Abstract
The sensitive detection of bone marrow involvement is crucial for tumor staging at diagnosis and for monitoring of the therapeutic response in the patient’s follow-up. In neuroblastoma, only conventional cytomorphological techniques are presently accepted for the detection of bone marrow involvement, yet since the therapeutic consequences of the bone marrow findings may be far-reaching, the need for highly reliable detection methods has become evident. For this purpose, we developed an automatic immunofluorescence plus FISH (AIPF) device which allows the exact quantification of disseminated tumor cells and the genetic verification in critical cases. In this study, the power of the immunofluorescence technique is compared with conventional cytomorphology. 198 samples from 23 neuroblastoma patients (stages 4 and 4s) at diagnosis and during follow-up were investigated. At diagnosis, 45.6% of the samples (26 of 57) which were positive by AIPF investigation were negative by cytomorphology. During follow-up, 74.2% (49 of 66) of AIPF-positive samples showed no cytological signs of tumor cell involvement. False negative morphological results were found in up to 10% of tumor cell content. A tumor cell infiltrate below 0.1% was virtually not detectable by conventional cytomorphology. Using the sensitive immunofluorescence technique, the analysis of only two instead of four puncture sites did not lead to false negative results. Thus, the immunofluorescence technique offers an excellent tool for reliable detection and quantification of disseminated tumor cells at diagnosis and during the course of the disease.
Unambiguous demonstration of tumor cell dissemination to the bone marrow at diagnosis of neuroblastoma is of crucial importance for clinical staging. Prerequisite for this procedure and for all other studies on disseminated tumor cells is a reliable and sensitive detection system. We therefore developed a detection system fulfilling all these requirements. 1,2 Beside diagnostic information, the clearance rate of the metastatic infiltration as a response to induction therapy was suggested to have an important prognostic impact in advanced disease. 3,4-6 The initial cytomorphological examination of Wright-Giemsa-stainedsmears for tumor cells from two bone marrow aspirates is incorporated in the recommendations of the International Neuroblastoma Staging System (INSS). 7 However, neuroblastoma cells may escape microscopic detection because of their relatively unspecific morphological appearance, especially when present as single cells. Moreover, the detection of disseminated disease is furthermore complicated by the uneven distribution of tumor cells in the body, which may result in false negative findings by classical cytology or histology. 8,9 Therefore, in contrast to the INSS recommendations, analysis of four iliac crest punctures for more precise assessment of BM infiltration has been emphasized.
The demonstration of minimal tumor cell amounts is certainly important, not only initially but also for judging responses to specific therapeutic applications. During the last decade, numerous alternative approaches using immunological 10-13 and molecular biological methods 14-18 were described with the intention of improving the detection of minimal neuroblastoma involvement in bone marrow, peripheral blood, and stem cell products. However, the reliability of tumor cell detection and quantification by these methods is still controversial. For routine detection of minimal amounts of tumor cells in clinical samples, high sensitivity and specificity as well as the ability for tumor cell quantification are vital prerequisites.
In our laboratory, a novel microscopic device for automatic immunofluorescence plus FISH analysis (AIPF) was developed (Metafer4/RCDetect, MetaSystems, Altlussheim, Germany) 19 which enables fast analysis and exact quantification of immunofluorescence-labeled tumor cells in hematopoietic samples at the sensitivity of 1/106 and below. 20 By the use of this device, automatic search for disseminated neuroblastoma cells was routinely performed in bone marrow samples following immunofluorescence labeling of the neuroblastoma specific ganglioside GD2. The cytogenetic aberrations found in the primary tumor were sequentially demonstrated in case of questionable morphology by fluorescence in situ hybridization (FISH), providing further evidence for the neoplastic nature of the target cell. Molecular cytogenetic verification allowed an observer independent identification of tumor cells which turned out to be essential in samples with low tumor cell infiltration, as false positive immunological reactions accounted for 38.5% of all analyzed bone marrow samples from localized neuroblastoma patients. 2
The purpose of this paper is to show to what extent the immunofluorescence-based automatic bone marrow analysis improves the detection of low-level tumor cell infiltration in comparison to cytomorphological examination of Wright-Giemsa-stained slides. The new approach was applied independent of, but parallel to, classical cytomorphological examinations of bone marrow smears from neuroblastoma patients at the St. Anna Children’s Hospital, enabling the comparison of results gained via both techniques. By the ability to automatically determine absolute tumor cell quantities, we demonstrate the limitations of bone marrow cytology for the analysis of minimally disseminated neuroblastoma cells. Furthermore, we analyzed whether punctures from more than two BM sites significantly improve the efficacy of tumor cell detection at elevated tumor cell detection sensitivity.
Materials and Methods
The extent of initial bone marrow involvement and the effect of chemotherapy was determined in 17 stage 4 and 6 stage 4s neuroblastoma patients. For this purpose, diagnostic bone marrow aspirates were taken from one to four iliac crest puncture sites in general anesthesia following a written consent signed by the parents of the patients. Bone marrow punctures were performed at diagnosis for initial staging and in the course of the disease to control response to cytotoxic therapy. The first drops of the aspirate were placed on microscopic slides to prepare 10 smears for conventional cytomorphological analysis following the Wright-Giemsa staining. 2- to 5-ml of bone marrow were then aspirated into a syringe supplied with heparin using the same needle from each puncture site. For further details of the handling of the bone marrow and laboratory and interpretation guidelines see Ambros and Ambros. 21 The mononuclear fractions were immediately isolated by density gradient centrifugation (Lymphoprep; Nycomed, Oslo, Norway) and the cells were processed for immunofluorescence analysis as described earlier. 19,20 Briefly, approx. 0.5 × 106 to 106 mononuclear cells were cytospun to microscopic slides (Hettich, Tuttlingen, Germany) and reacted with a GD2-specific monoclonal antibody following drying and fixation of the slides in buffered 3.7% formaldehyde solution for at least 1 hour, but ideally overnight at 4°C. The GD2-specific mab 14.18 (kindly provided by R.A. Reisfeld, The Scripps Research Institute, La Jolla, CA), previously described as highly specific to neuroblastoma cells, 22 was used for immunolabeling. The specific binding was detected by a FITC-conjugated anti-mouse secondary antibody (DAKO, Glostrup, Denmark) and the slides were covered with an antifade medium containing the nuclear stain DAPI (Vectashield/DAPI; Vector Laboratories, Burlingame, CA). 0.8 to 3 × 106 MNCs were analyzed per puncture site.
Cells with FITC immunofluorescence are selected by the Metafer4/RCDetect automated image analysis system (MetaSystems). This system enables automatic search and quantification of immunofluorescence-labeled cells according to their fluorescence pattern and intensity. During the scanning procedure, FITC-positive cells are automatically digitally photographed and the slide positions are recorded. The automatic scanning procedure per slide containing 106 MNCs takes 15 to 25 minutes depending on the number of positive cells. Cells which fulfill the search criteria (FITC and DAPI positivity, size and contour of the FITC-fluorescent object, and ratio of FITC and DAPI fluorescence area) are automatically captured by the system. The parameters for the search criteria were carefully set and verified in a number of control experiments to avoid false positive and false negative results. These search criteria are usually specific for a certain tumor type but have to be newly defined for most tumor entities. The fluorochromes can be chosen according to the laboratory settings and also three color analyses (eg, FITC+/TRITC−/DAPI+ fluorescence) are possible. After these fully automatic steps the observer can have a quick look at the cells on the screen or can have a close look directly in the microscope as every positive cell is precisely relocated by the automatic microscope. However, this microscopical analysis following the repositioning of isolated cells frequently does not support tumor-typical cell morphology. Features such as irregular broad cytoplasms, kidney-shaped small nuclei, condensed chromatin, and no prominent nucleoli could refer to damaged tumor cells due to eg, the cytotoxic treatment but also to falsely positive macrophages or other hematopoietic cells. Therefore, such cells are defined as “ambiguous”. To achieve a high degree of reliability one can either use another antibody or subject the cells to sequential FISH analysis directed to tumor typical cytogenetic aberrations found in the primary tumor. To enable the sequential genetic testing by FISH, slides are taken out of the stage and are re-inserted after successful FISH. The automatic relocation function allows the repositioning of all previously immunologically-positive cells. In this way every questionable cell can be re-evaluated and the genetic make-up can be studied in detail. In neuroblastomas 17q gain, MYCN amplification and 1p deletion are the most common aberrations, whereas stage 4 tumors show at least one of these aberrations, with very few exceptions. MYCN amplification was studied using the Oncor probe or an FITC-labeled BAC clone (RP11–355H10, provided by Dr. Pieter de Jong, BACPAC Resource Center at Children’s Hospital, Oakland Research Institute, Oakland, CA) in combination with a subtelomeric sequence from chromosome 2p (08–103-1, kindly provided by Dr. Rocchi, Bari, Italy), deletions or imbalances at 1p36.3 were monitored using probes D1Z2/D1Z1, 23,24 gain of 17q material was visualized using 17p/17q probes (411G7, 22G12, and 388C12, 516M14, kindly provided by Dr. Rocchi). Most of the stage 4s tumors, however, do not display the above mentioned aberrations, but usually display trisomies or polysomies of different chromosomes therefore centromere and subtelomere specific probes were used (eg, D1Z1, 411G7, 22G12). GD2-positive cells with the expected cytogenetic aberrations were interpreted as “unambiguous” tumor cells. GD2-positive cells lacking the expected tumor-typical cytogenetic aberrations were excluded from the evaluation as neuroblastoma cells but were also reported. FISH was performed in all 94 bone marrow samples, which contained less than 100 GD2+ cells.
The cytomorphological examination of Wright-Giemsa-stained bone marrow smears was performed by the hematology specialists of the St. Anna Children′s Hospital and was reported independently of the immunofluorescence results. The GD2 immunofluorescence was examined in a blinded fashion. Both the number of GD2-positive tumor cells and the number of analyzed MNCs were reported. For better comparison of the relative tumor cell contents of samples from different patients, tumor cell numbers were extrapolated to absolute cell numbers of 106 analyzed MNCs.
Results
Tumor Cell Detection by Cytomorphology
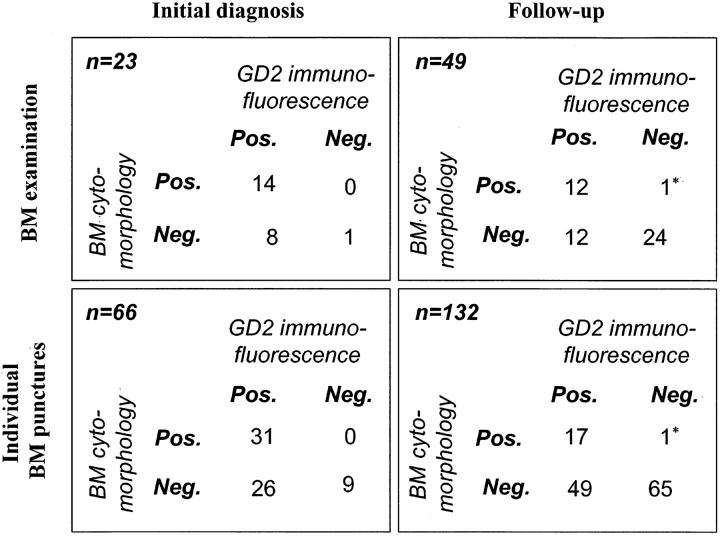
The initial bone marrow status was analyzed in all 23 patients (66 puncture samples), whereas disease monitoring (follow-up analysis) was performed at 49 time points (132 puncture samples). Bone marrow positivity appeared as a spectrum from massive tumorous infiltration to isolated cell nests in the smear. The cytomorphological analysis in Wright-Giemsa-stained slides revealed positivity in 14 of 23 (60.9%) initial examinations, 31 of 66 initial (46.9%) samples and 12 of 49 (24.5%) follow-up examinations, and 18 of 132 follow-up (13.6%) samples. Altogether, 49 of 198 positive results (24.7%) were obtained (Table 1) ▶ .
Table 1.
BM Cytomorphology and Results of GD2 Immunofluorescence
*Cells lacking tumor-typical MYCN amplification.
Tumor Cell Detection by GD2 Immunofluorescence Plus FISH Technique
Automated analysis of GD2 immunofluorescence-detected tumor cells in the range from 3 cells in 3.1 × 106 MNCs to 80% tumor cell content. However, the number of total cells provided for the analysis was also in a wide range (0.1 to 3.1 × 106, mean 0.88 × 106 MNCs). Positivity was shown in 57 of 66 initial samples (86.4%), while 66 of 132 (50%) bone marrow samples contained GD2+ tumor cells at follow-up bone marrow examinations. Altogether, 123 of 198 positive samples were detected by GD2 immunofluorescence. In 94 (24 initial and 70 follow-up) samples less than 100 GD2+ cells per 106 mononuclear cells were found, which showed, by original definition, “ambiguous” positivity. In these samples, the specificity of the test was achieved by sequential FISH analysis visualizing tumor-typical cytogenetic aberrations.
Correlation of BM Cytomorphology and GD2 Immunofluorescence
The correlation of the findings obtained by classical cytomorphology and automated search for GD2 immunofluorescence is presented in Table 1 ▶ . 34.8% (8 of 23) of the initial examinations (26 of 66, 39.4% of the individual punctions) and 24.5% (12 of 49) of the follow-up examinations (49 of 132, 37.1% of the individual punctions) showed divergent findings when comparing the two methods. Moreover, the discrepancy represented 45.6% (26 of 57) and 74.2% (49 of 66) of all positive samples in the respective sample group. While the overall rate of corresponding samples, which was found to be positive by GD2 immunofluorescence/verified by FISH and negative by cytomorphology, was 37.9% in this study (75 of 198), there was only one sample in which a single tumor cell nest was described in the Wright-Giemsa-stained BM smear, but no evidence of GD2-positive tumor cells could be detected by immunofluorescence. However, the targeted molecular cytogenetic analysis of the MYCN oncogene, which was carried out subsequently on the questionable Wright-Giemsa-stained cell nest, revealed two MYCN copies per cell. The primary tumor from this patient showed, in contrast, a well demonstrable 20-fold amplification of the MYCN oncogene in all three tumor pieces analyzed.
Sensitivity of BM Cytomorphology and GD2 Immunofluorescence/FISH
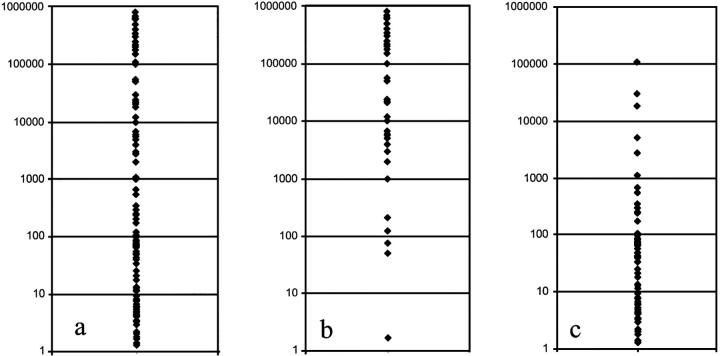
The comparison of the AIPF and cytomorphology data suggested a significantly lower sensitivity limit for cytomorphological analyses as compared to the immunofluorescence approach. This could be further supported by the quantitative evaluation of results based on AIPF search data. The AIPF enables the simultaneous determination of both the number of immunologically positive tumor cells and all mononuclear cells present in the sample, according to the nuclear DAPI staining. For a better comparison of the quantity of tumor cells in different samples, the number of positive cells was extrapolated to 106 MNCs. The distribution of quantitative GD2-immunofluorescence findings per 106 MNCs are presented in Figure 1a ▶ . The quantitative results of those samples which were found to be positive for tumor cells by both methods (cytomorphology and GD2 immunofluorescence) are displayed separately (Figure 1b) ▶ . Figure 1c ▶ , on the other hand, displays relative tumor cell quantities of those bone marrow samples which were found to be positive only by GD2 immunofluorescence, but not by cytomorphology. The detectability of tumor cells by the latter method became increasingly inconsistent at tumor cell concentrations between 10−2 and 10−4; 78.1% of the positive samples at 10−2, 88.6% of the positive samples at 10−3, and 92.8% of the positive samples at 10−4 tumor cell concentrations were missed. On the other hand, automatic search for GD2 immunofluorescence and subsequent FISH gave reproducible findings even when only a single tumor cell per one million MNCs was present (1/106 concentration).
Figure 1.
Quantitative distribution of neuroblastoma cells detected by cytomorphology and/or automatic GD2 immunofluorescence plus FISH analysis. a: Distribution of tumor cell numbers in samples which were found to be positive by automatic GD2 immunofluorescence analysis. b: Number of tumor cells in samples positive both for cytomorphology and GD2 immunofluorescence. c: Distribution of tumor cell numbers in samples negative by cytomorphology and positive by GD2 immunofluorescence. The number of tumor cells was extrapolated to 106 analyzed MNCs. y axis: number of tumor cells/106 MNCs
Influence of the Number of BM Punctures on Bone Marrow Positivity
The influence of the number and the site of punctures on the result of the BM examination was tested by comparing the AIPF with cytomorphology results of four iliac crest punctures with the analyzed BM samples. From 11 cytomorphological examinations with a positive finding, 3 (27.2%) and 5 (45.4%) cases displayed side-specific positivity in the antero-posterior or left-right relation, respectively (Table 2) ▶ . On the other hand, despite significant differences in the amount of tumor cells in individual puncture samples, no side-specific difference concerning sample positivity or negativity was observed in any of the 20 positive examinations analyzed by AIPF for four puncture sites.
Table 2.
Distribution of Cytomorphological and GD2-AIPF Positive Punctures
| Distribution of cytomorphological positive punctures in 11 bone marrows | ||||||
| A only | 1 | R only | 2 | |||
| P only | 2 | L only | 3 | |||
| A and P | 8 | R and L | 6 | |||
| Total | 11 | 11 | ||||
| Distribution of GD2-AIPF positive punctures in 20 bone marrows | ||||||
| A only | 0 | R only | 0 | |||
| P only | 0 | L only | 0 | |||
| A and P | 20 | R and L | 20 | |||
| Total | 20 | 20 | ||||
A, anterior; P, posterior; R, right; L, left.
Discussion
Cytomorphological examination to detect rare tumor cells is limited by numerous factors, eg, the small round-cell morphology of the tumor cells, lack of tumor cell nests, and lack of possibilities for quantification. Many immunological and RT-PCR applications, directed to detect rare solid tumor cells in hematological samples, were described to reach extremely high sensitivity levels (10−6 to 10−7) through demonstration of tissue-specific target molecules. However, they are often obscured by artificial immunological staining of non-tumorous cells, especially in alkaline phosphatase or peroxidase-stained slides or by the elevated probability to amplify non-specific targets. 25,26 Immunological demonstration of the neuroblastoma-specific cell-surface gangliosid GD2, otherwise a reliable cell-surface marker in all analyzed neuroblastomas 27,28 highlighting all neuroblastoma cells in the BM, 8 was also found to be associated with false positive reactions. 20 Fortunately, fluorescence microscopic visualization enables further analysis and verification of every questionable positivity by sequential molecular cytogenetic analysis or by further immunological characterization. In a recent report we could unambiguously show by sequential FISH analyses of GD2-positive bone marrow cells that rare GD2-positive cells, occasionally present in the BM of patients with localized/regional neuroblastoma, were not of tumorous nature. 2 These findings are contrasting with previous reports demonstrating tumor cells in 34% of the patients with localized disease using light microscopical detection of the immunological target. 12 Limitations in the sensitivity of the AIPF system cannot explain the differences because we are also able to unambiguously recognize very rare disseminated GD2-positive tumor cells with verified tumor typical genetic aberrations (down to 1/106) in bone marrow samples from stage 4 and 4s neuroblastomas. 2 Sampling error can also cause some false negative results. Statistical models indicate that at least 3 × 106 cells need to be analyzed to achieve a 95% probability to recognize 1 tumor cell in 106 MNCs. 29 Accordingly, the theoretical chance to detect 1 tumor cell in 106 MNCs lies, in our study, between 63% and 95%.
In the present study, 94 of 199 samples were processed for FISH analysis. These cases mostly contained few immuno-positive cells requiring further genetic verification. The remaining samples were either negative or displayed convincing numbers (>100) of immunologically positive cells which could be identified as tumor cells on morphological criteria. The genetic verification was undertaken in all these cases to make sure that no false positive reaction was overlooked. By doing this stringent comparison between immunologically positive cells with the genetic make-up we learned to better discriminate between tumor cells and false positive reactions solely on the basis of immunological staining pattern and morphological features. Current studies are undertaken to find immunological markers to further discriminate between tumor cells and non-tumor cells, to facilitate the unambiguous and simple identification of disseminated tumor cells (DTCs).
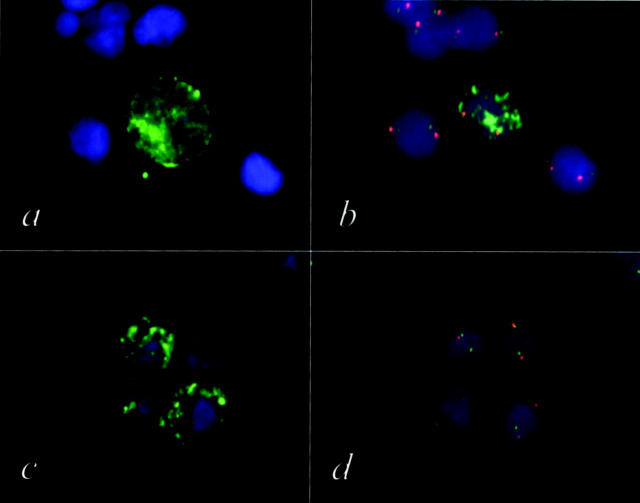
By the use of a computer-supported automatic device, the sensitivity of the bone marrow examination in neuroblastoma could be significantly enhanced. Using this method, an increase in the number of positive bone marrow examinations by more than 40% (41.3%) was achieved (overall increase of positive punctures by AIPF was 60.2%), as compared with classical BM cytology. On the contrary, only one sample negative by AIPF displayed a nest of possible tumor cells by classical cytological examination. Interestingly, none of the cells within this single cell group showed MYCN amplification by FISH, whereas the primary tumor displayed a 20-fold MYCN amplification (Figure 2) ▶ . Thus, we can conclude that this particular cell nest was not composed of tumor cells.
Figure 2.
a and c: Bone marrow cells from a neuroblastoma patient stained with FITC labeled GD2 antibody. b and d: After automatic relocation of the GD2-positive cells these cells were subsequently analyzed by FISH using a MYCN specific probe (FITC) and a chromosome 2-specific probe (TRITC). Only the GD2-positive cell shown in a and b displays the tumor typical MYCN amplification (b). The three GD2-positive cells shown in c show a normal MYCN copy number (d). Applying this genetic verification procedure (AIPF), tumor cells can be unambiguously discriminated from normal cells.
The vast majority of stage 4 tumors show genetic aberrations, ie, MYCN amplification, del1p36, and gain 17q, the latter representing the most frequent cytogenetic aberration in disseminated stage 4 patients as it has been described to be present in 85% of the analyzed tumors 30 (Stock et al, unpublished). In particular, gain of chromosomal regions and gene amplifications are ideal FISH targets because the risk of wrong interpretation of the FISH signals is reduced, as compared to the detection of deletions or imbalances. In theory the possibility of cytogenetic heterogeneity in disseminated tumor cells also exists in DTCs as it has been shown to occur at low frequency in primary tumors. 31 Our data, however, demonstrate that DTC always showed the same genetic aberration(s) as it was present in the primary tumor. In case of genetically heterogeneous primary tumors, not all aberrations found in the primary tumor are necessarily present in the DTCs. Because the genetic make-up of the primary tumor is usually known, the adequate DNA probes recognizing these aberrations can be applied. In cases where the genetic composition is not known, gain of 17q and MYCN amplification are ideal targets for tumor cell verification.
To prove whether the genetic aberrations are lost in DTCs or not, we analyzed the three different cytogenetic markers sequentially in the identical disseminated tumor cells from 7 neuroblastoma patients; no loss of genetic aberrations was found. 2 The improbability of obtaining false negative results by the automatic immunofluorescence/FISH approach is supported by mixing experiments. 20
The combination of two or possibly even more methods seems to be the best strategy to overcome the problems of false positive and/or false negative results in the detection of rare tumor cells. The example given in this paper where we used the high sensitivity of immunological detection systems and combined this method with the high specificity of the FISH technique can be extrapolated to other techniques. For example the combination of immunological and PCR techniques can also be used to reduce the chance of detecting false positive or false negative results.
Disseminated tumor cells below 1/103 MNC concentration were not detected reliably by cytomorphological examination. A significant part of the BM samples, especially samples from patients undergoing chemotherapy, displayed less tumor cells than this number, as demonstrated by the immunofluorescence technique. 76.8% of the positive samples from follow-up patients could only be detected by the latter method, but not by classical cytology. Accordingly, a 3 log (103) improvement in the sensitivity was reached by the AIPF method. We conclude that classical BM cytology was not sufficient for monitoring the response to cytotoxic therapy in stage 4 disease.
So far, evaluation of four puncture sites within one bone marrow examination was emphasized to increase the sensitivity of bone marrow analysis and avoid false negativity. When comparing the AIPF and CM results of the BM examinations from four individual puncture sites, we found that separate analyses of bone marrow samplings from four different sites were only of benefit to bone marrow cytomorphology, but did not change the number of positive samples when applying the AIPF approach. With the enhanced detection sensitivity of the latter method, the likelihood of false negativity by analysis of only two puncture sites was virtually eliminated. In the light of these data it seems sufficient to analyze, in agreement with INSS, samples from two puncture sites (left and right), provided that the detection method used guarantees a high sensitivity.
These data demonstrate the superior sensitivity and specificity of the automatic immunofluorescence detection technique compared to conventional bone marrow cytology for the detection of disseminated neuroblastoma cells, enabling a 3 log increase in the detection sensitivity. The less risky as well as time- and cost-saving analysis of bone marrow samples from two puncture sites provides reliable quantitative data at initial diagnosis and for disease monitoring. Future studies will have to illuminate the clinical value of quantitative bone marrow analysis with elevated sensitivity for staging and monitoring of advanced neuroblastoma but also of other solid tumors.
Acknowledgments
The help of M. Zavadil is greatly acknowledged.
Footnotes
Address reprint requests to Dr. Peter F. Ambros, CCRI, St. Anna Kinderspital, Kinderspitalgasse 6, A-1090 Vienna, Austria. E-mail: ambros@ccri.univie.ac.at.
Supported by the Children’s Cancer Research Institute, St. Anna Children’s Hospital, Vienna, Austria.
References
- 1.Ambros PF, Mehes G, Hattinger C, Ambros IM, Luegmayr A, Ladenstein R, Gadner H: Unequivocal identification of disseminated tumor cells in the bone marrow by combining immunological and genetic approaches: functional and prognostic information. Leukemia 2001, 15:275-277 [DOI] [PubMed] [Google Scholar]
- 2.Mehes G, Luegmayr A, Ambros IM, Ladenstein R, Ambros PF: Combined automatic immunological and molecular cytogenetic analysis allows exact identification and quantification of tumor cells in the bone marrow. Clin Cancer Res 2001, 7:1969-1975 [PubMed] [Google Scholar]
- 3.Hartmann O, Valteau-Couanet D, Vassal G, Lapierre V, Brugieres L, Delgado R, Couanet D, Lumbroso J, Benhamou E: Prognostic factors in metastatic neuroblastoma in patients over 1 year of age treated with high-dose chemotherapy and stem cell transplantation: a multivariate analysis in 218 patients treated in a single institution. Bone Marrow Transplant 1999, 23:789-795 [DOI] [PubMed] [Google Scholar]
- 4.Ladenstein R, Philip T, Lasset C, Hartmann O, Garaventa A, Pinkerton R, Michon J, Pritchard J, Klingebiel T, Kremens B, Pearson A, Coze C, Paolucci P, Frappaz D, Gadner H, Chauvin F: Multivariate analysis of risk factors in stage 4 neuroblastoma patients over the age of 1 year treated with megatherapy and stem-cell transplantation: a report from the European Bone Marrow Transplantation Solid Tumor Registry. J Clin Oncol 1998, 16:953-965 [DOI] [PubMed] [Google Scholar]
- 5.Seeger RC, Reynolds CP, Gallego R, Stram DO, Gerbing RB, Matthay KK: Quantitative tumor cell content of bone marrow and blood as a predictor of outcome in stage IV neuroblastoma: a Children’s Cancer Group study. J Clin Oncol 2000, 18:4067-4076 [DOI] [PubMed] [Google Scholar]
- 6.Ambros PF, Ambros IM, Ladenstein R, Luegmayr A, Rumpler S, Kovar H, Gadner H: Tumor cell clearing in the bone marrow of neuroblastoma patients: predictor for the outcome? Med Pediatr Oncol 1998, 31:195 [Google Scholar]
- 7.Brodeur GM, Seeger RC, Barrett A, Berthold F, Castleberry RP, D’Angio G, De BB, Evans AE, Favrot M, Freeman AI, Haase G, Hartmann O, Hayes FA, Helson L, Kemshead J, Lampert F, Ninane J, Ohkawa H, Philip T, Pinkerton CR, Pritchard J, Sawada T, Siegel S, Smith EI, Tsuchida Y, Voute PA: International criteria for diagnosis, staging, and response to treatment in patients with neuroblastoma. J Clin Oncol 1988, 6:1874-1881 [DOI] [PubMed] [Google Scholar]
- 8.Cheung NK, Heller G, Kushner BH, Liu C, Cheung IY: Detection of metastatic neuroblastoma in bone marrow: when is routine marrow histology insensitive? J Clin Oncol 1997, 15:2807-2817 [DOI] [PubMed] [Google Scholar]
- 9.Reid MM: Detection of bone marrow infiltration by neuroblastoma in clinical practice: how far have we come? Eur J Cancer 1994, 30A:134-135 [DOI] [PubMed] [Google Scholar]
- 10.Cheung NK, Von HD, Strandjord SE, Coccia PF: Detection of neuroblastoma cells in bone marrow using GD2 specific monoclonal antibodies. J Clin Oncol 1986, 4:363-369 [DOI] [PubMed] [Google Scholar]
- 11.Berthold F, Schneider A, Schumacher R, Bosslet K: Detection of minimal disease in bone marrow of neuroblastoma patients by immunofluorescence. Pediatr Hematol Oncol 1989, 6:73-83 [DOI] [PubMed] [Google Scholar]
- 12.Moss TJ, Reynolds CP, Sather HN, Romansky SG, Hammond GD, Seeger RC: Prognostic value of immunocytologic detection of bone marrow metastases in neuroblastoma. N Engl J Med 1991, 324:219-226 [DOI] [PubMed] [Google Scholar]
- 13.Faulkner LB, Tintori V, Tamburini A, Paoli A, Garaventa A, Viscardi E, Tucci F, Lippi AA, De Bernardi B, Bernini G: High-sensitivity immunocytologic analysis of neuroblastoma cells in paired blood and marrow samples. J Hematother 1998, 7:361-366 [DOI] [PubMed] [Google Scholar]
- 14.Burchill SA, Bradbury FM, Selby P, Lewis IJ: Early clinical evaluation of neuroblastoma cell detection by reverse transcriptase-polymerase chain reaction (RT-PCR) for tyrosine hydroxylase mRNA. Eur J Cancer 1995, 31A:553-556 [DOI] [PubMed] [Google Scholar]
- 15.Lode HN, Handgretinger R, Schuermann U, Seitz G, Klingebiel T, Niethammer D, Beck J: Detection of neuroblastoma cells in CD34+ selected peripheral stem cells using a combination of tyrosine hydroxylase nested RT-PCR and anti-ganglioside GD2 immunocytochemistry. Eur J Cancer 1997, 33:2024-2030 [DOI] [PubMed] [Google Scholar]
- 16.Miyajima Y, Horibe K, Fukuda M, Matsumoto K, Numata S, Mori H, Kato K: Sequential detection of tumor cells in the peripheral blood and bone marrow of patients with stage IV neuroblastoma by the reverse transcription-polymerase chain reaction for tyrosine hydroxylase mRNA. Cancer 1996, 77:1214-1219 [DOI] [PubMed] [Google Scholar]
- 17.Lo Piccolo MS, Cheung NK, Cheung IY: GD2 synthase: a new molecular marker for detecting neuroblastoma. Cancer 2001, 92:924-931 [DOI] [PubMed] [Google Scholar]
- 18.Hoon DS, Kuo CT, Wen S, Wang H, Metelitsa L, Reynolds CP, Seeger RC: Ganglioside GM2/GD2 synthetase mRNA is a marker for detection of infrequent neuroblastoma cells in bone marrow. Am J Pathol 2001, 159:493-500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehes G, Lorch T, Ambros PF: Quantitative analysis of disseminated tumor cells in the bone marrow by automated fluorescence image analysis. Cytometry 2000, 42:357-362 [DOI] [PubMed] [Google Scholar]
- 20.Mehes G, Luegmayr A, Hattinger CM, Lorch T, Ambros IM, Gadner H, Ambros PF: Automatic detection and genetic profiling of disseminated neuroblastoma cells. Med Pediatr Oncol 2001, 36:205-209 [DOI] [PubMed] [Google Scholar]
- 21.Ambros PF, Ambros IM: Pathology and biology guidelines for resectable and unresectable neuroblastic tumors and bone marrow examination guidelines. Med Pediatr Oncol 2001, 37:492-504 [DOI] [PubMed] [Google Scholar]
- 22.Mujoo K, Kipps TJ, Yang HM, Cheresh DA, Wargalla U, Sander DJ, Reisfeld RA: Functional properties and effect on growth suppression of human neuroblastoma tumors by isotype switch variants of monoclonal antiganglioside GD2 antibody 14.18. Cancer Res 1989, 49:2857-2861 [PubMed] [Google Scholar]
- 23.Cooke HJ, Hindley J: Cloning of human satellite III DNA: different components are on different chromosomes. Nucleic Acids Res 1979, 6:3177-3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buroker N, Bestwick R, Haight G, Magenis RE, Litt M: A hypervariable repeated sequence on human chromosome 1p36. Hum Genet 1987, 77:175-181 [DOI] [PubMed] [Google Scholar]
- 25.Borgen E, Beiske K, Trachsel S, Nesland JM, Kvalheim G, Herstad TK, Schlichting E, Qvist H, Naume B: Immunocytochemical detection of isolated epithelial cells in bone marrow: non-specific staining and contribution by plasma cells directly reactive to alkaline phosphatase. J Pathol 1998, 185:427-434 [DOI] [PubMed] [Google Scholar]
- 26.Zippelius A, Lutterbuse R, Riethmuller G, Pantel K: Analytical variables of reverse transcription-polymerase chain reaction-based detection of disseminated prostate cancer cells. Clin Cancer Res 2000, 6:2741-2750 [PubMed] [Google Scholar]
- 27.Wu ZL, Schwartz E, Seeger R, Ladisch S: Expression of GD2 ganglioside by untreated primary human neuroblastomas. Cancer Res 1986, 46:440-443 [PubMed] [Google Scholar]
- 28.Sariola H, Terava H, Rapola J, Saarinen UM: Cell-surface ganglioside GD2 in the immunohistochemical detection and differential diagnosis of neuroblastoma. Am J Clin Pathol 1991, 96:248-252 [DOI] [PubMed] [Google Scholar]
- 29.Funke I, Schraut W: Meta-analyses of studies on bone marrow micrometastases: an independent prognostic impact remains to be substantiated. J Clin Oncol 1998, 16:557-566 [DOI] [PubMed] [Google Scholar]
- 30.Bown N, Cotterill S, Lastowska M, O’Neill S, Pearson AD, Plantaz D, Meddeb M, Danglot G, Brinkschmidt C, Christiansen H, Laureys G, Speleman F: Gain of chromosome arm 17q and adverse outcome in patients with neuroblastoma. N Engl J Med 1999, 340:1954-1961 [DOI] [PubMed] [Google Scholar]
- 31.Ambros PF, Ambros IM, Kerbl R, Luegmayr A, Rumpler S, Ladenstein R, Amann G, Kovar H, Horcher E, De Bernardi B, Michon J, Gadner H: Intratumoural heterogeneity of 1p deletions and MYCN amplification in neuroblastomas. Med Pediatr Oncol 2001, 36:1-4 [DOI] [PubMed] [Google Scholar]