Abstract
Lymphocyte homing to secondary lymphoid tissue and lesions of chronic inflammation is directed by multi-step interactions between the circulating cells and the specialized endothelium of high endothelial venules (HEVs). In this study, we used the PCR-based method of suppression subtractive hybridization (SSH) to identify novel HEV genes by comparing freshly purified HEV endothelial cells (HEVECs) with nasal polyp-derived microvascular endothelial cells (PMECs). By this approach, we cloned the first nuclear factor preferentially expressed in HEVECs, designated nuclear factor from HEVs (NF-HEV). Virtual Northern and Western blot analyses showed strong expression of NF-HEV in HEVECs, compared to human umbilical vein endothelial cells (HUVECs) and PMECs. In situ hybridization and immunohistochemistry revealed that NF-HEV mRNA and protein are expressed at high levels and rather selectively by HEVECs in human tonsils, Peyers’s patches, and lymph nodes. The NF-HEV protein was found to contain a bipartite nuclear localization signal, and was targeted to the nucleus when ectopically expressed in HUVECs and HeLa cells. Furthermore, endogenous NF-HEV was found in situ to be confined to the nucleus of tonsillar HEVECs. Finally, threading and molecular modeling studies suggested that the amino-terminal part of NF-HEV (aa 1–60) corresponds to a novel homeodomain-like Helix-Turn-Helix (HTH) DNA-binding domain. Similarly to the atypical homeodomain transcription factor Prox-1, which plays a critical role in the induction of the lymphatic endothelium phenotype, NF-HEV may be one of the key nuclear factors that controls the specialized HEV phenotype.
The endothelium serves as a critical interface between blood and tissue, but exhibits a remarkable heterogeneity among different vascular beds despite certain common features. 1,2 Thus, the endothelium adapts to the local demands by regulating the flow of nutrients, numerous biologically active molecules, and also the circulating blood cells themselves. This gate-keeping role of endothelial cells (ECs) is governed by their differential gene expression pattern, which depends on the type of blood vessel and underlying tissue.
One of the most striking examples of EC differentiation is the post-capillary high endothelial venules (HEVs) found in organized secondary lymphoid tissue. 3,4 Such vessels are particularly abundant in the T-cell zones that surround the B-cell follicles, and serve as entry sites for extravasating T and B lymphocytes. HEV-like vessels also occur in chronically inflamed non-lymphoid tissue and may mediate aberrant lymphocyte influx at such sites. In rheumatoid arthritis, HEV-like vessels are seen close to the joint cavity, surrounded by dense lymphoid infiltrates. 5 Furthermore, in Crohn’s disease and ulcerative colitis, collectively called inflammatory bowel disease (IBD), HEVs are found associated with extensive accumulations of lymphocytes. 6 Recently, HEV-like vessels were also found in nasal allergy and various chronic skin diseases, including lesions of cutaneous T-cell lymphomas. 7-9 Finally, endothelium in rejecting heart transplants also exhibit HEV-like characteristics that correlate with the severity of the rejection. 10 All these observations suggest that aberrant development of HEV-like vessels might mediate abnormal lymphocyte recruitment to the target tissue, thereby contributing to intensification and maintenance of chronic inflammation.
Lymphocyte recruitment in HEVs depends on sequential multi-step interactions between lymphocytes and HEVECs, 11 and is initiated by transient interactions between L-selectin on the lymphocyte microvilli and glycosylated and sulfated ligands on the HEV surface. This step is followed by chemokine activation of lymphocyte integrins via G protein-coupled chemokine receptors, resulting in firm adhesion mediated through interactions with their HEV ligands intercellular adhesion molecule (ICAM)-1/ICAM-2. Much progress has recently been made in the molecular understanding of this adhesion cascade, including the identification of the unique HEV-expressed sulfated carbohydrate ligands for L-selectin 12 and the contribution by HEVECs to lymphocyte integrin activation by luminal presentation of endogenous or perivascularly derived chemokines. 13,14
Although a few genes preferentially expressed in HEVECs have been identified, including the L-selectin ligand N-acetyl-glucosamine-6-O-sulfotransferase (LSST), 15-17 the fucosyltransferase FucTVII, 18,19 the chemokine CCL21 (SLC/6Ckine/TCA-4/exodus-2), 20 and the SPARC-like antiadhesive matricellular protein hevin, 21,22 extensive molecular characterization of the HEVEC phenotype has become possible only with recently developed protocols for the isolation 21 and culture of human and mouse primary HEVECs. 23,24 Nevertheless, such analysis is still hampered by the low number of cells available after purification, thereby ruling out traditional subtraction cloning techniques, which typically require several micrograms of mRNA. 25 To circumvent this problem, we previously adapted the PCR-based method of suppression subtractive hybridization (SSH) 26 to identify genes preferentially expressed in human tonsillar HEVECs compared with human umbilical vein endothelial cells (HUVECs). 27 With this method we generated a subtracted HEVEC cDNA library from 1 μg of total RNA, and were able to clone several HEV-expressed cDNAs, including the promiscuous chemokine receptor DARC, mitochondrial genes, and secreted extracellular matrix (ECM) proteins, such as mac25/IGFBP7/angiomodulin. 27 Thus we showed that SSH could be applied for cloning of differentiation-specific genes from a very limited starting material. This strategy has since been applied for characterization of ECs from several other vascular beds. 28-30 SSH was also recently used to clone the novel vascular endothelial junction-associated molecule (VE-JAM) from an HEVEC cDNA library. 31
To be reliable, SSH requires a low but significant enrichment of genes in the cells of interest compared with those used for subtraction. Therefore, to identify differentiation-specific genes from HEVECs, subtraction was not performed with HUVECs but with the more closely related and truly microvascular nasal polyp-derived microvascular endothelial cells (PMECs). 32 This strategy allowed us to identify, in addition to the matricellular protein hevin (which validated our approach), a nuclear factor preferentially expressed in HEVECs, designated nuclear factor from HEV (NF-HEV). NF-HEV mRNA was detected by in situ hybridization in HEVs from several human lymphoid tissues, including tonsils, Peyer’s patches, and mesenteric lymph nodes. Virtual Northern and Western blot analysis revealed preferential expression of NF-HEV in HEVECs, compared to two other types of ECs, namely microvascular PMECs and macrovascular HUVECs. NF-HEV exhibits a consensus bipartite nuclear localization sequence and localized to the nucleus when ectopically expressed in HUVECs. Immunohistochemistry performed on human tonsil sections showed a similar in situ nuclear localization of endogenous NF-HEV in HEVECs. Finally, threading and molecular modeling analyses indicated that NF-HEV contains an homeodomain-like Helix-Turn-Helix (HTH) motif in its amino-terminal part. Together, our results characterized the first nuclear factor preferentially expressed in HEVECs that may play a key role in the control of the specialized HEV phenotype.
Materials and Methods
Suppression Subtractive Hybridization
SSH was performed as described 27 with some modifications. Total RNA was isolated from highly purified HEVECs 23 cultured for 2 days with an RNeasy kit (Qiagen, Courtaboef, France). PMECs were prepared from nasal polyps as described, 32 stained with anti-CD34-FITC (Diatec, Oslo, Norway), and purified by cell sorting (FACSVantage, Becton Dickinson, San Jose, CA). PMEC mRNA was isolated by μMACS mRNA Isolation kit (Miltenyi Biotech, Bergisch Gladbach, Germany). To obtain sufficient amounts of double-stranded (ds) cDNA for subtraction, both PMEC and HEVEC cDNAs were preamplified with the SMART PCR cDNA Synthesis kit (Clontech, Palo Alto, CA). cDNAs synthesized from 1 μg of total RNA (HEVECs) or 0.15 μg mRNA (PMECs) with Advantage KlenTaq polymerase (22 cycles, Clontech) were used with the PCR Select cDNA Subtraction kit (Clontech). Briefly, PCR-generated HEVEC and PMEC cDNAs were digested with RsaI (New England Biolabs, Beverly, MA) and ligated to ds cDNA adaptors. For the first hybridization, the mixtures of HEVEC and PMEC cDNAs were incubated for 8 hours at 68°C. For the second hybridization, excess PMEC cDNA was added and incubated for 22 hours at 68°C. Differentially expressed cDNAs were then selectively amplified by two successive PCR (27 cycles) and nested PCR (10 cycles) reactions.
T/A cloning libraries of the subtracted cDNAs were prepared as described. 33 Briefly, the HEVEC-PMEC and PMEC-HEVEC subtracted mixtures (200 ng) were cloned directly into pCR2.1-TOPO (TA Cloning kit, Invitrogen, Carlsbad, CA) and introduced into One Shot Competent TOP10 cells (Invitrogen) according to the manufacturer. The bacteria were plated on agar plates containing 100 μg/ml ampicillin, 100 μmol/L isopropyl-β-D-thiogalactoside (IPTG), and 50 μg/ml X-Gal, and then grown until blue/white colonies appeared.
Differential Hybridization Screening with Subtracted Probes
A total of 960 individual recombinant (white) colonies were picked and used to inoculate ten 96-well micotiter plates with LB medium and 100 μg/ml ampicilin, which was incubated overnight and diluted 1:4 with H2O. This diluted bacterial culture (1 μl) was used to PCR amplify cloned inserts in 25 μl reactions with M13rev and M13for (-20) primers flanking the vector cloning site under the following conditions: 95°C for 5 minutes; 30 cycles each at 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 1 minute. The PCR reaction products (12 μl) were then loaded onto duplicate agarose gels (1.6% w/v), denatured, and blotted onto nylon membranes. The filters were hybridized with equivalent amounts of 32P-labeled cDNA of similar specific activity derived from HEVEC and PMEC total RNA as described. 27 Miniprep DNA of the differentially hybridizing clones was prepared and sequenced at Medigenomix (Martinsried, Germany) with the plasmid-specific TOPO1 and TOPO2 oligonucleotides.
Virtual Northern Blot Analysis
SMART PCR generated cDNAs from HEVECs, PMECs, first passage HUVECs, and placenta total RNA (0.5 μg per lane) were electrophoresed on 1.6% agarose gels, transferred onto nylon filters, and hybridized as described 27 with a 32P-labeled cDNA probe corresponding to the coding region of NF-HEV.
In Situ Hybridization
In situ mRNA hybridization was performed as described. 34 Briefly, digoxigenin-labeled riboprobes were generated from the NF-HEV cDNA with the DIG RNA Labeling kit according to the manufacturer’s directions (Boehringer Mannheim, Mannheim, Germany). Frozen tissue sections (8 μm) from human palatine tonsils, Peyer’s patches, and mesenteric lymph nodes were fixed in 4% paraformaldehyde (PFA)/DEPC-treated phosphate-buffered saline (PBS) and subsequently washed in PBS containing 0.1% active DEPC (Sigma, St. Louis, MO). After equilibration in 5X SSC, sections were prehybridized (2 hours at 59°C) in hybridization solution (50% formamide, 5X SSC, 50 μg/ml yeast tRNA, 100 μg/ml heparin, 1X Denhardt solution, 0.1% Tween 20, 0.1% CHAPS, and 5 mmol/L EDTA). Sections were subsequently hybridized overnight at 59°C with 250 ng/ml of riboprobe in hybridization solution. High stringency wash was performed, and the sections were next incubated (45 minutes) with horseradish peroxidase (HRP)-conjugated rabbit anti-DIG (1:50; DAKO, Glostrup, Denmark) in blocking buffer (0.1% Boehringer Blocking agent dissolved in Tris-HCl 100 mmol/L, NaCl 150 mmol/L, pH 7.5), followed by signal amplification with biotin-tyramide deposition (GenPoint kit; DAKO). Subsequently, sections were incubated (20 minutes) with HRP-conjugated rabbit anti-biotin (1:50 in blocking buffer; DAKO), followed by an additional cycle of biotin-tyramide deposition. Signal was detected by incubation (20 minutes) with alkaline phospahatase (AP)-conjugated rabbit anti-biotin (1:50 in blocking buffer; DAKO), followed by the AP substrate Fast Red (Ventana Medical Systems, Tucson, AZ). Finally, the sections were counterstained with hematoxylin.
Epitope Tagging
The coding region of NF-HEV was cloned into the PCDNA3.1A/myc-his (Invitrogen) by PCR amplification of the NF-HEV open reading frame with primers 5′-GAATTCTGAAAAATGAAGCCTAAAATGAAGTATTCAAC-3′ and 5′-GGGCCCAGTTTCAGAGAGCTTAAACAAGATATTTTCAG-3′, digested with EcoRI and ApaI, and cloned in frame with the myc tag of the PCDNA3.1A.
Cell Culture, Transfection, and Immunofluorescence Studies
HUVECs (kindly provided by A. Bouloumié, Toulouse, France) were grown in ECGM medium (Promocell, Heidelberg, Germany) and transfected in RPMI medium. HeLa cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal calf serum (FCS) and 1% penicillin-streptomycin (all from Invitrogen). HUVECs were plated on coverslips and transiently transfected with 0.7 μg PCDNA3.1A-NF-HEV-myc-his expression vector and Genejammer Transfection reagent according to the manufacturer’s instructions (Stratagene, La Jolla, CA). HeLa cells were plated on coverslips and transiently transfected with 2 μg PCDNA3.1A-NF-HEV-myc-his expression vector by the calcium phosphate method. After medium change, transfected cells were incubated for 48 hours to allow gene expression and then washed twice with PBS, fixed for 15 minutes at room temperature in PBS containing 3.7% PFA, and washed again with PBS before neutralization with 50 mmol/L NH4Cl in PBS for 5 minutes at room temperature. Cells were permeabilized for 5 minutes at room temperature in PBS containing 0.1% Triton-X100, and washed twice with PBS. Permeabilized cells were then incubated for 2 hours at room temperature with an anti-myc monoclonal antibody (IgG1, 7 μg/ml, Clontech) in PBS with 1% (w/v) bovine serum albumin (BSA). Cells were then washed three times for 5 minutes at room temperature in PBS-BSA, and incubated for 1 hour with FITC-labeled rabbit anti-mouse IgG (1:40; Amersham Pharmacia Biotech, Paris, France) or Cy3-conjugated goat anti-mouse IgG (1:1000; Amersham Pharmacia Biotech). After extensive washing in PBS, samples were air-dried and mounted in Mowiol (Hoechst Pharmaceuticals, Frankfurt, Germany). Fluorescence of fixed immunostained cells was viewed with a Leica confocal laser-scanning microscope.
Antibody production, Immunohistochemistry, and Western Blotting
Rabbit polyclonal antibodies were raised against the peptides DKVLLSYYESQHPSNC and CYFRRETTKRPSLKTG, corresponding to amino acids 157–171 and 58–73 of the human NF-HEV sequence, respectively, using multiple antigen peptides technology (Eurogentec, Seraing, Belgium). The antisera wereapplied in immunohistochemistry as previously described. 27 In brief, acetone-fixed sections (4 μm) of human palatine tonsils were first incubated with amixture of mAb MECA-79 (rat IgM, 1:30; courtesy of E.C. Butcher, Stanford, CA) and anti-NF-HEV rabbit antiserum (1:1000), followed by a mixture of Cy3-conjugated goat anti-rat IgM (1:200; Jackson ImmunoResearch, West Grove, PA) and Alexa Fluor 488-conjugated goat anti-rabbit IgG (Molecular Probes, Eugene, OR). The sections were mounted with 4′6-diamidino-2-phenylindole (DAPI)-containing Vectashield (Vector, Burlingame, CA). Negative controls were tissue sections incubated with concentration-matched irrelevant rat IgM and preimmune rabbit serum. Lysates from purified HEVECs and primary cultures of PMECs and HUVECs (each corresponding to ∼105 cells) were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10%). Detection was performed with rabbit antiserum to NF-HEV (1:500), followed by HRP-conjugated donkey anti-rabbit Ig (1:1000; Amersham), and finally an enhanced chemiluminescence kit (Pierce, Rockford, IL).
Threading and Molecular Modeling Analyses
We used the InsightII, SeqFold, Homology and Discover modules from the Accelrys (San Diego, CA) molecular modeling software (version 98), run on a Silicon Graphics O2 workstation. Structural homologs of human NF-HEV were searched with the SeqFold threading program, 35 which combines sequence and secondary structure alignment. Optimal secondary structure prediction of the query protein domains was ensured by the DSC method 36 within SeqFold. The engrailed homeodomain (PDB code: 1DU0) was identified as the best structural template of the NF-HEV amino-terminal domain (NF-HEV aa 1–65). We used the threading-derived secondary structure alignments as input for homology-modeling, which was performed according to a previously described protocol. 37 The validity of the models was checked both by Ramachandran analysis and folding consistency verification as previously reported. 37
Results
Identification of the NF-HEV cDNA as a cDNA Preferentially Expressed in HEVECs
To identify cDNAs preferentially expressed in HEVEC, we generated a PCR Select library from HEVEC cDNA subtracted against PMEC cDNA (HEVEC-PMEC). 27 MECA-79-positive HEVECs were purified from human tonsils 23 and PMECs were isolated from nasal polyps as described. 32 A total of 960 clones were obtained in the PCR-select HEVEC-PMEC cDNA library. Differential screening of these 960 clones with radioactive probes generated from HEVEC or PMEC cDNAs, revealed 49 cDNAs preferentially expressed in HEVECs. Sequencing of these cDNAs showed that the most abundant family of genes was mitochondrial enzymes (12 clones), particularly transcripts for cytochrome c oxidase 1. This was in line with our previous report 27 that HEVECs express higher levels of these enzymes than other ECs. Our screen also identified three independent clones corresponding to the secreted matricellular protein hevin, one of the known markers of tonsillar HEVECs. 21,27 Using two distinct polyclonal antisera, we confirmed preferential expression of hevin in MECA-79-positive-HEVECs from human tonsils, as well as MadCAM-1-positive-HEVECs from human Peyer’s patches (data not shown). In addition to the hevin clones, which validated our HEVEC-PMEC SSH approach, we identified several other cDNAs corresponding to previously characterized genes, including endothelial multimerin (four clones), which is a secreted homomultimeric factor V-binding protein, 38 the complement inhibitor CD59 (two clones), and the Nck adaptor protein NCK1 (two clones). Abundant expression of multimerin and CD59 in HEVECs was confirmed by immunohistochemistry on human tonsil sections (data not shown).
Among the sequences corresponding to human genes not yet characterized, we focused on one that was represented by four distinct cDNA clones within the HEVEC-PMEC library. To assess the tissue distribution of this gene, we prepared riboprobes corresponding to the open reading frame, which were applied for in situ mRNA hybridization (Figure 1) ▶ . Strikingly, the antisense riboprobe hybridized strongly to HEVs in the T-cell zones of human tonsil (Figure 1A) ▶ , Peyer’s patch (Figure 1B) ▶ , and mesenteric lymph node (Figure 1C) ▶ . Indeed, higher magnification clearly revealed hybridization signals within HEVECs as well as in scattered cells in the T-and B-cell zones. Hybridization with a sense probe produced no signal (Figure 1, A–C) ▶ . Based on these in situ results, which confirmed preferential expression of this gene in human HEVs in vivo, and the localization of the corresponding protein in the cell nucleus (see below), we designated this gene nuclear factor from HEV (NF-HEV).
Figure 1.
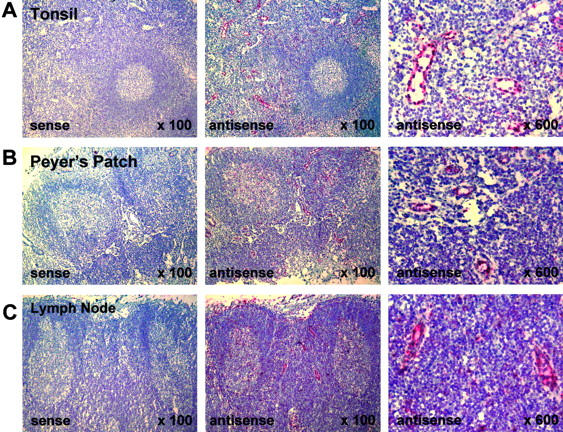
NF-HEV mRNA expression in HEVs of human tonsil, Peyer’s patch, and mesenteric lymph node. In situ hybridization was performed on paraformaldehyde-fixed sections with an RNA probe complementary to NF-HEV mRNA (antisense), and hybridization signal (red) occurs in HEVs of the T-cell zone around lymphoid follicles in tonsil (A), Peyer’s patch (B), and mesenteric lymph node (C). Higher magnification (×600, right panels) reveals that the signal is confined to HEVECs and scattered cells in the T- and B-cell zones. Hybridization with a sense probe produced no signal (left panels).
Sequence of the NF-HEV cDNA and Genomic Structure of the NF-HEV Gene
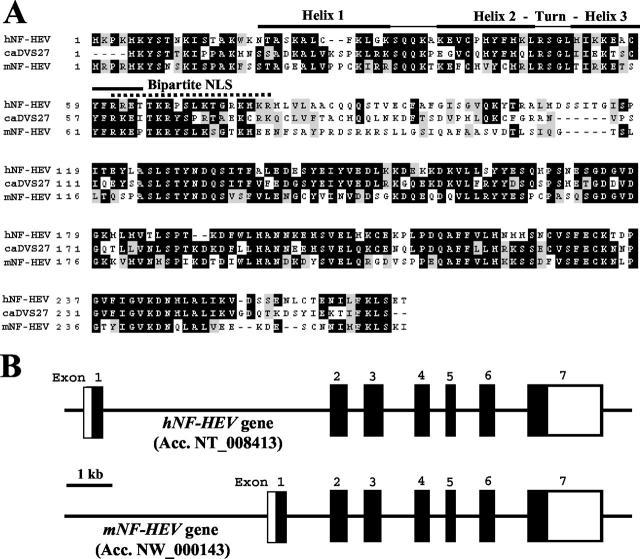
Sequencing of the four NF-HEV cDNA clones isolated from the PCR-select HEVEC-PMEC cDNA library revealed a sequence identical to that of a human cDNA deposited in GenBank with the annotation “Homo sapiens mRNA for DVS27-related protein” (GenBank Accession No.AB024518). This cDNA appears to encode a putative human ortholog of the canine DVS27 protein, previously identified in a screen for genes differentially expressed in canine vasospastic cerebral arteries after subarachnoid hemorrhage. 39 Database searches with both the nucleotide and amino acid sequences of canine DVS27 (GenBank Accession No. AB024517), using the programs BLASTN, TBLASTN, and BLASTP (GenBank non-redundant, human htgs, and human EST databases at National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/), failed to reveal any other human cDNA or protein more closely related to DVS27 than NF-HEV. This further suggested that human NF-HEV is an ortholog of canine DVS27. Two murine cDNAs encoding a putative mouse ortholog of human NF-HEV (GenBank Accession Nos. XM 123362 and NM 133775) were also identified by searching GenBank databases with the human NF-HEV sequence. Alignment of the human and mouse NF-HEV proteins (48% identity over 270 residues) with the canine DVS27 sequence (56% identity between hNF-HEV and canine DVS27) revealed that the NF-HEV/DVS27 protein is composed of two evolutionary conserved regions separated by a highly divergent linker region in the central part (Figure 2A) ▶ .
Figure 2.
Human and mouse NF-HEV proteins and genes. A: Amino acid sequence alignment of human NF-HEV (hNF-HEV) with its mouse ortholog (mNF-HEV) and canine DVS27 (caDVS27). Conserved residues are boxed. Black boxes indicate identical residues, whereas shaded boxes show similar amino acids. Dashed lines represent gaps introduced to align sequences. Sequence alignment was performed with ClustalW (http://www2.ebi.ac.uk/clustalw) and colored with Boxshade (http://www.ch.embnet.org/software/BOX_form.html). The bipartite NLS and the three helices of the homeodomain-like HTH putative DNA-binding motif are indicated. B: Genomic structure of the human and mouse NF-HEV genes. Open boxes indicate non-translated exon sequence and black boxes coding exon sequence. The two genes share a similar organization with seven exons. A major difference is the size of the first intron, which is >9 kb in the human gene but only ∼2 kb in its mouse ortholog.
A BLAST search of the non-redundant sequence database at NCBI with human NF-HEV cDNA or protein sequences as baits, revealed a genomic hit from the Homo sapiens chromosome 9 sequence (GenBank Accession No. NT 008413) that covered the whole NF-HEV cDNA. This genomic contig contains three independent UniSTS (UniSTS entries: SHGC-15129, stSG27179, and RH101248) that have been previously mapped at 9p24.1, between microsatellite markers D9S178 and D9S168. This suggested that the human NF-HEV gene is located on the short arm of chromosome 9 at 9p24.1. Alignment between the NF-HEV cDNA and genomic sequences revealed that there are seven exons that span more than 16 kb of genomic DNA (Figure 2B) ▶ . All of the exon-intron boundaries followed the GT-AG rule. The human NF-HEV gene shared a similar organization with its mouse ortholog (Figure 2B) ▶ , that we identified in a Mus musculus genomic contig (GenBank Accession No. NW 000143). The size of exons were found to be strictly conserved between the two species, with the exception of exon 3 that contains 15 additional nucleotides in the human sequence, corresponding to an insertion of five residues in the middle of the human NF-HEV protein (Figure 2A) ▶ .
NF-HEV Is a Nuclear Protein
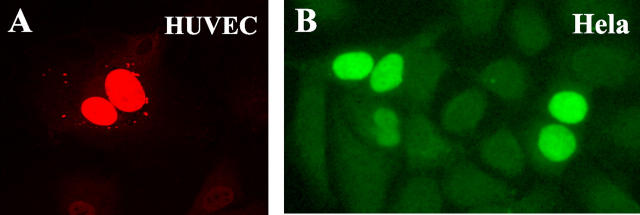
Because the predicted NF-HEV amino acid sequence contains a consensus bipartite nuclear localization sequence (NLS, Prosite PS00015), near the linker region (Figure 2A) ▶ , we decided to investigate whether NF-HEV could localize to the nucleus of ECs. For that purpose, we designed an expression construct with NF-HEV fused to the c-myc epitope tag, which was transfected into primary HUVECs and detected by indirect immunofluorescence staining with antibodies to myc. Confocal immunofluorescence microscopy revealed a strict intranuclear localization of the epitope-tagged NF-HEV (Figure 3A) ▶ . The myc-tagged-NF-HEV protein also localized to the nucleus when ectopically expressed in HeLa cells (Figure 3B) ▶ , NIH 3T3, and COS-7 cells (data not shown), suggesting that the nuclear localization of NF-HEV is not a specific property of ECs.
Figure 3.
NF-HEV encodes a nuclear protein. A–B: Nuclear localization of epitope-tagged NF-HEV ectopically expressed in primary HUVECs or immortalized HeLa epithelial cells. HUVECs (A) and HeLa cells (B) transfected with myc-tagged NF-HEV expression vector were stained by indirect immunofluorescence with antibodies to myc and analyzed by confocal laser scanning microscopy. Original magnification, ×1000.
Preferential Expression of NF-HEV in HEVECs Compared with Other EC Types
Preferential expression of NF-HEV in human HEVECs was confirmed by virtual Northern and Western blot analyses. Virtual Northern blot analysis of PCR-generated full-length cDNA from human HEVECs, PMECs, HUVECs, and placenta with a cDNA probe corresponding to the NF-HEV open reading frame, revealed a prominent band of ∼2.6 kb in HEVECs (Figure 4A) ▶ ; it agreed well with the size of NF-HEV mRNA. Conversely, this 2.6-kb signal was detected at only very low levels in PMECs and HUVECs and was almost undetectable in placenta (Figure 4A) ▶ . To confirm such preferential expression of NF-HEV in HEVECs at the protein level, we raised rabbit antibodies against NF-HEV peptides. By immunoblotting, these antibodies recognized a ∼30-kd protein in lysates from tonsil stroma and purified HEVECs, but not in PMECs or HUVECs (Figure 4B) ▶ . The apparent molecular weight of ∼30 kd for endogenous NF-HEV was in agreement with the predicted Mw of 31 kd and the size of a recombinant NF-HEV protein produced in Escherichia coli (data not shown).
Figure 4.
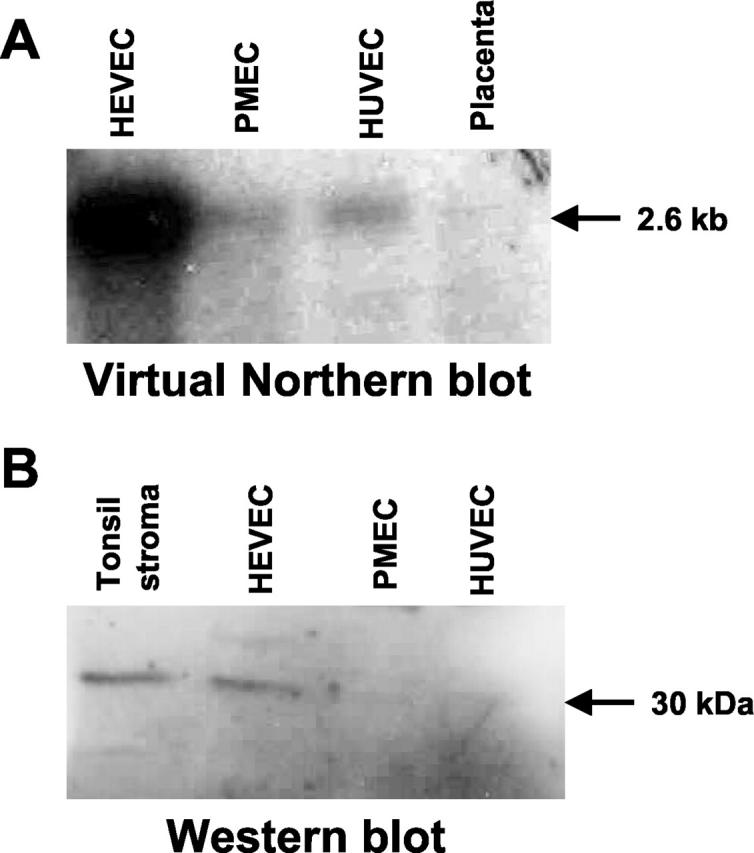
Virtual Northern and Western blot analyses demonstrating preferential expression of NF-HEV in HEVECs. A: Virtual Northern blot analysis of NF-HEV expression in HEVECs, PMECs, HUVECs, or placenta tissue. PCR-generated full-length cDNAs from the various types of ECs were electrophoresed on a 1% agarose gel, transferred to nylon filters, and hybridized under high-stringency conditions with a 32P-labeled human NF-HEV cDNA probe. B: Western blot analysis of extracts of tonsillar stroma, HEVECs, PMECs, or HUVECs with rabbit antibodies to NF-HEV. A single band of ∼30 kd was detected in extracts of tonsillar stroma and HEVECs.
Endogenous NF-HEV Localizes to the Nucleus of HEVECs in Situ
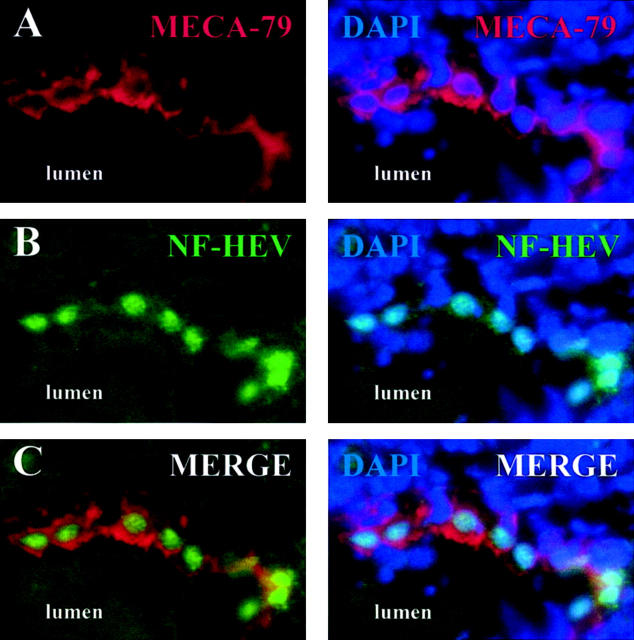
To determine the subcellular localization of NF-HEV in HEVECs in situ, we performed immunohistochemistry with the rabbit antibodies raised against NF-HEV peptides. Immunostaining of human tonsil sections with these reagents demonstrated strong NF-HEV expression in MECA-79-positive HEVs (Figure 5A–C) ▶ . NF-HEV and MECA-79 immunostainings overlapped and NF-HEV appeared to be expressed in all MECA-79-positive HEVs but not in other vessels. Costaining of nuclear DNA with DAPI further showed that NF-HEV was concentrated in the nucleus of HEVECs (Figure 5 ▶ , right panels). Lower magnification also revealed that the antibodies to NF-HEV decorated, scattered single cells (the identity of which remains unknown) in the T- and B-cell zones (data not shown), in addition to the MECA-79-positive-HEVs. This result was consistent with the in situ hybridization results described above (Figure 1) ▶ . In conclusion, although NF-HEV expression did not appear to be strictly HEV-specific, our immunohistochemistry data clearly revealed abundant in vivo expression of NF-HEV in the nucleus of HEVECs.
Figure 5.
In situ expression of NF-HEV protein in the nucleus of tonsillar HEVECs. Cryosections of human tonsils (4 μm, acetone-fixed) were double-stained with HEV-specific rat mAb MECA-79 (A) or antibodies to NF-HEV peptides (B). C: Two-color overlays reveal that NF-HEV immunoreactivity is associated with MECA-79-positive HEVECs. Counterstaining with the nuclear dye DAPI showed a clear nuclear localization of NF-HEV in MECA-79-positive HEVECs (right panels). No nuclear staining was observed with preimmune rabbit serum (not shown). Original magnification, ×600.
NF-HEV Contains a Putative Homeodomain-Like Helix-Turn-Helix DNA-Binding Domain
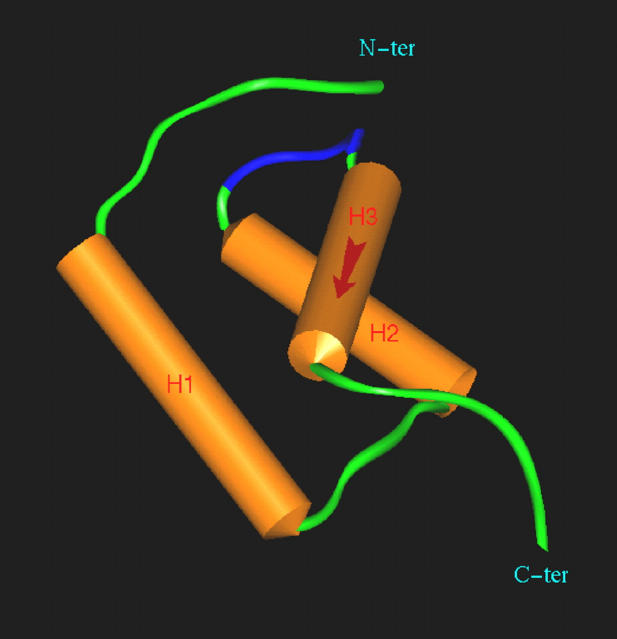
Searches in Prosite and Pfam databases with the NF-HEV sequence failed to reveal significant similarities to previously characterized protein sequence motifs, except for a low level of homology of the NF-HEV amino acids 28 to 68 with prokaryotic HTH DNA-binding domains (HTH ARAC, Prosite PS00041). Because detection of sequence homology is more sensitive and selective when aided by secondary structure information, we searched for structural homologs of human NF-HEV in the PDB crystallographic database, with the SeqFold threading program 35 which combines sequence and secondary structure alignment. This search revealed significant structural homologies between the first 65 amino-terminal residues of NF-HEV and the DNA-binding homeodomains of several drosophila (engrailed, fushi-tarazu) and vertebrate (POU) transcription factors. The crystallographic structure (PDB No. 1DU0) of the drosophila transcription factor engrailed homeodomain 40 provided the best score of the search. We used the resulting threading-derived secondary structure alignment, to generate a homology-based model for the amino-terminal domain of human NF-HEV (Figure 6 ▶ ; see the Materials and Methods section for a detailed account of the model-building and structural check protocols). Similarly to the homeodomain 41 and various other eukaryotic HTH DNA-binding domains (human centromere protein CENP-B, human Myb transcription factor, and yeast telomere binding protein RAP1), 42 NF-HEV was predicted to contain a homeodomain-like HTH motif that could be described as a right-handed three-helical bundle, 40,41 composed of an hydrophobic core of two α-helices (helices 2 and 3 corresponding to the HTH motif) completed by another N-terminal α-helix (helix 1). A conserved characteristic of this HTH motif is the packing of α-helices 2 and 3 at nearly a right angle to each other (Figure 6) ▶ ; the turn between α-helices 2 and 3 offsets α-helix 3 so that the N-terminal part of α-helix 3, which is predicted to bind to the target DNA major groove, is packed against the middle of α-helix 2. Together, our threading and modeling results suggested that the amino-terminal part of NF-HEV (aa 1–65) corresponds to a novel homeodomain-like HTH DNA-binding domain.
Figure 6.
NF-HEV contains a homeodomain-like HTH motif. Model of the three-dimensional structure of the homeodomain-like HTH motif of human NF-HEV (aa 1–65), based on its threading-derived homology with the crystallographic structure of the homeodomain from drosophila transcription factor engrailed (PDB code: 1DU0). The α-helices have been numbered in order and color-coded in brown. The potential DNA recognition helix (α-helix 3) is marked by a red arrow. The turn of the HTH motif is coded in blue. Molecular modeling was performed as described in the Materials and Methods.
Discussion
Despite intensive efforts, the specialized HEV endothelium remains poorly characterized at the molecular level, and only a few genes preferentially expressed in HEVECs have been identified. 15,16,18,20,21,43 Molecular characterization of HEVECs has been hampered by the fact that these cells, like other specialized ECs, are not abundantly available for analysis and lose their specialized phenotype rapidly when grown in vitro isolated from their natural tissue environment. 3,44 In this study, we used the PCR-based method of SSH to compare freshly purified HEVECs with nasal microvascular PMECs. This strategy allowed us to identify NF-HEV, the first nuclear factor preferentially expressed in HEVECs. In situ hybridization revealed high levels of NF-HEV mRNA in HEVs from human tonsils, mesenteric lymph nodes, and Peyer’s patches. Virtual Northern and Western blot analyses confirmed preferential expression of NF-HEV mRNA and protein in HEVECs compared with another type of microvascular ECs (PMECs) and ECs from large vessels (HUVECs). Finally, immunohistochemistry on human tonsillar sections with antibodies to NF-HEV peptides demonstrated that the NF-HEV protein accumulates at high levels in the nucleus of MECA-79-positive HEVECs in situ. This nuclear localization of NF-HEV in vivo is in agreement with the presence of a consensus bipartite NLS in the amino-terminal part of the protein. Also notably, we observed nuclear targeting of an epitope-tagged NF-HEV protein when ectopically expressed in primary HUVECs and immortalized HeLa epithelial cells.
In addition to the consensus NLS, NF-HEV contains another protein motif (homeodomain-like HTH motif) that exhibits structural similarity with several DNA-binding domains, including the HTH motifs of prokaryotic regulators and the homeodomains of vertebrate POU transcription factors as well as the drosophila transcription factors fushi-tarazu and engrailed. 41 Our molecular modeling studies predict that this putative NF-HEV DNA-binding domain may fold, similarly to the homeodomain, as a right-handed three-helical bundle, composed of an hydrophobic core of two α-helices corresponding to the HTH motif, which is completed by another N-terminal α-helix. To this end, we used a threading method that can reliably detect structural homology between protein domains that show only some 20% sequence homology. 45 The critical issue is then to determine whether the two domains found to be structurally related, are distant parents from the same phylogenetic superfamily or are the products of convergent evolution without common ancestry. Further studies will therefore be required to determine whether the NF-HEV homeodomain-like HTH motif is a distant relative of the homeodomain, which possesses similar sequence-specific DNA-binding properties.
NF-HEV may, in fact, be one of the nuclear factors that participates in the control of the specialized HEV phenotype. Thus, NF-HEV may play a role in HEV differentiation analogous to the role of the atypical homeodomain transcription factor Prox-1 in the induction of the lymphatic endothelium phenotype. 46-48 Recent studies have shown that ectopic expression of this transcription factor in blood vascular ECs is sufficient for expression of lymphatic endothelium-specific genes such as VEGFR-3. 48 These results suggest that Prox-1 is a major regulator of the lymphatic EC phenotype. In contrast, none of the nuclear factors likely to play important roles in the induction of the HEV phenotype have yet been identified. It has been known for a long time that the specialized features of HEVs are under the control of the local tissue environment. 3 In contrast to non-lymphoid post-capillary venules, HEVs are present in a microenvironment with many cytokines generated during activation of abundant lymphoid cells. Evidence for a key role (either direct or indirect) of these cytokines in the induction and maintenance of the HEV specialized phenotype has been provided by in vivo studies in transgenic mice, which have revealed that structural and functional characteristics of HEVs are induced on HEV-like vessels in the pancreas of transgenic mice whose pancreatic β cells express the chemokines CCL21 49 or CXCL13, 50 or the cytokines IFN-γ, IL-10, 3 or lymphotoxin. 51 The induction of HEV-specific genes by these cytokines is likely to be mediated by several distinct nuclear transcription factors. Because we showed that NF-HEV is a putative DNA-binding protein preferentially expressed in HEVECs, it is an attractive candidate for mediation of the cytokine effects that induce HEV-specific genes.
NF-HEV did not appear to be a strictly HEV-specific factor because human lymphoid organs (tonsils, mesenteric lymph nodes and Peyer’s patches) showed NF-HEV expression both at the mRNA and protein level in scattered single cells of the T-cell zone. In addition, the probable canine ortholog (DVS27) of human NF-HEV has previously been found to be expressed at the mRNA level in cultured smooth muscle cells stimulated with IL-1 or IFN-γ, and in vasospastic cerebral arteries after subarachnoid hemorrhage in a canine cerebral vasospasm model. 39 Similarly to NF-HEV, the lymphatic endothelium transcription factor Prox-1 is expressed in some non-EC types such as intestinal and pancreatic epithelial cells, hepatocytes, and cardiomyocytes. 52 However, Prox-1 is able to induce the lymphatic endothelium marker VEGFR3 in ECs but not in epithelial cells, 48 suggesting that the role of Prox-1 as a regulator of the lymphatic endothelium phenotype most likely requires the presence of other EC-specific transcriptional co-activators. A similar scenario may apply to NF-HEV; despite its expression in some non-EC types, NF-HEV may nevertheless be one of the key nuclear factors for the induction of the HEV phenotype.
Even though the nuclear localization and structural homology with DNA-binding domains is consistent with the idea that NF-HEV functions as a transcription factor, NF-HEV might have other functions in the nucleus. For instance, NF-HEV may play a role in chromatin dynamics, RNA or DNA metabolism (including DNA replication and repair), nuclear signaling pathways, or nucleo-cytoplasmic trafficking of nuclear components. Future studies should aim at investigating the biological role of NF-HEV in the nucleus and its functional role in HEV differentiation.
Acknowledgments
We thank Dr. Hege S. Carlsen for the generous gift of Peyer’s patch and mesenteric lymph node specimens. We also thank Dr. A. Bouloumié (Toulouse, France) and the staff at the Departments of Obstetrics and ENT surgery of the Rikshospitalet for kindly providing primary human endothelial cells or relevant material. Furthermore, Inger Johanne Ryen is gratefully acknowledged for expert technical assistance. Special thanks go to other members of the LIIPAT staff (Oslo, Norway) and the Laboratory of Vascular Biology (IPBS, Toulouse, France) for stimulating discussions.
Footnotes
Address reprint requests to Dr. Guttorm Haraldsen, Department and Institute of Pathology, University of Oslo, Rikshospitalet, Oslo, Norway. E-mail: guttorm.haraldsen@labmed.uio.no or to Dr. Jean-Philippe Girard, IPBS-CNRS UMR 5089, 205 Route de Narbonne, 31077 Toulouse, France. E-mail: Jean-Philippe.Girard@ipbs.fr.
Supported by grants from the Research Council of Norway (139324/300), the Norwegian Cancer Society (B02085), Centre National de la Recherche Scientifique “Puces à Acide DésoxyriboNucléique,” Ministère de la Recherche Action Concertée Incitative “Jeunes chercheurs” et “Génopôle-Toulouse Midi-Pyrénées-Santé,” Fondation de France, Région Midi-Pyrénées, and Association pour la Recherche sur le Cancer. G.H. is a Career Investigator of the Research Council of Norway
References
- 1.Cines DB, Pollak ES, Buck CA, Loscalzo J, Zimmerman GA, McEver RP, Pober JS, Wick TM, Konkle BA, Schwartz BS, Barnathan ES, McCrae KR, Hug BA, Schmidt AM, Stern DM: Endothelial cells in physiology and in the pathophysiology of vascular disorders. Blood 1998, 91:3527-3561 [PubMed] [Google Scholar]
- 2.Risau W: Differentiation of endothelium. EMBO J 1995, 9:926-933 [PubMed] [Google Scholar]
- 3.Girard JP, Springer TA: High endothelial venules (HEVs): specialized endothelium for lymphocyte migration. Immunol Today 1995, 16:449-457 [DOI] [PubMed] [Google Scholar]
- 4.Kraal G, Mebius RE: High endothelial venules: lymphocyte traffic control and controlled traffic. Adv Immunol 1997, 65:347-395 [PubMed] [Google Scholar]
- 5.Freemont AJ: Molecules controlling lymphocyte-endothelial interactions in lymph nodes are produced in vessels of inflamed synovium. Ann Rheum Dis 1987, 46:924-928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salmi M, Granfors K, MacDermott R, Jalkanen S: Aberrant binding of lamina propria lymphocytes to vascular endothelium in inflammatory bowel diseases. Gastroenterology 1994, 106:596-605 [DOI] [PubMed] [Google Scholar]
- 7.Jahnsen FL, Lund-Johansen F, Dunne JF, Farkas L, Haye R, Brandtzaeg P: Experimentally induced recruitment of plasmacytoid (CD123high) dendritic cells in human nasal allergy. J Immunol 2000, 165:4062-4068 [DOI] [PubMed] [Google Scholar]
- 8.Lechleitner S, Kunstfeld R, Messeritsch-Fanta C, Wolff K, Petzelbauer P: Peripheral lymph node addressins are expressed on skin endothelial cells. J Invest Dermatol 1999, 113:410-414 [DOI] [PubMed] [Google Scholar]
- 9.Farkas L, Beiske K, Lund-Johansen F, Brandtzaeg P, Jahnsen FL: Plasmacytoid dendritic cells (natural interferon-α/β-producing cells) accumulate in cutaneous lupus erythematosus lesions. Am J Pathol 2001, 159:237-243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toppila S, Paavonen T, Nieminen MS, Hayry P, Renkonen R: Endothelial L-selectin ligands are likely to recruit lymphocytes into rejecting human heart transplants. Am J Pathol 1999, 155:1303-1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.von Andrian UH, Mackay CR: T-cell function and migration: two sides of the same coin. N Engl J Med 2000, 343:1020-1034 [DOI] [PubMed] [Google Scholar]
- 12.Rosen SD: Endothelial ligands for L-selectin: from lymphocyte recirculation to allograft rejection. Am J Pathol 1999, 155:1013-1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stein JV, Rot A, Luo Y, Narasimhaswamy M, Nakano H, Gunn MD, Matsuzawa A, Quackenbush EJ, Dorf ME, von Andrian UH: The CC chemokine thymus-derived chemotactic agent 4 (TCA-4, secondary lymphoid tissue chemokine, 6Ckine, exodus-2) triggers lymphocyte function-associated antigen 1-mediated arrest of rolling T lymphocytes in peripheral lymph node high endothelial venules. J Exp Med 2000, 191:61-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baekkevold ES, Yamanaka T, Palframan RT, Carlsen HS, Reinholt FP, von Andrian UH, Brandtzaeg P, Haraldsen G: The CCR7 ligand elc (CCL19) is transcytosed in high endothelial venules and mediates T cell recruitment. J Exp Med 2001, 193:1105-1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bistrup A, Bhakta S, Lee JK, Belov YY, Gunn MD, Zuo FR, Huang CC, Kannagi R, Rosen SD, Hemmerich S: Sulfotransferases of two specificities function in the reconstitution of high endothelial cell ligands for L-selectin. J Cell Biol 1999, 145:899-910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiraoka N, Petryniak B, Nakayama J, Tsuboi S, Suzuki M, Yeh JC, Izawa D, Tanaka T, Miyasaka M, Lowe JB, Fukuda M: A novel, high endothelial venule-specific sulfotransferase expresses 6-sulfo sialyl Lewis(x), an L-selectin ligand displayed by CD34. Immunity 1999, 11:79-89 [DOI] [PubMed] [Google Scholar]
- 17.Hemmerich S, Bistrup A, Singer MS, van Zante A, Lee JK, Tsay D, Peters M, Carminati JL, Brennan TJ, Carver-Moore K, Leviten M, Fuentes ME, Ruddle NH, Rosen SD: Sulfation of L-selectin ligands by an HEV-restricted sulfotransferase regulates lymphocyte homing to lymph nodes. Immunity 2001, 15:237-247 [DOI] [PubMed] [Google Scholar]
- 18.Smith PL, Gersten KM, Petryniak B, Kelly RJ, Rogers C, Natsuka Y, Alford JA, III, Scheidegger EP, Natsuka S, Lowe JB: Expression of the α(1, 3)fucosyltransferase Fuc-TVII in lymphoid aggregate high endothelial venules correlates with expression of L-selectin ligands. J Biol Chem 1996, 271:8250-8259 [DOI] [PubMed] [Google Scholar]
- 19.Maly P, Thall A, Petryniak B, Rogers CE, Smith PL, Marks RM, Kelly RJ, Gersten KM, Cheng G, Saunders TL, Camper SA, Camphausen RT, Sullivan FX, Isogai Y, Hindsgaul O, von Andrian UH, Lowe JB: The α(1, 3)fucosyltransferase Fuc-TVII controls leukocyte trafficking through an essential role in L-, E-, and P-selectin ligand biosynthesis. Cell 1996, 86:643-653 [DOI] [PubMed] [Google Scholar]
- 20.Gunn MD, Tangemann K, Tam C, Cyster JG, Rosen SD, Williams LT: A chemokine expressed in lymphoid high endothelial venules promotes the adhesion and chemotaxis of naive T lymphocytes. Proc Natl Acad Sci USA 1998, 95:258-263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Girard JP, Springer TA: Cloning from purified high endothelial venule cells of hevin, a close relative of the antiadhesive extracellular matrix protein SPARC. Immunity 1995, 2:113-123 [DOI] [PubMed] [Google Scholar]
- 22.Girard JP, Springer TA: Modulation of endothelial cell adhesion by hevin, an acidic protein associated with high endothelial venules. J Biol Chem 1996, 271:4511-4517 [DOI] [PubMed] [Google Scholar]
- 23.Baekkevold ES, Jahnsen FL, Johansen FE, Bakke O, Gaudernack G, Brandtzaeg P, Haraldsen G: Culture characterization of differentiated high endothelial venule cells from human tonsils. Lab Invest 1999, 79:327-336 [PubMed] [Google Scholar]
- 24.Izawa D, Tanaka T, Saito K, Ogihara H, Usui T, Kawamoto S, Matsubara K, Okubo K, Miyasaka M: Expression profile of active genes in mouse lymph node high endothelial cells. Int Immunol 1999, 11:1989-1998 [DOI] [PubMed] [Google Scholar]
- 25.Byers RJ, Hoyland JA, Dixon J, Freemont AJ: Subtractive hybridization: genetic takeaways and the search for meaning. Int J Exp Pathol 2000, 81:391-404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, Siebert PD: Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci USA 1996, 93:6025-6030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Girard JP, Baekkevold ES, Yamanaka T, Haraldsen G, Brandtzaeg P, Amalric F: Heterogeneity of endothelial cells: the specialized phenotype of human high endothelial venules characterized by suppression subtractive hybridization. Am J Pathol 1999, 155:2043-2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stier S, Totzke G, Grunewald E, Neuhaus T, Fronhoffs S, Sachinidis A, Vetter H, Schulze-Osthoff K, Ko Y: Identification of syntenin and other TNF-inducible genes in human umbilical arterial endothelial cells by suppression subtractive hybridization. FEBS Lett 2000, 467:299-304 [DOI] [PubMed] [Google Scholar]
- 29.Wang H, Zhan Y, Xu L, Feuerstein GZ, Wang X: Use of suppression subtractive hybridization for differential gene expression in stroke: discovery of CD44 gene expression and localization in permanent focal stroke in rats. Stroke 2001, 32:1020-1027 [DOI] [PubMed] [Google Scholar]
- 30.Kirsch T, Wellner M, Luft FC, Haller H, Lippoldt A: Altered gene expression in cerebral capillaries of stroke-prone spontaneously hypertensive rats. Brain Res 2001, 910:106-115 [DOI] [PubMed] [Google Scholar]
- 31.Palmeri D, van Zante A, Huang CC, Hemmerich S, Rosen SD: Vascular endothelial junction-associated molecule, a novel member of the immunoglobulin superfamily, is localized to intercellular boundaries of endothelial cells. J Biol Chem 2000, 275:19139-19145 [DOI] [PubMed] [Google Scholar]
- 32.Jahnsen FL, Brandtzaeg P, Haye R, Haraldsen G: Expression of functional VCAM-1 by cultured nasal polyp-derived microvascular endothelium. Am J Pathol 1997, 150:2113-2123 [PMC free article] [PubMed] [Google Scholar]
- 33.von Stein OD, Thies WG, Hofmann M: A high throughput screening for rarely transcribed differentially expressed genes. Nucleic Acids Res 1997, 25:2598-2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.St Croix B, Rago C, Velculescu V, Traverso G, Romans KE, Montgomery E, Lal A, Riggins GJ, Lengauer C, Vogelstein B, Kinzler KW: Genes expressed in human tumor endothelium. Science 2000, 289:1197-1202 [DOI] [PubMed] [Google Scholar]
- 35.Olszewski KA, Yan L, Edwards DJ: SeqFold - fully automated fold recognition and modeling software - evaluation and application. Theor Chem Acc 1999, 101:57-61 [Google Scholar]
- 36.King RD, Sternberg MJ: Identification and application of the concepts important for accurate and reliable protein secondary structure prediction. Protein Sci 1996, 5:2298-2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manival X, Ghisolfi-Nieto L, Joseph G, Bouvet P, Erard M: RNA-binding strategies common to cold-shock domain- and RNA recognition motif-containing proteins. Nucleic Acids Res 2001, 29:2223-2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hayward CP, Cramer EM, Song Z, Zheng S, Fung R, Masse JM, Stead RH, Podor TJ: Studies of multimerin in human endothelial cells. Blood 1998, 91:1304-1317 [PubMed] [Google Scholar]
- 39.Onda H, Kasuya H, Takakura K, Hori T, Imaizumi T, Takeuchi T, Inoue I, Takeda J: Identification of genes differentially expressed in canine vasospastic cerebral arteries after subarachnoid hemorrhage. J Cereb Blood Flow Metab 1999, 19:1279-1288 [DOI] [PubMed] [Google Scholar]
- 40.Grant RA, Rould MA, Klemm JD, Pabo CO: Exploring the role of glutamine 50 in the homeodomain-DNA interface: crystal structure of engrailed (Gln50>ala) complex at 2.0 A. Biochemistry 2000, 39:8187-8192 [DOI] [PubMed] [Google Scholar]
- 41.Kissinger C, Liu B, Martin-Blanco E, Kornberg T, Pabo C: Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions. Cell 1990, 63:579-590 [DOI] [PubMed] [Google Scholar]
- 42.Iwahara J, Kigawa T, Kitagawa K, Masumoto H, Okazaki T, Yokoyama S: A helix-turn-helix structure unit in human centromere protein B (CENP-B). EMBO J 1998, 17:827-837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lasky LA, Singer MS, Dowbenko D, Imai Y, Henzel WJ, Grimley C, Fennie C, Gillett N, Watson SR, Rosen SD: An endothelial ligand for L-selectin is a novel mucin-like molecule. Cell 1992, 69:927-938 [DOI] [PubMed] [Google Scholar]
- 44.Mebius RE, Bauer J, Twisk AJ, Breve J, Kraal G: The functional activity of high endothelial venules: a role for the subcapsular sinus macrophages in the lymph node. Immunobiology 1991, 182:277-291 [DOI] [PubMed] [Google Scholar]
- 45.Madej T, Boguski MS, Bryant SH: Threading analysis suggests that the obese gene product may be a helical cytokine. FEBS Lett 1995, 373:13-18 [DOI] [PubMed] [Google Scholar]
- 46.Wigle JT, Oliver G: Prox1 function is required for the development of the murine lymphatic system. Cell 1999, 98:769-778 [DOI] [PubMed] [Google Scholar]
- 47.Wigle JT, Harvey N, Detmar M, Lagutina I, Grosveld G, Gunn MD, Jackson DG, Oliver G: An essential role for Prox1 in the induction of the lymphatic endothelial cell phenotype. EMBO J 2002, 21:1505-1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petrova TV, Makinen T, Makela TP, Saarela J, Virtanen I, Ferrell RE, Finegold DN, Kerjaschki D, Yla-Herttuala S, Alitalo K: Lymphatic endothelial reprogramming of vascular endothelial cells by the Prox-1 homeobox transcription factor. EMBO J 2002, 21:4593-4599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fan L, Reilly CR, Luo Y, Dorf ME, Lo D: Ectopic expression of the chemokine TCA4/SLC is sufficient to trigger lymphoid neogenesis. J Immunol 2000, 164:3955-3959 [DOI] [PubMed] [Google Scholar]
- 50.Luther SA, Lopez T, Bai W, Hanahan D, Cyster JG: BLC expression in pancreatic islets causes B cell recruitment and lymphotoxin-dependent lymphoid neogenesis. Immunity 2000, 12:471-481 [DOI] [PubMed] [Google Scholar]
- 51.Kratz A, Campos-Neto A, Hanson MS, Ruddle NH: Chronic inflammation caused by lymphotoxin is lymphoid neogenesis. J Exp Med 1996, 183:1461-1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilting J, Papoutsi M, Christ B, Nicolaides KH, Von Kaisenberg CS, Borges J, Stark GB, Alitalo K, Tomarev SI, Niemeyer C, Rossler J: The transcription factor Prox1 is a marker for lymphatic endothelial cells in normal and diseased human tissues. EMBO J 2002, 16:1271-1273 [DOI] [PubMed] [Google Scholar]