Abstract
Overexpression of CD30 is the hallmark of Hodgkin and Reed-Sternberg (H-RS) cells and drives constitutive nuclear factor-κB activation that is the molecular basis for the pathophysiology of Hodgkin’s lymphoma. Transcription of the CD30 gene is controlled by the core promoter that is driven by Sp-1 and the microsatellite sequences (MSs) that represses core promoter activity. To understand the mechanism(s) of CD30 overexpression in H-RS cells, we structurally and functionally characterized the CD30 MSs. Although the CD30 MS of H-RS cell lines was polymorphic, it was not truncated compared with that of control cells. A strong core promoter activity and constitutive Sp-1 binding were revealed in all cell lines examined irrespective of the levels of CD30 expression. In transient reporter gene assays, all MS clones derived from H-RS cell lines repressed the core promoter activity in unrelated cell lines, but not in the H-RS cell lines. An AP-1-binding site was found in the MS at nucleotide position of −377 to −371, the presence of which was found to relieve repression of the core promoter in H-RS cell lines but not in other tumor cell lines. H-RS cell lines showed constitutive and strong AP-1-binding activity, but other cell lines did not. The AP-1 complex contained JunB, whose overexpression activated reporter constructs driven by the CD30 promoter including the MSs, and was dependent on the AP-1 site. JunB expression was detected in H-RS cells in vitro and in vivo, but not in reactive cells or tumor cells of non-Hodgkin’s lymphoma of diffuse large B-cell type. Transduction of JunB small interfering RNAs suppressed CD30 promoter activity in L428 cells but not in control cells. Taken together, overexpression and binding of JunB to the AP-1 site appear to relieve the repression of the core promoter by the CD30 MS in H-RS cells, which provide one basis for the constitutive overexpression of CD30 in Hodgkin’s lymphoma.
Hodgkin’s lymphoma (HL), which accounts for approximately one-third of all malignant lymphomas, is characterized by the presence of only a small fraction of malignant cells. Neoplastic cells represented as mononucleated Hodgkin and multinucleated Reed-Sternberg (H-RS) cells are embedded in a varying infiltrate of reactive cells including B and T lymphocytes, eosinophils, plasma cells, and fibroblasts. 1 In most cases, H-RS cells carry clonal immunoglobulin (Ig) gene rearrangements and thus derive from germinal center B cells. 2-6
H-RS cells are characterized by constitutive expression of CD30, which is a member of the tumor necrosis factor receptor superfamily. 7-10 CD30 was originally discovered as a marker protein overexpressed on the surface of H-RS cells. 11 Other investigators and our group have shown that CD30 signals leading to nuclear factor-κB activation are mediated by interactions with tumor necrosis factor receptor-associated factor (TRAF) 2 and TRAF5. 12-15
Recently, it was found that constitutively activated nuclear factor-κB (p50/p65) is the hallmark of H-RS cells and is considered to be a molecular basis for aberrant growth and cytokine gene expression. This, in turn, provides an explanation for the clinical and histological characteristics of HL. 16-21
We recently reported that overexpression of CD30 results in ligand-independent constitutive signaling that activates nuclear factor-κB. 22 Overexpression of CD30 in H-RS cells leads to self-aggregation and recruitment of TRAF2 and TRAF5, the result being constitutive nuclear factor-κB activation. Thus, constitutive CD30 signaling is the basis for the growth of H-RS cells as well as for characteristic clinical and histological features of HL. 22,23 Consequently, understanding of the mechanisms for constitutive overexpression of CD30 has pivotal importance in delineating the pathogenesis of HL.
Recent cloning and characterization of the promoter region of the CD30 gene enabled us to understand the regulation of CD30 expression. 24,25 The CD30 promoter is composed of three regions: a microsatellite sequence (MS) containing CCAT repeats, a core promoter with Sp-1-binding sites, and a downstream promoter element (DPE), which is essential for start site selection. The core CD30 promoter contains three Sp-1-binding sites (−43 to −38, −96 to −91, and −206 to −201) and two ETS binding sites (−238 to −233 and −158 to −153). The Sp-1 site at −43 to −38 is responsible for maximum promoter activity. 25 Sp-1 and ETS family transcription factors can induce CD30 transcription. 26 On the other hand, the CCAT motif repeated in CD30 MS represses the core CD30 promoter activity. Because the MS is highly polymorphic in length, it has been hypothesized that polymorphic changes truncating the length of CD30 MS may lead to relief of transcriptional repression, resulting in the overexpression of CD30 in H-RS cells of HL. 25,27
To examine the above hypothesis and better understand the mechanism(s) underlying CD30 overexpression in H-RS cells, we cloned the CD30 MS from H-RS cells and characterized their structure and function. The results suggest that the AP-1 site in the MS mediates relief of suppression of the CD30 promoter through interaction with JunB, which appears to be one mechanism for the constitutive and strong expression of CD30 in H-RS cells.
Materials and Methods
Cell Cultures and DNA Extraction
Jurkat, K562, and HEK293 cell lines were obtained from the Japanese Cancer Research Resources Bank (Tokyo, Japan) and Fujisaki Cell Biology Center (Okayama, Japan). HDLM-2, KMH-2, L428, and L540 cell lines were purchased from the German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany). Nonadherent cell lines were cultured in RPMI 1640 and adherent cells in Dulbecco’s modified Eagle’s medium with supplementation of recommended concentrations of fetal calf serum and antibiotics. Peripheral blood mononuclear cells (PBMCs) were separated from peripheral blood samples obtained from normal volunteers by Lymphoprep (Nycomed Pharma AS, Oslo, Norway) according to manufacturer’s instructions. High-molecular genomic DNA samples were prepared from cell lines and control PBMCs using the standard method as described. 28
Cloning of the CD30 MS
The region containing the CD30 MS was amplified from genomic DNA samples by polymerase chain reaction (PCR). The reactions contained 1.5 mmol/L MgCl2 1× PCR buffer, 0.2 mmol/L dNTPs, and 2.5 U TaqDNA polymerase (Takara, Kyoto, Japan) and 25 pmol each of forward and reverse primers in 50 μl of cocktail. The nucleotide sequences of the primers flanking the CD30 MS were as follows: the forward primer, 30MSF 5′-ACCCATTTACCCACTCACCTGC-3′ and the reverse primer, 30MSR 5′-CAACTGGCCTAGGGAGACTGC-3′. The reaction condition was as follows: initial denature at 94°C for 30 seconds, 30 cycles of 95°C for 30 seconds, 60°C for 30 seconds, 72°C for 1 minute, and the final extension at 72°C for 7 minutes. PCR-amplified products were cloned using pGEM-T easy vector (Promega, Madison, WI) for sequencing. DNA sequences were determined by an ABI Prism 377 DNA sequencer and the dye terminator cycle-sequencing kit (Perkin Elmer Corp., Foster City, CA). The Genetix Mac ver. 10 program (Software Development, Co., Ltd., Tokyo, Japan) was used for nucleotide sequence analysis.
Northern Blotting
Northern blot analysis was done essentially as described. 29 Briefly, 2 μg per lane of poly(A)-selected RNA was size-fractionated on 1% formalin agarose gel electrophoresis and subsequently blotted onto nylon membranes (Micron Separations, Inc., Westborough, MA). Filters were hybridized in 4× standard saline citrate, 1× Denhardts, 0.5% sodium dodecyl sulfate, 0.1 mol/L NaPO4 (pH7.0), 10% dextran Na at 65°C with 1.0 × 106 cpm/ml of random prime-labeled probes. After washings to a final stringency of 0.2× standard saline citrate and 0.1% sodium dodecyl sulfate at 65°C, filters were exposed to XAR-5 films (Eastman Kodak, Rochester, NY) at −80°C. Reverse transcriptase-PCR-amplified fragments of human CD30 and human GAPDH were used as probes.
Reporter Gene Assays
The CD30 core promoter region from −295 to +198 was amplified from the control PBMC DNA by PCR using a primer pair having a HindIII site at the end. The fragment was cloned into pGL3 basic plasmid (Promega). The resultant plasmid was named pGL-CD30c. MS regions were obtained by PCR from H-RS cell lines and control PBMCs and inserted into the SmaI site upstream of the CD30 core promoter and resultant constructs were named pGL-CD30cMS. To prepare an MS region having a mutant AP-1 site, a reverse primer having a mutated AP-1 sequence was used to amplify the cloned MS fragment (KMH-2, no. 2 in Figure 2 ▶ ). The sequences of the primers used are as follows: 30MSF 5′-CATTTGCTGTAAAACAAATCAGTGAG-3′ and the reverse primer, 30MSR 5′-CATTTGCTGTAAATGAATCAGTGAG-3′. The amplified fragment was inserted at the SmaI site upstream of the CD30 core promoter sequence of the pGL-CD30c and the resultant plasmid was named pGL-CD30cMSmA. Activities of the CD30 core promoter with or without MS were studied by transient reporter gene assays. Renilla luciferase expression vector driven by the herpes simplex virus thymidine kinase promoter, pRL-TK (Promega), was co-transfected to standardize the transfection efficiency in each experiment. Luciferase activities were measured by the Dual Luciferase assay kit (Promega). Transfection was done using Lipofectamine 2000 Reagent (Invitrogen, Tokyo, Japan) according to the manufacturer’s instructions.
Figure 2.
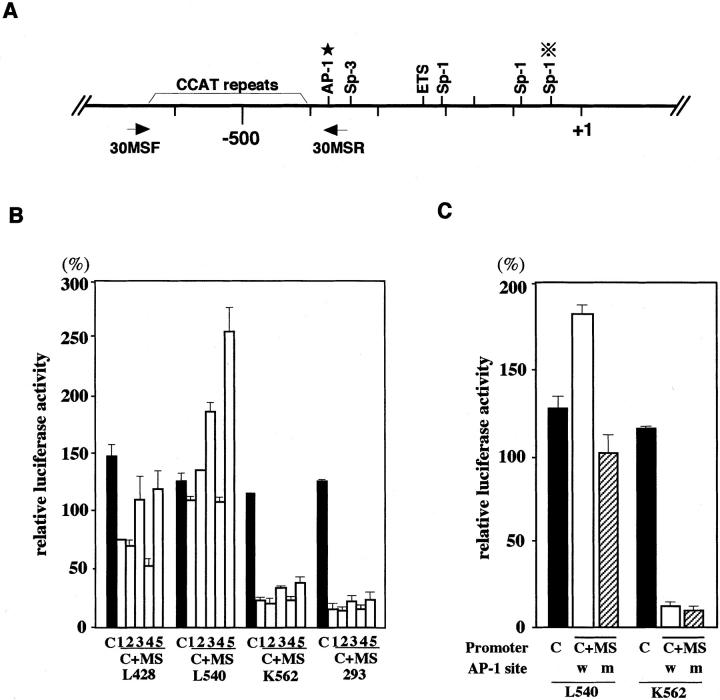
Schematic representation of the structure of the CD30 MS. Nucleotide sequences of more than four clones from each cell line or a PBMC sample were determined. The CCAT repeat in the MS is indicated by a filled circle and the 5′-CCACTTATGCAT-3′ and 5′-CCACCTATGCAT-3′ motifs are indicated by hatched squares. The number of the CCAT repeats in each clone is indicated on the right.
Electrophoretic Mobility Shift Analysis (EMSA)
EMSA was done as described previously. 22,30 Double-stranded oligonucleotides containing the Sp-1 and AP-1 consensus sites (5′-ATTCGATCGGGGCGGGGCGAGC-3′ and 5′-CGCTTGATGAGTCAGCCGGAA-3′, respectively) were purchased from Promega. The sequence of the CD30 MS AP-1 site probe is as follows: 5′-CACTCACTGATTCATTTTACA-3′ (nucleotide position −384 to −364). Nuclear extracts were prepared basically according to the method reported by Andrews and Faller. 31 Briefly, cells were washed in cold phosphate-buffered saline, and suspended in buffer A [10 mmol/L HEPES-KOH (pH 7.9 at 48°C), 1.5 mmol/L MgCl2, 10 mmol/L KCl, 0.5 mmol/L dithiothreitol, 0.2 mmol/L phenylmethyl sulfonyl fluoride]. After brief vortexing, samples were centrifuged and the pellet was resuspended in 50 μl of cold buffer C [20 mmol/L HEPES-KOH (pH 7.9 at 48°C), 25% glycerol, 420 mmol/L NaCl, 1.5 mmol/L MgCl2, 0.2 mmol/L ethylenediaminetetraacetic acid, 0.5 mmol/L dithiothreitol, 0.2 mmol/L phenylmethyl sulfonyl fluoride] and incubated for 20 minutes for high-salt extraction. Cellular debris was removed by centrifugation and the supernatant fraction was used as the nuclear extract. For binding assays, samples of 2-μg nuclear extract were incubated with ∼20,000 cpm of γ-32P end-labeled probe at room temperature for 30 minutes in a buffer containing 20 mmol/L HEPES-KOH (pH 7.9), 50 mmol/L KCl, 0.5 mmol/L ethylenediaminetetraacetic acid, 5% glycerol, 1 mmol/L dithiothreitol, 0.1% Nonidet P-40, 0.5 mmol/L phenylmethyl sulfonyl fluoride, 1 mg/ml bovine serum albumin, 2 μg Poly (dI-dC). DNA-protein complexes were resolved on 5% polyacrylamide gels in 0.5× Tris-borate-ethylenediaminetetraacetic acid buffer. The gels were then dried and autoradiographed. For competition assays, 2 μg of nuclear extracts were incubated for 30 minutes with the unlabeled probe before addition of γ-32P end-labeled probe. Antibody used for supershift assays were as follows: anti-Sp-1 rabbit polyclonal antibody (Upstate Biotechnology, Lake Placid, NY), rabbit polyclonal antibodies against c-Fos (H-125), c-Jun (H-79), JunD (329), Fos B (C-11), Fra-1 (R-20), Fra-2 (L-15), CREB-1 (C-21), and mouse monoclonal antibodies against JunB (C-11) and ATF2 (F2BR-1) (all from Santa Cruz Biotechnology, Inc. Santa Cruz, CA).
Immunoblot Analysis
Immunoblotting experiments were done, as described previously. 22,23 A JunB expression vector pME-JunB was prepared using the cDNA amplified from L540 mRNA by reverse transcriptase-PCR and the pME vector. The entire coding region of JunB cDNA was amplified using primers that have a restriction enzyme site. The nucleotide sequence of the primers are as follows: forward primer, 5′-TATACGCGTCGCCTGGGCCGCCCGGATG-3′, reverse primer, 5′-ATAGCGGCCGCTTATCAGAAGGCGTGTCCCTTGACC-3′. pME-JunB was co-transfected into K562 cells with a reporter construct such as pGL-CD30cMS or pGL-CD30cMSmA for reporter gene assays. For immunoblot analysis, cell lysates were prepared from a part of the transfected cells (2 × 106 cells) in a lysis buffer (10 mmol/L Tris-HCl, pH 7.4, 1% sodium dodecyl sulfate, 1 mmol/L sodium orthovanadate, 0.1 mmol/L sodium molybdate, 1 mmol/L phenylmethyl sulfonyl fluoride). Twenty mg of cell lysates were separated by 10% sodium dodecyl sulfate-polyacrylamide gel, and transferred to the polyvinylidene difluoride membrane (Bio-Rad Laboratories Hercules, CA). Blocking of the membrane was done with 1% albumin fraction V in Tris-buffered saline at 4°C for 12 hours. After washing with Tris-buffered saline with 0.05% Tween 20, membranes were incubated with 2 μg/ml of the JunB antibody for 1 hour at room temperature. Alkaline phosphatase-conjugated anti-mouse antibody was used to detect immunoreactive proteins by Western blue stabilized substrate for alkaline phosphatase (Promega).
Immunohistochemistry
Immunostaining of JunB was performed on paraffin-embedded specimens of H-RS cell lines and control cell lines as well as lymph nodes affected with HL or diffuse large B-cell non-Hodgkin’s lymphoma (NHL). Before incubation of the anti-JunB monoclonal antibody C-11 (Santa Cruz), a heat-induced antigen retrieval was performed. Immunodetection was done with biotinylated goat anti-mouse IgG, followed by peroxidase-labeled streptavidin (DAKO Japan, Kyoto, Japan). A Histofine kit (Nichirei, Tokyo, Japan) was used to detect the color reaction for peroxidase, according to the manufacturer’s instructions.
Inhibition of JunB Expression by siRNA
Target sequences of siRNA 32 for JunB were selected as follows: JunB1 5′-AATGGAACAGCCCTTCTACCA-3′; JunB2 5′-AAGATGAACCACGTGACACCC-3′; JunB3 5′-AAACAGAAGGTCATGACCCAC-3′. siRNAs were designed and synthesized by Silencer siRNA construction kit (Ambion Inc., Austin, TX) according to the manufacture’s instruction manual. The resultant siRNAs were named as JunB siRNA1, JunB siRNA2 and JunB siRNA3, respectively. Twenty pmol of siRNAs were transduced into 2 × 105 of L428 and 1 × 105 of HEK 293 cells by Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instruction. Cells were plated and maintained in appropriate conditions. Among above three JunB siRNAs, JunB siRNA1 was most effective at silencing JunB expression, and used in the following experiments. The scrambled oligonucleotide 5′-CATGCTTAAATGGGCCCATGA-3′ served as a control.
Results
Expression of CD30 mRNA and the CD30 MS Length
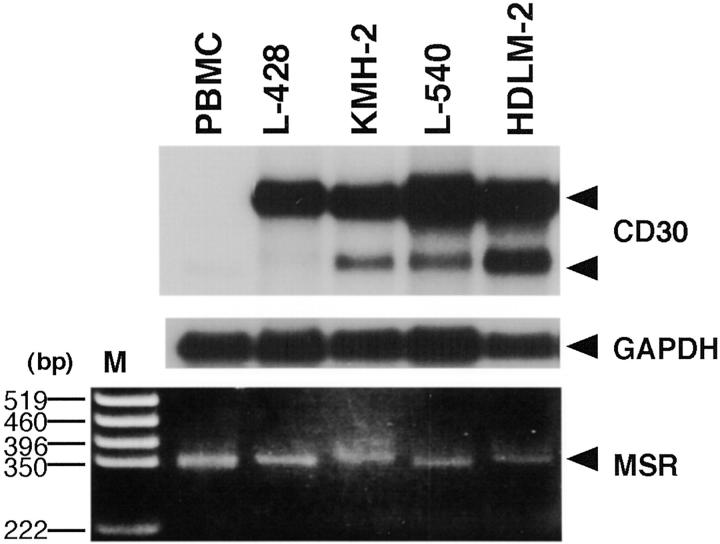
Earlier reports demonstrated a suppressive activity of the CD30 MS that contains a CCAT repeat on the core promoter activity of CD30. 24-26,33 To understand the roles of CD30 MS in overexpression of CD30 in H-RS cells, we first studied the correlation between the length of CD30 MS and levels of CD30 expression. Using H-RS cell lines, L428, KMH-2, L540, and HDLM-2, that abundantly express CD30 transcripts (Figure 1 ▶ , top), we analyzed the size of the PCR-amplified MS of the CD30 promoter (Figure 1 ▶ , bottom). Sizes of the amplified fragments of H-RS cells were generally not decreased compared to those of the control PBMCs. These results suggested that CD30 overexpression does not result simply from a reduction in the number of repressive CCAT repeats in the CD30 MS.
Figure 1.
CD30 mRNA expression and the length of MSs. Poly(A)-selected RNA (2 μg/lane) was analyzed by Northern blotting using cDNA probes of CD30 (top) and GAPDH (middle). Arrowheads indicate positions for two sizes of CD30 transcripts and GAPDH. PCR-amplified fragments of the CD30 MS were analyzed by agarose gel electrophoresis and ethidium bromide staining (bottom). The position for the PCR products of the MS is indicated by an arrowhead on the right. DNA size markers (M) are shown on the left.
Structure of the CD30 MS in H-RS Cell Lines
Next we determined the nucleotide sequences of the amplified CD30 MS. In addition to four H-RS cell lines, two samples of PBMCs were included as controls. At least four clones of the PCR product from each cell line or PBMC sample were analyzed. The amplified clones showed different sizes except for those amplified from L-428 that had exactly the same sequences. In KMH-2 and L540 H-RS cell lines, amplified clones included two kinds of clones, whereas HDLM-2 had one major clone and two other minor ones. These results indicate that the sizes of the CD30 MS in these H-RS cell lines are mostly heterozygous (Figure 2) ▶ . As indicated in the figure, the number of the repeats in the CD30 MS were as follows: L428, all four clones had 42 repeats; KMH-2, five clones with 43 repeats and four clones with 46 repeats; L540, seven clones with 43 repeats and two clones with 42 repeats; HDLM-2, four clones with 46 repeats and one clone each with 40, 47, and 49 repeats. All four clones from one PBMC sample had 40 repeats, and the other PBMC sample had two kinds of clones with 39 and 41 repeats. Thus, the numbers of CCAT repeats of H-RS cells are greater than those of PBMC samples (Figure 2 ▶ , right column).
The results obtained for the PBMC controls suggest that the PCR was amplifying the genomic copies without error. However, the HDLM-2 cell line in particular produced multiple PCR products indicating heterogeneity in target MSs that may be explained by hyperploidy as often seen in cancer cells lines including HDLM-2.
Suppression of the CD30 Core Promoter Activity by the CD30 MS
Sequence analysis suggested that CD30 MS of H-RS cell lines were also polymorphic as was previously reported. 25,33 Repressive effects of the CCAT repeat on the core promoter have been documented. 25 On the other hand, the H-RS cells that overexpress CD30 have larger MS regions 33 as we confirmed above. Thus, the role of the CCAT repeat unit in the overexpression of CD30 remains to be clarified. Therefore, we next examined the regulatory effects of cloned CD30 MS fragments against the CD30 core promoter using H-RS cell lines and unrelated cell lines such as K562 and HEK293 by transient reporter gene assays. A schematic representation of the CD30 promoter region is shown in Figure 3A ▶ . When the reporter plasmid driven by the core promoter was used, the results showed high-basal promoter activity in all cell lines irrespective of CD30 expression levels (Figure 3B ▶ , black bars). In contrast, a significant difference was observed in the suppressive activity of the MS sequence on the core promoter between H-RS cell lines and unrelated cell lines. The MS sequence significantly suppressed the core promoter activity in K562 and HEK293, whereas the suppression was less evident or absent in H-RS cell lines (Figure 3B ▶ , open bars). K562 cells express CD30 at a low level and HEK293 cells do not. Various CD30 MS fragments cloned from H-RS cell lines as well as PBMCs showed basically the same effects on the core promoter activity (Figure 3B) ▶ . These results suggest that regulatory effects of the CD30 MS on the core promoter are independent of the structure of each MS and may represent differences in the conditions of transcriptional and/or signaling controls between H-RS cell lines and unrelated cell lines.
Figure 3.
Regulation of the CD30 promoter by the MS. A: Schematic representation of the CD30 promoter. Binding sites for transcription factors are indicated above the line. An asterisk indicates the position of the AP-1 site in the MS. Positions of the primers used to amplify the MS are shown below the line. B: Effects of the CD30 MS on the core promoter activity in various cell lines. Luciferase reporter constructs driven by the CD30 core promoter (−295 to + 198), with or without the CD30 MS, were transiently transfected with Renilla luciferase vector (pRL-TK). The promoter activities among different cell lines were compared with the results of parallel experiments with a firefly luciferase construct driven by the SV40 promoter and enhancer (pSV-Luc) and pRL-TK. The relative luciferase activities are expressed as percentages of those of pSV-Luc. C, a construct driven by the core promoter from PBMCs; C+MS, a construct driven by the core promoter with CD30 MS. The fragments used are as follows: clone number 1 of L428, number 2 of KMH-2, number 2 of L540, number 2 of HDLM-2, and number 1 of the control PBMCs in Figure 2 ▶ . The numbers 1 to 5 indicate luciferase constructs containing a CD30 core promoter with MS fragment derived from L428, KMH-2, L540, HDLM-2, and the control PBMCs, respectively. C: Effects of a mutation in the AP-1 site on the core promoter activities. Luciferase constructs having the CD30 core promoter alone (C) or with an MS fragment with a mutation in the AP-1 (C+MS, m) or without a mutation in the AP-1 (C+MS, w) were transfected into a H-RS cell line L540 and an unrelated cell line, K562. Luciferase activities were standardized using an SV40-driven luciferase construct as described above and expressed as a percentage. C, a core promoter-driven luciferase construct; C+MS, a construct driven by the core promoter with CD30 MS; w, the MS without a mutation; m, the MS with a mutation.
AP-1-Binding Sequence within the CD30 MS Is Involved in the Regulation of the Core Promoter Activity
Sequence analysis of the CD30 MS region searching for putative transcription binding sites revealed an AP-1-binding site located at nucleotide position of −377 to −371 (5′-TGATTCA-3′) (Figure 3A) ▶ . To examine the functional roles of this site, activities of the CD30 promoter with or without a mutation in the AP-1-binding site were studied by transient transfection assays. Introduction of the MS sequence with a wild-type AP-1 site resulted in enhancement of the core promoter activity in L540 cells, whereas it showed a marked suppression of the core promoter activity in K562 cells as was reported previously. 25 A mutation in the putative AP-1 site (5′-TGATTCA-3′ to 5′-TGATTTG-3′) of the MS abolished the enhancing effect in L540 cells, although it did not result in a marked suppression of the core promoter activity as observed in K562 and HEK293 cells. The mutation did not show any effects on the core promoter activities in K562 cells (Figure 3C) ▶ .
These results were basically reproduced when we used other H-RS cell lines and HEK293 cells, although the levels were different (data not shown). These results clearly demonstrated differences in the regulatory function of the MS sequence depending on the cell lines used. Furthermore, they indicated that the enhancement of the core promoter activity by the MS sequence observed in H-RS cell lines is mediated by the putative AP-1 site.
Constitutive AP-1-Binding Activity in H-RS Cells Involves JunB
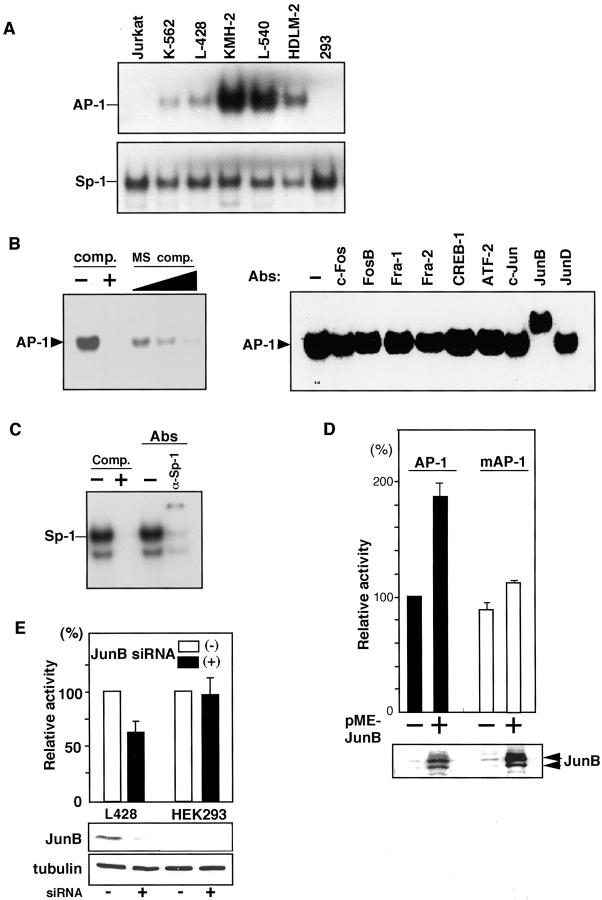
Because the above results suggested regulatory roles of the putative AP-1 site in the MS sequence, we examined binding of nuclear factors to this site by EMSA using a probe prepared from the sequence containing this AP-1 site and nuclear extracts of various cell lines. Results revealed strong binding activities of nuclear factors from H-RS cell lines but not those from other cell lines, except for weak binding observed in K562 nuclear extracts (Figure 4A ▶ , top). In contrast, Sp-1-binding activity was ubiquitously observed in all cell lines (Figure 4A ▶ , bottom), which is in accordance with the constitutive core promoter activities observed in all cell lines tested (Figure 3A) ▶ . The specificity of the binding was confirmed by competition assays using unlabeled oligomers of the consensus AP-1 probe and an oligonucleotide containing the putative AP-1-binding site (Figure 4B ▶ , left). To clarify the components of the AP-1-binding activity, super shift analysis was done using the L540 nuclear extract and antibodies against AP-1 family transcription factors. The results demonstrated a complete shift when the anti-JunB antibody was used. However, no band shift was observed with antibodies reactive to the other members of the AP-1 family such as FosB, Fra1, Fra2, as well as those of the CREB/ATF family such as CREB-1, CREM1, ATF1, and ATF2 because the antibody against CREB-1 recognizes CREM1 and ATF1 as well as CREB-1 (Figure 4B ▶ , right).
Figure 4.
AP-1 activities in H-RS cells. A: AP-1- and Sp-1-binding activities. Nuclear extracts of H-RS cell lines and unrelated ones were studied by EMSA. Probes used are indicated on the left. B: Competition and supershift analyses of AP-1. Competition assays were done using a 50-fold molar excess of the unlabeled consensus Ap-1 probe or the unlabeled oligomer containing the AP-1 site in the CD30 MS at 50- to 150-fold molar excess (left). Comp., the consensus AP-1 oligomer; MS comp., oligomer containing the AP-1 site in the CD30 MS. AP-1 supershift assays were done using L-540 nuclear extracts (right). Antibodies used are indicated above the lanes. C: Competition and supershift analyses of Sp-1-binding complex using L540 nuclear extract. Comp., competition assay using 50-fold molar excess of the unlabeled consensus Sp-1 oligomer; Abs, supershift assay with anti-Sp-1 antibody. D: Responses to transduced JunB. Luciferase constructs driven by the CD30 promoter with or without a mutation in AP-1 site (pGL-CD30cMS and pGL-CD30cMSmA, respectively) was co-transfected with a JunB expression plasmid (pME-JunB) into K562 cells. pRL-TK was co-transfected to standardize transfection efficiency. AP-1, pGL-CD30cMS; mAP-1, pGL-CD30cMSmA. E: Effects of siRNA-mediated repression of endogenous JunB in H-RS cells on CD30 promoter activity. Twenty pmol of siRNA for JunB, 0.5 μg of reporter construct containing CD30 promoter (pGL-CD30cMS), and 0.2 μg of pRL-TK vector were transfected with 2.5 μl of Lipofectamine (Invitrogen). CD30 promoter activities were measured by Dual Luciferase assay kit (Promega) after 24 hours. Bottom, immunoblot analysis of JunB and tubulin expression.
The specificity of Sp-1-binding activities was confirmed by the competition analysis using an unlabeled consensus Sp-1 probe (Figure 4C) ▶ . Components of the Sp-1-binding activity of the L540 nuclear extracts were examined by the supershift assay using an antibody against Sp-1. Most of the binding activities were supershifted by pretreatment of the nuclear extracts with the antibody (Figure 4C) ▶ , indicating the constitutive Sp-1 activity in L540 cells. These results clearly indicate that the AP-1-binding complex of L540 nuclear extracts contains JunB. Thus, it was suggested that the suppression of the Sp-1-driven core promoter activity by the CD30 MS sequence might be counteracted by the AP-1 activity in H-RS cell lines.
To test the possibility that JunB drives the CD30 promoter through binding to the AP-1 site in the MS and opposes the suppressive activity of the MS on the core promoter, transient transfection assays were done using K562 cells and a JunB expression vector. The luciferase activity driven by the CD30 promoter having the MS with a wild-type AP-1 site was enhanced approximately twofold by transduction of JunB, whereas that driven by the promoter with a mutated AP-1 site was not enhanced significantly (Figure 4D) ▶ . Transduction of JunB siRNAs into L428 cells resulted in suppression of CD30 promoter activity to approximately two-thirds of the control. This suppressive effect was not observed in HEK293 cells (Figure 4E) ▶ , or when we used CD30 promoter with mutated AP-1 site within the MS (data not shown). The repression of endogenous JunB by transduction of JunB siRNAs indicated that the AP-1 site in the MS of L428 Hodgkin’s cells is functionally responsive to JunB and plays a role in the relief of suppression by the MS sequence.
Constitutive Expression of JunB in H-RS Cells in Vitro and in Vivo
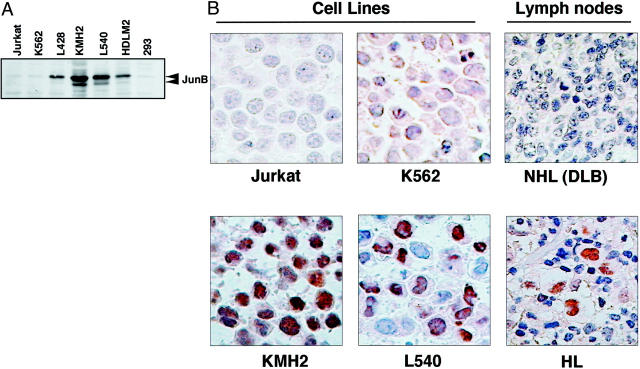
The above results suggested that binding of JunB to the AP-1 site in the MS counteracts the suppressive activity of the MS in H-RS cells, which explain the constitutive overexpression of CD30. To test this hypothesis, we next examined expression of JunB in H-RS cell lines and H-RS cells in the lymph node of the patients with classical HL. Immunoblot analysis revealed expression of JunB in H-RS cell lines, but not in other tumor cell lines (Figure 5A) ▶ , suggesting that JunB expression may be a common and unique characteristic of H-RS cells. To examine this possibility, we next studied expression of JunB in H-RS cell lines and that in the biopsied lymph nodes of patients with classical HL and diffuse large B-cell NHL. Eight samples of classical HL and five diffuse large B-cell NHL samples were studied by immunohistochemistry using an anti-JunB antibody. JunB expression was clearly detected in the H-RS cell lines and H-RS cells, but not reactive cells in the lymph nodes from HL patients but not in Jurkat cells, K562 cells, or in lymph nodes from patients with diffuse large B-cell NHL (Figure 5B) ▶ . The results clearly demonstrated specific expression of JunB in H-RS cells but not in any other cells including reactive cells in HL lymph nodes and not in tumor cells of diffuse large B-cell NHL. Thus, we have shown that constitutive expression of JunB may be a unique and common characteristic of H-RS cells that is involved in the constitutive overexpression of CD30, counteracting the suppressive CD30 MS activity against the CD30 core promoter.
Figure 5.
Constitutive expression of JunB in H-RS cells in vitro and in vivo. A: Immunoblot analysis of Jun B. Whole cell lysates of four H-RS cell lines and three unrelated cell lines were examined. Positions of JunB are indicated on the right. B: Immunohistochemistry of JunB. Results of H-RS cell lines and control ones, and representative results of biopsied samples of classical HL and diffuse large B-cell NHL.
Discussion
In this study we structurally characterized the CD30 MS of H-RS cell lines and found that, although they are polymorphic, the average length of the MS is not shorter than that of control PBMCs. We identified an AP-1-binding site in the CD30 MS, by which suppressive activity of the MS against the CD30 core promoter was counteracted in the H-RS cell lines but not in other tumor cell lines. The AP-1 site was found to be a distinct complex that includes JunB as determined by EMSA when we used nuclear extracts of H-RS cell lines but not those of other cell lines. The AP-1 site was responsive to transduced JunB in transient reporter gene assays. We further demonstrated constitutive overexpression of JunB in H-RS cells in vitro and in vivo. These results suggest that constitutively active JunB causes overexpression of CD30 in H-RS cells through acting on the AP-1 site of the CD30 MS and relieving its suppressive activity on the Sp-1-derived core promoter.
Overexpression of CD30 mRNA in H-RS cells is well documented. However, recent reports demonstrated that the high activity of the SP-1 driven core promoter is repressed by CCAT elements in the CD30 MS. 25 Thus, it has been hypothesized that the size of the CD30 MS may be shortened in H-RS cells, possibly because of the microsatellite instability that has been reported in various kinds of hematopoietic malignancies including HL. 34-37 However, we demonstrated here that the number of CCAT repressive elements in H-RS cells was generally not decreased compared to normal cells. Furthermore, the CD30 MS from H-RS cells showed a strong repressive activity on the core promoter in transient reporter gene assays in control cells (Figure 3B) ▶ . These results provide strong evidence against the hypothesis and indicate that differences in the promoter activity are independent of the repeat number of the CCAT-repressive element. Our results suggest that not the structural changes but transcriptional regulations are responsible for the high activity of the CD30 promoter in H-RS cell lines.
We demonstrated that the CD30 MS repressive activity is not evident in H-RS cells compared to that in other tumor cell lines (Figure 3B) ▶ . In some experiments, addition of the CD30 MS resulted in enhancement of the core promoter activity. Searching for putative binding sites of transcription factors in the MS, we identified an AP-1 site in the CD30 MS at nucleotide position of −377 to −371 (5′-TGATTCA-3′). This AP-1 site counteracted repression by the MS in H-RS cells through binding to JunB, whose constitutive expression in H-RS cells was demonstrated in the present study. Taken together, these results indicate that the AP-1 site can counteract the repressive activity of the CCAT repeat in the CD30 promoter MS of H-RS cells but not in other cell lines, and in H-RS cells specific relief of repression can be explained by constitutive expression of JunB in that it binds to the AP-1 site at −377 to −371 (5′-TGATTCA-3′).
Little is known about roles of the AP-1 in the biology of H-RS cells or HL. Previous reports suggested that c-Fos/c-Jun expression is involved in the differentiation of H-RS cells. 38 Constitutive expression of c-fos and c-jun in H-RS cell lines (KMH2 and HDLM2) was also reported. 39 Another report showed that the JNK pathway is critically involved in the formation of RS cell morphology. 40 Expression of JunB and JunD in an H-RS cell line was documented previously. 2 Very recently, aberrant expression of JunB as well as c-Jun was reported to be a characteristic of H-RS cells and tumor cells of anaplastic large-cell lymphoma and contributes to the proliferation of H-RS cells. 41 In the present study, we showed involvement of JunB in the relief of suppressive activity of the CD30 MS through binding to the AP-1 site. The result provides clear evidence for another important biological function of JunB in H-RS cells. The relevance of our findings in primary H-RS cells will be confirmed by analysis of MS sequences derived from primary H-RS cells by laser microdissection and micromanipulation. We are currently investigating involvement of JunB in regulation of the CD30 promoter in anaplastic large-cell lymphoma cells.
JNK represents one subgroup of MAP kinases that is activated by cytokines and exposure to environmental stress. A major target of the JNK signaling pathway is activation of the AP-1 transcriptional factor that is mediated in part by the phosphorylation of c-Jun and related molecules. 42 The AP-1 family of transcription factors that are activated by the JNK pathway is involved in cellular proliferation and differentiation. 43 However, a previous report suggested that JNK is not involved in the activation of JunB. 44 This is in accordance with our observation that expression of JNK-1 did not further enhance relief of suppressive CD30 MS activity by JunB (data not shown). Although c-Jun and JunB were considered to have antagonistic functions in biological processes such as oncogenic transformation and cell proliferation, a recent report suggested a potential of JunB to substitute for c-Jun. Deregulated JunB expression could rescue the expression of target genes of Jun. 45 Involvement of JunB in activation of the AP-1-driven promoter was also reported. 46 The CD30 promoter appears to be another example of JunB target genes, because the promoter was activated by JunB in reporter gene assays. Furthermore, we have shown that the AP-1 complex of H-RS cells includes activated JunB and repression of endogenous JunB expression by siRNAs leads to down-regulation of CD30 promoter activity.
In the present study, an MS fragment with a mutated AP-1 site did not completely abrogate the relief of suppression in H-RS cell lines (Figure 3C) ▶ . This implies that as yet unknown transcription factors and/or mechanisms are likely involved in the constitutive overexpression of CD30. Thus, studies of transcription factor(s) that may interact with the suppressive CCAT element in the CD30 MS will provide clues to understanding the high promoter activity in H-RS cells.
In summary, the high activity of the CD30 promoter in H-RS cells appears to be determined by the balance among 1) Sp-1-driven core promoter activity, 2) suppressive activity mediated by the CCAT motif in the CD30 MS, and 3) AP-1 site-mediated relief of the MS suppressive activity. Because there appears to be no significant differences in Sp-1-driven core promoter activity among cell lines with or without CD30 expression, relief of the MS suppression by the JunB-binding AP-1 site appears to play a key role in the high CD30 promoter activity of H-RS cells.
Acknowledgments
We thank the staff in the Department of Molecular Biology and DNA Center at School of Medicine, Kitasato University.
Footnotes
Address reprint requests to Ryouichi Horie, M.D., Ph.D., Fourth Department of Internal Medicine, Kitasato University, School of Medicine, 1-15-1 Sagamihara, Kanagawa, 228-8555, Japan. E-mail: rhorie@med.kitasato-u.ac.jp.
Supported in part by grants from the Ministry of Education, Culture, Sports, Science, and Technology; the Japan Society of Promotion of Science (to T. W., R. H.), and by the Mochida Memorial Foundation for Medical and Pharmaceutical Research (to R. H.).
References
- 1.Gruss HJ, Kadin ME: Pathophysiology of Hodgkin’s disease: functional and molecular aspects. Baillieres Clin Haematol 1996, 9:417-446 [DOI] [PubMed] [Google Scholar]
- 2.Cossman J, Annunziata CM, Barash S, Staudt L, Dillon P, He WW, Ricciardi-Castagnoli P, Rosen CA, Carter KC: Reed-Sternberg cell genome expression supports a B-cell lineage. Blood 1999, 94:411-416 [PubMed] [Google Scholar]
- 3.Kanzler H, Kuppers R, Hansmann ML, Rajewsky K: Hodgkin and Reed-Sternberg cells in Hodgkin’s disease represent the outgrowth of a dominant tumor clone derived from (crippled) germinal center B cells. J Exp Med 1996, 184:1495-1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuppers R, Rajewsky K: The origin of Hodgkin and Reed/Sternberg cells in Hodgkin’s disease. Annu Rev Immunol 1998, 16:471-493 [DOI] [PubMed] [Google Scholar]
- 5.Kuppers R, Rajewsky K, Zhao M, Simons G, Laumann R, Fischer R, Hansmann ML: Hodgkin disease: Hodgkin and Reed-Sternberg cells picked from histological sections show clonal immunoglobulin gene rearrangements and appear to be derived from B cells at various stages of development. Proc Natl Acad Sci USA 1994, 91:10962-10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marafioti T, Hummel M, Foss HD, Laumen H, Korbjuhn P, Anagnostopoulos I, Lammert H, Demel G, Theil J, Wirth T, Stein H: Hodgkin and Reed-Sternberg cells represent an expansion of a single clone originating from a germinal center B-cell with functional immunoglobulin gene rearrangements but defective immunoglobulin transcription. Blood 2000, 95:1443-1450 [PubMed] [Google Scholar]
- 7.Durkop H, Latza U, Hummel M, Eitelbach F, Seed B, Stein H: Molecular cloning and expression of a new member of the nerve growth factor receptor family that is characteristic for Hodgkin’s disease. Cell 1992, 68:421-427 [DOI] [PubMed] [Google Scholar]
- 8.Falini B, Pileri S, Pizzolo G, Durkop H, Flenghi L, Stirpe F, Martelli MF, Stein H: CD30 (Ki-1) molecule: a new cytokine receptor of the tumor necrosis factor receptor superfamily as a tool for diagnosis and immunotherapy. Blood 1995, 85:1-14 [PubMed] [Google Scholar]
- 9.Horie R, Watanabe T: CD30: expression and function in health and disease. Semin Immunol 1998, 10:457-470 [DOI] [PubMed] [Google Scholar]
- 10.Smith CA, Gruss HJ, Davis T, Anderson D, Farrah T, Baker E, Sutherland GR, Brannan CI, Copeland NG, Jenkins NA, Grabstein KH, Gliniak B, McAlister IB, Fanslow W, Alderson M, Falk B, Gimpel S, Gillis S, Din WS, Goodwin RG, Armitage RJ: CD30 antigen, a marker for Hodgkin’s lymphoma, is a receptor whose ligand defines an emerging family of cytokines with homology to TNF. Cell 1993, 73:1349-1360 [DOI] [PubMed] [Google Scholar]
- 11.Schwab U, Stein H, Gerdes J, Lemke H, Kirchner H, Schaadt M, Diehl V: Production of a monoclonal antibody specific for Hodgkin and Sternberg-Reed cells of Hodgkin’s disease and a subset of normal lymphoid cells. Nature 1982, 299:65-67 [DOI] [PubMed] [Google Scholar]
- 12.Gedrich RW, Gilfillan MC, Duckett CS, Van Dongen JL, Thompson CB: CD30 contains two binding sites with different specificities for members of the tumor necrosis factor receptor-associated factor family of signal transducing proteins. J Biol Chem 1996, 271:12852-12858 [DOI] [PubMed] [Google Scholar]
- 13.Lee SY, Kandala G, Liou ML, Liou HC, Choi Y: CD30/TNF receptor-associated factor interaction: NF-kappa B activation and binding specificity. Proc Natl Acad Sci USA 1996, 93:9699-9703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aizawa S, Nakano H, Ishida T, Horie R, Nagai M, Ito K, Yagita H, Okumura K, Inoue J, Watanabe T: Tumor necrosis factor receptor-associated factor (TRAF) 5 and TRAF2 are involved in CD30-mediated NFkappaB activation. J Biol Chem 1997, 272:2042-2045 [DOI] [PubMed] [Google Scholar]
- 15.Duckett CS, Gedrich RW, Gilfillan MC, Thompson CB: Induction of nuclear factor kappaB by the CD30 receptor is mediated by TRAF1 and TRAF2. Mol Cell Biol 1997, 17:1535-1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bargou RC, Leng C, Krappmann D, Emmerich F, Mapara MY, Bommert K, Royer HD, Scheidereit C, Dorken B: High-level nuclear NF-kappa B and Oct-2 is a common feature of cultured Hodgkin/Reed-Sternberg cells. Blood 1996, 87:4340-4347 [PubMed] [Google Scholar]
- 17.Bargou RC, Emmerich F, Krappmann D, Bommert K, Mapara MY, Arnold W, Royer HD, Grinstein E, Greiner A, Scheidereit C, Dorken B: Constitutive nuclear factor-kappaB-RelA activation is required for proliferation and survival of Hodgkin’s disease tumor cells. J Clin Invest 1997, 100:2961-2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wood KM, Roff M, Hay RT: Defective IkappaBalpha in Hodgkin cell lines with constitutively active NF-kappaB. Oncogene 1998, 16:2131-2139 [DOI] [PubMed] [Google Scholar]
- 19.Cabannes E, Khan G, Aillet F, Jarrett RF, Hay RT: Mutations in the IkBa gene in Hodgkin’s disease suggest a tumour suppressor role for IkappaBalpha. Oncogene 1999, 18:3063-3070 [DOI] [PubMed] [Google Scholar]
- 20.Emmerich F, Meiser M, Hummel M, Demel G, Foss HD, Jundt F, Mathas S, Krappmann D, Scheidereit C, Stein H, Dorken B: Overexpression of I kappa B alpha without inhibition of NF-kappaB activity and mutations in the I kappa B alpha gene in Reed-Sternberg cells. Blood 1999, 94:3129-3134 [PubMed] [Google Scholar]
- 21.Krappmann D, Emmerich F, Kordes U, Scharschmidt E, Dorken B, Scheidereit C: Molecular mechanisms of constitutive NF-kappaB/Rel activation in Hodgkin/Reed-Sternberg cells. Oncogene 1999, 18:943-953 [DOI] [PubMed] [Google Scholar]
- 22.Horie R, Watanabe T, Morishita Y, Ito K, Ishida T, Kanegae Y, Saito I, Higashihara M, Mori S, Kadin ME: Ligand-independent signaling by overexpressed CD30 drives NF-kappaB activation in Hodgkin-Reed-Sternberg cells. Oncogene 2002, 21:2493-2503 [DOI] [PubMed] [Google Scholar]
- 23.Horie R, Watanabe T, Ito K, Morisita Y, Watanabe M, Ishida T, Higashihara M, Kadin M, Watanabe T: Cytoplasmic aggregation of TRAF2 and TRAF5 proteins in the Hodgkin-Reed-Sternberg cells. Am J Pathol 2002, 160:1647-1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Croager EJ, Muir TM, Abraham LJ: Analysis of the human and mouse promoter region of the non-Hodgkin’s lymphoma-associated CD30 gene. J Interferon Cytokine Res 1998, 18:915-920 [DOI] [PubMed] [Google Scholar]
- 25.Croager EJ, Gout AM, Abraham LJ: Involvement of Sp1 and microsatellite repressor sequences in the transcriptional control of the human CD30 gene. Am J Pathol 2000, 156:1723-1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Durkop H, Oberbarnscheidt M, Latza U, Bulfone-Paus S, Hirsch B, Pohl T, Krause H, Hummel M, Stein H: The restricted expression pattern of the Hodgkin’s lymphoma-associated cytokine receptor CD30 is regulated by a minimal promoter. J Pathol 2000, 192:182-193 [DOI] [PubMed] [Google Scholar]
- 27.Kadin ME: Regulation of CD30 antigen expression and its potential significance for human disease. Am J Pathol 2000, 156:1479-1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aizawa S, Satoh H, Horie R, Ito K, Choi SH, Takeuchi H, Watanabe T: Cloning and characterization of a cDNA for rat CD30 homolog and chromosomal assignment of the genomic gene. Gene 1996, 182:155-162 [DOI] [PubMed] [Google Scholar]
- 29.Horie R, Ito K, Tatewaki M, Nagai M, Aizawa S, Higashihara M, Ishida T, Inoue J, Takizawa H, Watanabe T: A variant CD30 protein lacking extracellular and transmembrane domains is induced in HL-60 by tetradecanoylphorbol acetate and is expressed in alveolar macrophages. Blood 1996, 88:2422-2432 [PubMed] [Google Scholar]
- 30.Horie R, Aizawa S, Nagai M, Ito K, Higashihara M, Ishida T, Inoue J, Watanabe T: A novel domain in the CD30 cytoplasmic tail mediates NFkappaB activation. Int Immunol 1998, 10:203-210 [DOI] [PubMed] [Google Scholar]
- 31.Andrews NC, Faller DV: A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res 1991, 19:2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hammond SM, Caudy AA, Hannon GJ: Post-transcriptional gene silencing by double-stranded RNA. Nat Rev Genet 2001, 2:110-119 [DOI] [PubMed] [Google Scholar]
- 33.Durkop H, Oberbarnscheidt M, Latza U, Bulfone-Paus S, Krause H, Pohl T, Stein H: Structure of the Hodgkin’s lymphoma-associated human CD30 gene and the influence of a microsatellite region on its expression in CD30(+) cell lines. Biochim Biophys Acta 2001, 1519:185-191 [DOI] [PubMed] [Google Scholar]
- 34.Baccichet A, Benachenhou N, Couture F, Leclerc JM, Sinnett D: Microsatellite instability in childhood T cell acute lymphoblastic leukemia. Leukemia 1997, 11:797-802 [DOI] [PubMed] [Google Scholar]
- 35.Hatta Y, Yamada Y, Tomonaga M, Miyoshi I, Said JW, Koeffler HP: Microsatellite instability in adult T-cell leukaemia. Br J Haematol 1998, 101:341-344 [DOI] [PubMed] [Google Scholar]
- 36.Mark Z, Toren A, Amariglio N, Schiby G, Brok-Simoni F, Rechavi G: Instability of dinucleotide repeats in Hodgkin’s disease. Am J Hematol 1998, 57:148-152 [DOI] [PubMed] [Google Scholar]
- 37.Tanosaki S, Inokuchi K, Shimada T, Dan K: Relation between microsatellite instability and N-ras mutation and duration of disease free survival in patients with acute leukemia. Cancer 1998, 83:475-481 [PubMed] [Google Scholar]
- 38.Hsu SM, Xie SS, el-Okda MO, Hsu PL: Correlation of c-fos/c-jun expression with histiocytic differentiation in Hodgkin’s Reed-Sternberg cells. Examination in HDLM-1 subclones with spontaneous differentiation. Am J Pathol 1992, 140:155-165 [PMC free article] [PubMed] [Google Scholar]
- 39.Gruss HJ, Brach MA, Drexler HG, Bonifer R, Mertelsmann RH, Herrmann F: Expression of cytokine genes, cytokine receptor genes, and transcription factors in cultured Hodgkin and Reed-Sternberg cells. Cancer Res 1992, 52:3353-3360 [PubMed] [Google Scholar]
- 40.Knecht H, Berger C, McQuain C, Rothenberger S, Bachmann E, Martin J, Esslinger C, Drexler HG, Cai YC, Quesenberry PJ, Odermatt BF: Latent membrane protein 1 associated signaling pathways are important in tumor cells of Epstein-Barr virus negative Hodgkin’s disease. Oncogene 1999, 18:7161-7167 [DOI] [PubMed] [Google Scholar]
- 41.Mathas S, Hinz M, Anagnostopoulos I, Krappmann D, Lietz A, Jundt F, Bommert K, Mechta-Grigoriou F, Stein H, Dorken B, Scheidereit C: Aberrantly expressed c-Jun and JunB are a hallmark of Hodgkin lymphoma cells, stimulate proliferation and synergize with NF-kappaB. EMBO J 2002, 21:4104-4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weston CR, Davis RJ: The JNK signal transduction pathway. Curr Opin Genet Dev 2002, 12:14-21 [DOI] [PubMed] [Google Scholar]
- 43.Shaulian E, Karin M: Stress-induced JNK activation is independent of Gadd45 induction. J Biol Chem 1999, 274:29595-29598 [DOI] [PubMed] [Google Scholar]
- 44.Gupta S, Barrett T, Whitmarsh AJ, Cavanagh J, Sluss HK, Derijard B, Davis RJ: Selective interaction of JNK protein kinase isoforms with transcription factors. EMBO J 1996, 15:2760-2770 [PMC free article] [PubMed] [Google Scholar]
- 45.Passegue E, Jochum W, Behrens A, Ricci R, Wagner EF: JunB can substitute for Jun in mouse development and cell proliferation. Nat Genet 2002, 30:158-166 [DOI] [PubMed] [Google Scholar]
- 46.Shoda T, Fukuda K, Uga H, Mima H, Morikawa H: Activation of mu-opioid receptor induces expression of c-fos and junB via mitogen-activated protein kinase cascade. Anesthesiology 2001, 95:983-989 [DOI] [PubMed] [Google Scholar]