Abstract
The clinically important melanoma diagnostic antibodies HMB-45, melan-A, and MITF (D5) recognize gene products of the melanocyte-lineage genes SILV/PMEL17/GP100, MLANA/MART1, and MITF, respectively. MITF encodes a transcription factor that is essential for normal melanocyte development and appears to regulate expression of several pigmentation genes. In this report, the possibility was examined that MITF might additionally regulate expression of the SILV and MLANA genes. Both genes contain conserved MITF consensus DNA sequences that were bound by MITF in vitro and in vivo, based on electrophoretic mobility shift assay and chromatin-immunoprecipitation. In addition, MITF regulated their promoter/enhancer regions in reporter assays, and up- or down-regulation of MITF produced corresponding modulation of endogenous SILV and MLANA in melanoma cells. Expression patterns were compared with these factors in a series of melanoma cell lines whose mutational status of the proto-oncogene BRAF was also known. SILV and MLANA expression correlated with MITF, while no clear correlation was seen relative to BRAF mutation. Finally, mRNA expression array analysis of primary human melanomas demonstrated a tight correlation in their expression levels in clinical tumor specimens. Collectively, this study links three important melanoma antigens into a common transcriptional pathway regulated by MITF.
MITF, a bHLHZip (basic/helix-loop-helix/leucine-zipper) transcription factor, is essential for the development and maintenance of the melanocyte lineage. 1 MITF binds the canonical E-box promoter sequence CACGTG as well as the non-palindromic sequence CACATG. 2,3 The three major pigmentation enzymes tyrosinase, TYRP1, and DCT all contain the MITF consensus binding site in their promoters and are thought to be transcriptional targets of MITF. 2,4,5 In humans, germline heterozygous MITF mutation produces, with variable penetrance, Waardenburg Syndrome IIA, 6 and Tietz syndrome, 7,8 manifesting pigmentation disturbances and deafness, the latter due to inner ear melanocyte deficiency. 9 In mice, the allele Mitfmi leads to complete absence of melanocytes in the homozygous state, 10 while mice homozygous for Mitfvit (vitiligo) are born with a nearly normal coat color, but turn white over the first few months of life as melanocytes are lost, suggesting a role for MITF in both melanocyte development and in postnatal survival. 11,12 In contrast to various melanocytic markers such as melanin or c-kit whose expression may be lost or difficult to detect in malignant melanocytic lesions, 13-15 MITF expression is usually maintained in human melanoma specimens, 16-19 and MITF is increasingly used as a histopathological marker for melanoma diagnosis. 20
PMEL17 is the human homologue of murine silver, a locus whose disruption produces a significant pigmentation phenotype which resembles a silver color. 21 Although the precise function of this gene (also known as GP100/SILV, hereafter referred to as SILV) remains to be ascertained, SILV localizes to an early melanosome trafficking compartment 22 and may function in melanosome structure, 23 biosynthesis of the melanin intermediate 5,6-dihydroxyindole-2-carboxylic acid (DHICA), 24,25 or morphogenesis of premelanosomes. 26
MLANA/MART1 (hereafter referred to as MLANA) was cloned by two separate groups using melanoma reactive cytotoxic T lymphocytes to screen a cDNA library derived from melanoma cells. 27,28 MLANA expression is restricted to melanocytes, melanomas, and retinal pigment epithelium. 27 Its function remains to be fully elucidated, but it has recently been found to localize to vesicles, including melanosomes, suggesting a role in melanosome biogenesis. 29 Although near the loci for two murine coat color mutants, MLANA has been excluded as a candidate for either, 30 and its loss of function phenotype is currently unknown.
Aside from its role in pigmentation, SILV encodes antigenic epitopes which are recognized by multiple melanoma diagnostic antibodies including HMB-45, currently one of the most commonly used melanocytic markers for clinical melanoma diagnosis in humans. 31,32 The Melan-A and MART1 antibodies both recognize the MLANA gene product and are increasingly being used as markers for the diagnosis 33-36 and prognosis of melanoma. 37 Due to its early vertical (deep) growth phase, cutaneous melanomas exhibit a high propensity for local invasion and metastasis. Furthermore, melanoma may present as “metastasis of unknown primary” or relapse after considerable time intervals, often lacking differentiated features (such as pigmentation). The diagnosis of melanoma may therefore rely on combinations of immunohistochemical markers for diagnosis. Despite the common loss of many melanocyte-specific markers in melanoma, HMB-45 positivity has been seen in a high fraction of human melanoma specimens 38,39 and antibody staining for the melanocyte-melanoma lineage-specific antigen MLANA was shown to be positive in all primary malignant melanomas and nevi tested. 40,41 In subsequent studies, MLANA reactivity has been shown to largely overlap with tyrosinase 42 and HMB-45. 33,43 MITF (antibody D5) was also found to stain a high fraction of melanomas, with a pattern that included all HMB45-positive specimens. 16 A recent side-by-side examination of MITF, HMB-45, and Melan-A revealed high sensitivity and specificity for all three in melanoma diagnosis of cytologic specimens. 44
Both SILV and MLANA have been identified as antigens recognized by tumor infiltrating lymphocytes, 28,45-47 leading to their use in vaccine protocols aimed at immunotherapy for melanoma. 48-51 Dramatic melanoma regressions were recently reported for patients undergoing intensive immunotherapy directed against SILV and MLANA. 52 In addition, it has been suggested that expression patterns of MITF and MLANA may correlate with specific clinical behaviors of melanoma, with higher expression of these markers associated with better prognosis. 17,53-55
In this study, we demonstrate evidence of linkage between the expression patterns of the three melanoma diagnostic markers, MITF, SILV and MLANA. MITF consensus recognition sites were identified in the promoters/enhancers of both SILV and MLANA, and were found to be occupied by endogenous MITF using chromatin immunoprecipitation. Reporter assays, quantitative-PCR, Northern, and Western analysis all suggested that MITF either regulates reporters or the endogenous genes within melanoma cells and melanocytes. Protein and mRNA levels of the three antigens were found to correlate in a panel of melanoma cell culture lines as well as human primary melanocytes. Finally, we show MITF, SILV and MLANA mRNA levels are highly correlated in a primary human melanoma gene expression data set. Together, the results suggest that MITF transcriptionally regulates SILV and MLANA, placing three commonly used diagnostic markers within a common transcriptional pathway.
Materials and Methods
Cell Culture and Media
501mel was a gift from Dr. Ruth Halaban (Yale University School of Medicine) and maintained in Ham’s F10 medium (Mediatech, Herndon, VA) supplemented with 10% fetal bovine serum (FBS) (Sigma, St. Louis, MO) or RPMI (Mediatech) with 10% FBS. SKMEL5 and A375 melanoma cells were purchased from American Type Culture Collection (ATCC; Manassas, VA). MALME-3M, SKMEL2, SKMEL28, UACC62, UACC257 and M14 melanoma lines were all obtained from NIH (Frederick, MD). All melanoma cell lines were maintained in RPMI with 10% FBS. 293 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Mediatech) with 10% FBS. Human primary melanocytes were isolated as described previously 56 and maintained in TICVA medium (F10 with penicillin/streptomycin/glutamine (Invitrogen, Carlsbad, CA), 7.5% FBS, 50 ng/ml TPA (Sigma), 225 μmol/L IBMX (Sigma), 1 μmol/L Na3VO4 (Sigma), and 1 mmol/L dbcAMP; Sigma).
Chromatin Immunoprecipitation
Chromatin immunoprecipitation assays (ChIPs) were performed with cells grown in logarithmic phase. Cells were harvested by scraping and homogenized in a hypotonic buffer (10 mmol/L Tris-HCl, pH 7.4, 15 mmol/L NaCl, 60 mmol/L KCl, 1 mmol/L EDTA, 0.1% Nonidet P40, 5% sucrose, and 1X Complete proteinase inhibitor cocktail (Roche, Indianapolis, IN)) on ice using a Dounce homogenizer. The nuclei were isolated by centrifugation onto a 10% sucrose pad and then cross-linked with 1% formaldehyde in phosphate-buffered saline (PBS) for 20 minutes at room temperature with gentle shaking. Nuclei were then spun down and resuspended in ChIPs buffer (10 mmol/L Tris-HCl, pH 7.4, 100 mmol/L NaCl, 60 mmol/L KCl, 0.1% Nonidet P40, and 1X Complete proteinase inhibitor cocktail (Roche)) and sonicated by two 1-minute pulses on ice using a Fisher dismembranator fitted with a microtip. Rabbit antibodies against human MITF, anti-GST (Roche), and anti-Acetylated-Histone3 (Upstate, Waltham, MA) were then added to a 10-fold ChIPs buffer diluted sample and incubated on a nutator for 3 hours at room temperature. Ultralink protein-A/G-beads (Pierce, New York, NY) were added to the sample and a control sample and incubated for an additional hour. Immunoprecipitates were then washed twice with ChIPs buffer, twice with 500 mmol/L NaCl ChIPs buffer, and once with TE, pH 8.0. The immunoprecipitates were released from the beads by incubating at 65°C for 20 minutes in 1%SDS/TE and protein digested by proteinase K treatment side-by-side with an additional unprecipitated sample as input control. Cross-links were released by heating at 70°C for 10 hours and DNA recovered by extraction with phenol and chloroform at high salt (0.6 mol/L Na acetate, pH 8.0) followed by ethanol precipitation. Semi-quantitative PCR was then performed on samples to amplify a fragment containing the intronic E-box (E1) or the upstream E-box (E2) in the human SILV transcription initiation region. The forward and reverse primers for the E1 region are 5′- CAT AAG ATA CCC CAT TCT TTC TCC ACT T-3′ and 5′-GAG AAT GTG GTA TTG GGT AAG AAC AC-3′, respectively. The primers for the E2 region are 5′-CAT GGA GAA CTT CCA AAA GGT GG-3′ and 5′-TAC TCT CCC CAG GGA GTA TAA GT-3′. The MLANA promoter and control regions were analyzed similarly. The primers for MLANA promoter region are: 5′-TTG GAA TAA ATT GGG CTA CGA ACT T and 5′-TGG CAG GAT CTC AGC TCA CTA CAA C. The primers for MLANA downstream control regions are: 5′- ATG CCT GGC CTC TAT CCA CT and 5′- GGC GAC AGA GTG AGT GAG AT.
EMSA (Electrophoretic Mobility Shift Assay)
Nuclear extracts were prepared as previously described. 57 Probe/competitor double-stranded oligonucleotides were 30 bp and spanned the individual E-boxes. A MLANA-specific double-stranded probe spanning the E2 site was prepared using the following sequences for sense oligos: wild-type probe, 5′-TTTCCATGTTCACGTGTGAGATATGC; mutant probe, 5′-TTTCCATGTTGAGGTGTGAGATATG. The SILV probe was prepared using the following sequences for sense oligos: wild-type probe, 5′- CCCAGAGCCCTTTCATGTGATGCTCAGCT; mutant probe, 5′- CCCAGAGCCCTTTGAGGTGATGCTCAGCT. Probe labeling was performed using T4 polynucleotide kinase (New England BioLabs, Beverly, MA) and α-[32P]-ATP (PerkinElmer, Boston, MA) to label the 5′ oligo before annealing. Binding reactions contained 25 fmol (∼100,000 cpm)-labeled probe, 1 μg poly (dI-dC) (Amersham Biosciences, Piscataway, NJ), 5% glycerol, 0.1 mol/L KCl, 10 mmol/L Tris (pH 7.6), 0.2 mmol/L DTT, 1 μl (5 mg/ml) human 501 melanoma nuclear extract, and 4 to 8 μl D5 anti-human Mitf mAb (Neomarkers, Fremont, CA) in a 20 μl reaction volume. Reactions were incubated for 30 minutes at room temperature and loaded on 6% 0.5X TBE non-denaturing gels. For competition studies, indicated amounts of molar excess double-stranded mutant probes were included in the binding reactions.
Construction of SILV and MLANA Reporters
A 2.8-kb fragment containing the first exon of SILV and flanking sequences was amplified from human genomic DNA (Roche) using primers 5′-GCA ATC CTA TGA CCA CGG CCT CCC AAA G-3′ and 5′-GTT TTC CAT GGA TTC CAC ACC ACC CTA TAC-3′. A smaller fragment was subsequently amplified from the 2.8-kb fragment with primers 5′-CCG GTA CCC ACA TAA CTC CAC TCC ATG GAG-3′ and 5′-CCC CAT GGC CAG CTA ATT TTT GTA CTT TTA-3′, digested with KpnI and NcoI, and inserted into pGL3-basic vector (Promega, Madison, WI). Subsequently, the splice acceptor of SILV exon 4 was amplified with primers 5′-ttC CAT GGc ctc aac ccc caa cta ml-3′ and 5′-ccC CAT GGg gct ccc tga aag ata a-3′ and inserted into the NcoI site. The initiation ATG of luciferase was cloned into the comparable location of the MLANA gene to generate a luciferase reporter including 1.9 kb of the MLANA promoter upstream of the transcriptional start site. MLANA promoter was amplified with primers: 5′- ACT GTT TGG TGG TCT CTG CTG GTC TGA T and 5′-GCT GGC TGG CCG CGT GTA TGA, and inserted into the pCR4-TOPO vector (Invitrogen). The vector was then digested with EcoRI, the promoter fragment was filled in with Klenow, and inserted into pGL3-basic digested with SmaI.
SILV and MLANA Reporter Construct Mutagenesis
Site-directed mutagenesis was performed using the QuickChange-method (Stratagene, La Jolla, CA) according to the supplier’s recommendations. Mutagenesis primers were designed as follows (using the following sense strands with their corresponding complementary oligonucleotides):
SILV-E1: 5′-CCCAGAGCCCTTTGAGGTGATGCTCAGCT-3′
SILV-E2: 5′-AAATCCGCCTGGAGAGGTGAGTGGCCTCT-3′
MLANA-E1: 5′-CAGCACCTAACCACCTCTCACACAACCAATG-3′
MLANA-E2: 5′-GTTTCCATGTTCACCTCTGAGATATGCCTCC-3′
Transfection and Dual-Luciferase Reporter Assay
For all reporter assays triplicate transfections of 293 cells were carried out in 24-well plates using Fugene-6 (Roche) according to manufacturer’s recommendations. SILV reporter constructs were transfected together with either pRC-CMV/MITF (wt) (encoding full-length M-form human MITF) 57-59 or pcDNA3.1 (Invitrogen) and pRL-null (Promega). A total of 1 μg DNA was used for each transfection. Cell lysates were prepared 48 hours after transfection using 100 μl of passive lysis buffer as per manufacturer’s recommendations (Promega). The MLANA promoter-luciferase fragment was cut from pGL3-Basic backbone with KpnI and BamHI, column-purified, and transfected with either pcDNA3/MITF (wt) (encoding full-length M-form human MITF) or pcDNA3.1 (Invitrogen) and pRL-TK (Promega). A total of 0.5 μg of DNA was used for each transfection. Cell lysates were prepared 20 hours after transfection using 100 μl of passive lysis buffer as per manufacturer’s recommendations (Promega). All lysates were incubated for 15 minutes at room temperature. An aliquot was used to perform luciferase assays using the Dual-Luciferase kit (Promega) according to the manufacturer’s recommendations, and signals were normalized for transfection efficiency by the internal Renilla controls.
Adenovirus Infection and RNA Preparation
Adenoviruses encoding wild-type MITF, dominant-negative mutant MITF, or vector control encoding a fusion of GFP-wee1 for nuclear localization, were generated as previously described. 57 Briefly, 106 human primary melanocytes were plated per 100-mm plate. On the second day, cells were overlaid with 2 ml of serum-free F10 media containing 10 mmol/L MgCl2 and concentrated adenovirus was added at MOI 300. The cells were incubated at 37°C for one-half hour after which virus was removed and fresh full media was added. Total RNA was isolated with RNAqueous-4PCR kit (Ambion, Austin, TX) at 68 and 80 hours after infection according to manufacturer’s instructions. SKMEL5 melanoma cells were infected at MOI 100 and the RNA was harvested at 52, 68, and 80 hours post-infection. UACC62 cells were infected at MOI 200 and RNA was harvested at 72 hours post-infection.
Real-Time/Quantitative PCR
The real-time PCR primers for human SILV were 5′-TCT GGG CTG AGC ATT GGG-3′ and 5′AGA CAG TCA CTT CCA TGG TGT GTG-3′. The probe for human SILV was 5′- 6-FAM-CAGGCAGGGCAATGCTGGGC-TAMRA-3′ (Applied Biosystems, Foster City, CA). The total volume of each reaction was 25 μl including 12.5 μl 2X Master Mix without UNG (uracil-N-glycosylase), 0.625 μl MultiScribe Reverse Transcriptase and RNase inhibitor (Applied Biosystems), 0.5 μl of each primer (10 μmol/L stock), 0.25 μl of the probe (5 μmol/L stock) and 1 μl of the template at 100 ng/μl. Reverse transcription proceeded at 48°C for 30 minutes. Then, 40 cycles of PCR reaction were carried out at 95°C for 15 seconds and at 60°C for 1 minute. Real-time PCR was carried out using the ABI PRISM 7700 Sequence Detection System (Applied Biosystems) with analysis using the integrated Sequence Detection System Software Version 1.7. Standard curves were generated for all primer sets to confirm linearity of signals over the experimentally measured ranges.
Northern Analysis
Total RNA was isolated from human UACC-62 melanoma cells using the Trizol reagent (Invitrogen). Ten μg of total RNA was resolved on a 1% agarose gel and blotted onto Hybond-N membrane (Amersham Biosciences) and cross-linked in a GS Gene Linker (Bio-Rad, Hercules, CA). The MLANA probe was prepared by random priming of the full-length reverse transcription-PCR-derived CDS for human MLANA. A probe for human GAPDH was prepared by a restriction digestion of the full-length GAPDH cDNA, followed by random priming of the purified fragment.
Immunoblots
Melanoma cells and adenovirus-infected primary melanocytes were collected and lysed in 2X lysis/loading buffer (125 mmol/L Tris, pH 6.8, 4.6% SDS, 20% glycerol, and 0.04% pyronin Y), resolved by electrophoresis in 12% SDS-polyacrylamide gels, and transferred to nitrocellulose membranes (Protran from Schleicher & Schuell, Keene, NH). All proteins were detected using chemiluminescence and antibodies to SILV (anti-PEP13h, a gift from Dr. Vincent Hearing), MLANA (NeoMarkers AB-2, Fremont, CA), MITF (C5), SOX10 (Active Motif, Carlsbad, CA), BRN2 (Santa Cruz Biotechnology, Santa Cruz, CA), CREB/ATF1 (25C10G; Santa Cruz Biotechnology), phospho-CREB/ATF1 (Cell Signaling Technologies, Beverly, MA), or α-tubulin (Sigma).
Preparation of Genomic DNA and Analysis of BRAF Status (V599E Mutation) in Melanoma Cell Lines
Genomic DNA from various melanoma cell lines was isolated with the DNeasy Tissue kit (Qiagen, Valencia, CA) according to manufacturer’s instructions. Fragments spanning the T1796A mutation were amplified with primers 5′- AAG CAT CTC ACC TCA TCC TAA CAC ATT T and 5′- CTT TCT AGT AAC TCA GCA GCA TCT CAG G from the genomic DNA. These fragments were subsequently sequenced with primer 5′- ATA GCC TCA ATT CTT ACC ATC CAC AAA to determine the mutation status.
Microarray and Pearson Correlation Coefficient Analyses
Expression data on NCI 60 cell lines were collected as described elsewhere. 60 Expression profiling data on 190 human primary tumors were collected using Affymetrix Hu6800 high-density oligonucleotide microarrays as described elsewhere. 61 Expression values were given a lower threshold of 20 units and a ceiling of 16,000 units. The data set was filtered to eliminate genes whose expression levels did not vary more than fivefold or at least 100 absolute units across the data set. Of the 7129 genes on the microarray, 6466 genes passed this filter and were subjected to further analysis. The raw expression value for each gene was normalized to a mean of zero and SD of one across all samples in the data set. Degree of similarity analysis was performed using the Pearson correlation coefficient to identify genes with similar patterns of expression to MITF across the melanoma and non-melanoma collections, resulting in a sorting of the 6466 genes by their degree of correlation.
Results
It has been previously shown that MITF modulates the transcription of several key enzymes in the pigment pathway and might regulate other melanocyte-specific differentiation-associated genes in melanocytes. 62,63 Of the melanocyte-specific melanoma-associated antigens, tyrosinase, TYRP1, and DCT are widely considered to be transcriptional targets of MITF. 2,4,5 Two remaining genes are SILV and MLANA, which encode proteins recognized by the widely used diagnostic antibodies, HMB45 and Melan-A, respectively. Based on the role of MITF in the regulation of melanocyte-specific genes, we asked whether MITF might also modulate the expression of these genes.
Promoter/Enhancer Structures of SILV and MLANA
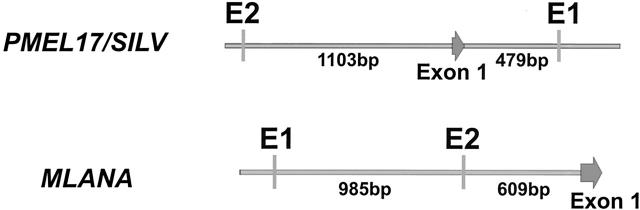
Since MITF modulates its targets through association with E-box elements in their promoters/enhancers (Figure 1) ▶ , we examined human genome sequences surrounding SILV and MLANA transcriptional initiation sites. For SILV, one E-box (CACGTG) was found 1103 bp upstream its start and a second E-box (CATGTG) was identified in a region within the first intron. This intronic E-box contains a 5′-flanking T which was previously reported to contribute to the recognition of CATGTG by MITF. 64 For MLANA, two E-boxes were identified upstream of its transcriptional start site at −609 and −1594 bp, respectively. The distal site matches the consensus CACATG while the proximate site is a canonical (CACGTG) E-box that has been previously reported to be responsible for melanocyte specificity of the promoter, though the responsible transcriptional regulator was not known. 65 Murine homologues of SILV and MLANA were also examined, and were found to contain E-boxes in similar positions as the human genes (data not shown).
Figure 1.
The SILV and MLANA promoters/enhancers. Initial transcribed exons are denoted by large arrows and E-boxes are represented by vertical bars.
In Vitro and in Vivo Binding of MITF to the SILV E1 Region and MLANA Promoter Region
To examine in vivo chromatin occupancy by MITF and help establish the directness of potential transcriptional regulation, chromatin immunoprecipitation (ChIP) was carried out from human melanoma cells (Figure 2) ▶ . Primers were designed which span the upstream and intronic E-boxes located within the SILV transcription initiation region. Immunoprecipitation with antibody against MITF did reveal occupancy by MITF of the E1 enhancer region (SILV enhancer), but not a control region located near the E2 site (Figure 1) ▶ located further upstream. Additional specificity controls included no antibody or control polyclonal antibody (directed against GST protein). An antibody directed against acetylated histone H3 represented a positive control in each reaction. For MLANA, ChIP revealed occupancy by MITF of the E-box containing promoter region. No occupancy was seen for MITF at the 3′ UTR (“control region”), and additional controls included no antibody or comparable polyclonal anti-GST antibodies. Although ChIP does not permit accurate assessments of precise sequence elements being contacted by the transcription factor, it does suggest that MITF is bound to promoter/enhancer elements near the conserved E-boxes for both genes.
Figure 2.
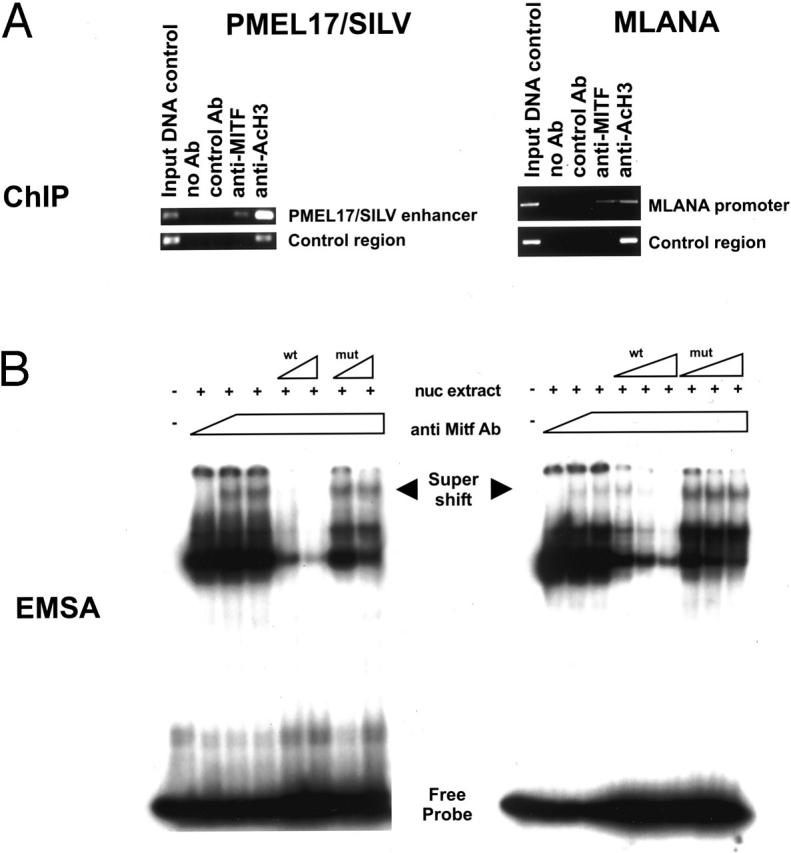
Binding of MITF to SILV enhancer and MLANA promoter in vitro and in vivo. A: Chromatin immunoprecipitations were performed on materials isolated from melanoma cells. DNA from lysates before immunoprecipitation was used as positive input control. The SILV enhancer primer set amplifies the intronic regulatory region which is important for gene expression, while the control primer set amplifies the region containing the upstream E-box which is dispensable for gene expression (negative control), based on reporter assays (see below and data not shown). The MLANA promoter primer set spanning the upstream E-boxes, while the control primer set amplifies a portion of the 3′ UTR (negative control). B: EMSA assays were performed on melanoma lysates using probes containing SILV intronic E-box (E1) and MLANA proximate E-box (E2), respectively. Similar binding of MITF was seen at the E1 site of MLANA as well (data not shown).
To specifically examine DNA binding by MITF of the relevant E-boxes, EMSA assays were performed using DNA probes containing these E-box elements plus melanoma nuclear extracts (as a source of MITF) and a monoclonal anti-MITF antibody (D5) for supershift analysis. Supershifted complexes were observed on addition of wild-type radiolabeled probes to the SILV E1 or MLANA E2 and E1 (data not shown) boxes in the presence of anti-MITF antibody (Figure 2) ▶ . More importantly, the supershifted bands were specifically competed away with increasing titration of wild-type unlabeled probe, but not with identical titration of point mutated cold probes (Figure 2) ▶ . Together, these results suggest that MITF directly binds the intronic E-box element in the SILV enhancer and the proximate E-box upstream of the MLANA transcriptional start site.
E-Box-Dependent Modulation of SILV and MLANA Reporters by MITF
To further test the role of specific promoter/enhancer elements in the regulation of SILV, luciferase reporters were constructed which recapitulate the intronic location of the E-box element for its transcription. As shown in Figure 3 ▶ , the intact genomic region was used as a template for the reporter. Transcription initiation should occur at the SILV exon 1 site, and the intron-containing transcript should be spliced to a downstream SILV-derived splice acceptor (which is fused to luciferase). To verify that the template was correctly initiated and spliced, RT-PCR was used together with 5′ RACE of transfected cells. Sequencing of the resulting products confirmed that the luciferase reporter did correctly initiate and splice the luciferase transcript to exon 2 (data not shown). Using this reporter, wild-type MITF was found to stimulate luciferase activity (Figure 3 ▶ , pSILV-E1E2), and point mutagenesis revealed an essential role for the intronic E-box element (E1) in mediating MITF-dependent transactivation. These data are consistent with the notion that the conserved E1 element may function as an intronic enhancer for MITF-regulated SILV expression.
Figure 3.
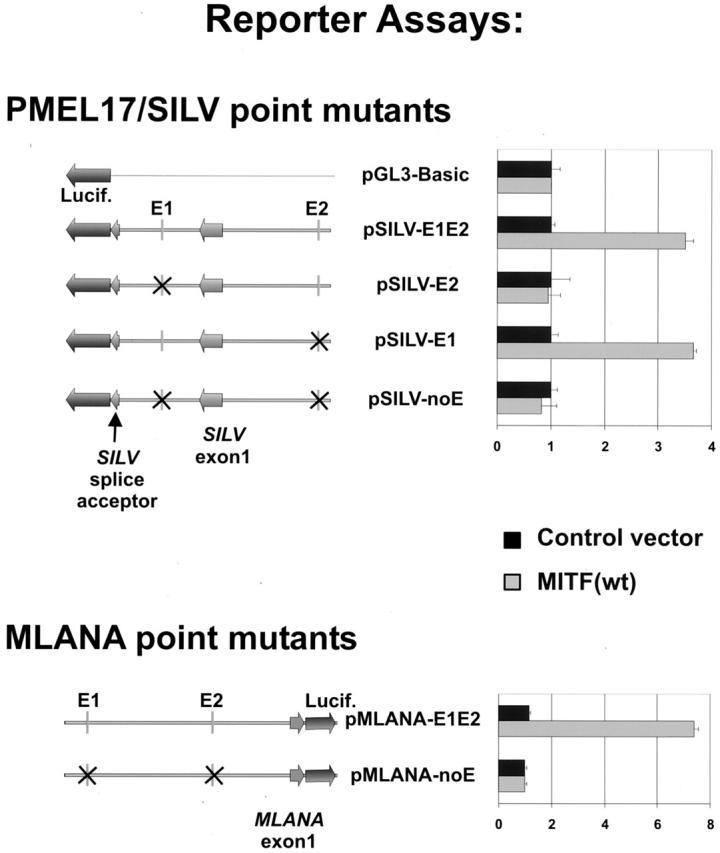
Reporter assays of SILV and MLANA point mutants. SILV and MLANA reporters were transfected into 293 cells. Firefly luciferase activities in samples were normalized to Renilla luciferase activities in the same specimens. Relative luciferase activities are shown. To recapitulate the putative SILV intronic enhancer, the genomic locus was incorporated into the pGL3 reporter with luciferase placed downstream of a SILV splice acceptor sequence. As described in Results, correct splicing of the reporter transcript was separately confirmed.
MLANA promoter activity was also assessed using luciferase reporters. 1.9 kb of the MLANA gene upstream of its transcriptional start fused to the luciferase ATG was used in reporter assays. Wild-type MITF significantly up-regulated the wild-type reporter (pMLANA-E1E2) (Figure 3) ▶ . Point mutations of both E-boxes completely abolished responsiveness to MITF (Figure 3) ▶ . These data suggest that the modulation of MLANA expression by MITF is E-box-dependent and further supports the directness of this regulation.
Transcriptional Regulation of Endogenous SILV and MLANA by MITF
To investigate whether MITF regulates endogenous SILV and MLANA mRNA levels in the melanocyte lineage, early passage human primary melanocytes and multiple human melanoma cell lines (501mel, SKMEL5, UACC-62 and A375) were infected with adenoviruses overexpressing either control, wild-type MITF or dominant-negative MITF (R218del). The R218 deletion mutant of MITF is derived from a dominantly inherited basic domain mutant mouse allele which preserves dimerization, but ablates DNA binding and thus sequesters wild-type dimerization partners in non-functional heterodimers. This allele has thus been extensively used as a specific dominant-negative mutant. 2,56,57 Adenoviral infection efficiency approached 100% in these cells, based on GFP signal (data not shown) and was shown to produce strong expression of wild-type or mutant MITF by Western blot (data not shown). Enhanced or disrupted DNA binding by MITF in nuclear extracts occurred by about 48 hours post-infection, lasting until at least 96 hours post-infection, as measured by EMSA analyses (data not shown). Total RNA was isolated after 72 hours. Real-time (quantitative) PCR was performed for SILV on the RNA samples. Wild-type MITF significantly stimulated SILV mRNA expression while dominant-negative MITF suppressed SILV mRNA levels (Figure 4A) ▶ . MLANA expression was examined by Northern blot analysis (due to unknown complications with quantitative PCR). Wild-type MITF up-regulated, while dominant-negative MITF suppressed MLANA mRNA levels (Figure 4B) ▶ relative to vector control adenovirus (normalized to GAPDH as a control).
Figure 4.
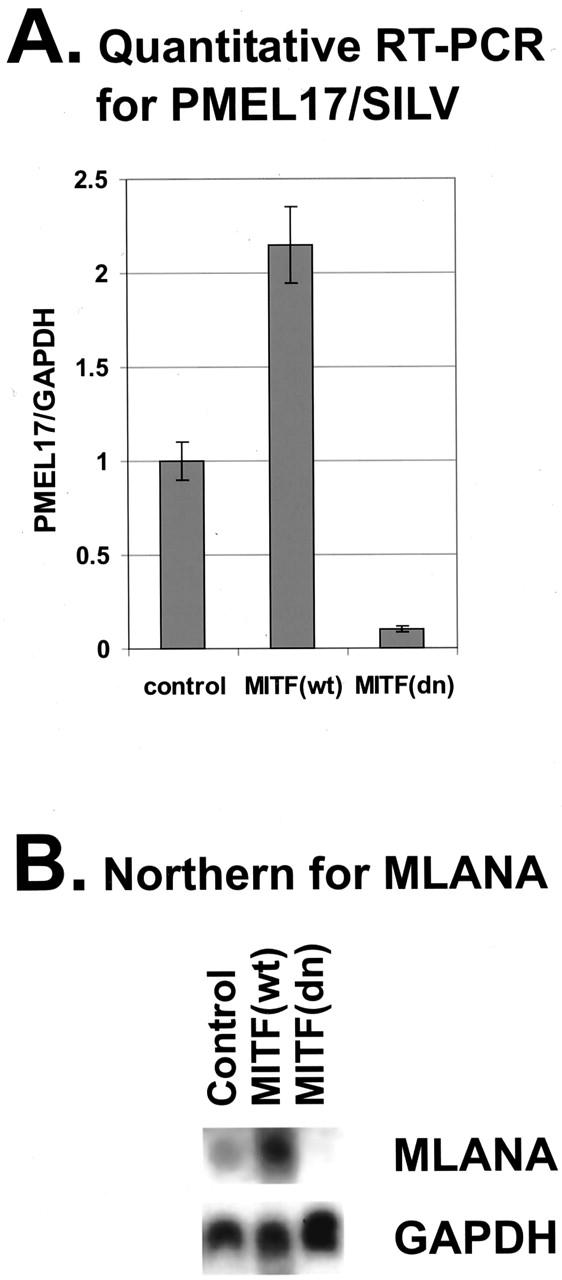
Regulation of endogenous SILV and MLANA mRNA levels by MITF. A: SILV mRNA levels were assayed after adenovirus infection (top panel). Total RNA was isolated from melanoma cells infected with adenoviruses 72 hours after infection. Quantitative RT-PCR was performed and data were normalized to endogenous GAPDH levels. Expression levels for MITF (wt) and MITF (dn)-virus infected samples were normalized to the control samples. B: MLANA mRNA levels were assayed after adenovirus infection (bottom panel). Total RNA was harvested 72 hours after adenovirus infection and Northern analysis was performed.
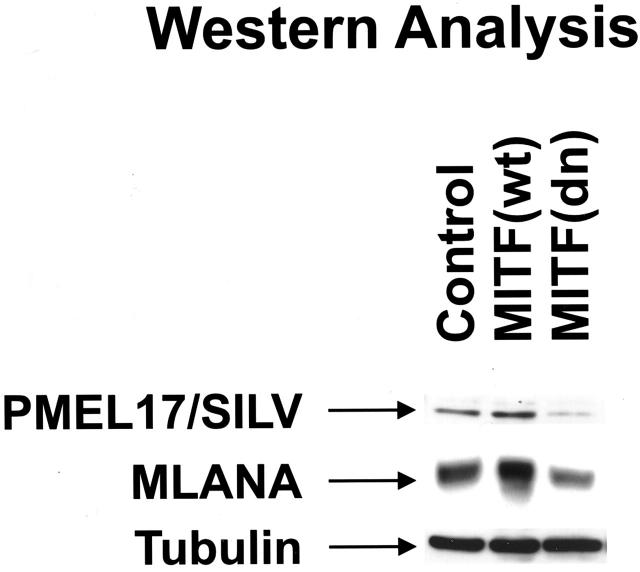
Protein levels of SILV and MLANA were also examined following up- or down-regulation of endogenous MITF using adenoviruses, as above. Western blotting for SILV and MLANA (Figure 5) ▶ revealed increases of both in the presence of wild-type MITF and decreases of both in the presence of dominant-negative MITF. These data are consistent with the hypothesis that MITF regulates expression of SILV and MLANA in the melanocyte lineage.
Figure 5.
Regulation of endogenous SILV and MLANA protein levels by MITF. Western analysis was performed on melanoma lysates harvested 72 hours after adenovirus infection.
Comparison of MITF, SILV, and MLANA Expression in Cultured Melanomas and Melanocytes
Next, we examined protein levels of these genes in a panel of 11 human melanoma cell lines as well as primary early passage human melanocytes. As shown in Figure 6 ▶ , a general pattern became apparent when comparing expression of MLANA, SILV, and MITF. A relatively clear pattern of correlated expression was seen for MITF, SILV, and MLANA in these specimens.
Figure 6.
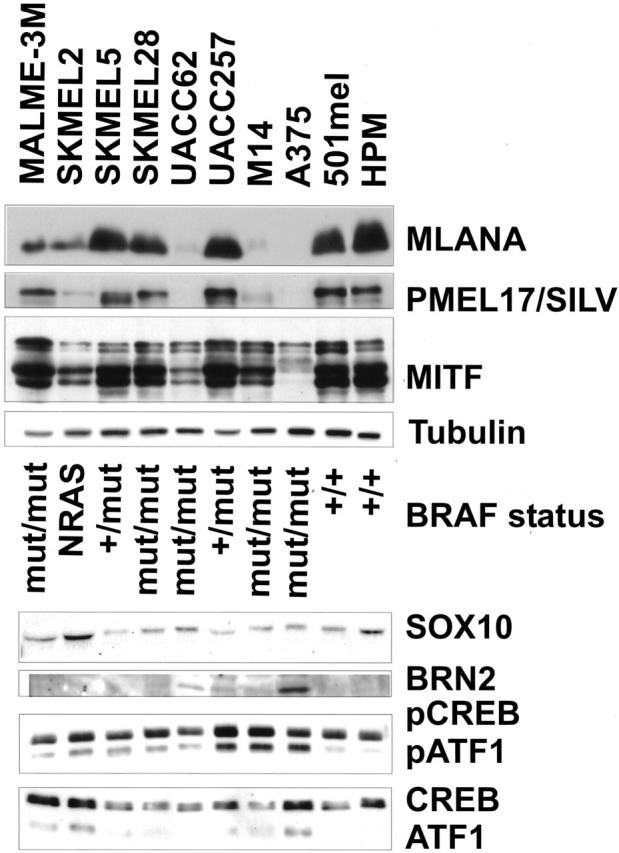
Correlation between MITF, MLANA, and SILV expression and BRAF mutation status. Western analysis were performed on cell lysates harvested from various melanoma cell lines and human primary melanomas with antibodies specified in Materials and Methods. BRAF mutation status was assessed by PCR and subsequent sequencing on corresponding genomic DNA. Whereas MITF and SILV expression correlated quite closely, no simple correlation was seen between MITF and BRAF mutational state or the various transcriptional regulators shown (which have been suggested to regulate MITF expression in specific contexts).
Overall, considerable variability was seen in MITF expression levels among the various cell sources. Yet, MITF is clearly expressed in all of these samples, including A375, which contains significantly less visible protein by Western blot. To determine whether other potential upstream factors may similarly correlate with the pattern of MITF, SILV, and MLANA expression (and therefore be candidate regulators of MITF), we examined the expression of several transcription factors that have been previously demonstrated to modulate MITF expression. SOX10 binds multiple sites in the MITF promoter in melanocytes, and its mutation in humans is associated with a similar pigmentation/deafness phenotype as MITF deficiency. 9,66-68 The BRN2 transcription factor has been suggested to regulate or interact with MITF. 69,70 CREB/ATF1 have been shown to modulate MITF expression in melanocytes through recognition of a consensus CRE element, in response to cAMP signaling which is downstream of the melanocyte stimulating hormone. 71-73 As shown in the bottom panel of Figure 6 ▶ , the expression levels of these factors did not correlate in a simple fashion with MITF or the other melanocytic markers.
In addition, post-translational modifications of MITF have also been observed. One of these is mediated by the MAP-kinase pathway, which leads to the phosphorylation of Ser73 in MITF by ERK1 and ERK2. 58 This post-translational modification leads to recruitment of the p300 coactivator and concomitant degradation of MITF protein. 57 Recently, it was found that oncogenic mutant BRAF(V599E) occurs in the majority of human melanomas studied. 74 Since BRAF activates the MAP kinase pathway, it might modulate MITF expression (either post-translationally or possibly even transcriptionally by unknown intermediates). This prompted us to analyze the mutational frequency of BRAF in the panel of melanomas and see whether this could explain the varied levels of MITF protein found (Figure 6 ▶ , middle). A weak trend suggested that most abundant expression of MITF, MLANA, and SILV occurs in the setting of wild-type BRAF, but expression was otherwise variable and did not correlate clearly with BRAF status. Quantitative RNA measurements similarly demonstrated no simple correlation between BRAF mutational status and MITF mRNA levels (data not shown). Thus, no simple relationship emerged from these studies of other transcription factors and regulatory pathways that might explain the variable expression of MITF, SILV, and MLANA among melanomas.
Analysis of SILV, MLANA and MITF Expression in Tumor Microarrays
Several of the cell lines used in our analysis of protein levels are part of an NCI database on which transcriptional profiling has been performed. 60 Since protein expression patterns reflect both transcriptional and post-transcriptional regulation, we sought verification at the mRNA level that MITF, SILV, and MLANA expression also correlated. Indeed, it has been demonstrated that at least one mechanism which likely confers amelanotic (unpigmented) phenotype to certain melanomas is the post-translational degradation of pigmentation factors, specifically tyrosinase. 15 As shown in Figure 7A ▶ , the levels of MITF, SILV, and MLANA fluctuate together in a similar pattern among the cell lines available for analysis.
Figure 7.
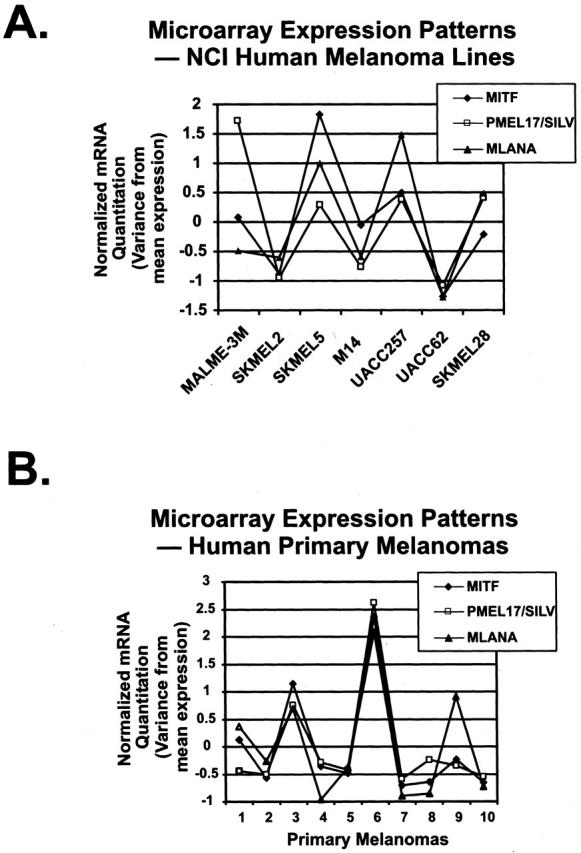
Correlations between MITF, SILV, and MLANA mRNA levels in cultured melanoma cell lines and human primary melanomas. NCI human melanoma cell lines (A) and human primary melanomas (B). Quantitative mRNA levels for all three genes were acquired from Affymetrix microarrays as described in Materials and Methods, and deviations from mean expression are plotted across each series.
Finally, we wished to examine whether mRNA expression levels of MITF, SILV, and MLANA correlate within primary clinical melanoma specimens (rather than cultured cell lines which have been subjected to selective pressures of unknown clinical relevance). We therefore examined microarray expression profile data from a large unbiased data set of primary human cancers, including a series of 10 melanoma clinical specimens. 61 To find genes highly correlated with endogenous MITF expression in melanomas, we identified the genes best matching the MITF expression pattern using a Pearson correlation coefficient ranking of similarity. 75 Of the 6466 genes analyzed, both SILV and MLANA were among the top 20 genes correlating with levels of MITF in the primary melanomas of this series (Figure 7) ▶ . Importantly, neither gene was significantly correlated with MITF expression in non-melanoma tumors due to lack of expression of SILV and MLANA (data not shown). Although this correlation does not prove a functional relationship between these factors, it utilizes a human primary tumor data set to help validate the model that MITF transcriptionally regulates the SILV and MLANA genes.
Discussion
The results of the present work demonstrate that the essential melanocyte-specific transcription factor MITF regulates expression of the genes encoding the melanoma tumor markers MLANA and SILV. Evidence in support of this model includes the presence of conserved MITF consensus DNA binding elements within enhancer/promoter elements of both genes that are shown to be bound by endogenous MITF protein in melanoma cells. Furthermore MITF regulates luciferase reporters for these genes in a manner dependent on those DNA elements. MITF up- or down-regulation is seen to correspondingly modulate expression of MLANA and SILV in parallel directions, at both mRNA and protein levels. Across several melanoma cell lines, MITF, SILV, and MLANA correlate in expression at the protein level. Finally, expression microarray data reveal that MITF, MLANA, and SILV behave similarly in cultured cell lines as well as in primary melanoma specimens.
The MITF gene contains multiple alternative promoters which are capable of contributing unique initial coding exons to the common body of the gene. One of the alternative promoters is melanocyte-specific and subject to MSH/cAMP regulation. 71-73 However, isoforms expressed from alternative promoters are present in other tissues, including osteoclasts and mast cells. As melanocyte-restricted target genes, MLANA and SILV will likely provide an opportunity to study whether their melanocyte restricted expression arises through unique activity of the melanocyte isoform of MITF or whether other transcription factors might contribute (together with MITF) to confer melanocyte-specific expression.
Levels of the Three Major Melanoma Markers Are Tightly Correlated with Each Other
Despite the generally correlated protein expression levels for MLANA, SILV, MITF, no clear transcriptional regulator upstream of MITF expression could be identified on the basis of matching expression levels. For certain transcription factors, key post-translational events (such as phosphorylation of CREB) may modulate activity more than expression levels. Another important possibility is that MITF is transcriptionally regulated in a combinatorial fashion by multiple upstream regulators, whose correlations with MITF expression would thus appear complex. This model is particularly attractive as a means of understanding how ubiquitous factors, such as CREB, can regulate tissue-restricted target genes. Through cooperative linkages with other upstream regulators at the MITF promoter, such combinatorial networks might produce highly restricted target gene expression, using relatively ubiquitous upstream factors.
We are also interested in exceptions to the simple correlations in MITF, SILV, and MLANA protein expression patterns, and have found a single melanoma cell line (not among the NCI cancer cell data set) for which MLANA expression appears under-represented relative to MITF and SILV (data not shown). Multiple models could account for this discrepancy, including the possibility that transcriptional regulators other than MITF may also be rate-limiting in specific contexts. Alternatively post-translational differences could account for such discrepancies. For example, Halaban and colleagues 15 have previously examined the mechanism underlying lack of pigment production in amelanotic melanomas, and discovered aberrant tyrosinase protein degradation. It is plausible that components of the pigmentation machinery as well as byproducts of pigmentation chemistry may be toxic to melanoma cells and therefore a selective pressure may exist to disrupt components of the pathway in selected tumors. Of note, previous studies have demonstrated that oncogenic transformation associated with dysregulation of E2F may be connected to down-regulation of MITF. 76 In fact, these studies led to the previous suggestion that MITF may transcriptionally regulate the SILV gene, as is mechanistically demonstrated here.
Melanoma Markers and Prognosis
It has been observed that low levels of MLANA and MITF correlate with a worse prognosis. 17,37,53-55 It is likely that in many of these tumors there is not a selective down-regulation of a single antigen, but rather a coordinated decrease of many target genes, governed largely by MITF and its central regulation of pigmentation as the most observable of differentiation pathways. Thus, higher levels of antigen expression represent a more differentiated state, while lower levels represent a less differentiated state. Such regulation could be of potential therapeutic value if MITF expression were modulated.
Modulating Antigen Expression via MITF
Notably absent from the literature are cases of melanoma antigen up-regulation in metastatic disease. The trend appears to be toward an expected state of a less differentiated phenotype with loss of many markers of differentiation. While previous work has previously linked MITF to the regulation of pigment genes and work presented here shows MITF regulation of SILV and MLANA, we have shown in an earlier study that MITF transcriptionally modulates the apoptosis modulator, Bcl-2. In addition, suppression of MITF through a dominant-negative mutant triggers apoptosis in melanoma cells while overexpression of Bcl-2 at least partially rescues this effect. 56 Bcl-2 expression also correlates closely with MITF in primary melanoma expression profiles. 56 Therefore, the worsened prognosis which has been suggested for melanomas expressing lower levels of MITF, occurs despite lower expression of the anti-apoptotic Bcl-2 factor. One model to explain this observation is that lower levels of MITF expression might select for tumor cells that are less differentiated (rather than expressing higher levels of survival genes). Additional studies will be needed to examine this question. Even though MITF expression fluctuates among melanomas, some level of staining is retained in nearly all melanomas and has been demonstrated to be a specific and sensitive diagnostic marker. Together, these observations underlie MITF’s apparent dual roles in regulating both survival and differentiation in the melanocyte lineage.
The cytokine α-MSH up-regulates MITF expression via a well-studied signal transduction pathway that also leads to increased pigment enzyme expression. 71-73 This pathway would therefore be expected to up-regulate MLANA and SILV expression as well. The combination of α-MSH treatment with immunotherapy directed against antigens such as MLANA and SILV could potentially lead to therapeutic benefit both by increasing tumor cell differentiation and increasing the expression of melanoma-associated antigens that have been associated with therapeutically meaningful responses.
Acknowledgments
We thank members of the Fisher lab for useful discussions and comments as well as Dr. Todd Golub for assistance with microarrays. A.J.M. is supported by a National Institutes of Health Medical Scientist Training Program grant and H.R.W. is a Swedish Wenner-Gren Foundation postdoctoral fellow. S.R. is supported by a medical fellowship from the National Institutes of Health.
Footnotes
Address reprint requests to David E. Fisher, M.D., Ph.D., Dana-Farber Cancer Institute and Children’s Hospital, Department of Pediatric Hematology/Oncology, Harvard Medical School, 44 Binney Street, Boston, MA 02115. E-mail: David_Fisher@dfci.harvard.edu.
Supported by National Institutes of Health grant AR43369 to D.E.F.
J.D. and A.J.M. contributed equally to this work.
Present address for M.A.H. is Universitäts Klinikum Eppendorf, Hamburg, Germany
References
- 1.Hodgkinson CA, Moore KJ, Nakayama A, Steingrimsson E, Copeland NG, Jenkins NA, Arnheiter H: Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell 1993, 74:395-404 [DOI] [PubMed] [Google Scholar]
- 2.Hemesath TJ, Steingrimsson E, McGill G, Hansen MJ, Vaught J, Hodgkinson CA, Arnheiter H, Copeland NG, Jenkins NA, Fisher DE: Microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev 1994, 8:2770-2780 [DOI] [PubMed] [Google Scholar]
- 3.Tachibana M, Perez-Jurado LA, Nakayama A, Hodgkinson CA, Li X, Schneider M, Miki T, Fex J, Francke U, Arnheiter H: Cloning of MITF, the human homolog of the mouse microphthalmia gene and assignment to chromosome 3p14.1-p12.3. Hum Mol Genet 1994, 3:553-557 [DOI] [PubMed] [Google Scholar]
- 4.Bentley NJ, Eisen T, Goding CR: Melanocyte-specific expression of the human tyrosinase promoter: activation by the microphthalmia gene product and role of the initiator. Mol Cell Biol 1994, 14:7996-8006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yasumoto K, Yokoyama K, Shibata K, Tomita Y, Shibahara S: Microphthalmia-associated transcription factor as a regulator for melanocyte-specific transcription of the human tyrosinase gene. Mol Cell Biol 1994, 14:8058-8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tassabehji M, Newton VE, Read AP: Waardenburg syndrome type 2 caused by mutations in the human microphthalmia (MITF) gene. Nat Genet 1994, 8:251-255 [DOI] [PubMed] [Google Scholar]
- 7.Amiel J, Watkin PM, Tassabehji M, Read AP, Winter RM: Mutation of the MITF gene in albinism-deafness syndrome (Tietz syndrome). Clin Dysmorphol 1998, 7:17-20 [PubMed] [Google Scholar]
- 8.Smith SD, Kelley PM, Kenyon JB, Hoover D: Tietz syndrome (hypopigmentation/deafness) caused by mutation of MITF. J Med Genet 2000, 37:446-448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Price ER, Fisher DE: Sensorineural deafness and pigmentation genes: melanocytes and the Mitf transcriptional network. Neuron 2001, 30:15-18 [DOI] [PubMed] [Google Scholar]
- 10.Silvers WK: Microphthalmia and other considerations. Silvers WK eds. The Coat Colors of Mice: A Model for Mammalian Gene Action and Interaction. 1979:pp 268-291 Springer-Verlag, New York
- 11.Lerner AB, Shiohara T, Boissy RE, Jacobson KA, Lamoreux ML, Moellmann GE: A mouse model for vitiligo. J Invest Dermatol 1986, 87:299-304 [DOI] [PubMed] [Google Scholar]
- 12.Steingrimsson E, Moore KJ, Lamoreux ML, Ferre-D’Amare AR, Burley SK, Zimring DC, Skow LC, Hodgkinson CA, Arnheiter H, Copeland NG, Jenkins NA: Molecular basis of mouse microphthalmia (mi) mutations helps explain their developmental and phenotypic consequences. Nat Genet 1994, 8:256-263 [DOI] [PubMed] [Google Scholar]
- 13.Carrel S, Rimoldi D: Melanoma-associated antigens. Eur J Cancer 1993, 29A:1903-1907 [DOI] [PubMed] [Google Scholar]
- 14.Zakut R, Perlis R, Eliyahu S, Yarden Y, Givol D, Lyman SD, Halaban R: KIT ligand (mast cell growth factor) inhibits the growth of KIT-expressing melanoma cells. Oncogene 1993, 8:2221-2229 [PubMed] [Google Scholar]
- 15.Halaban R, Cheng E, Zhang Y, Moellmann G, Hanlon D, Michalak M, Setaluri V, Hebert DN: Aberrant retention of tyrosinase in the endoplasmic reticulum mediates accelerated degradation of the enzyme and contributes to the dedifferentiated phenotype of amelanotic melanoma cells. Proc Natl Acad Sci USA 1997, 94:6210-6215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King R, Weilbaecher KN, McGill G, Cooley E, Mihm M, Fisher DE: Microphthalmia transcription factor: a sensitive and specific melanocyte marker for melanoma diagnosis. Am J Pathol 1999, 155:731-738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salti GI, Manougian T, Farolan M, Shilkaitis A, Majumdar D, Das Gupta TK: Micropthalmia transcription factor: a new prognostic marker in intermediate-thickness cutaneous malignant melanoma. Cancer Res 2000, 60:5012-5016 [PubMed] [Google Scholar]
- 18.Miettinen M, Fernandez M, Franssila K, Gatalica Z, Lasota J, Sarlomo-Rikala M: Microphthalmia transcription factor in the immunohistochemical diagnosis of metastatic melanoma: comparison with four other melanoma markers. Am J Surg Pathol 2001, 25:205-211 [DOI] [PubMed] [Google Scholar]
- 19.Granter SR, Weilbaecher KN, Quigley C, Fisher DE: Role for microphthalmia transcription factor in the diagnosis of metastatic malignant melanoma. Appl Immunohistochem Mol Morphol 2002, 10:47-51 [DOI] [PubMed] [Google Scholar]
- 20.Chang KL, Folpe AL: Diagnostic utility of microphthalmia transcription factor in malignant melanoma and other tumors. Adv Anat Pathol 2001, 8:273-275 [DOI] [PubMed] [Google Scholar]
- 21.Kwon BS, Halaban R, Ponnazhagan S, Kim K, Chintamaneni C, Bennett D, Pickard RT: Mouse silver mutation is caused by a single base insertion in the putative cytoplasmic domain of Pmel 17. Nucleic Acids Res 1995, 23:154-158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raposo G, Tenza D, Murphy DM, Berson JF, Marks MS: Distinct protein sorting and localization to premelanosomes, melanosomes, and lysosomes in pigmented melanocytic cells. J Cell Biol 2001, 152:809-824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi T, Urabe K, Orlow SJ, Higashi K, Imokawa G, Kwon BS, Potterf B, Hearing VJ: The Pmel 17/silver locus protein: characterization and investigation of its melanogenic function. J Biol Chem 1994, 269:29198-29205 [PubMed] [Google Scholar]
- 24.Chakraborty AK, Platt JT, Kim KK, Kwon BS, Bennett DC, Pawelek JM: Polymerization of 5, 6-dihydroxyindole-2-carboxylic acid to melanin by the pmel 17/silver locus protein. Eur J Biochem 1996, 236:180-188 [DOI] [PubMed] [Google Scholar]
- 25.Solano F, Martinez-Esparza M, Jimenez-Cervantes C, Hill SP, Lozano JA, Garcia-Borron JC: New insights on the structure of the mouse silver locus and on the function of the silver protein. Pigment Cell Res 2000, 13:118-124 [DOI] [PubMed] [Google Scholar]
- 26.Berson JF, Harper DC, Tenza D, Raposo G, Marks MS: Pmel17 initiates premelanosome morphogenesis within multivesicular bodies. Mol Biol Cell 2001, 12:3451-3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawakami Y, Eliyahu S, Delgado CH, Robbins PF, Sakaguchi K, Appella E, Yannelli JR, Adema GJ, Miki T, Rosenberg SA: Identification of a human melanoma antigen recognized by tumor-infiltrating lymphocytes associated with in vivo tumor rejection. Proc Natl Acad Sci USA 1994, 91:6458-6462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coulie PG, Brichard V, Van Pel A, Wolfel T, Schneider J, Traversari C, Mattei S, De Plaen E, Lurquin C, Szikora JP: A new gene coding for a differentiation antigen recognized by autologous cytolytic T lymphocytes on HLA-A2 melanomas. J Exp Med 1994, 180:35-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Maziere AM, Muehlethaler K, van Donselaar E, Salvi S, Davoust J, Cerottini JC, Levy F, Slot JW, Rimoldi D: The melanocytic protein Melan-A/MART-1 has a subcellular localization distinct from typical melanosomal proteins. Traffic 2002, 3:678-693 [DOI] [PubMed] [Google Scholar]
- 30.Wright A, Kawakami Y, Pavan W: Mart1 is located on mouse chromosome 19 and is excluded as a candidate for ep and ru. Mamm Genome 1997, 8:377-378 [DOI] [PubMed] [Google Scholar]
- 31.Adema GJ, de Boer AJ, van ’t Hullenaar R, Denijn M, Ruiter DJ, Vogel AM, Figdor CG: Melanocyte lineage-specific antigens recognized by monoclonal antibodies NKI-beteb, HMB-50, and HMB-45 are encoded by a single cDNA. Am J Pathol 1993, 143:1579-1585 [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner SN, Wagner C, Hofler H, Atkinson MJ, Goos M: Expression cloning of the cDNA encoding a melanoma-associated Ag recognized by mAb HMB-45: identification as melanocyte-specific Pmel 17 cDNA. Lab Invest 1995, 73:229-235 [PubMed] [Google Scholar]
- 33.Fetsch PA, Marincola FM, Filie A, Hijazi YM, Kleiner DE, Abati A: Melanoma-associated antigen recognized by T cells (MART-1): the advent of a preferred immunocytochemical antibody for the diagnosis of metastatic malignant melanoma with fine-needle aspiration. Cancer 1999, 87:37-42 [PubMed] [Google Scholar]
- 34.Fetsch PA, Marincola FM, Abati A: The new melanoma markers: mART-1 and Melan-A (the NIH experience). Am J Surg Pathol 1999, 23:607-610 [DOI] [PubMed] [Google Scholar]
- 35.Orchard GE: Comparison of immunohistochemical labelling of melanocyte differentiation antibodies melan-A, tyrosinase, and HMB 45 with NKIC3 and S100 protein in the evaluation of benign naevi and malignant melanoma. Histochem J 2000, 32:475-481 [DOI] [PubMed] [Google Scholar]
- 36.Orchard GE: Melan A (MART-1): a new monoclonal antibody for malignant melanoma diagnosis. Br J Biomed Sci 1998, 55:8-9 [PubMed] [Google Scholar]
- 37.Berset M, Cerottini JP, Guggisberg D, Romero P, Burri F, Rimoldi D, Panizzon RG: Expression of Melan-A/MART-1 antigen as a prognostic factor in primary cutaneous melanoma. Int J Cancer 2001, 95:73-77 [DOI] [PubMed] [Google Scholar]
- 38.Gown AM, Vogel AM, Hoak D, Gough F, McNutt MA: Monoclonal antibodies specific for melanocytic tumors distinguish subpopulations of melanocytes. Am J Pathol 1986, 123:195-203 [PMC free article] [PubMed] [Google Scholar]
- 39.Bacchi CE, Gown AM: Specificity of antibody HMB-45. Arch Pathol Lab Med 1992, 116:899-900 [PubMed] [Google Scholar]
- 40.Kageshita T, Kawakami Y, Hirai S, Ono T: Differential expression of MART-1 in primary and metastatic melanoma lesions. J Immunother 1997, 20:460-465 [DOI] [PubMed] [Google Scholar]
- 41.Nicotra MR, Nistico P, Mangoni A, Di Filippo F, Marincola FM, Natali PG: Melan-A/MART-1 antigen expression in cutaneous and ocular melanomas. J Immunother 1997, 20:466-469 [DOI] [PubMed] [Google Scholar]
- 42.Kaufmann O, Koch S, Burghardt J, Audring H, Dietel M: Tyrosinase, melan-A, and KBA62 as markers for the immunohistochemical identification of metastatic amelanotic melanomas on paraffin sections. Mod Pathol 1998, 11:740-746 [PubMed] [Google Scholar]
- 43.Blessing K, Sanders DS, Grant JJ: Comparison of immunohistochemical staining of the novel antibody melan-A with S100 protein and HMB-45 in malignant melanoma and melanoma variants. Histopathology 1998, 32:139-146 [DOI] [PubMed] [Google Scholar]
- 44.Sheffield MV, Yee H, Dorvault CC, Weilbaecher KN, Eltoum IA, Siegal GP, Fisher DE, Chhieng DC: Comparison of five antibodies as markers in the diagnosis of melanoma in cytologic preparations. Am J Clin Pathol 2002, 118:930-936 [DOI] [PubMed] [Google Scholar]
- 45.Jager E, Ringhoffer M, Karbach J, Arand M, Oesch F, Knuth A: Inverse relationship of melanocyte differentiation antigen expression in melanoma tissues and CD8+ cytotoxic-T-cell responses: evidence for immunoselection of antigen-loss variants in vivo. Int J Cancer 1996, 66:470-476 [DOI] [PubMed] [Google Scholar]
- 46.Kirkin AF, Dzhandzhugazyan K, Zeuthen J: Melanoma-associated antigens recognized by cytotoxic T lymphocytes. APMIS 1998, 106:665-679 [DOI] [PubMed] [Google Scholar]
- 47.Kawakami Y, Eliyahu S, Delgado CH, Robbins PF, Rivoltini L, Topalian SL, Miki T, Rosenberg SA: Cloning of the gene coding for a shared human melanoma antigen recognized by autologous T cells infiltrating into tumor. Proc Natl Acad Sci USA 1994, 91:3515-3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kirkin AF, thor Straten P, Hansen MR, Barfoed A, Dzhandzhugazyan KN, Zeuthen J: Establishment of gp100 and MART-1/Melan-A-specific cytotoxic T lymphocyte clones using in vitro immunization against preselected highly immunogenic melanoma cell clones. Cancer Immunol Immunother 1999, 48:239-246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhai Y, Yang JC, Kawakami Y, Spiess P, Wadsworth SC, Cardoza LM, Couture LA, Smith AE, Rosenberg SA: Antigen-specific tumor vaccines: development and characterization of recombinant adenoviruses encoding MART1 or gp100 for cancer therapy. J Immunol 1996, 156:700-710 [PubMed] [Google Scholar]
- 50.Kawakami Y, Rosenberg SA: Immunobiology of human melanoma antigens MART-1 and gp100 and their use for immuno-gene therapy. Int Rev Immunol 1997, 14:173-192 [DOI] [PubMed] [Google Scholar]
- 51.Rosenberg SA, Zhai Y, Yang JC, Schwartzentruber DJ, Hwu P, Marincola FM, Topalian SL, Restifo NP, Seipp CA, Einhorn JH, Roberts B, White DE: Immunizing patients with metastatic melanoma using recombinant adenoviruses encoding MART-1 or gp100 melanoma antigens. J Natl Cancer Inst 1998, 90:1894-1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dudley ME, Wunderlich JR, Robbins PF, Yang JC, Hwu P, Schwartzentruber DJ, Topalian SL, Sherry R, Restifo NP, Hubicki AM, Robinson MR, Raffeld M, Duray P, Seipp CA, Rogers-Freezer L, Morton KE, Mavroukakis SA, White DE, Rosenberg SA: Cancer regression and autoimmunity in patients after clonal repopulation with antitumor lymphocytes. Science 2002, 298:850-854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Selzer E, Wacheck V, Lucas T, Heere-Ress E, Wu M, Weilbaecher KN, Schlegel W, Valent P, Wrba F, Pehamberger H, Fisher D, Jansen B: The melanocyte-specific isoform of the microphthalmia transcription factor affects the phenotype of human melanoma. Cancer Res 2002, 62:2098-2103 [PubMed] [Google Scholar]
- 54.Marincola FM, Hijazi YM, Fetsch P, Salgaller ML, Rivoltini L, Cormier J, Simonis TB, Duray PH, Herlyn M, Kawakami Y, Rosenberg SA: Analysis of expression of the melanoma-associated antigens MART-1 and gp100 in metastatic melanoma cell lines and in in situ lesions. J Immunother Emphasis Tumor Immunol 1996, 19:192-205 [DOI] [PubMed] [Google Scholar]
- 55.Hofbauer GF, Kamarashev J, Geertsen R, Boni R, Dummer R: Melan A/MART-1 immunoreactivity in formalin-fixed, paraffin-embedded primary and metastatic melanoma: frequency and distribution. Melanoma Res 1998, 8:337-343 [DOI] [PubMed] [Google Scholar]
- 56.McGill GG, Horstmann M, Widlund HR, Du J, Motyckova G, Nishimura EK, Lin YL, Ramaswamy S, Avery W, Ding HF, Jordan SA, Jackson IJ, Korsmeyer SJ, Golub TR, Fisher DE: Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell 2002, 109:707-718 [DOI] [PubMed] [Google Scholar]
- 57.Wu M, Hemesath TJ, Takemoto CM, Horstmann MA, Wells AG, Price ER, Fisher DZ, Fisher DE: c-Kit triggers dual phosphorylations, which couple activation and degradation of the essential melanocyte factor Mi. Genes Dev 2000, 14:301-312 [PMC free article] [PubMed] [Google Scholar]
- 58.Hemesath TJ, Price ER, Takemoto C, Badalian T, Fisher DE: MAP kinase links the transcription factor microphthalmia to c-Kit signalling in melanocytes. Nature 1998, 391:298-301 [DOI] [PubMed] [Google Scholar]
- 59.Motyckova G, Weilbaecher KN, Horstmann M, Rieman DJ, Fisher DZ, Fisher DE: Linking osteopetrosis and pycnodysostosis: regulation of cathepsin K expression by the microphthalmia transcription factor family. Proc Natl Acad Sci USA 2001, 98:5798-5803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Staunton JE, Slonim DK, Coller HA, Tamayo P, Angelo MJ, Park J, Scherf U, Lee JK, Reinhold WO, Weinstein JN, Mesirov JP, Lander ES, Golub TR: Chemosensitivity prediction by transcriptional profiling. Proc Natl Acad Sci USA 2001, 98:10787-10792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramaswamy S, Tamayo P, Rifkin R, Mukherjee S, Yeang CH, Angelo M, Ladd C, Reich M, Latulippe E, Mesirov JP, Poggio T, Gerald W, Loda M, Lander ES, Golub TR: Multiclass cancer diagnosis using tumor gene expression signatures. Proc Natl Acad Sci USA 2001, 98:15149-15154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kawakami Y, Suzuki Y, Shofuda T, Kiniwa Y, Inozume T, Dan K, Sakurai T, Fujita T: T-cell immune responses against melanoma and melanocytes in cancer and autoimmunity. Pigment Cell Res 2000, 13(Suppl 8):163-169 [DOI] [PubMed] [Google Scholar]
- 63.Castelli C, Rivoltini L, Andreola G, Carrabba M, Renkvist N, Parmiani G: T-cell recognition of melanoma-associated antigens. J Cell Physiol 2000, 182:323-331 [DOI] [PubMed] [Google Scholar]
- 64.Aksan I, Goding CR: Targeting the microphthalmia basic helix-loop-helix-leucine zipper transcription factor to a subset of E-box elements in vitro and in vivo. Mol Cell Biol 1998, 18:6930-6938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Butterfield LH, Stoll TC, Lau R, Economou JS: Cloning and analysis of MART-1/Melan-A human melanoma antigen promoter regions. Gene 1997, 191:129-134 [DOI] [PubMed] [Google Scholar]
- 66.Verastegui C, Bille K, Ortonne JP, Ballotti R: Regulation of the microphthalmia-associated transcription factor gene by the Waardenburg syndrome type 4 gene, SOX10. J Biol Chem 2000, 275:30757-30760 [DOI] [PubMed] [Google Scholar]
- 67.Lee M, Goodall J, Verastegui C, Ballotti R, Goding CR: Direct regulation of the microphthalmia promoter by Sox10 links Waardenburg-Shah syndrome (WS4)-associated hypopigmentation and deafness to WS2. J Biol Chem 2000, 275:37978-37983 [DOI] [PubMed] [Google Scholar]
- 68.Bondurand N, Pingault V, Goerich DE, Lemort N, Sock E, Caignec CL, Wegner M, Goossens M: Interaction among SOX10, PAX3, and MITF, three genes altered in Waardenburg syndrome. Hum Mol Genet 2000, 9:1907-1917 [DOI] [PubMed] [Google Scholar]
- 69.Eisen T, Easty DJ, Bennett DC, Goding CR: The POU domain transcription factor Brn-2: elevated expression in malignant melanoma and regulation of melanocyte-specific gene expression. Oncogene 1995, 11:2157-2164 [PubMed] [Google Scholar]
- 70.Thomson JA, Murphy K, Baker E, Sutherland GR, Parsons PG, Sturm RA, Thomson F: The brn-2 gene regulates the melanocytic phenotype and tumorigenic potential of human melanoma cells. Oncogene 1995, 11:691-700 [PubMed] [Google Scholar]
- 71.Price ER, Horstmann MA, Wells AG, Weilbaecher KN, Takemoto CM, Landis MW, Fisher DE: α-Melanocyte-stimulating hormone signaling regulates expression of microphthalmia, a gene deficient in Waardenburg syndrome. J Biol Chem 1998, 273:33042-33047 [DOI] [PubMed] [Google Scholar]
- 72.Bertolotto C, Abbe P, Hemesath TJ, Bille K, Fisher DE, Ortonne JP, Ballotti R: Microphthalmia gene product as a signal transducer in cAMP-induced differentiation of melanocytes. J Cell Biol 1998, 142:827-835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Busca R, Ballotti R: Cyclic AMP a key messenger in the regulation of skin pigmentation. Pigment Cell Res 2000, 13:60-69 [DOI] [PubMed] [Google Scholar]
- 74.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA: Mutations of the BRAF gene in human cancer. Nature 2002, 417:949-954 [DOI] [PubMed] [Google Scholar]
- 75.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES: Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 1999, 286:531-537 [DOI] [PubMed] [Google Scholar]
- 76.Halaban R, Bohm M, Dotto P, Moellmann G, Cheng E, Zhang Y: Growth regulatory proteins that repress differentiation markers in melanocytes also down-regulate the transcription factor microphthalmia. J Invest Dermatol 1996, 106:1266-1272 [DOI] [PubMed] [Google Scholar]