neuroendocrine cells of the intestinal epithelium.
Figure 6.
Western blot analysis of COS cells transiently expressing RELP that contains a C-terminal FLAG epitope tag. T, COS cells transfected to express RELP-FLAG; C, mock transfected COS cells. The primary antibodies for protein detection were affinity-purified polyclonal anti-RELP C-terminal peptide antibodies (a) and anti-FLAG epitope antibodies (b).
Figure 7.
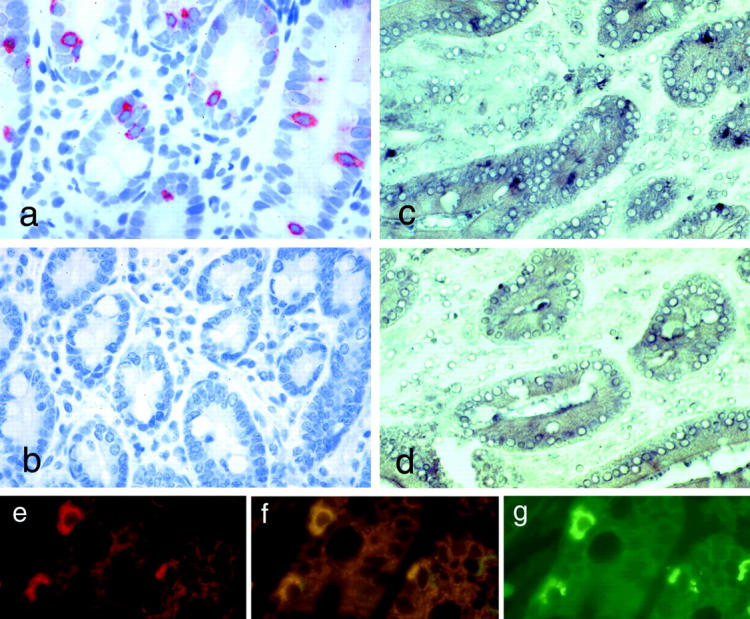
Immunohistochemical staining using polyclonal anti-RELP C-terminal peptide antibodies of normal duodenal mucosa showing solitary RELP positive cells (a), and abolishment of the staining in the presence of the peptide used for immunization (b). In situ hybridization with labeled anti-sense RELP RNA probe (c) confirms transcription in the same cells. No signal is detected when a sense RELP RNA probe is used (d). Double-immunofluorescence staining shows that chromogranin A (e) is co-expressed (f) with RELP (g). Magnifications, ×200.
Figure 8.
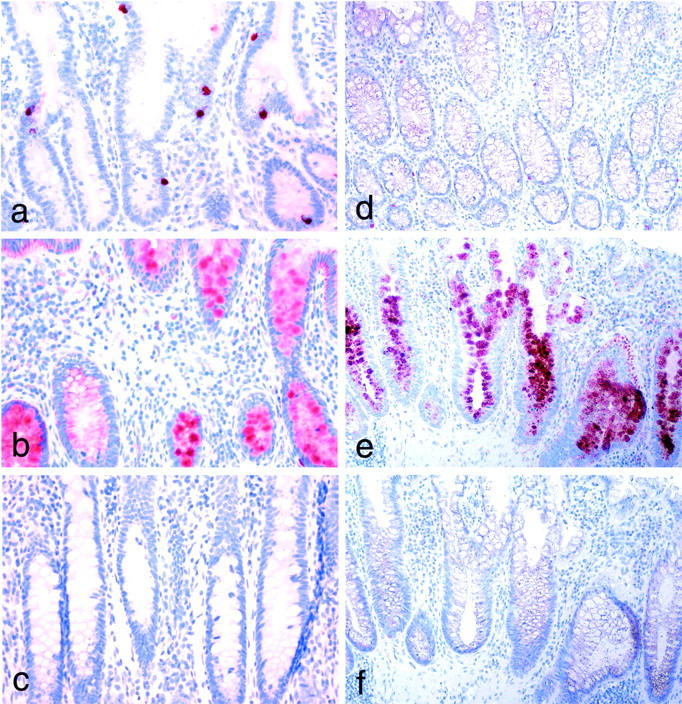
Immunohistochemical staining of normal ileal mucosa with monoclonal antibodies to RELP (a) where scattered epithelial cells display strong positivity. Staining of ileal mucosa from a patient with Crohn′s disease (b) shows robust reactivity in goblet cells in crypts of the inflammated lamina propria. c: Control staining with IgG fraction of normal rabbit serum. Immunohistochemical staining of colon mucosa. d: Weak staining for RELP in normal mucosa. e: A strong reactivity is seen in the crypts of mucosa from a patient with ulcerative colitis. f: Control stainings with IgG fraction of normal rabbit serum. Magnifications, ×200.
RELP mRNA was abundant in EST libraries derived from IBD. Immunohistochemical staining of sections representing Crohn’s disease in the small intestine (Figure 8b) ▶ showed a strong expression of RELP, particularly in the goblet cells in the affected mucosa.
Only a weak expression of RELP was seen in selected cells in normal colon mucosa (Figure 8d) ▶ whereas a strong reactivity was seen in the cryptal epithelium of mucosa from patients with ulcerative colitis (Figure 8e) ▶ . No specific staining was seen when the rabbit anti-RELP antibodies were replaced with the IgG fraction of preimmune rabbit serum (Figure 8, c and f) ▶ . Positive staining was also found in the parietal cells in the gastric corpus mucosa (Figure 9, a and b) ▶ . An up-regulated expression of the protein was present in epithelial cells at regenerating margins of peptic ulcers in the stomach and duodenum (not shown). Moreover, the expression of RELP was also induced in the goblet cells of glands representing intestinal metaplasia in the mucosa of the gastric antrum (Figure 9c) ▶ . A similar robust enhancement of RELP expression was seen in goblet cells in intestinal metaplasia of the esophagus (Barrett′s esophagus; not shown).
Figure 9.
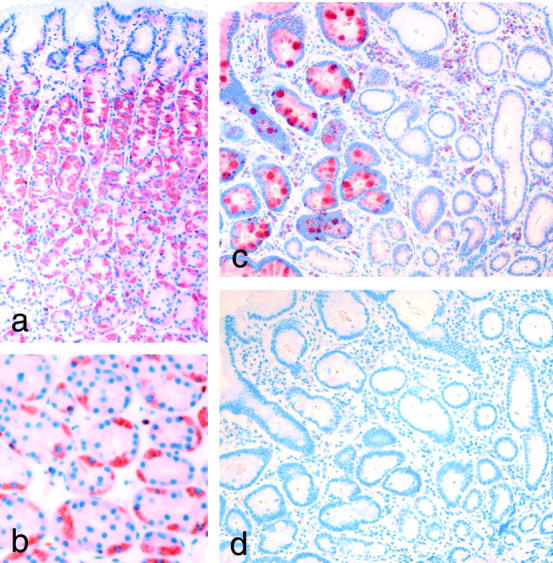
Immunohistochemical staining of gastric mucosa. a: RELP reactivity in parietal cells in a perpendicular section of corpus mucosa. b: A cross-section at higher magnification. c: Staining of antrum mucosa where the normal epithelium does not show positivity for RELP while the goblet cells in gland representing intestinal metaplasia show strong reactivity. d: Control staining with IgG fraction of normal rabbit serum of a section adjacent to c. Magnifications: a, c, and d, ×200; b, ×400.
Subcellular Localization of the RELP Protein
To define the subcellular localization of the RELP protein, gastric carcinoma cell line MKN45 cells were transfected to transiently express RELP with the FLAG epitope. The fusion protein was visualized by immunofluorescence, and the images were captured using a confocal microscope. RELP-FLAG epitope protein was localized in granules around the nucleus, and less prominently in the peripheral. RELP-FLAG protein was readily secreted into the medium, as indicated by Western blot analysis (data not shown).
Discussion
Crohn’s disease and ulcerative colitis are the major forms of idiopathic IBDs, which, although considered to be distinct clinical entities, share common pathogenetic mechanisms. A defective barrier function of the intestinal epithelium and an abnormal mucosal immune response appear to be the factors that characterize IBDs and facilitate the ongoing inflammation. Genetic susceptibility to Crohn’s disease is stronger than to ulcerative colitis. 1 The susceptibility loci identified by genome screening for Crohn’s disease comprise candidate genes primarily expressed in cells involved in immune responses, such as NOD2, CD11 integrins, CD19, IFN-γ, MHC, T cell receptor α/δ complex, ICAM1, C3, interleukin (IL)-3, IL-4, IL-5, IL-13, and CSF-2. 2 HLA-DR and IL-1RA, the polymorphisms of which are linked to the pathogenesis of ulcerative colitis, play a role in immune surveillance. 3 The elevated risk for cancer in IBDs is partially based on the damage that neutrophil-derived oxygen radicals cause on the DNA in the inflamed mucosa 7,8 ; however, the molecular events of the dysregulated regenerative responses of the inflammatory epithelium and its eventual transformation are poorly characterized. To understand the pathogenesis of IBDs and the underlying mechanisms leading to the development of cancer it is important to study the gene expression patterns of the mucosal epithelium. With that aim, we identified and characterized RELP. RELP is homologous to REG proteins, known to exert mitotic and motogenic activities in various cell types, 9-22 although the exact mechanism of function is elusive. REG proteins are primarily localized to the pancreas, 23,24 but are also expressed in the stomach 25,26 and the gut, 19,27 and in regenerating peripheral nerves. 21 Recently, the sequence of the major transcript of relp was published, and the predicted protein product was annotated Reg IV. 28 Because of the different chromosomal localization of relp and the human reg genes 29,30 and different sizes of the genes, 31,32 the relp gene could, however, be considered a separate entity. The RELP protein also displays lower homology with REG proteins than they share with each other. Relp appears to have segregated from the ancestral reg gene before the formation of the current reg gene cluster.
The proximal promoter region for relp could not be predicted, making the interpretation of the regulation of its transcription intangible. While the members of the REG protein family are highly represented in the pancreas, RELP message is the most abundant in the small intestine and in IBD-derived libraries. Immunofluorescence double staining and in situ hybridization indicated that RELP is selectively expressed in neuroendocrine cells of normal intestinal mucosa. This is reminiscent of rat REG1 protein expression in the enterochromaffin-like cells of the gastric corpus. 26 Given that RELP is also present in the parietal cells of the gastric mucosa, it might, analogously with REG1, act as a downstream effector for gastrin. 27,33
Inflammatory mediators, such as various cytokines, might be involved in the regulation of the relp gene expression. This could provide an explanation for the robust accumulation of RELP protein in the goblet cells of mucosa affected by ulcerative colitis or Crohn’s disease and also in goblet cells of intestinal metaplasia in the antrum and the esophagus. The levels of RELP in serum and/or urine may also provide a marker for non-invasive estimations of the severity or activity of IBDs.
Acknowledgments
We thank A. Harju and T. Arumäe for technical assistance.
Footnotes
Address reprint requests to Leif C. Andersson, Department of Pathology, Haartman Institute, University of Helsinki, Haartmaninkatu 3, FIN-00014, Helsinki, Finland. E-mail: leif.andersson@helsinki.fi.
Supported in part by grants from The Finnish Cancer Society, The Finnish Academy of Science, The Sigrid Jusélius Foundation, The Novo Nordisk Foundation, and R.W. Johnson Pharmaceutical Research and Development.
References
- 1.Podolsky DK: Inflammatory bowel disease. N Engl J Med 2002, 347:417-429 [DOI] [PubMed] [Google Scholar]
- 2.Shanahan F: Crohn’s disease. Lancet 2002, 359:62-69 [DOI] [PubMed] [Google Scholar]
- 3.Farrell RJ, Peppercorn MA: Ulcerative colitis. Lancet 2002, 359:331-340 [DOI] [PubMed] [Google Scholar]
- 4.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, Achkar JP, Brant SR, Bayless TM, Kirschner BS, Hanauer SB, Nunez G, Cho JH: A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 2001, 411:603-606 [DOI] [PubMed] [Google Scholar]
- 5.Ekbom A, Helmick C, Zack M, Adami HO: Ulcerative colitis and colorectal cancer: a population-based study. N Engl J Med 1990, 323:1228-1233 [DOI] [PubMed] [Google Scholar]
- 6.Rhodes JM, Campbell BJ: Inflammation and colorectal cancer: iBD-associated and sporadic cancer compared. Trends Mol Med 2002, 8:10-16 [DOI] [PubMed] [Google Scholar]
- 7.Knaapen AM, Schins RP, Polat D, Becker A, Borm PJ: Mechanisms of neutrophil-induced DNA damage in respiratory tract epithelial cells. Mol Cell Biochem 2002, 235:143-151 [PubMed] [Google Scholar]
- 8.Liu Q, Shimoyama T, Suzuki K, Umeda T, Nakaji S, Sugawara K: Effect of sodium butyrate on reactive oxygen species generation by human neutrophils. Scand J Gastroenterol 2001, 36:744-750 [DOI] [PubMed] [Google Scholar]
- 9.Chiba T, Fukui H, Kinoshita Y: REG protein: a possible mediator of gastrin-induced mucosal cell growth. J Gastroenterol 2000, 35(Suppl 12):52-56 [PubMed] [Google Scholar]
- 10.Levine JL, Patel KJ, Zheng Qh, Shuldiner AR, Zenilman ME: A recombinant rat regenerating protein is mitogenic to pancreatic derived cells. J Surg Res 2000, 89:60-65 [DOI] [PubMed] [Google Scholar]
- 11.Otonkoski T, Mally MI, Hayek A: Opposite effects of β-cell differentiation and growth on REG expression in human fetal pancreatic cells. Diabetes 1994, 43:1164-1166 [DOI] [PubMed] [Google Scholar]
- 12.Bernard-Perronese FR, Renaud WP, Guy-Crotte OM, Bernard P, Figarella CG, Okamoto H, Balas DC, Segenas-Balas FO: Expression of REG protein during cell growth and differentiation of two human colon carcinoma cell lines. J Histochem Cytochem 1999, 47:863-870 [DOI] [PubMed] [Google Scholar]
- 13.Watanabe T, Yonekura H, Terazono K, Yamamoto H, Okamoto H: Complete nucleotide sequence of human REG gene and its expression in normal and tumoral tissues: the REG protein, pancreatic stone protein, and pancreatic thread protein are one and the same product of the gene. J Biol Chem 1990, 265:7432-7439 [PubMed] [Google Scholar]
- 14.Lasserre C, Christa L, Simon MT, Vernier P, Brechot C: A novel gene (HIP) activated in human primary liver cancer. Cancer Res 1992, 52:5089-5095 [PubMed] [Google Scholar]
- 15.Bartoli C, Gharib B, Giorgi D, Sansonetti A, Dagorn JC, Berge-Lefranc JL: A gene homologous to the REG gene is expressed in the human pancreas. FEBS Lett 1993, 327:289-293 [DOI] [PubMed] [Google Scholar]
- 16.Rafaeloff R, Pittenger GL, Barlow SW, Qin XF, Yan B, Rosenberg L, Duguid WP, Vinik AI: Cloning and sequencing of the pancreatic islet neogenesis associated protein (INGAP) gene and its expression in islet neogenesis in hamsters. J Clin Invest 1997, 99:2100-2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katsumata N, Chakraborty C, Myal Y, Schroedter IC, Murphy LJ, Shiu RP, Friesen HG: Molecular cloning and expression of peptide 23, a growth hormone-releasing hormone-inducible pituitary protein. Endocrinology 1995, 136:1332-1339 [DOI] [PubMed] [Google Scholar]
- 18.Zenilman ME, Magnuson TH, Swinson K, Egan J, Perfetti R, Shuldiner AR: Pancreatic thread protein is mitogenic to pancreatic-derived cells in culture. Gastroenterology 1996, 110:1208-1214 [DOI] [PubMed] [Google Scholar]
- 19.Zenilman ME, Kim S, Levine BA, Lee C, Steinberg JJ: Ectopic expression of REG protein: a marker of colorectal mucosa at risk for neoplasia. J Gastrointest Surg 1997, 1:194-202 [DOI] [PubMed] [Google Scholar]
- 20.Zenilman ME, Chen J, Magnuson TH: Effect of REG protein on rat pancreatic ductal cells. Pancreas 1998, 17:256-261 [DOI] [PubMed] [Google Scholar]
- 21.Livesey FJ, O’Brien JA, Li M, Smith AG, Murphy LJ, Hunt SP: A Schwann cell mitogen accompanying regeneration of motor neurons. Nature 1997, 390:614-618 [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi S, Akiyama T, Nata K, Abe M, Tajima M, Shervani NJ, Unno M, Matsuno S, Sasaki H, Takasawa S, Okamoto H: Identification of a receptor for REG (regenerating gene) protein, a pancreatic β-cell regeneration factor. J Biol Chem 2000, 276:10723-10726 [DOI] [PubMed] [Google Scholar]
- 23.Terazono K, Yamamoto H, Takasawa S, Shiga K, Yonemura Y, Tochino Y, Okamoto H: A novel gene activated in regenerating islets. J Biol Chem 1988, 263:2111-2114 [PubMed] [Google Scholar]
- 24.Sanchez D, Figarella C, Marchand-Pinatel S, Bruneau N, Guy-Crotte O: Preferential expression of REG I β gene in human adult pancreas. Biochem Biophys Res Commun 2001, 284:729-737 [DOI] [PubMed] [Google Scholar]
- 25.Anastasi E, Ponte E, Gradini R, Bulotta A, Sale P, Tiberti C, Okamoto H, Dotta F, Mario UD: Expression of REG and cytokeratin 20 during ductal cell differentiation and proliferation in a mouse model of autoimmune diabetes. Eur J Endocrinol 1999, 141:644-652 [DOI] [PubMed] [Google Scholar]
- 26.Asahara M, Mushiake S, Shimada S, Fukui H, Kinoshita Y, Kawanami C, Watanabe T, Tanaka S, Ichikawa A, Uchiyama Y, Narushima Y, Takasawa S, Okamoto H, Tohyama M, Chiba T: REG gene expression is increased in rat gastric enterochromaffin-like cells following water immersion stress. Gastroenterology 1996, 111:45-55 [DOI] [PubMed] [Google Scholar]
- 27.Higham AD, Bishop LA, Dimaline R, Blackmore CG, Dobbins AC, Varro A, Thompson DG, Dockray GJ: Mutations of REGIα are associated with enterochromaffin-like cell tumor development in patients with hypergastrinemia. Gastroenterology 1999, 116:1310-1318 [DOI] [PubMed] [Google Scholar]
- 28.Hartupee JC, Zhang H, Bonaldo MF, Soares MB, Dieckgraefe BK: Isolation and characterization of a cDNA encoding a novel member of the human regenerating protein family: rEG IV. Biochim Biophys Acta 2001, 1518:287-293 [DOI] [PubMed] [Google Scholar]
- 29.Miyashita H, Nakagawara K, Mori M, Narushima Y, Noguchi N, Moriizumi S, Takasawa S, Yonekura H, Takeuchi T, Okamoto H: Human REG family genes are tandemly ordered in a 95-kilobase region of chromosome 2p12. FEBS Lett 1995, 377:429-433 [DOI] [PubMed] [Google Scholar]
- 30.Bartoli C, Dagorn JC, Fontes M, Berge-Lefrane JL: A limited genomic region contains the human REG and REG-related genes. Eur Hum Genet 1995, 3:344-350 [DOI] [PubMed] [Google Scholar]
- 31.Gharib B, Fox MF, Bartoli C, Giorgi D, Sansonetti A, Swallow DM, Dagorn JC, Berge-Lefranc JL: Human regeneration protein/lithostathine genes map to chromosome 2p12. Ann Hum Genet 1993, 57:9-16 [DOI] [PubMed] [Google Scholar]
- 32.Abe M, Nata K, Akiyama T, Shervani NJ, Kobayashi S, Tomioka-Kumagai T, Ito S, Takasawa S, Okamoto H: Identification of a novel REG family gene, REG III δ, and mapping of all three types of REG family gene in a 75 kilobase mouse genomic region. Gene 2000, 246:111-122 [DOI] [PubMed] [Google Scholar]
- 33.Chiba T, Fukui H, Kinoshita Y: Reg protein: a possible mediator of gastrin-induced mucosal cell growth. J Gastroenterol 2000, 35(Suppl 12):52-56 [PubMed] [Google Scholar]