Abstract
The cytokine tumor necrosis factor (TNF)-α has previously been shown to prime hepatocytes to a state of replicative competence, but has not been shown to act as a complete mitogen for these cells. In the present study we have altered our previously described long-term dimethyl sulfoxide culture system to exclude all known hepatocyte mitogens from the culture media and enable us to directly examine the effects of TNF-α on primary rat hepatocytes. We have shown that cells maintained under these culture conditions retain the biochemical and morphological features of well-differentiated hepatocytes. Treatment with TNF-α induced DNA synthesis relative to control, to a level not significantly different from that induced by the known hepatocyte mitogen, epidermal growth factor (EGF). Maximal DNA synthesis was induced by treatment with 250 U/ml TNF-α for 24 hours. Mitotic figures were observed in cultures treated with TNF-α or EGF but not in untreated controls. Treatment of cultures with TNF-α, but not EGF, induced activation of both nuclear factor-κB p50 homodimers and p50/p65 heterodimers. DNA synthesis induced by TNF-α was inhibited by treatment with transforming growth factor-β. Based on the results of our studies, we conclude that TNF-α acts as a complete mitogen for rat hepatocytes.
The proinflammatory cytokine tumor necrosis factor (TNF)-α is secreted systemically by activated macrophages and in liver by biliary epithelial and venous endothelial cells. 1 TNF-α has been shown to be an important mediator in both normal and pathological processes in liver. 2-7 TNF-α levels are dramatically increased during liver regeneration and in patients with chronic hepatitis B and hepatitis C viral infections, alcoholic hepatitis, and inflammatory liver disease. 6-9
The TNF-α molecule exists as a homotrimer 10 and interacts with two homotrimerized transmembrane receptors located on the cell surface, TNF receptor 1 (TNF-R1) and TNF receptor 2 (TNF-R2). 11 Both receptors contain three highly conserved cysteine-rich domains in their extracellular regions, characteristic of members of the TNF receptor superfamily. 12,13 Binding of TNF-α to the extracellular domains of these receptors leads to receptor activation and downstream signal transduction via divergent intracellular domains, capable of inducing an array of responses including apoptosis and cell proliferation. 14-18 Activated TNF-R1 recruits adapter molecules including Fas-associated death domain and TNF receptor-associated death domain to its clustered cytosolic domains and leads either to activation of the caspase cascade and ultimately death of the cell, or activation of the transcription factors nuclear factor (NF)-κB and AP-1, promoting cell survival and proliferation. Activation of TNF-R2 results in recruitment of TNF receptor associated factors 1 and 2 (TRAFs 1 and 2) and leads to activation of NF-κB.
Hepatocytes exist in G0 in vivo 19,20 but retain the capacity to enter the cell cycle in response to specific stimuli. A limited number of growth factors including transforming growth factor (TGF)-α, 21,22 acidic fibroblast growth factor, 23,24 basic fibroblast growth factor, 25 epidermal growth factor (EGF), 26,27 hepatocyte growth factor (HGF), 28,29 and keratinocyte growth factor 30 have been shown to induce hepatocytes to enter the cell cycle without additional stimulation and, therefore, have been classified as complete mitogens.
Evidence suggests that the cytokine transforming growth factor β1 (TGF-β1) may function as a negative regulator of hepatocyte replication during liver regeneration. 31-33 TGF-β1 has been previously shown to inhibit DNA synthesis induced by the hepatocyte mitogens EGF and HGF. 34 In long-term hepatocyte culture, Serra and Isom 35 have demonstrated that TGF-β1 inhibits DNA synthesis induced by both EGF and TGF-α. Carr and colleagues 36 have demonstrated that while TGF-β1 was able to inhibit mitogen-induced DNA synthesis, it was unable to inhibit DNA synthesis induced by co-mitogens such as insulin and glucagon.
TNF-α has been shown to promote cell growth in fibroblasts. 37-39 The ability of TNF-α to act as a priming agent for hepatocytes has been addressed 3,20,40-42 and indirect evidence suggesting that TNF-α may function as a complete mitogen exists. 1,2,4,5 The ability of TNF-α to act as a complete mitogen for rat hepatocytes has never been directly examined, in part because there has never been an appropriate in vitro model system in which this question can be addressed.
Our laboratory has previously described a system in which primary rat hepatocytes can be maintained in long-term culture in a chemically defined, serum-free media supplemented with dimethyl sulfoxide (DMSO). 43,44 These cells resemble hepatocytes at the ultrastructural level, 44 maintain expression of numerous paracellular junction proteins, 44-46 have functional gap junctions, 45,46 and continue to express liver-specific transcription factors and genes for several months. 43
Hepatocytes in long-term DMSO culture do not synthesize DNA or divide but they retain the capacity to do so. One problem with using this cell culture system to test the mitogenic capability of a growth factor or cytokine is that the medium in which the cells are cultured is supplemented with EGF. In the present study, we addressed the question of whether TNF-α could act as a complete mitogen for primary rat hepatocytes in long-term culture, and, if so, whether this mitogenic effect was sensitive to inhibition by TGF-β. To accomplish this goal it was first necessary to modify the long-term rat hepatocyte DMSO culture system so that it was deficient in the mitogen EGF and characterize the cells under these novel culture conditions to determine whether they retain the morphological and biochemical properties of well-differentiated hepatocytes.
Materials and Methods
Hepatocyte Cultures
Primary rat hepatocytes were isolated by collagenase perfusion of adult male Fischer 344 rat livers (180 to 200 g) as described previously. 44 Hepatocytes were plated on rat tail collagen-coated 60-mm tissue culture dishes at a density of 1.0 × 106 cells per dish. Hepatocytes were initially plated in a Dulbecco’s modified Eagle’s medium (DMEM)-F12-based chemically defined medium containing insulin (0.06 μg/mL), glucagon (0.04 μg/mL), dexamethasone (0.4 μg/mL), transferrin (100 μg/mL) (designated modified DMEM-F12) (Life Technologies, Gaithersburg, MD) supplemented with 3% fetal calf serum (HyClone, Logan, UT). Cultures were refed this media 4 hours after plating. Twenty-four hours after plating, cells were fed DMEM-F12-based media deficient in fetal calf serum supplemented with 2% DMSO (Sigma, St. Louis, MO) and 1 μmol/L [+]-α-tocopherol (Sigma) with or without the addition of 25 ng/ml of EGF (Sigma). Cells were maintained under these conditions for 10 to 30 days before the start of all experiments. Cultures were treated with murine TNF-α (Roche Applied Science, Indianapolis, IN) with concentrations between 100 and 5000 U/ml as described (TNF-α concentrations: 1 ng = 10 U/ml).
Hematoxylin and Eosin (H&E) Staining
Cultures of primary rat hepatocytes were fixed in 10% neutral buffered formalin for 15 minutes at room temperature. Cells were stained with hematoxylin (Shandon Lipshaw, Pittsburgh, PA) for 7 minutes and incubated in 0.2% ammonia water until color change was observed. Cultures were dipped in Eosin-Y (Shandon Lipshaw) and visualized by transmissive light microscopy.
Electron Microscopy
Primary rat hepatocytes plated on rat tail collagen-coated Permanox dishes (Electron Microscopy Sciences, Fort Washington, PA) were fixed with 1% paraformaldehyde and 0.5% gluteraldehyde in 0.1 mol/L of sodium cacodylate buffer (pH 7.3) for 1 hour at 4°C. The cells were postfixed in 1% osmium tetroxide plus 1.5% potassium ferrocyanide overnight at 4°C, dehydrated in graded ethanols, and embedded in Epon 812 (Tousimis Research Corp., Rockville, MD). Ultrathin (∼70 nm) sections were cut from the embedded cells and stained with uranyl acetate and lead citrate and visualized on a transmission electron microscope (EM400; Phillips, Einhoven, The Netherlands). 47
RNase Protection Assay
Probes for rat albumin, α1-antitrypsin (A1AT), GAPDH, and cyclophilin were generated by polymerase chain reaction as previously described. 48 The sizes for each of the polymerase chain reaction products were A1AT, 513 nucleotides; albumin, 314 nucleotides; cyclophilin, 290 nucleotides; and GAPDH, 269 nucleotides. Probes were labeled with T7 RNA polymerase and 32P-CTP. RNase protection assays were performed as previously described. 48 Labeled probes were hybridized against 20 μg of total genomic RNA isolated from primary rat hepatocytes cultured in the presence or absence of EGF. After RNase digestion, protected probes were separated on a 4% urea-polyacrylamide gel electrophoresis sequencing gel with 1× Tris-borate-EDTA running buffer and quantitated using a Phosphorimager and ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
DNA Synthesis Assay
The 5-bromo-2′-deoxy-uridine (BrdU) staining kit was obtained from Roche Applied Science (Indianapolis, IN). Unless otherwise indicated, cultures were incubated with BrdU for the final 24 hours of each treatment period. Cells were then fixed in 30% glycine-buffered ethanol and developed according to the manufacturer’s instructions. For all BrdU incorporation experiments a minimum of 24 fields of cells were counted for each experimentalcondition. The percentage of BrdU-positive cells for each field was calculated and the average of these percentages represented graphically. This corresponds to ∼4000 cells per condition.
DNA Fragmentation Analysis
Low molecular weight DNA was isolated as previously described. 49-52 For each experimental condition, one 60-mm plate of hepatocytes was trypsinized and cells pelleted by centrifugation at 1500 × g for 10 minutes. The cell pellet was resuspended in phosphate-buffered saline (PBS) and centrifuged at 14,000 × g for 2 minutes. The resulting cell pellet was resuspended in lysis buffer (10 mmol/L Tris-HCl, pH 8.0, 10 mmol/L EDTA, 0.5% Triton X-100) and centrifuged at 14,000 × g for 20 minutes at 4°C. The supernatant was treated with Proteinase K and sodium dodecyl sulfate for 2 hours at 50°C. The samples were then extracted with phenol/chloroform (1:1) for 5 minutes. The DNA was precipitated overnight with isopropanol and 5 mol/L NaCl at −20°C. For each sample, 1.0 μg of DNA was end-labeled with Klenow polymerase and 32P-dCTP for 10 minutes. DNA samples were separated on Trevigel gels (Trevigen, Gaithersburg, MD), fixed overnight in 10% acetic acid, and dried. Gels were then analyzed by autoradiography.
Acridine Orange Staining
As previously described by Bour and colleagues, 52 cells were fixed in ethanol-acetic acid for 20 minutes at −20°C and subsequently fixed in 100% ethanol at −20°C for an additional 20 minutes. Cultures were rinsed in graded ethanols (50, 80, and 100% ethanol twice for 2 minutes), rinsed in 1% acetic acid, and stained with 1% acridine orange for 3 minutes. Hepatocytes were rinsed in 0.067 mol/L of phosphate buffer (0.067 mol/L Na2HPO4, 0.067 mol/L KH2PO4) and differentiated in 0.1 mol/L of CaCl2. Acridine orange staining was visualized on a Nikon florescence microscope equipped with a dual wavelength fluorescein isothiocyanate/tetramethyl-rhodamine isothiocyanate filter.
Electrophoretic Mobility Shift Assay and Supershift Analysis
Nuclear protein was extracted as described by Olnes and Kurl. 53 Primary rat hepatocytes were harvested in lysis buffer [10 mmol/L HEPES, 10 mmol/L KCl, 0.1 mmol/L EDTA, 0.1 mmol/L EGTA, 1 mmol/L dithiothreitol, and Protease Inhibitor Cocktail for Mammalian Cell Extracts (Sigma)] and lysed with the addition of Nonidet P-40 (Sigma). Samples were centrifuged for 5 minutes at 250 × g and the pellets resuspended in nuclear salt lysis buffer (20 mmol/L HEPES, 400 mmol/L KCl, 1.0 mmol/L EDTA, 1.0 mmol/L EGTA, 10% glycerol, and Protease Inhibitor Cocktail for Mammalian Cell Extracts). Samples were incubated with shaking for 30 minutes and subsequently centrifuged at 12,000 × g for 10 minutes. Oligonucleotide probes corresponding to the NF-κB consensus sequence (sense, 5′-TAG TTG AGG GGA CTT TCC CAG GCA-3′; anti-sense, 5′-TGC CTG GGA AAG TCC CCT CAA CTA-3′) 54 were annealed, gel-purified, and labeled with polynucleotide kinase (New England BioLabs, Beverly, MA) and γ32P-ATP. Five μg of nuclear protein was incubated with 7.0 × 104 cpm oligonucleotide probe at room temperature for 30 minutes. For supershift analysis, samples were subsequently incubated with PBS, rabbit preimmune sera (DAKO, Carpinteria, CA), and anti-p65 or anti-p50 antibodies (Santa Cruz Biotechnology, Santa Cruz, CA). Complexes were separated on 4% nonreducing gels.
Statistical Analyses
In all BrdU incorporation experiments, a minimum of 24 data points for each experimental condition were studied. Triplicate samples were analyzed for RNase protection experiments. Analysis of variance, Dunnett’s test (relative to control), and Tukey-Kramer tests were performed as appropriate using JMP 3.0.2 (SAS Institute, Cary, NC). The null hypothesis was rejected for P values less than or equal to 0.05.
Results
Primary Rat Hepatocytes Maintained in the Absence of EGF Retain Morphological and Biochemical Markers Characteristic of Differentiated Hepatocytes
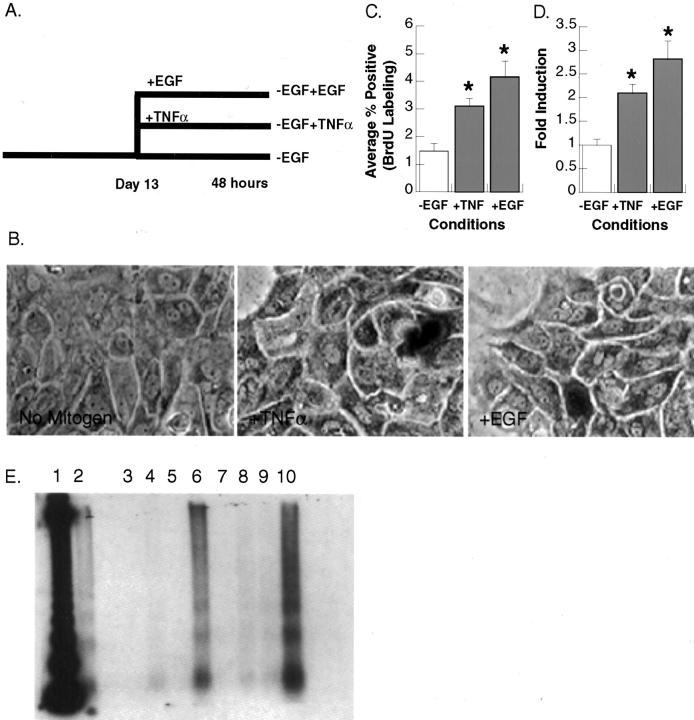
To determine whether TNF-α can act as a primary mitogen for hepatocytes in long-term DMSO culture, we developed a system in which all other known hepatocyte mitogens are excluded. In this system, the EGF normally present in the culture media at 25 ng/ml from day 1 after plating is excluded. These cultures can be maintained long term, through 40 days after plating, as has been previously established for cultures fed medium supplemented with EGF. 44 It should be noted that no fetal calf serum is present under either culture condition.
To determine whether cultures maintained in the absence of EGF (−EGF) retain intact hepatocyte morphology, primary rat hepatocytes maintained in long-term DMSO culture in the presence (+EGF) and absence (−EGF) of EGF were fixed and stained with H&E 13 days after plating (Figure 1A) ▶ . Hepatocytes cultured in the absence of EGF remained mono- and bi-nucleated and retained nuclear circularity as observed in +EGF cultures. A reduced number of cells were present in −EGF cultures relative to cultures maintained in the presence of EGF. On average, ∼282 cells per field were observed in +EGF cultures, compared with only 171 cells per field in −EGF cultures. This represents a 40% reduction in cell number in −EGF cultures of primary rat hepatocytes. Although a reduced number of cells was present in cultures maintained in the absence of EGF, no apparent differences in cell or island morphology were observed. Because this decrease in cell number occurs within days of plating, rather than continuously throughout time in long-term culture, it is most likely because of the absence of the growth factor EGF at early time points after plating. No spontaneous apoptosis occurs in these cultures after 10 days of plating (Figure 2E ▶ , compare lanes 3 and 7).
Figure 1.
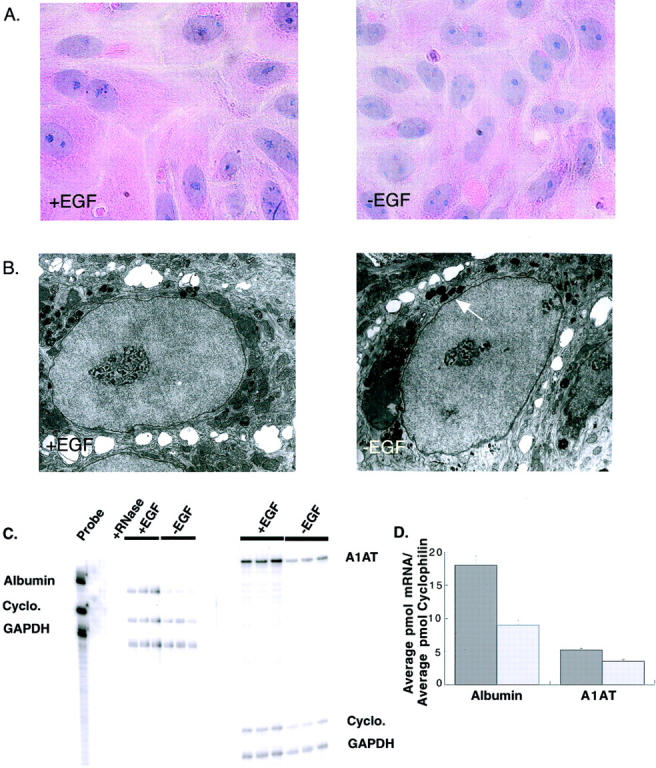
Primary rat hepatocytes cultured in the absence of EGF retain the morphological and biochemical markers of well-differentiated hepatocytes. A: H&E stain of primary rat hepatocytes cultured in the presence (+EGF) or absence (−EGF) of EGF. B: Electron micrographs of primary rat hepatocytes cultured in the presence or absence of EGF. Arrow indicates presence of peroxisomes. C: Phosphorimager scan of RPA for rat albumin, A1AT, cyclophilin, and GAPDH. Left: RPA for albumin, cyclophilin, and GAPDH. Lane 1, Probe, no RNase (positive control); lane 2, probe, +RNase (negative control); lanes 3 to 5, +EGF cultures of hepatocytes; lanes 6 to 8, −EGF cultures of hepatocytes. Right: RPA for A1AT, cyclophilin, and GAPDH. Lanes 1 to 3, +EGF cultures of hepatocytes; lanes 4 to 6, −EGF cultures of hepatocytes. D: Graphical representation of RPA results after ImageQuant analysis. Liver-specific gene expression was normalized to cyclophilin expression to control for equal RNA loading. Original magnifications: ×400 (A); ×11,500 (B).
Figure 2.
TNF-α and EGF act as primary mitogens for rat hepatocytes. A: Protocol for the induction of DNA synthesis by TNF-α and EGF. Hepatocytes were maintained for 13 days in the absence of EGF and for an additional 48 hours in control media (−EGF), media containing TNF-α (2500 U/ml) (−EGF +TNF-α), or media containing EGF (25 ng/ml) (−EGF +EGF). BrdU was added to the media of all cultures for the final 24 hours of the treatment period. B: BrdU labeling of primary rat hepatocytes. Cells were fixed and stained for BrdU incorporation. C and D: DNA synthesis in TNF-α- or EGF-treated hepatocytes. Percentages of BrdU-labeled nuclei were calculated for each condition. Fold induction in BrdU incorporation was calculated by normalizing data to control cultures. Error bars indicate the SE for individual conditions. *, Values indicate that results are significantly increased relative to −EGF controls (one-tailed t-test, P < 0.05). E: Induction of apoptosis in primary rat hepatocytes. Samples in lanes 3to 6 were extracted from +EGF cultures and samples in lanes 7 to 10 from −EGF cultures. Lane 1, 123-bp ladder; lane 2, +TNF-α +cycloheximide (5 μmol/L) (positive control); lanes 3 and 7, untreated control; lanes 4 and 8, +cycloheximide (5 μmol/L); lanes 5 and 9, +TNF-α (2500 U/ml); lanes 6 and 10, +cycloheximide (5 μmol/L) +TNF-α. Original magnification, ×200 (B).
To further characterize the morphological features of cells maintained in the absence of EGF, control and −EGF cultures were examined by transmission electron microscopy at 21 days after plating. The hepatocytes retained the nuclear circularity and overall morphology of +EGF control hepatocytes (Figure 1B) ▶ . We did observe that hepatocytes in −EGF cultures had an increased number of peroxisomes relative to +EGF controls.
Primary rat hepatocytes in long-term DMSO culture maintain high-level expression of liver-specific genes including albumin and A1AT. 43 RNase protection assays (RPAs) were performed to determine whether expression of these hepatocyte differentiation markers was maintained in cells cultured in the absence of EGF. RNA was isolated from primary rat hepatocytes cultured in the presence and absence of EGF for 25 days after plating and analyzed for expression of albumin and A1AT (Figure 1C) ▶ . GAPDH and cyclophilin mRNA expressions were also monitored to ensure equal RNA loading. At 25 days after plating, A1AT and albumin mRNAs were expressed at levels 1.466- and 2.00-fold greater, respectively, in +EGF cultures than were observed in −EGF cultures (Figure 1D) ▶ . The experiment reported represents the maximum difference in albumin and A1AT mRNA we observed when comparing the two culture systems. In a separate experiment, RNA extracted 20 days after plating showed no significant difference in liver-specific gene expression between these culture systems (data not shown). It should be noted that although the levels of albumin and A1AT mRNA expressed by rat hepatocytes in long-term −EGF DMSO culture may be lower than for +EGF DMSO cultures, these levels are high relative to those observed for hepatocytes in short term in vitro culture systems.
TNF-α Is Able to Promote DNA Synthesis to Levels Similar to that Observed for the Known Hepatocyte Mitogen EGF
To determine whether TNF-α alone is able to induce DNA synthesis in primary rat hepatocytes, cultures were maintained in DMEM-F12 in the absence of EGF for 13 days. Cultures were then treated with murine TNF-α (2500 U/ml) or EGF (25 ng/ml) for 48 hours (Figure 2A) ▶ and labeled with BrdU for the last 24 hours of the treatment period. After treatment with TNF-α or EGF, no changes in cell morphology were observed in cultures (Figure 2B) ▶ . The percentage of cells positive for BrdU incorporation was determined and the results summarized in Figure 2C ▶ . Cultures of −EGF hepatocytes had low basal levels of BrdU incorporation, with 1% of the cells synthesizing DNA. Both TNF-α and EGF were able to produce significant increases in BrdU incorporation relative to untreated control cultures with ∼3 to 4% of cells synthesizing DNA by 48 hours after treatment. Further, neither the increase in labeling index nor the fold induction in DNA synthesis (Figure 2D) ▶ induced by TNF-α treatment differed significantly from that induced by the known hepatocyte mitogen EGF.
In addition to its potential as a mitogenic agent, TNF-α is known to induce apoptosis in many cell types. To rule out the possibility that the increase in BrdU incorporation we observed in TNF-α-treated cultures was because of an increase in DNA fragmentation rather than an increase in DNA synthesis, we monitored internucleosomal cleavage of DNA. No spontaneous apoptosis was observed in untreated cultures or cultures treated with TNF-α alone (Figure 2E) ▶ . As a positive control for apoptosis, hepatocytes were treated with both cycloheximide and TNF-α (conditions previously shown to induce apoptosis in hepatocytes in long-term DMSO culture; Iocca and Isom, unpublished data) and analyzed for ladder formation (Figure 2E ▶ , lanes 6 and 10). As expected apoptosis was induced only when the cells were sensitized with cycloheximide and treated with TNF-α.
Complete Mitogenic Potential of TNF-α Is Not Decreased with Increasing Time in Culture
We next wanted to determine whether the length of time primary hepatocytes are retained in the −EGF/DMSO culture system had any effect on the ability of the cells to respond to TNF-α. Hepatocytes, in culture for 27 days before the start of the study (compared to the 13 days used previously) were treated for 24, 48, or 96 hours with murine TNF-α at 5000 U/ml and labeled for the last 24 hours of the treatment period with BrdU (Figure 3A) ▶ . As observed previously, ∼1% of untreated control cells stained positive for BrdU incorporation (Figure 3B) ▶ . Maximal TNF-α-induced DNA synthesis occurred between 48 and 96 hours after treatment during which time, ∼3 to 4% of cells incorporated BrdU (Figure 3, B and C) ▶ . In contrast, the maximal DNA synthesis induced by EGF was observed at 48 hours after treatment, with 5 to 6% of cells staining positive for BrdU incorporation, and declined to levels not significantly different from untreated control by 96 hours after treatment. These results indicate that the length of time that hepatocytes are in culture does not alter the complete mitogenic effect of TNF-α on rat hepatocytes.
Figure 3.
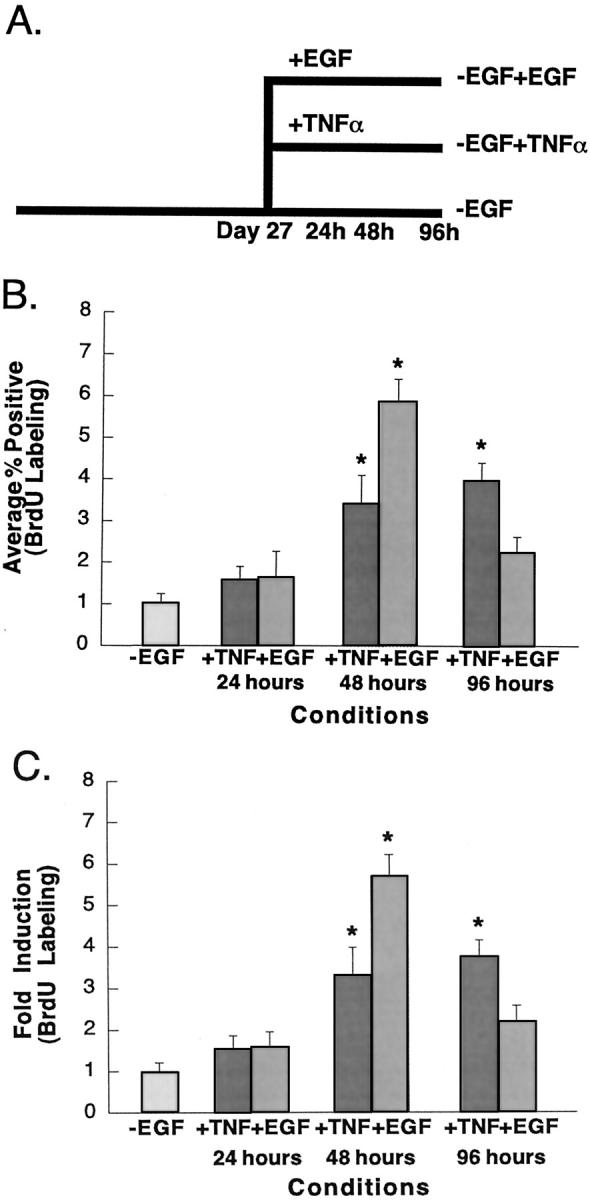
Time course of TNF-α- and EGF-induced DNA synthesis. A: Protocol for the induction of DNA synthesis by EGF and TNF-α. Hepatocytes were maintained for 27 days in the absence of EGF. Cultures were either maintained in this media during the experiment to serve as controls (−EGF); treated with TNF-α (5000 U/ml) for 24, 48, or 96 hours (−EGF +TNF-α); or treated with EGF (25 ng/ml) for 24, 48, or 96 hours (−EGF +EGF). BrdU was added to the media of all cultures for the final 24 hours of treatment. B and C: DNA synthesis in TNF-α- and EGF-treated hepatocytes. Percentages of BrdU-labeled nuclei were calculated for the conditions described previously. Fold induction in BrdU incorporation was calculated by normalizing data to control cultures. Error bars indicate the SE for individual conditions. *, Values indicate results that were significantly increased relative to −EGF controls (one-tailed t-test, P < 0.05).
Low Doses of TNF-α Produce the Greatest Increases in DNA Synthesis
To determine the concentration at which murine TNF-α induces the maximal number of hepatocytes to enter S phase, cultures were maintained in modified DMEM-F12 in the absence of EGF for 16 days. Cells were then treated for 48 hours with 100, 250, 500, 1500, 2500, or 5000 U of murine TNF-α/ml and labeled with BrdU for the final 24 hours of the treatment period. Significant increases in DNA synthesis occurred at concentrations ranging from 250 to 5000 U TNF-α/ml. Induction was maximal (fourfold relative to control cultures) after treatment with 250 U TNF-α/ml (Figure 4A) ▶ . Higher TNF-α concentrations resulted in two- to threefold inductions of DNA synthesis relative to control. To further refine the dose response of TNF-α-induced DNA synthesis, we repeated this experiment with 100, 150, 200, 250, 375, 500, and 2500 U TNF-α/ml. The lowest concentration of TNF-α able to produce a significant increase in DNA synthesis in primary rat hepatocytes was 250 U/ml, which also was the dose that generated the greatest increase in DNA synthesis (data not shown).
Figure 4.
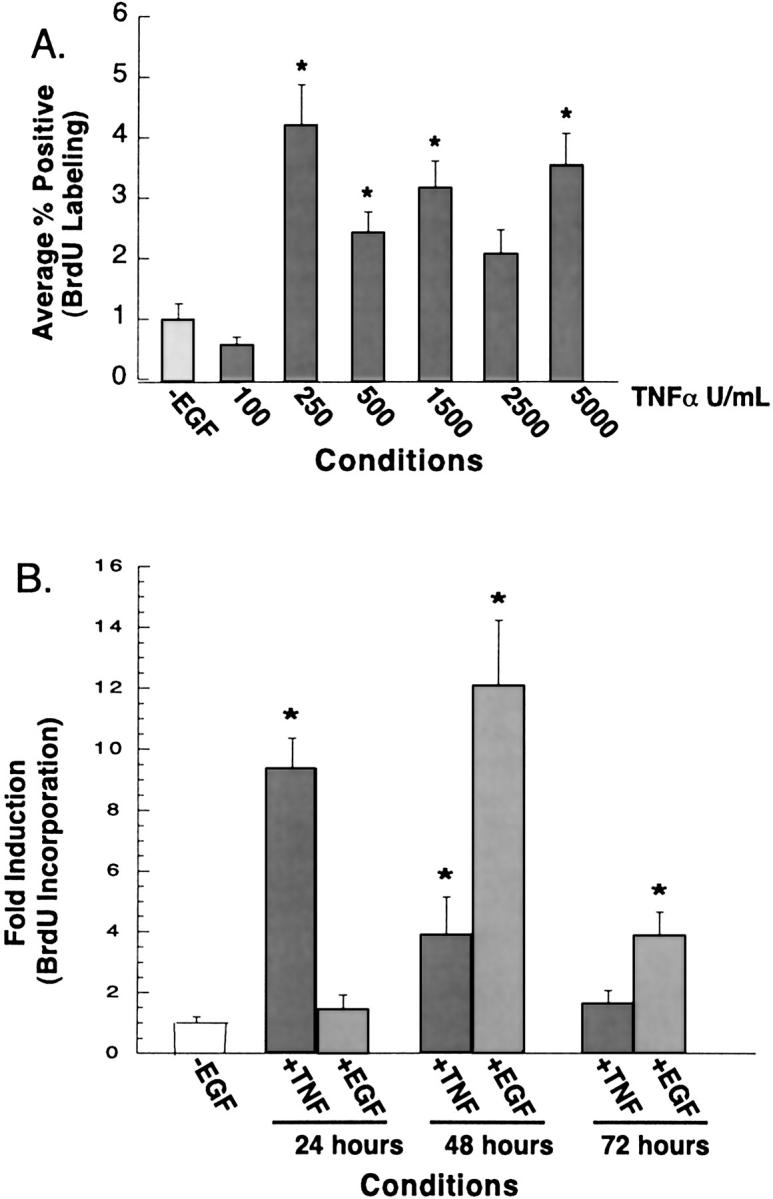
Concentration curve of TNF-α-induced DNA synthesis. Hepatocytes were maintained for 16 days in the absence of EGF. Cultures were either maintained in this media during the 48-hour experiment to serve as controls (−EGF) or treated with TNF-α (100, 250, 500, 1500, 2500, or 5000 U/ml). BrdU was added to the media of all cultures 24 hours after treatment. A: DNA synthesis in TNF-α-treated hepatocytes. Percentages of BrdU-labeled nuclei were calculated for each condition. B: Time courses of TNF-α- and EGF-induced DNA synthesis. Hepatocytes were maintained for 13 days in the absence of EGF. Cultures were either maintained in this media during the experiment to serve as controls (−EGF), treated with TNF-α (250 U/ml) or EGF (25 ng/ml) for 24, 48, or 72 hours. BrdU was added to the media of all cultures 24 hours before harvest. Percentages of BrdU-labeled nuclei were calculated for each condition. Fold induction in BrdU incorporation was calculated by normalizing data to control cultures. Error bars indicate the SE for individual conditions. *, Values indicate results that were significantly increased relative to −EGF controls (one-tailed t-test, P < 0.05).
To determine the time course of TNF-α-induced DNA synthesis at the optimal TNF-α concentration, cells cultured in the absence of EGF for 13 days after plating were treated with TNF-α or EGF for 24, 48, or 72 hours and labeled with BrdU for the last 24 hours of the treatment period. In contrast to the time course obtained for TNF-α at 5000 U/ml in −EGF cultures, in which maximal DNA synthesis occurred 48 hours after treatment, the greatest induction of DNA synthesis by 250 U/ml of TNF-α occurred at 24 hours after treatment (Figure 4B) ▶ . At this time point, a ninefold induction in the number of cells positive for BrdU incorporation relative to control was observed. In cultures maintained in the absence of EGF, the time course for DNA synthesis induced by low-dose TNF-α treatment differed from that observed for high-dose TNF-α treatment in −EGF cultures.
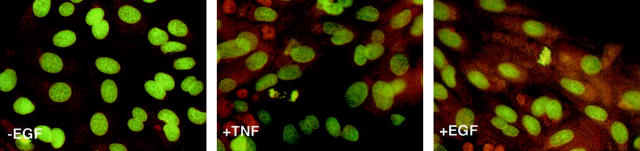
An Increase in Mitotic Figure Formation Occurs after Treatment with TNF-α or EGF
We next examined whether the observed increase in BrdU incorporation correlates with an increase in mitotic figure formation. To address this question, cells maintained in the absence of EGF for 13 days after plating were treated with either murine TNF-α or EGF for 48, 72, or 96 hours; fixed; and stained with acridine orange. Mitotic figures were not observed in the untreated −EGF cultures at any of the time points examined. This was the expected result because hepatocytes in long-term DMSO culture remain in G0. However, a low number of mitotic figures were observed in TNF-α- and EGF-treated cultures (Figure 5) ▶ . The mitotic indices after 96 hours of treatment with TNF-α or EGF were 0.05% and 0.09%, respectively. Thus, although a small percentage of cells synthesizing DNA appear to be continuing through the cell cycle, the majority of the cells induced to enter S phase by TNF-α or EGF do not undergo mitosis. Therefore, we believe that although TNF-α and EGF are able to induce DNA synthesis in primary rat hepatocytes, additional signals are necessary to induce cells to proceed through the cell cycle.
Figure 5.
Acridine orange staining of primary rat hepatocytes after TNF-α or EGF treatment. Cells were maintained for 13 days after plating in the absence of EGF. Cultures were then either maintained under control conditions for the duration of the experiment (−EGF), treated with TNF-α (250 U/ml) (+TNF) or EGF (25 ng/ml) (+EGF) for 48, 72, or 96 hours. Cells were fixed and stained with acridine orange. Original magnification, ×200.
DNA Synthesis Is Induced to the Same Degree by Both Murine and Human TNF-α
In many systems, cells respond differently to TNF-αderived from different species. To determine whether species specificity played a role in TNF-α-induced DNA synthesis, cells cultured in the absence of EGF for 13 days after plating were treated with murine or human TNF-α for 24 hours at 250 U/ml, the previously determined optimal conditions for induction of DNA synthesis. Both mitogens produced significant increases in DNA synthesis (three- to fourfold) compared to control cultures and the inductions were not statistically different from one another (Figure 6) ▶ .
Figure 6.
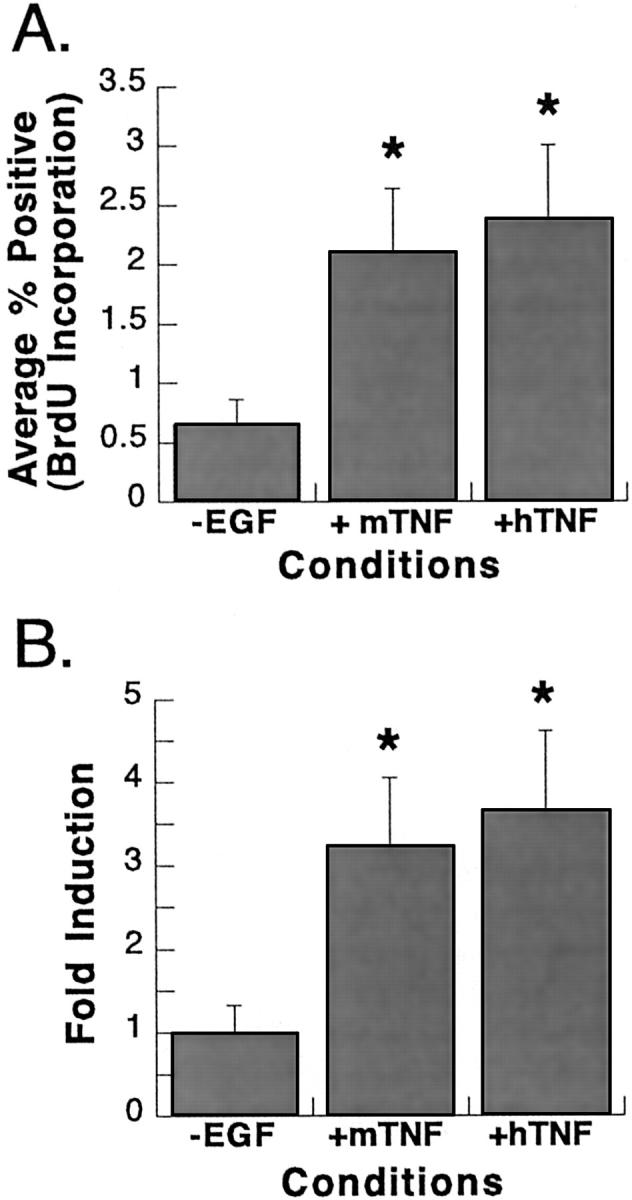
BrdU labeling of primary rat hepatocytes after treatment with mouse or human TNF-α. Cultures were maintained in the absence of EGF for 13 days before the start of the experiment. Cells were then maintained under the following conditions for an additional 48 hours: DMEM-F12 lacking EGF (−EGF), DMEM-F12 containing mouse TNF-α (250 U/ml) (+mTNF), or DMEM-F12 containing human TNF-α (250 U/ml) (+hTNF). BrdU was added to the media for the last 24 hours of the treatment period. A: Percentages of BrdU-labeled nuclei were calculated for each condition. B: Fold induction in BrdU incorporation was calculated by normalizing data to control cultures. Error bars indicate the SE for individual conditions. *, Values indicate results that are significantly increased relative to −EGF controls (one-tailed t-test, P < 0.05).
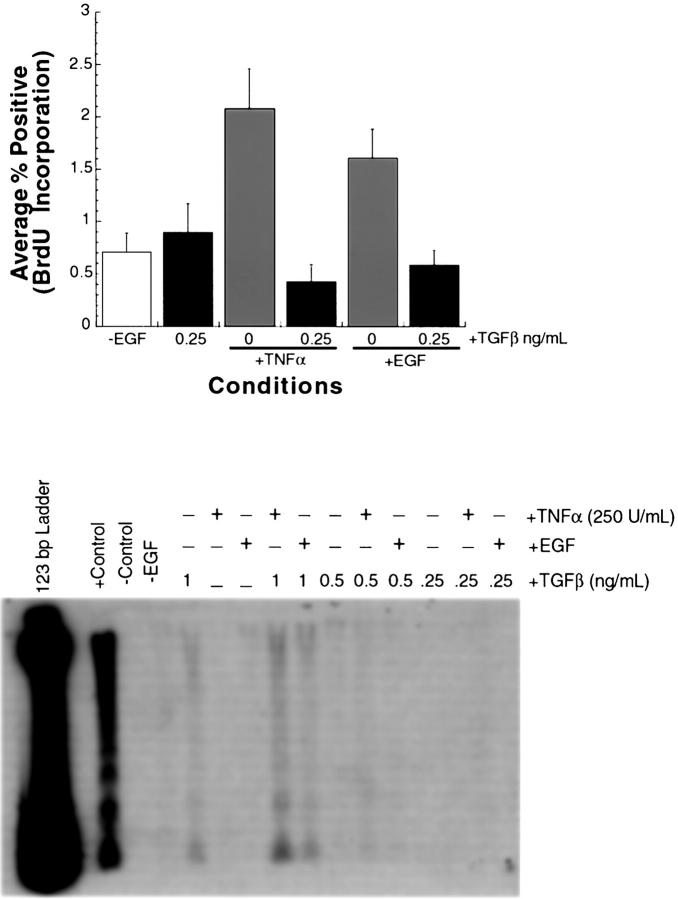
TGF-β Abrogates the Induction of DNA Synthesis by TNF-α and EGF in Primary Rat Hepatocytes
Carr and colleagues 36 have demonstrated that hepatocyte DNA synthesis induced by the mitogen EGF can be abrogated by co-treatment of cultures with EGF and TGF-β. It was determined that this effect was due neither to direct competition between the cytokines nor to toxicity. To determine whether TGF-β was able to inhibit DNA synthesis induced by the mitogen TNF-α, cells maintained in the absence of EGF were treated with TNF-α (250 U/ml), EGF (25 ng/ml), TNF + TGF-β (0.25 ng/ml), or EGF + TGF-β (0.25 ng/ml) and labeled with BrdU for 24 hours. When TGF-β was added in combination with either mitogen the number of cells incorporating BrdU was suppressed to control levels (Figure 7A) ▶ . Thus, DNA synthesis induced by TNF-α is abrogated by TGF-β as previously described for the known mitogen EGF. 35,36
Figure 7.
TGF-β abrogates the induction of DNA synthesis by TNF-α and EGF in primary rat hepatocytes. Hepatocytes were maintained for 16 days in the absence of EGF. A: Hepatocytes were either maintained under these conditions for the duration of the experiment (−EGF) treated with TGF-β alone (0.25 ng/ml), TNF-α alone (250 U/ml), EGF alone (25 ng/ml), TNF-α in combination with TGF-β, or EGF in combination with TGF-β. Cultures were treated for 24 hours and BrdU was added to the culture media at the time of treatment. Percentages of BrdU-labeled nuclei were calculated for each condition. Error bars indicate the SE for individual conditions. B: DNA fragmentation after treatment with TNF-α and TGF-β. Hepatocytes were either maintained in media lacking EGF for the duration of the experiment (−EGF) and treated with TGF-β alone (0.25, 0.5, or 1.0 ng/ml), TNF-α alone (250 U/ml), EGF alone (25 ng/ml), TNF-α in combination with TGF-β, or EGF in combination with TGF-β for 24 hours. Untreated primary rat hepatocytes or hepatocytes treated with cycloheximide (5 μmol/L) and TNF-α (2500 U/ml) in combination were used as negative and positive controls, respectively. Low-molecular weight DNA was isolated and analyzed for internucleosomal cleavage.
In addition to its ability to inhibit cell proliferation, TGF-β is well documented as a proapoptotic agent. Because TNF-α can also function in this capacity, we wanted to determine whether the combination of these factors may be inducing apoptosis in hepatocytes, thus accounting for the change in BrdU incorporation. Cultures of primary rat hepatocytes were treated as described above. Low-molecular weight DNA was isolated and analyzed for internucleosomal DNA fragmentation (Figure 7B) ▶ . No spontaneous apoptosis was detected in cultures of primary hepatocytes. At the concentration of TGF-β (0.25 ng/ml) that abrogated stimulation of DNA synthesis by either TNF-α or EGF, no ladder formation was detected when treatment was with TGF-β alone or in combination with TNF-α or EGF (Figure 7B) ▶ . When hepatocytes were treated with TGF-β at 1 ng/ml alone or in combination with TNF-α or EGF, low levels of ladder formation were detectable. We conclude that the decrease in BrdU incorporation after treatment with TGF-β in combination with TNF-α was not because of increased apoptosis of the cultures.
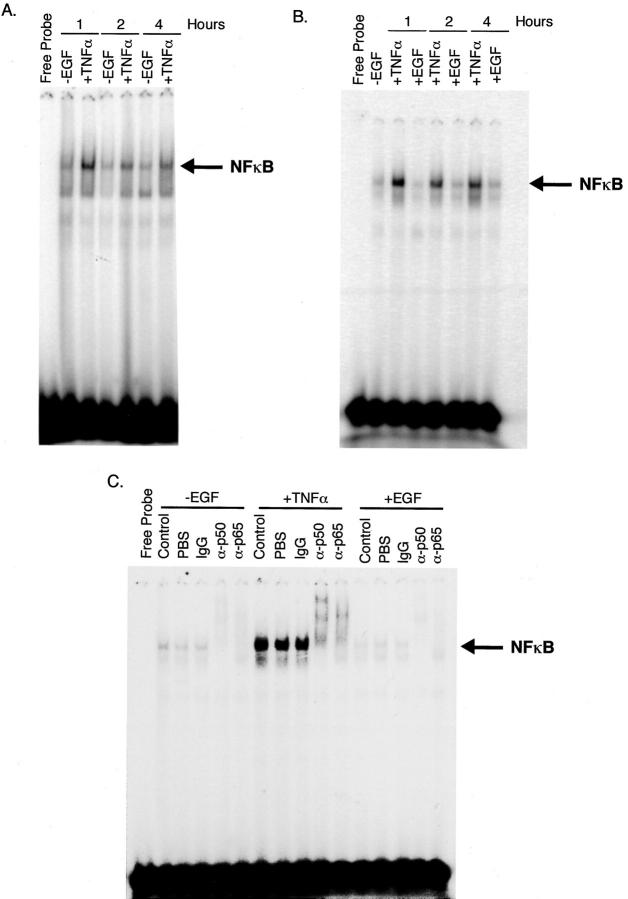
TNF-α Treatment Leads to an Increase in NF-κB Activation
The ability of TNF-α to induce DNA synthesis and cell proliferation is commonly mediated by the transcription factor NF-κB. To determine whether this was the case in primary rat hepatocytes, cells were maintained in control media (−EGF) or treated with TNF-α (+TNF-α) for 1, 2, or 4 hours. Nuclear protein was harvested from these cultures and subjected to electrophoretic mobility shift assay. TNF-α treatment caused an induction of NF-κB activity within 1 hour of treatment (Figure 8A) ▶ . This induction was most dramatic at early time points after treatment (1 to 4 hours) but was sustained through 24 hours after TNF-α treatment (data not shown). To determine whether this effect was specific to TNF-α, or was a common factor in hepatocyte cell-cycle progression after mitogen treatment, primary rat hepatocytes were maintained in control media for the duration of the experiment (−EGF), or treated with TNF-α (+TNF-α) or EGF (+EGF) for 1, 2, or 4 hours (Figure 8B) ▶ . No increase in NF-κB activity was observed after treatment with the known hepatocyte mitogen EGF at any of the three time points, indicating that activation of this transcription factor is specifically induced after TNF-α stimulation.
Figure 8.
NF-κB is activated after TNF-α treatment. Hepatocytes were maintained for 16 days in the absence of EGF. Arrows indicate the position of the NF-κB complex. A: Cells were either maintained in −EGF media (−EGF) or treated with TNF-α (250 U/ml) (+TNF-α) for an additional 1, 2, or 4 hours. Nuclear protein was incubated with 32P-labeled oligonucleotides corresponding to the consensus binding sequence for NF-κB. B: Cells were maintained in the absence of EGF for 15 days before the start of the experiment. Cells were maintained in this media for the duration of the experiment (−EGF) or treated with TNF-α (+TNF-α) or EGF (+EGF) for 1, 2, or 4 hours. Nuclear protein was incubated with 32P-labeled oligonucleotides corresponding to the consensus binding site for NF-κB. C: Supershift analysis of NF-κB activity. Extracts were incubated as described for gel shifts and subsequently incubated an additional half hour with no addition (control), addition of PBS, normal rabbit IgG (IgG), anti-p50 antibody (α-p50), or anti-p65 antibody (α-p65).
To confirm these results and identify the components of the NF-κB dimer, supershift analysis was performed (Figure 8C) ▶ . Hepatocytes were treated with TNF-α (or EGF as a control) for 1 hour and nuclear protein extracted. Neither PBS nor IgG lead to altered complex mobility. Incubation with antibody to the p50 component of NF-κB produced two supershifted complexes whereas incubation with antibody against p65 produced a single supershifted complex, suggesting that both p50 homodimers and p50/p65 heterodimers are activated after TNF-α treatment of primary rat hepatocytes.
Discussion
We have shown in this study, that primary hepatocytes maintained in long-term DMSO culture in the absence of EGF (−EGF) retain the properties of differentiated hepatocytes. Use of the −EGF rat hepatocyte long-term DMSO culture system is advantageous to examine the primary mitogenic potential of TNF-α for several reasons. In contrast to in vivo models for liver regeneration, the −EGF culture system enables us to examine the effects of TNF-α alone without interference from growth factors, cytokines, and hepatocyte mitogens. Hepatocytes maintained in the −EGF long-term DMSO culture system express high levels of the liver-specific genes albumin and A1AT relative to hepatocytes in short-term in vitro culture. Furthermore, these cultures maintain the morphological characteristics of hepatocytes at the ultrastructural level. An increased number of peroxisomes were present in cultures of rat hepatocytes maintained in the absence of EGF as compared to +EGF cultures, indicating that these cells are under stress as a result of the EGF deprivation. Despite this growth factor deficiency, no spontaneous apoptosis occurs in this culture system. We conclude that cells maintained in the −EGF culture system retain the morphological and biochemical markers of well-differentiated hepatocytes and that this system is a valuable tool for the study of the primary mitogenic potential of TNF-α or other factors.
In the present study, we have demonstrated that TNF-α serves as a complete mitogen for rat hepatocytes in long-term DMSO culture. TNF-α induced DNA synthesis to levels not statistically different from that of the known hepatocyte mitogen EGF and did not induce apoptosis. The maximal induction of DNA synthesis in primary rat hepatocytes by TNF-α was observed 24 hours after treatment with 250 U of TNF-α/ml. A small number of hepatocytes induced to enter S phase by TNF-α treatment proceeded through the cell cycle as evidenced by the detection of mitotic figures 96 hours after treatment. TNF-α activated NF-κB in the form of both p50 homodimers and p65/p50 heterodimers within 1 hour of treatment. As previously described for EGF, 35,36 TNF-α-induced DNA synthesis was inhibited by TGF-β.
It has been suggested that for hepatocytes to respond to proliferative stimuli in vivo, they must first undergo a period of priming during which the cells exit G0 and enter a state of replicative competence. 3,20,40-42 Several stimuli capable of priming hepatocytes for replication have been identified to date including minimal hepatectomy (removal of 30% of the liver), 20 collagenase perfusion, 41 sham surgery, and growth hormone injections. 42 Webber and colleagues 3 demonstrated that while the complete hepatocyte mitogens TGF-α and HGF induced DNA synthesis after portal vein infusion, simultaneous treatment with either mitogen and TNF-α increased hepatocyte DNA synthesis fourfold above mitogen stimulation alone. The authors concluded from these data that TNF-α had the ability to prime hepatocytes to a state of replicative competence.
In addition to the ability of TNF-α to prime hepatocytes for replication, considerable evidence exists in the literature to suggest that TNF-α may also serve as a mitogen for these cells. Within 1 hour after two-thirds partial hepatectomy, TNF-α mRNA and protein levels increase dramatically. 1 Treatment of animals with TNF-α-neutralizing antibodies before two-thirds partial hepatectomy inhibits DNA synthesis and liver regeneration 2 indicating that TNF-α is necessary for the normal regenerative response in liver. TNF receptor 1 (TNF-R1) knockout mice showed severely impaired DNA synthesis as compared to their wild-type counterparts after two-thirds partial hepatectomy. This deficiency in liver regeneration lead to a 50% mortality rate in these animals whereas no mortality occurred in wild-type mice after partial hepatectomy. 4 In contrast, TNF-R2 knockout mice show no impairment of DNA synthesis or liver regeneration as compared to wild-type animals. 5 These results suggest that TNF-α and TNF-R1 play an important role in liver regeneration after injury.
Although hepatocytes in long-term DMSO culture exist in G0, they retain the capacity to reenter the cell cycle and synthesize DNA in response to a variety of stimuli. We have previously demonstrated that when hepatocytes in long-term DMSO culture are subjected to infection with SV40 virus or transfection with SV40 DNA, some of the hepatocytes replicated to yield epithelial cell colonies that were capable of being expanded to yield immortalized cell lines. 55 Serra and Isom 35 have demonstrated that hepatocytes cultured in the presence of DMSO and EGF for 20 days can be induced to respond to both EGF and TGF-α stimulation after a 7-day period of insulin, glucagon, and EGF deprivation. In addition, Cable and Isom 56 have shown that maintenance of primary rat hepatocytes in DMSO containing media with iron, copper, and zinc, not only induces hepatocytes to synthesize DNA but also results in a threefold increase in cell number in the cultures. Taken together these data showed that primary rat hepatocytes in long-term DMSO culture in the presence of EGF retained the capacity to both synthesize DNA and to proliferate. In the current study, we have shown that primary hepatocytes maintained in long-term DMSO culture in the absence of EGF also retain the capacity to re-enter the cell cycle.
It has been reported previously that TNF-α derived from different species can differ in signaling capacity. Specifically, Leist and colleagues 57 showed that in cultures of murine cells, human TNF-α was able to signal only through TNF-R1, whereas murine TNF-α was able to activate both TNF-R1 and TNF-R2. In the present study, no significant difference in the number of cells induced to enter S phase by murine compared to human TNF-α was observed, suggesting that TNF-R1 is the receptor used for TNF-α-induced DNA synthesis in rat hepatocytes in long-term culture. As discussed briefly in the Introduction, after interaction of TNF-α with its receptors numerous intracellular changes occur. A hallmark associated with TNF-α-induced cell proliferation is induction of NF-κB activity. Therefore, it was important to determine whether NF-κB was activated when primary hepatocytes in long-term culture in the absence of EGF were treated with TNF-α. Indeed, the expected result was obtained; a rapid and dramatic increase in NF-κB activation occurred after TNF-α treatment. It is also important to note that treatment with the mitogen EGF did not result in NF-κB activation indicating that the response was specific to TNF-α.
It has been shown using short-term cultures of primary rat hepatocytes, that TGF-β is able to inhibit induction of DNA synthesis by EGF and HGF. 34,36 In addition, TGF-β mRNA has been shown to be elevated in nonparenchymal cells between 4 and 72 hours after partial hepatectomy, 58 suggesting that TGF-β may be a key player in negative regulation of liver regeneration. Serra and Isom 35 previously demonstrated that TGF-β inhibited DNA synthesis induced by both EGF and TGF-α in hepatocytes in long-term DMSO culture. In the present study, we have shown that addition of low doses of TGF-β to TNF-α treated primary rat hepatocytes abrogates the induction of DNA synthesis by this mitogen. This finding supports the conclusion that TNF-α acts as a primary mitogen for these cells because DNA synthesis induced by co-mitogens or priming agents, including insulin and glucagon, is not inhibited by TGF-β treatment. 36
It has been demonstrated that serum TNF-α levels are significantly increased in patients with a variety of chronic liver diseases including hepatitis B virus infection, hepatitis C virus infection, hemochromatosis, Wilson’s disease, α1-antitrypsin deficiency, primary biliary cirrhosis, autoimmune hepatitis, primary hepatocellular carcinoma, and alcoholic liver disease. 59 In addition to modulation of TNF-α expression in cases of chronic liver disease, altered expression of the receptors TNF-R1 and TNF-R2 is also observed. The ability of TNF-α to act as a complete mitogen for primary rat hepatocytes and the up-regulation of TNF-α expression in patients with chronic liver disease raise the possibility that TNF-α may contribute to the progression of these disease states to cirrhosis and carcinogenesis. Our future studies will focus on the mechanism by which TNF-α exerts its mitogenic effect on primary rat hepatocytes and the interaction of TNF-α with other known growth factors for hepatocytes.
TNF-α has previously been identified as a priming factor for hepatocytes and has been shown to be instrumental in liver regeneration. 1-5,20,40-42 This study represents the first direct evidence that TNF-α is able to act as a complete mitogen for primary rat hepatocytes in long-term DMSO culture.
Acknowledgments
We thank Dr. Edward Cable for helpful discussions and assistance in the preparation of this manuscript, Roland Myers for assistance with electron microscopy, and Tom Miller for excellent technical assistance.
Footnotes
Address reprint requests to Dr. Harriet C. Isom, Milton S. Hershey Medical Center, Department of Microbiology and Immunology, H107, The Penn State College of Medicine, 500 University Dr., Hershey, PA, 17033. E-mail: hisom@psu.edu.
Supported by the National Cancer Institute (grants CA23931 and DK53430).
References
- 1.Loffreda S, Rai R, Yang SQ, Lin HZ, Diehl AM: Bile ducts and portal and central veins are major producers of tumor necrosis factor α in regenerating liver. Gastroenterology 1997, 112:2089-2098 [DOI] [PubMed] [Google Scholar]
- 2.Akerman P, Cote P, Yang SQ, McClain C, Nelson S, Bagby GJ, Diehl AM: Antibodies to tumor necrosis factor-α inhibit liver regeneration after partial hepatectomy. Am J Physiol 1992, 263:G579-G585 [DOI] [PubMed] [Google Scholar]
- 3.Webber EM, Bruix J, Pierce RH, Fausto N: Tumor necrosis factor primes hepatocytes for DNA replication in the rat. Hepatology 1998, 28:1226-1234 [DOI] [PubMed] [Google Scholar]
- 4.Yamada Y, Kirillova I, Peschon JJ, Fausto N: Initiation of liver growth by tumor necrosis factor: deficient liver regeneration in mice lacking type I tumor necrosis factor receptor. Proc Natl Acad Sci USA 1997, 94:1141-1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamada Y, Webber EM, Kirillova I, Peschon JJ, Fausto N: Analysis of liver regeneration in mice lacking type I or type II TNF receptor: requirement for type I but not type II receptor. Hepatology 1998, 28:906-913 [DOI] [PubMed] [Google Scholar]
- 6.Neumann MG, Benhamou JP, Malkiewicz IM, Ibrahim A, Valla DC, Martinot-Peignoux M, Asselah T, Bourliere M, Katz GG, Shear NH, Marcellin P: Kinetics of serum cytokines reflect changes in the severity of chronic hepatitis C presenting minimal fibrosis. J Viral Hepatol 2002, 9:134-140 [DOI] [PubMed] [Google Scholar]
- 7.Fang JW, Shen WW, Meager A, Lau JY: Activation of the tumor necrosis factor-alpha system in the liver in chronic hepatitis B virus infection. Am J Gastroenterol 1996, 91:748-753 [PubMed] [Google Scholar]
- 8.Bird GL, Sheron N, Goka AK, Alexander GJ, Williams RS: Increased plasma tumor necrosis factor in severe alcoholic hepatitis. Ann Intern Med 1990, 112:917-920 [DOI] [PubMed] [Google Scholar]
- 9.Khoruts A, Stahnke L, McClain CJ, Logan G, Allen JI: Circulating tumor necrosis factor, interleukin-1 and interleukin-6 concentrations in chronic alcoholic patients. Hepatology 1991, 13:267-276 [PubMed] [Google Scholar]
- 10.Chan FK-M, Chun HJ, Zheng L, Siegel RM, Bui KL, Lenardo MJ: A domain in TNF receptors that mediates ligand-independent receptor assembly and signaling. Science 2000, 288:2351-2353 [DOI] [PubMed] [Google Scholar]
- 11.Brockhaus M, Schoenfeld H-J, Schlaeger E-J, Hunziker W, Lesslauer W, Loetscher H: Identification of two types of tumor necrosis factor receptors on human cell lines by monoclonal antibodies. Proc Natl Acad Sci USA 1990, 87:3127-3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith CA, Farrah T, Goodwin RA: The TNF receptor superfamily of cellular and viral proteins: activation, costimulation and death. Cell 1994, 76:959-962 [DOI] [PubMed] [Google Scholar]
- 13.Banner DW, D’Arcy A, Janes W, Gentz R, Schoenfeld HJ, Broger C, Loetscher H, Lesslauer W: Crystal structure of the soluble human 55 kd TNF receptor-human TNF beta complex: implications for TNF receptor activation. Cell 1993, 73:431-445 [DOI] [PubMed] [Google Scholar]
- 14.Dembic Z, Loetscher H, Gubler U, Pan YC, Lahm HW, Gentz R, Brockhaus M, Lesslauer W: Two human TNF receptors have similar extracellular, but distinct intracellular, domain sequences. Cytokine 1990, 2:231-237 [DOI] [PubMed] [Google Scholar]
- 15.Lewis M, Tartaglia LA, Lee A, Bennett GL, Rice GC, Wong GHW, Chen EY, Goeddel DV: Cloning and expression of cDNAs for two distinct murine tumor necrosis factor receptors demonstrate one receptor is species specific. Proc Natl Acad Sci USA 1991, 88:2830-2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodwin RG, Anderson D, Jerzy R, Davis T, Brannan CI, Copeland NG, Jenkins NA, Smith CA: Molecular cloning and expression of the type 1 and type 2 murine receptors for tumor necrosis factor. Mol Cell Biol 1991, 11:3020-3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rothe M, Wong SC, Henzel WJ, Goeddel DV: A novel family of putative signal transducers associated with the cytoplasmic domain of the 75 kDa tumor necrosis factor receptor. Cell 1994, 78:681-692 [DOI] [PubMed] [Google Scholar]
- 18.Shu H-B, Takeuchi M, Goeddel DV: The tumor necrosis factor receptor 2 signal transducers TRAF2 and c-IAP1 are components of the tumor necrosis factor receptor 1 signaling complex. Proc Natl Acad Sci USA 1996, 93:13973-13978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Etienne PL, Baffet G, Desvergne B, Boisnard-Rissel M, Glaise D, Guguen-Guillouzo C: Transient expression of c-fos and constant expression of c-myc in freshly isolated hepatocytes. Oncogene Res 1988, 3:255-262 [PubMed] [Google Scholar]
- 20.Webber EM, Godowski PJ, Fausto N: In vivo response of hepatocytes to growth factors requires an initial priming stimulus. Hepatology 1994, 14:489-497 [PubMed] [Google Scholar]
- 21.Brenner DA, Koch KS, Leffert HL: Transforming growth factor α stimulates proto-oncogene c-jun expression and a mitogenic program in primary cultures of adult rat hepatocytes. DNA 1989, 8:279-285 [DOI] [PubMed] [Google Scholar]
- 22.Mead JE, Fausto N: Transforming growth factor α may be a physiological regulator of liver regeneration by means of an autocrine mechanism. Proc Natl Acad Sci USA 1989, 86:1559-1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kan M, Huang J, Mansson P-EYH, Carr B, McKeehan WL: Heparin-binding growth factor type I (acidic fibroblast growth factor): a potential biphasic autocrine and paracrine regulator of hepatocyte regeneration. Proc Natl Acad Sci USA 1989, 86:7432-7436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Houck KA, Zarnegar R, Muga SJ, Michalopoulos GK: Acidic fibroblast growth factor (HBGF-1) stimulates DNA synthesis in primary rat hepatocyte cultures. J Cell Physiol 1990, 143:129-132 [DOI] [PubMed] [Google Scholar]
- 25.Hoffmann B, Paul D: Basic fibroblast growth factor and transforming growth factor-alpha are hepatotrophic mitogens in vitro. J Cell Physiol 1990, 142:149-154 [DOI] [PubMed] [Google Scholar]
- 26.Richman RA, Claus TH, Pilkis SJ, Friedman DL: Hormonal stimulation of DNA synthesis in primary cultures of adult rat hepatocytes. Proc Natl Acad Sci USA 1976, 73:3589-3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGowan JA, Strain AJ, Bucher NLR: DNA synthesis in primary cultures of adult rat hepatocytes in a defined medium: effects of epidermal growth factor, insulin, glucagon, and cyclic-AMP. J Cell Physiol 1981, 108:353-363 [DOI] [PubMed] [Google Scholar]
- 28.Michalopoulos GK, Houck KA, Dolan ML, Leutteke NC: Control of hepatocyte replication by two serum factors. Cancer Res 1984, 44:4414-4419 [PubMed] [Google Scholar]
- 29.Nakamura T, Nawa K, Ichihara A: Partial purification and characterization of hepatocyte growth factor from serum of hepatectomized rats. Biochem Biophys Res Commun 1984, 122:1450-1459 [DOI] [PubMed] [Google Scholar]
- 30.Housley RM, Morris CF, Boyle W, Ring B, Biltz R, Tarpley JE, Aukerman SL, Devine PL, Whitehead RH, Pierce GF: Keratinocyte growth factor induces proliferation of hepatocytes and epithelial cells throughout the rat gastrointestinal tract. J Clin Invest 1994, 94:1764-1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bissell DM, Wang S-S, Jarnagin WR, Roll FJ: Cell-specific expression of transforming growth factor-β in rat liver. J Clin Invest 1995, 96:447-455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Russell WE: Transforming growth factor beta (TGF-β) inhibits hepatocyte DNA synthesis independently of EGF binding and EGF receptor autophosphorylation. J Cell Physiol 1988, 135:253-261 [DOI] [PubMed] [Google Scholar]
- 33.Strain AJ, Hill DJ, Milner RDG: Divergent action of transforming growth factor β on DNA synthesis in human foetal liver cells. Cell Biol Int Rep 1986, 10:855-860 [DOI] [PubMed] [Google Scholar]
- 34.Nakamura T, Tomita Y, Hirai R, Yamaoka K, Kaji K, Ichihara A: Inhibitory effect of transforming growth factor β on DNA synthesis of adult rat hepatocytes in primary culture. Biochem Biophys Res Commun 1985, 133:1042-1050 [DOI] [PubMed] [Google Scholar]
- 35.Serra R, Isom HC: Stimulation of DNA synthesis and protooncogene expression in primary rat hepatocytes in long-term DMSO culture. J Cell Physiol 1993, 154:543-553 [DOI] [PubMed] [Google Scholar]
- 36.Carr BI, Hayashi I, Branum EL, Moses HL: Inhibition of DNA synthesis in rat hepatocytes by platelet-derived type beta transforming growth factor. Cancer Res 1986, 46:2330-2334 [PubMed] [Google Scholar]
- 37.Cornelius P, Marlowe M, Lee MD, Pekala PH: The growth factor-like effects of tumor necrosis factor-α. J Biol Chem 1990, 265:20506-20516 [PubMed] [Google Scholar]
- 38.Vilcek J, Palombella VJ, Henriksen-DeStefano D, Swenson C, Feinman R, Hirai M, Tsujimoto M: Fibroblast growth enhancing activity of tumor necrosis factor and its relationship to other polypeptide growth factors. J Exp Med 1986, 163:632-643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sugarman BJ, Aggarwal BB, Hass PE, Figari IS, Palladino MA, Shepard HM: Recombinant human tumor necrosis factor-α: effects on proliferation of normal and transformed cells in vitro. Science 1985, 230:943-945 [DOI] [PubMed] [Google Scholar]
- 40.Mead JE, Braun L, Martin DA, Fausto N: Induction of replicative competence (“priming”) in normal liver. Cancer Res 1990, 50:7023-7030 [PubMed] [Google Scholar]
- 41.Liu M-L, Mars WM, Zarnegar R, Michalopoulos GK: Collagenase pretreatment and the mitogenic effects of hepatocyte growth factor and transforming growth factor-α in adult rat liver. Hepatology 1994, 19:1521-1527 [PubMed] [Google Scholar]
- 42.Moolten FL, Oakman NJ, Bucher NL: Accelerated response of hepatic DNA synthesis to partial hepatectomy in rats pretreated with growth hormone or surgical stress. Cancer Res 1970, 30:2353-2357 [PubMed] [Google Scholar]
- 43.Isom H, Georgoff I, Salditt-Georgieff M, Darnell JE: Persistence of liver-specific messenger RNA in cultured hepatocytes: different regulatory events for different genes. J Cell Biol 1987, 105:2877-2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Isom HC, Secott T, Georgoff I, Woodworth C, Mummaw J: Maintenance of differentiated rat hepatocytes in primary culture. Proc Natl Acad Sci USA 1985, 82:3252-3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bilello JP, Cable EE, Myers RL, Isom HC: Role of paracellular junction complexes in baculovirus-mediated gene transfer to non-dividing rat hepatocytes. Gene Ther 2003, 10:733-749 [DOI] [PubMed] [Google Scholar]
- 46.Bilello JP, Cable EE, Isom HC: Expression of E-cadherin and other paracellular junction genes is decreased in iron-loaded hepatocytes. Am J Pathol 2003, 162:1323-1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cable EE, Connor JR, Isom HC: Accumulation of iron by primary rat hepatocytes in long term culture. Am J Pathol 1998, 152:781-792 [PMC free article] [PubMed] [Google Scholar]
- 48.Cable EE, Miller TG, Isom HC: Regulation of heme metabolism in rat hepatocytes and hepatocyte cell lines: δ-aminolevulinic acid synthase and heme oxygenase are regulated by different heme-dependent mechanisms. Arch Biochem Biophys 2000, 384:280-295 [DOI] [PubMed] [Google Scholar]
- 49.Bissonnette RP, Echeverri F, Mahboubi A, Green DR: Apoptotic cell death induced by c-myc is inhibited by bcl-2. Nature 1992, 359:552-554 [DOI] [PubMed] [Google Scholar]
- 50.Wyllie AH, Morris RG, Smith AL, Dunlop D: Chromatin cleavage in apoptosis: association with condensed chromatin morphology and dependence on macromolecular synthesis. J Pathol 1984, 142:67-77 [DOI] [PubMed] [Google Scholar]
- 51.Hoang AT, Cohen KJ, Barrett JF, Bergstrom DA, Dang CV: Participation of cyclin A in Myc induced apoptosis. Proc Natl Acad Sci USA 1994, 91:6875-6879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bour ES, Ward LK, Cornman GA, Isom HC: Tumor necrosis factor-α-induced apoptosis in hepatocytes in long-term culture. Am J Pathol 1996, 148:485-495 [PMC free article] [PubMed] [Google Scholar]
- 53.Olnes MI, Kurl RN: Isolation of nuclear extracts from fragile cells: a simplified procedure applied to thymocytes. Biotechniques 1994, 17:828-829 [PubMed] [Google Scholar]
- 54.Plumpe J, Malek NP, Bock CT, Rakemann T, Manns MP, Trautwein C: NFκB determines between apoptosis and proliferation in hepatocytes during liver regeneration. Am J Physiol 2000, 278:G173-G183 [DOI] [PubMed] [Google Scholar]
- 55.Woodworth C, Secott T, Isom HC: Transformation of rat hepatocytes by transfection of simian virus 40 DNA to yield proliferating cells. Cancer Res 1986, 46:4018-4026 [PubMed] [Google Scholar]
- 56.Cable EE, Isom HC: Exposure of primary rat hepatocytes in long-term DMSO culture to selected transition metals induces hepatocyte proliferation and formation of duct-like structures. Hepatology 1997, 26:1444-1457 [DOI] [PubMed] [Google Scholar]
- 57.Leist M, Gantner F, Jilg S, Wendel A: Activation of the 55 kDa TNF receptor is necessary and sufficient for TNF-induced liver failure, hepatocyte apoptosis, and nitrite release. J Immunol 1995, 154:1307-1316 [PubMed] [Google Scholar]
- 58.Braun L, Mead JE, Panzica M, Mikumo R, Bell GI, Fausto N: Transforming growth factor β mRNA increases during liver regeneration: a possible paracrine mechanism of growth regulation. Proc Natl Acad Sci USA 1988, 85:1539-1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tilg H, Wilmer A, Vogel W, Herold M, Nölchen B, Judmaier G, Huber C: Serum levels of cytokines in chronic liver diseases. Gastroenterology 1992, 103:264-274 [DOI] [PubMed] [Google Scholar]