Abstract
The proliferation of keratinocytes in the hair follicle varies from slowly cycling, intermittently proliferating stem cells in the bulge to rapidly proliferating, transient cells in the bulb. To better understand the biological differences between these two compartments, we sought to identify differentially expressed genes using cDNA macroarray analysis. Cyclin D1 was one of 13 genes increased in the bulge compared to the bulb, and its differential expression was corroborated by quantitative real-time polymerase chain reaction (PCR) on the original samples. Using immunohistochemical staining, laser-capture microdissection (LCM) and quantitative real-time PCR, we localized cyclin D1 to the suprabasal cells of the telogen bulge and anagen outer root sheath (ORS). Surprisingly, cyclin D1, D2, and D3 were not detectable by immunohistochemistry in the rapidly proliferating hair-producing cells of the anagen bulb (matrix cells), while these cells were strongly positive for Ki-67 and retinoblastoma protein. In contrast, pilomatricoma, a tumor thought to be derived from matrix cells, was positive for cyclin D1, D2, and D3. Our results suggest that cyclin D1 may mediate the proliferation of stem cells in the bulge to more differentiated transient amplifying cells in the suprabasal ORS. In contrast, non-cyclin D1-proteins appear to control cell division of the highly proliferative bulb matrix cells. This non-cyclin D1-mediated proliferation may provide a protective mechanism against tumorigenesis, which is overridden in pilomatricomas. Our data also demonstrate that the combination of DNA macroarray, LCM and quantitative real-time PCR is a powerful approach for the study of gene expression in defined cell populations with limited starting material.
Hair grows in a regulated cyclical process consisting of three distinct phases: anagen, catagen, and telogen. In human scalp follicles, anagen, the growing phase, lasts several years; catagen, the involution phase, lasts approximately 1 to 3 weeks, and telogen, the resting phase, lasts for approximately 3 months. The cellular mechanisms involved in the maintenance of the phases and the transitions between them are poorly understood. The lack of progress in this area reflects the complicated structure and physiology of the hair follicle. In addition, human hair follicles grow in an asynchronous fashion; the majority (∼90%) of hair follicles are in the anagen phase and less than 10% of follicles are in telogen, thus making it difficult to study the hair follicle cycle in humans. However, understanding what controls the proliferation of the follicle could have wide-ranging implications for carcinogenesis and hair disorders.
One important aspect of the transition from the telogen phase to the anagen phase is the mechanism which promotes stem cell proliferation. The stem cells of the bulge region are normally quiescent throughout catagen, telogen and most of the anagen phase but proliferate briefly at anagen onset. In contrast, the anagen matrix cells, which are considered “transient amplifying” (TA) cells, proliferate constantly during anagen. In fact, these cells display one of the highest proliferative rates of any mammalian tissue, with a growth fraction of almost 100%, even outranking most malignant tumors. As in many other highly proliferative tissues, such as bone marrow and gastrointestinal epithelium, this may represent a risk for mutagenesis. But malignant tumors derived from matrix keratinocytes are exceedingly rare; although benign tumors, such as pilomatricomas, occasionally arise from hair matrix cells.
In mammalian cells, proliferation is under the control of factors that regulate the transitions between different cell-cycle stages at two main checkpoints. The better-characterized checkpoint is at the G1-S transition, which initiates DNA replication in S phase. The other checkpoint is at the G2-M transition, which controls mitosis and cell division. The cyclins are a family of key cell-cycle regulators that function by association with and activation of cyclin-dependent kinases (CDKs) at specific points in the cell cycle to phosphorylate various proteins that are important during cell cycle progression. A key substrate for G1 cyclins/CDK complexes is the retinoblastoma tumor suppressor protein (RB). The phosphorylation of RB then releases E2F, which is important in the initiation of DNA replication and in the G1-S phase transition. The levels of cyclins are regulated at the level of transcription as well as by targeted degradation via the ubiquitin pathway.
β-catenin plays an intricate role in Wnt signaling. β-catenin regulates gene expression by direct interaction with Lef-1, providing a molecular mechanism for the transmission of signals, from cell-adhesion components or Wnt protein to the nucleus. Lef1/β-catenin has been identified as a key regulator in hair follicle differentiation and development. The cyclin D1 gene is a direct target for transactivation by the β-catenin/LEF-1 pathway through a LEF-1 binding site in the cyclin D1 promoter and therefore a direct downstream molecule in the β-catenin pathway.
To better characterize the transition from quiescent stem cells to proliferative daughter TA cells, we used DNA macroarrays to compare gene expression in the bulge cells to the bulb matrix cells. We identified cyclin D1 as one of the genes up-regulated in the telogen bulge compared to the anagen bulb. Localization of cyclin D1 expression in the hair follicle using laser-capture microdissection (LCM), real-time polymerase chain reaction (PCR) and immunohistochemical staining suggests that it may play a role in conversion of stem cells to transient amplifying cells, but not in the maintenance of the continual hair follicle proliferation required for hair production.
Materials and Methods
Tissue Preparation
Adult human scalp skin (obtained from the Cooperative Human Tissue Network) was fixed in PBS-buffered 10% formalin; unfixed tissue samples were embedded in tissue-Tek OCT compound (Miles, Naperville, IL), snap-frozen in liquid nitrogen, and stored at −80°C until used for frozen sectioning.
Dissection of Anagen Bulb and Telogen Bulge
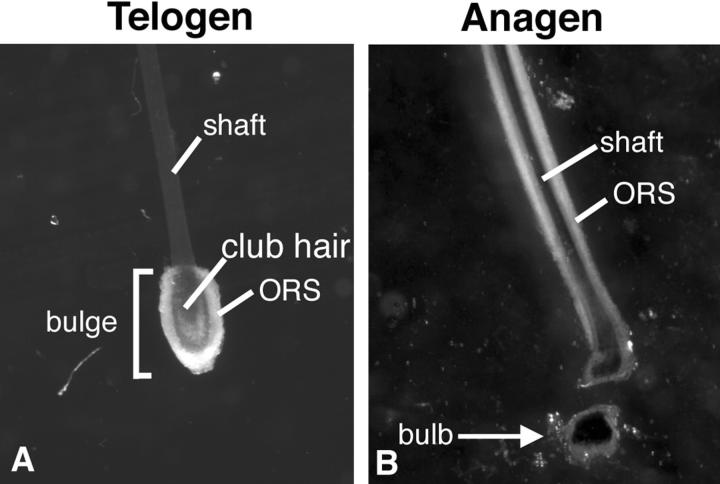
Hair follicles were isolated from dispase-treated (Sigma, St. Louis, MO) fresh skin as previously described. The anagen bulb and telogen bulge were isolated using a dissecting microscope (Figure 1) ▶ . The tissue was rapidly frozen on dry ice and stored at −80°C. Total RNA from telogen bulge and anagen bulb was extracted using the guanidine isothiocyanate method.
Figure 1.
Images of entire epithelial component of telogen (A) and anagen (B) hair follicles released from human scalp tissue at the basement membrane by incubation with the enzyme dispase (×20).
Macroarrays
The Atlas Human cDNA Expression Array I (Clontech, Palo Alto, CA) was used in the experiments. This array has been spotted in duplicate with 10 ng of PCR-amplified cDNA fragments which are 200- to 500-bp long and represent 588 genes. Several plasmid and bacteriophage DNAs are included as negative controls to confirm hybridization specificity, along with ten housekeeping genes for normalizing mRNA abundance. We first reverse-transcribed 2 μg of total RNA from each sample using the reagents provided and [α-32P]dCTP. These radioactively labeled cDNA probes were then hybridized overnight to the membranes following the manufacturer’s instructions. After washing three times with 2X SSC, 1% SDS at 68°C for 30 minutes, then washing twice with 0.1X SSC, 0.5% SDS at 68°C for 30 minutes, the hybridization pattern was analyzed by autoradiography. To ensure accurate comparisons, signals were obtained exposing the array to a phosphoimager (Bio-Rad, Hercules, CA). Duplicate blots were used to compare the expression patterns of telogen and anagen RNA populations.
The phosphoimager blot was analyzed using IPLab Spectrum software. Individual spots were measured by densitometry. The average densitometry signals of the duplicate spots, minus background, were calculated. The signals were then normalized against an average of the signals from the control housekeeping genes ubiquitin and 23-kd highly basic protein. The ratio of expression between telogen and anagen signals was then calculated (Table 1) ▶ .
Table 1.
Genes Differentially Expressed in the Telogen Bulge versus Anagen Bulb
| Gene name | GenBank # | Ratio (telogen/anagen) |
|---|---|---|
| Chaperonin (HSP60) | M34664 | 3.1 |
| CLK-1 | L29222 | 2.9 |
| c-myc | V00568 | 8.1 |
| Cyclin D1 | X59798 | 5.4 |
| DAD-1 | D15057 | 1.5 |
| Ezrin (cytovillin 2) | X51521 | 4.6 |
| Gs-α subunit | M14631 | 4.3 |
| MLK-3 | L32976 | 2.7 |
| MRP-8 | X06234 | 22.2 |
| NET-1 | U02081 | 6.1 |
| NF-E1 | M76541 | 13.7 |
| RhoA | L25080 | 3.7 |
| TR3 orphan receptor | L13740 | 5.2 |
| Ubiquitin* | M26880 | 0.9 |
| 23 kd highly basic protein* | X56932 | 1 |
Housekeeping genes.
The approximate ratios of the signals after normalization to housekeeping genes are listed in the third column.
Quantitative Real-Time PCR
Due to the limited quantity of total RNA (1.5 μg from telogen follicle and 2.6 μg from anagen follicle) that was available for further studies, we adopted a quantitative real-time PCR approach to verify the DNA array results. We used TaqMan probes and the Applied Biosystems Prism 7700 Sequencing Detection System (Applied Biosystems, Foster City, CA) in our experiments. The cyclin D1, GAPDH, and ribosomal 18s TaqMan probe and primer mix, as well as the TaqMan Universal PCR Master Mix, were purchased from Applied Biosystems. The cyclin D1 probe was labeled at the 5′ end with a reporter fluorochrome (6-carboxyfluorescein [6-FAM]), and at the 3′ end with a quencher fluorochrome (6-carboxy-tetramethyl-rhodamine [TAMRA]). The 5′ end of the GAPDH or ribosomal 18s probes were labeled with VIC (ABI). The cyclin D1 and GAPDH primers and probes were designed to span one intron so that genomic DNA would not amplify. Other important controls were reactions without reverse transcriptase and reactions without template, which test for contamination of RNA by genomic DNA, and for contamination of the reagents, respectively. These controls were included in the same runs as the experimental reactions.
The PCR conditions were 50°C for 2 minutes and 94°C for 10 minutes, repeated for 40 cycles. After 40 cycles, data were processed using the software accompanying the Applied Biosystems Prism 7700 Sequencing Detection System. The linearity of the fluorescence response for each sample at each cycle and at baseline was checked for each tube to generate an accurate threshold cycle (Ct). The Ct is determined by identifying the cycle number at which the reporter dye emission intensities rises above background noise.
Total RNA extracted from human prostate tissue with serial 1/10 dilutions was run in duplicate to generate the concentration curves for cyclin D1 and the housekeeping genes GAPDH and ribosomal 18s. Cyclin D1, GAPDH and/or ribosomal 18s samples were run in triplicate and concentrations of each were determined comparing the Ct to the concentration curves. The amount of cyclin D1 was normalized to the amount of housekeeping gene in the each sample.
Immunohistochemistry
Six-micron thick paraffin sections were cut and stained with hematoxylin and eosin (H&E). Consecutive sections were immunostained with mouse monoclonal antibodies against cyclin D1, D3, retinoblastoma protein, Ki-67 (all from DAKO, Carpinteria, CA), and cyclin D2 (Research Diagnostics Inc., Flanders, NJ). Tissue sections were boiled in 1X Tris buffer (pH 8.1) for 20 minutes, washed and incubated with cyclin D1 antibody at room temperature for 2 hours; for cyclin D2 and D3, incubation was for 20 minutes at 4°C overnight. Tissue sections were boiled in Target Retrieval Solution (DAKO) for 20 minutes, and incubated with Ki67 or pRB antibody at ROOM TEMPERATURE for 2 hours. The standard avidin-biotin peroxidase (ABC) complex method was used to visualize the antigen antibody complex.
LCM and Gene Expression Analysis
To compare the differential gene expression among different compartments within the hair follicle, we performed LCM of basal and suprabasal cells of the ORS using the Arcturus PixCell II and CapSure Transfer film carrier (Arcturus Engineering, Inc., Mountain View, CA). Briefly, frozen tissue was embedded in OCT compound (Tissue-Tek) and snap-frozen in liquid nitrogen. The blocks were wrapped in foil and kept at −80°C until sectioning. Eight-μm sections were placed onto plain uncoated glass slides. Routine H&E staining was performed and after xylene dehydration, the slides were allowed to air dry for 20 minutes. Sections were inspected to ensure no tissue folds or errant hair shafts were present on the surface of the section. Basal layer and suprabasal layer ORS cells were captured on CapSure LCM caps (caps) by LCM using a 7.5-mm laser beam at 50 to 100 mV. Multiple captures (2 to 3) from similar areas of different follicles were performed using one cap. Cells on the caps were lysed and total RNA was extracted using a Cells-to-cDNA Kit from Ambion (Austin, TX) following the manufacturer’s instructions with slight modifications. Briefly, the cell lysis buffer was pre-warmed to over 75°C. Cell lysis buffer was added to cover the cap, and then vigorously vortexed, followed by incubation for 5 minutes at 75°C. The tube was cooled and 1 μl of DNase I for every 50 μl of Cell Lysis Buffer was added. We then incubated the sample at 37°C for 30 minutes and 75°C for 5 minutes to inactivate the DNase I. A portion of the cell lysate was used as template for the first-strand cDNA synthesis in a reverse transcription (RT) reaction with random primers. A control reaction without reverse transcriptase, to demonstrate that the template for the PCR product was cDNA and not genomic DNA, was always used. RT was performed by heating the mixture of 10 μl cell lysate (RNA), 4 μl dNTP mix, and 2 μl first-strand random decamers for 3 minutes at 70°C, placing the reaction on ice for 1 minute, centrifuging the tube briefly and then placing it on ice. The remaining RT reagents were added, centrifuged briefly and then placed at 42°C for 1 hour. This cDNA was subsequently used for quantitative real-time PCR.
Data Analysis
The correlation coefficients of the concentration curves were calculated using Excel (Microsoft).
Results
Differential Expression of Cyclin D1 in the Telogen Bulge by cDNA Macroarray
We identified a total of 36 genes on the cDNA macroarray after hybridization with probe from telogen follicle bulge RNA, and 68 genes on the duplicate macroarray blot using probe generated from anagen bulb RNA. Cyclin D1 was one of 13 genes differentially expressed in the stem-cell-rich telogen follicle and not in the anagen bulb (Table 1) ▶ . The housekeeping genes ubiquitin, glyceraldehyde 3-phosphate dehydrogenase (GAPDH), 23kDa highly basic protein and ribosomal protein S9 were present along the bottom of the blot and allowed for proper alignment. All negative control spots for phage and plasmid DNA were negative (data not shown).
Differential CyclinD1 Gene Expression in Telogen Bulge Cells was Confirmed by Quantitative Real-Time PCR
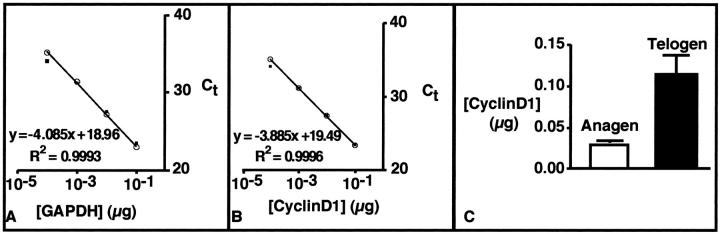
Quantitative real-time PCR using TaqMan probes is a sensitive and precise technique for quantitating mRNA levels. High specificity is conferred by the requirement of three oligonucleotides to anneal to the DNA before any data are collected. The precision is a function of the Ct, which is calculated during the exponential phase of the PCR reaction. In this study, the concentration curves were constructed by serial dilution (1:10) of total RNA from prostate tissue, which was assayed for cyclin D1 and GAPDH expression (Figure 2) ▶ . Duplicate samples yielded nearly identical results (Figure 2) ▶ with coefficient of determination (R2) of the concentration curves greater than 0.99. These standard curves were used later to quantify the amount of cyclin D1 in each sample.
Figure 2.
Quantitative real-time PCR demonstrates increased expression of cyclin D1 in telogen follicle. A: Concentration curve of GAPDH. B: Concentration curve of cyclin D1. C: Comparison of cyclin D1 gene expression in telogen bulge and anagen bulb.
To quantify expression levels of cyclin D1, we performed quantitative real-time PCR on RNA extracted from anagen bulb and telogen bulge cells, respectively. The amount of cyclin D1 mRNA in the tissue was normalized to the amount of GAPDH in the same sample. Similar to the macroarray results, cyclin D1 was more than threefold increased in the mRNA from the telogen bulge (Figure 2) ▶ ; therefore, two different methods (DNA blot and quantitative real-time PCR) confirmed the up-regulation of cyclin D1 in telogen bulge cells.
Cyclin D1 Protein Localized to the Suprabasal Cells of the Bulge ORS of Both Telogen and Anagen Hair Follicles
The finding of elevated cyclin D1 gene expression in the telogen bulge compared to the anagen bulb was surprising, since bulge cells are quiescent and bulb cells are highly proliferative. To study this further, we immunostained follicles at different stages of the hair follicle cycle to localize the cyclin D1 protein. In telogen follicles, immunohistochemical detection of cyclin D1 showed distinct nuclear staining in the suprabasal layers of the ORS (Figure 3A) ▶ . In anagen follicles, cyclin D1-positive cells were mostly confined to the suprabasal layers of the ORS as well (Figure 3C) ▶ . Rare staining of the basal cells was also observed, which is similar to the staining pattern of Ki67 (Figure 3, B and D) ▶ . In the hair bulb, only rare isolated cells were positive for cyclin D1 at the periphery. The matrix cells around the lower half of the dermal papilla, which is the most proliferative area in the hair follicle, were negative for cyclin D1 (Figure 4B) ▶ . In hair follicles at anagen onset, cyclin D1-positive cells were present in the suprabasal layer of the residual telogen follicle, but the cells in the newly developing anagen matrix were negative for cyclin D1 (Figure 4G) ▶ . The anagen matrix and telogen bulge cells were also negative for cyclin D2 and D3 by immunohistochemistry (data not shown). Rare cyclin D2-positive cells were located in the suprabasal layer of the ORS and cyclin D3 immunostaining was negative in hair follicles. Reactive lymph nodes were used as positive controls and appropriate staining was observed for cyclins D1, D2, and D3.
Figure 3.
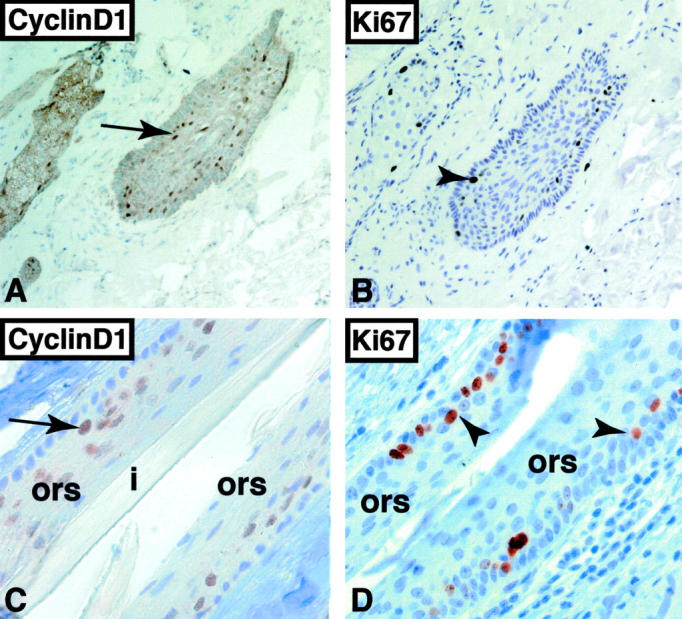
Immunohistochemical detection of cyclin D1 and Ki67 in telogen bulge (A and B) and anagen ORS (C and D). A: Cyclin D1-positive cells are located in the suprabasal layer of the telogen bulge (arrow, ×100). B: Majority of Ki67-positive cells are located in the suprabasal layer of the telogen bulge (arrowhead, ×100). Cyclin D1-positive (arrow, C) and Ki67-positive cells (arrowheads, D) are located in the suprabasal layer of the anagen ORS (×630).
Figure 4.
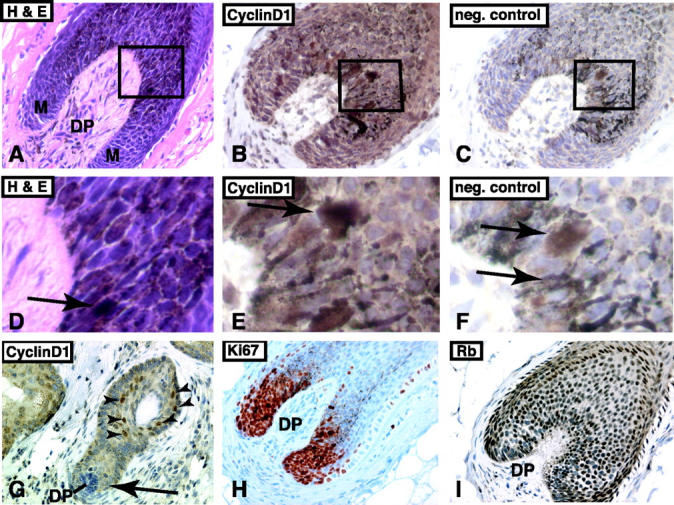
Immunohistochemical detection of cyclin D1, Ki67, and RB in the anagen hair bulb. A: H&E-stained tissue section of anagen hair bulb (DP, dermal papilla; M, matrix; ×200). Higher magnification view of boxed area is shown in D. B: Immunohistochemical staining for cyclin D1 in anagen bulb (×400). The anagen bulb is rich in pigment-laden melanocytes. No true nuclear staining is present. The boxed area is shown in E. C: Negative control of consecutive section of anagen bulb shown in B. The pigment-laden melanocytes are easily identified (×400). The boxed area is shown in F. Arrows in D, E, and F point to pigment-laden cells. G: At anagen onset, the suprabasal cells of the telogen bulge are positive for cyclin D1 (arrowheads). However, the newly formed anagen bulb is negative for cyclin D1 (arrow, ×200). H: Ki67 staining shows that cells around the dermal papilla are strongly positive and proliferating (×200). E: Immunostaining for Rb shows that the matrix cells retain expression of Rb (×200). Immunohistochemical sections are counterstained with hematoxylin.
To confirm the proliferative status of bulb matrix cells, we performed immunohistochemical staining for Ki67, a marker of cell proliferation. Serial sections of human scalp stained for cyclin D1 were also stained for Ki67. The results were similar to prior studies and showed that matrix cells surrounding the dermal papilla stained intensely with Ki67 (Figure 4H) ▶ . Since expression of cyclin D1 is regulated by the RB protein, we also performed immunohistochemistry for RB protein, which showed that matrix cells and bulge cells were positive for RB protein (Figure 4I) ▶ .
Gene Expression in Anagen ORS by Microdissection and Real-Time PCR
Immunohistochemical staining for cyclin D1 revealed that there was a distinct difference of cyclin D1 protein expression between the suprabasal and basal layers of the ORS. This observation prompted us to study cyclin D1 gene expression in these two regions using quantitative real-time PCR and LCM. We collected microdissected tissue from the suprabasal and basal layers of the anagen ORS in multiple follicles (Figure 5) ▶ . The relative concentration of cyclin D1 was measured in the two populations by quantitative real-time PCR (Figure 6) ▶ .
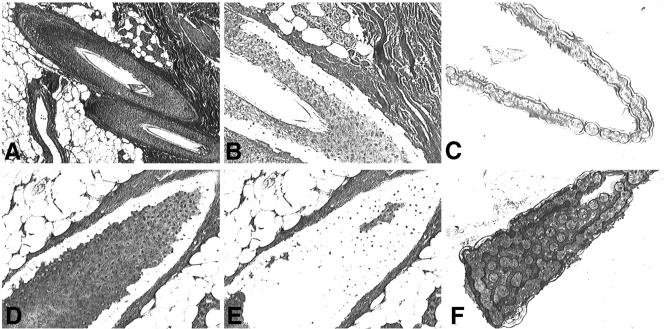
Figure 5.
Laser-capture microdissection for isolation of hair follicle basal cells (A to C) and suprabasal cells (D to F). A and D: Overview of the tissue section before the dissection (×40 and ×200). B and E: Overview of the tissue section after the microdissection (×200). C and F: Microdissected tissue on the transfer film carrier (×200). The sections were stained with H&E.
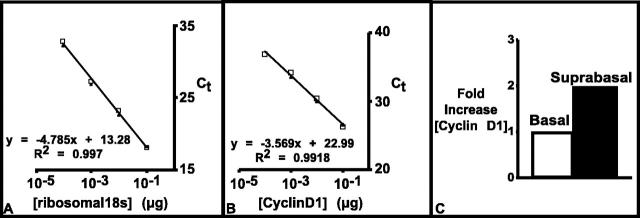
Figure 6.
Differential expression of cyclin D1 mRNA in ORS. A: Concentration curve of ribosomal 18s. B: Concentration curve of cyclin D1. C: Concentration of cyclin D1 in basal and suprabasal cells relative to concentration in basal layer cells.
Serial 1:10 dilutions of total RNA from prostate tissue were used to construct concentration curves for cyclin D1 and ribosomal 18s. In preliminary studies, (data not shown), microdissected tissues from basal and suprabasal layers of ORS after a single capture (approximately 50 to 200 cells) were analyzed for cyclin D1 concentration. The results showed that although the variance between duplicates was very small, the concentration of cyclin D1 varied considerably from different ORS areas, indicating that cells with a similar morphological appearance may have different gene expression levels. Therefore, multiple captures (2 to 3) of cells from the ORS of different hair follicles were performed and tissues from 4 to 5 captures were pooled to obtain an overall estimation of cyclin D1 expression. The results showed that cyclin D1 mRNA was up-regulated approximately twofold in the suprabasal layer compared to the basal layer (Figure 6) ▶ . This is in line with the results of the immunohistochemical staining.
Pilomatricoma Cells Express Cyclins D1, D2, and D3
Pilomatricomas are thought to originate from bulb matrix cells because of their morphological appearance. To determine whether cyclin Ds play a role in proliferation of these tumors, we immunostained tissue sections from five of these tumors (Figure 7) ▶ . We found that cyclin D1, D2, and D3 all were expressed in the basaloid cells within pilomatricomas.
Figure 7.
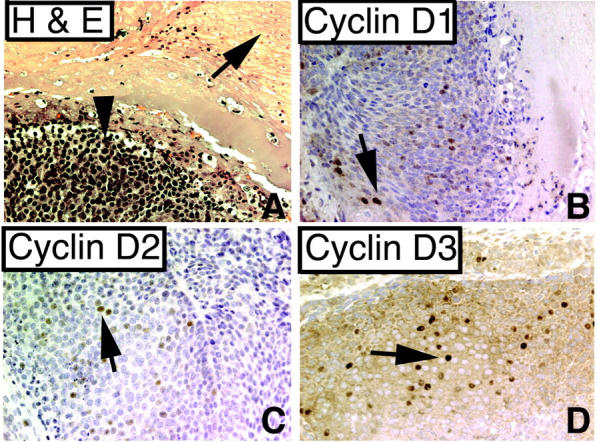
Pilomatricomas express cyclins D1, D2, and D3. A: H&E stain of pilomatricoma. Arrow indicates shadow cells; arrowhead indicates basoloid cells (×200), Tumor cells are focally positive for cyclin D1 (B), cyclin D2 (C), and cyclin D3 (D) (arrows, ×200).
Discussion
DNA array technology allows for expression analysis of multiple genes on the same blot or array. Presumably, the combination of genes that are expressed is a major determinant of cellular phenotype and function, and changes in gene expression patterns reflect alterations in both cell morphology and function. DNA arrays are powerful tools but verification of positive clones can be time-consuming and difficult, especially if only small amounts of starting material are available. To address this restraint, we used quantitative real-time PCR. The combination of DNA blot and quantitative real-time PCR allowed us to successfully identify and confirm that cyclin D1 was differentially expressed within different subpopulations of the follicle.
Advances in gene sequencing and amplification techniques now allow researchers to quantitatively measure single-digit gene copy number. However, the efficacy of these sophisticated methods depends on the purity of the cell populations being analyzed. Simply homogenizing biopsy sample material results in a mixture of cell populations, which may bias results. LCM provides a means for obtaining DNA, RNA, and protein from specific cells within heterogeneous cell populations from tissue biopsies and cytological smears. Using LCM, we were able to separately microdissect out basal and suprabasal cells of the ORS and, with quantitative real-time PCR, measure the relative cyclin D1 gene expression in cells from a single hair follicle. The relative gene expression in these samples varied, indicating that differences in cyclin D1 expression between basal and suprabasal cells could be greater in one particular area within a follicle, as was indicted by the immunostaining results. Variable cyclin D1 levels among different follicles could also be a function of their different stages within the hair follicle cycle. Uneven distribution of antigen is common in normal and especially in tumor tissues. Therefore, in our case, we sampled the basal and suprabasal areas of several follicles by performing multiple captures and then pooled these samples. The results showed that cyclin D1 mRNA was increased overall in the suprabasal layer compared to the basal layer, and this was in line with the immunohistochemical staining. These results suggest that cyclin D1-mediated proliferation predominantly occurs in the immediate progeny of the stem-cell-rich bulge cells, indicating that cyclin D1 may be important for cells to exit the stem cell compartment.
Epithelial stem cells in the hair follicle bulge rarely proliferate, but their progeny are thought to generate the lower hair follicle, including the rapidly proliferating bulb matrix cells, the suprabasal cells of the ORS, and in times of wounding, the overlying epidermis as well. The lef1/β-catenin complex is thought to regulate key cell-fate decisions that control whether bulge cells become hair follicle or epidermal cells. The absence of β-catenin signaling favors formation of epidermis rather than hair follicle. 21 The lef1/β-catenin complex can bind to the cyclin D1 promoter and activate cyclin D1 gene expression and cell proliferation; however, the expression pattern and function of cyclin D1 during human hair follicle growth has not been studied previously. The expression patterns of cyclin D1 that we observed suggest that cyclin D1 is important for proliferation of the suprabasal hair follicle ORS cells, but may not play a role in bulb matrix cell proliferation. This is in line with evidence that indicates strong lef1/β-catenin activity in differentiated cells of the hair follicle rather than in the proliferating matrix cells.
Three D-type cyclins are expressed in the G1 phase of the cell cycle, and depending on cell lineage, various combinations of D-type cyclins are induced by mitogens in the middle of G1. Cyclin D1 is believed to drive cell-cycle progression by associating with its catalytic partners CDK4 and CDK6 and guiding these kinases to RB. Besides promoting growth, cyclin D1 also acts as an oncogene. Overexpression of D-type cyclins or CDK4 and inactivation of CDK4 inhibitors are common in human tumors. Dysregulation of cyclin D1 synthesis allows cell-cycle progression in the absence of growth factors and may contribute to the initiation of oncogenesis. For example, overexpression of cyclin D1 has been reported in various human malignant tumors, including 27% of esophageal cancers. In breast cancers, the cyclin D1 gene is amplified in 23% of cases, and also in 58% of hepatocellular carcinomas. Cyclin D1 gene over-expression is associated with mantle-cell lymphoma and various other tumors. Cyclin D2 overexpression is associated with gastric carcinoma cells and seems to play a role in tumor progression. Cyclin D3 is the least studied and a few reports suggest that cyclin D3 also plays a role in tumorigenesis. However, overexpression of cyclin D1, D2, or D3 in the epidermis of transgenic mice causes epidermal hyperplasia but not skin tumor formation, suggesting that cyclin D activation alone is not sufficient to induce tumorigenesis.
Cyclin D1 knockout mice show normal development of most mouse tissues, such as kidney, salivary glands and seminal vesicles, which normally have readily detectable cyclin D1 transcript levels. Hepatocytes from these mice still have proliferative responses to mitogenic stimuli and have increased expression of cyclin E. Lack of cyclin D1 expression does result in significant reduction in mouse skin and mammary tumor development. However, complete elimination of tumor development was not observed in these models, suggesting that other cyclin/cdk complexes may compensate for the loss of function of cyclin D, and play an important role in growth and tumorigenesis. Cyclin D1 knockout mice appear to also have normal hair production.
Our results, together with the knockout studies, suggest that the normal proliferation of hair matrix cells is not mediated through cyclin D1 and interestingly, despite the high proliferation-rate of these cells, malignant tumors arising from hair matrix are exceedingly rare. On the other hand, we found that pilomatricoma, a benign tumor arising from the hair matrix, expresses cyclin D1, D2, and D3. In this respect, pilomatricomas may be analogous to mantle-cell lymphomas, which express cylinD1, but which arise from lymph node mantle cells that normally do not normally express cyclin D1. Together, these results suggest that non-cyclin D1-mediated proliferation might offer protection against tumorigenesis in highly proliferative tissues.
In conclusion, we have shown that the combination of DNA array, LCM and quantitative real-time PCR is a powerful combination of research tools for studying gene expression within different cell populations of the same tissue. In addition, the complex anatomical structure and physiology of the human hair follicle is reflected in the highly regulated distribution of gene expression. Cyclin D1 likely is one of many genes which show such an intricate pattern of expression, and the analysis of additional genes should help unravel the mysteries of hair growth and cutaneous carcinogenesis.
Acknowledgments
We thank Dr. James Eberwine for the use of his LCM apparatus, Dorothy Campbell and the histology staff of the Departments of Pathology and Dermatology at the University of Pennsylvania for technical assistance. The Cooperative Human Tissue Network, which is funded by the National Cancer Institute, provided tissue specimens.
Footnotes
Address reprint requests to George Cotsarelis, M.D., Department of Dermatology, Hospital of University of Pennsylvania, M8 Stellar Chance Building, 422 Curie Boulevard, Philadelphia, PA 19104. E-mail: cotsarel@mail.med.upenn.edu.
Supported by National Institutes of Health grants R29-AR-44038 and R01-AR46837, and grants from the National Alopecia Areata Foundation.
The abstract of this paper received the Stowell-Orbison Award from the United States and Canadian Academy of Pathology.
References
- 1.Paus R, Cotsarelis G: The biology of hair follicles. N Engl J Med 1999, 341:491-497 [DOI] [PubMed] [Google Scholar]
- 2.Kligman AM: The human hair cycle. J Invest Dermatol 1959, 33:307-316 [DOI] [PubMed] [Google Scholar]
- 3.Cotsarelis G, Millar SE: Towards a molecular understanding of hair loss and its treatment. Trends Mol Med 2001, 7:293-301 [DOI] [PubMed] [Google Scholar]
- 4.Paus R: Control of the hair cycle and hair diseases as cycling disorders. Curr Opin Dermatol 1996, 3:248-258 [Google Scholar]
- 5.Stenn KS, Paus R: Controls of hair follicle cycling. Physiol Rev 2001, 81:449-494 [DOI] [PubMed] [Google Scholar]
- 6.Cotsarelis G, Sun TT, Lavker RM: Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell 1990, 61:1329-1337 [DOI] [PubMed] [Google Scholar]
- 7.Lyle S, Christofidou-Solomidou M, Liu Y, Elder DE, Albelda S, Cotsarelis G: The C8/144B monoclonal antibody recognizes cytokeratin 15 and defines the location of human hair follicle stem cells. J Cell Sci 1998, 111:3179-3188 [DOI] [PubMed] [Google Scholar]
- 8.Wilson C, Cotsarelis G, Wei ZG, Fryer E, Margolis-Fryer J, Ostead M, Tokarek R, Sun TT, Lavker RM: Cells within the bulge region of mouse hair follicle transiently proliferate during early anagen: heterogeneity and functional differences of various hair cycles. Differentiation 1994, 55:127-136 [DOI] [PubMed] [Google Scholar]
- 9.Ito M, Kizawa K, Toyoda M, Morohashi M: Label-retaining cells in the bulge region are directed to cell death after plucking, followed by healing from the surviving hair germ. J Invest Dermatol 2002, 119:1310-1316 [DOI] [PubMed] [Google Scholar]
- 10.Paus R: Principles of hair cycle control. J Dermatol 1998, 25:793-802 [DOI] [PubMed] [Google Scholar]
- 11.Van Scott EJ, Ekel T, Auerbach R: Determinants of rate and kinetics of cell division in scalp hair. J Invest Dermatol 1963, 41:269-273 [PubMed] [Google Scholar]
- 12.Chan EF, Gat U, McNiff JM, Fuchs E: A common human skin tumour is caused by activating mutations in β-catenin. Nat Genet 1999, 21:410-413 [DOI] [PubMed] [Google Scholar]
- 13.Kato J: Induction of S phase by G1 regulatory factors. Front Biosci 1999, 4:D787-792 [DOI] [PubMed] [Google Scholar]
- 14.O’Connell MJ, Walworth NC, Carr AM: The G2-phase DNA-damage checkpoint. Trends Cell Biol 2000, 10:296-303 [DOI] [PubMed] [Google Scholar]
- 15.Kato JY: Control of G1 progression by D-type cyclins: key event for cell proliferation. Leukemia 1997, 11(Suppl 3):347-351 [PubMed] [Google Scholar]
- 16.Muller H, Lukas J, Schneider A, Warthoe P, Bartek J, Eilers M, Strauss M: Cyclin D1 expression is regulated by the retinoblastoma protein. Proc Natl Acad Sci USA 1994, 91:2945-2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pagano M: Cell cycle regulation by the ubiquitin pathway. EMBO J 1997, 11:1067-1075 [DOI] [PubMed] [Google Scholar]
- 18.Behrens J, von Kries JP, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W: Functional interaction of β-catenin with the transcription factor LEF-1. Nature 1996, 382:638-642 [DOI] [PubMed] [Google Scholar]
- 19.Gat U, DasGupta R, Degenstein L, Fuchs E: De novo hair follicle morphogenesis and hair tumors in mice expressing a truncated β-catenin in skin. Cell 1998, 95:605-614 [DOI] [PubMed] [Google Scholar]
- 20.Millar SE: Molecular mechanisms regulating hair follicle development. J Invest Dermatol 2002, 118:216-225 [DOI] [PubMed] [Google Scholar]
- 21.Huelsken J, Vogel R, Erdmann B, Cotsarelis G, Birchmeier W: β-catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell 2001, 105:533-545 [DOI] [PubMed] [Google Scholar]
- 22.Shtutman M, Zhurinsky J, Simcha I, Albanese C, D’Amico M, Pestell R, Ben Ze’ev A: The cyclin D1 gene is a target of the β-catenin/LEF-1 pathway. Proc Natl Acad Sci USA 1999, 96:5522-5527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coombs LM, Pigott D, Proctor A, Eydmann M, Denner J, Knowles MA: Simultaneous isolation of DNA, RNA, and antigenic protein exhibiting kinase activity from small tumor samples using guanidine isothiocyanate. Anal Biochem 1990, 188:338-343 [DOI] [PubMed] [Google Scholar]
- 24.Xu X, Chu AY, Pasha TL, Elder DE, Zhang PJ: Immunoprofile of MITF, tyrosinase, melan-A, and MAGE-1 in HMB45-negative melanomas. Am J Surg Pathol 2002, 26:82-87 [DOI] [PubMed] [Google Scholar]
- 25.Klein D: Quantification using real-time PCR technology: applications and limitations. Trends Mol Med 2002, 8:257-260 [DOI] [PubMed] [Google Scholar]
- 26.Scholzen T, Gerdes J: The Ki-67 protein: from the known and the unknown. J Cell Physiol 2000, 182:311-322 [DOI] [PubMed] [Google Scholar]
- 27.Wollina U: Histochemistry of the human hair follicle with consideration of anagen phases I to VI. Acta Histochem 1992, 92:171-178 [DOI] [PubMed] [Google Scholar]
- 28.Eberwine J, Yeh H, Miyashiro K, Cao Y, Nair S, Finnell R, Zettel M, Coleman P: Analysis of gene expression in single live neurons. Proc Natl Acad Sci USA 1992, 89:3010-3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eberwine J: Single-cell molecular biology. Nat Neurosci 2001, 4 (Suppl):1155-1156 [DOI] [PubMed] [Google Scholar]
- 30.Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA, Liotta LA: Laser capture microdissection. Science 1996, 274:998-1001 [DOI] [PubMed] [Google Scholar]
- 31.Lehmann U, Kreipe H: Real-time PCR analysis of DNA and RNA extracted from formalin-fixed and paraffin-embedded biopsies. Methods 2001, 25:409-418 [DOI] [PubMed] [Google Scholar]
- 32.Czerniak B, Herz F, Wersto RP, Koss LG: Asymmetric distribution of oncogene products at mitosis. Proc Natl Acad Sci USA 1992, 89:4860-4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glockner S, Buurman H, Kleeberger W, Lehmann U, Kreipe H: Marked intratumoral heterogeneity of c-myc and cyclin D1 but not of c-erbB2 amplification in breast cancer. Lab Invest 2002, 82:1419-1426 [DOI] [PubMed] [Google Scholar]
- 34.Oshima H, Rochat A, Kedzia C, Kobayashi K, Barrandon Y: Morphogenesis and renewal of hair follicles from adult multipotent stem cells. Cell 2001, 104:233-245 [DOI] [PubMed] [Google Scholar]
- 35.DasGupta R, Fuchs E: Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development 1999, 126:4557-4568 [DOI] [PubMed] [Google Scholar]
- 36.Niemann C, Owens DM, Hulsken J, Birchmeier W, Watt FM: Expression of δNLef1 in mouse epidermis results in differentiation of hair follicles into squamous epidermal cysts and formation of skin tumours. Development 2002, 129:95-109 [DOI] [PubMed] [Google Scholar]
- 37.Sherr CJ: Cancer cell cycles. Science 1996, 274:1672-1677 [DOI] [PubMed] [Google Scholar]
- 38.Morgan DO: Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol 1997, 13:261-291 [DOI] [PubMed] [Google Scholar]
- 39.Weinstein IB: Disorders in cell circuitry during multistage carcinogenesis: the role of homeostasis. Carcinogenesis 2000, 21:857-864 [DOI] [PubMed] [Google Scholar]
- 40.Dong Y, Sui L, Sugimoto K, Tai Y, Tokuda M: Cyclin D1-CDK4 complex, a possible critical factor for cell proliferation and prognosis in laryngeal squamous cell carcinomas. Int J Cancer 2001, 95:209-215 [DOI] [PubMed] [Google Scholar]
- 41.Gorospe M, Liu Y, Xu Q, Chrest FJ, Holbrook NJ: Inhibition of G1 cyclin-dependent kinase activity during growth arrest of human breast carcinoma cells by prostaglandin A2. Mol Cell Biol 1996, 16:762-770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagasawa S, Onda M, Sasajima K, Makino H, Yamashita K, Takubo K, Miyashita M: Cyclin D1 overexpression as a prognostic factor in patients with esophageal carcinoma. J Surg Oncol 2001, 78:208-214 [DOI] [PubMed] [Google Scholar]
- 43.Takano Y, Takenaka H, Kato Y, Masuda M, Mikami T, Saegusa M, Okayasu I: Cyclin D1 overexpression in invasive breast cancers: correlation with cyclin-dependent kinase 4 and oestrogen receptor overexpression, and lack of correlation with mitotic activity. J Cancer Res Clin Oncol 1999, 125:505-512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joo M, Kang YK, Kim MR, Lee HK, Jang JJ: Cyclin D1 overexpression in hepatocellular carcinoma. Liver 2001, 21:89-95 [DOI] [PubMed] [Google Scholar]
- 45.Donnellan R, Chetty R: Cyclin D1 and human neoplasia. Mol Pathol 1998, 51:1-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takano Y, Kato Y, Masuda M, Ohshima Y, Okayasu I: Cyclin D2, but not cyclin D1, overexpression closely correlates with gastric cancer progression and prognosis. J Pathol 1999, 189:194-200 [DOI] [PubMed] [Google Scholar]
- 47.Moller MB, Nielsen O, Pedersen NT: Cyclin D3 expression in non-Hodgkin lymphoma: correlation with other cell cycle regulators and clinical features. Am J Clin Pathol 2001, 115:404-412 [DOI] [PubMed] [Google Scholar]
- 48.Rodriguez-Puebla ML, LaCava M, Miliani de Marval PL, Jorcano JL, Richie ER, Conti CJ: Cyclin D2 overexpression in transgenic mice induces thymic and epidermal hyperplasia whereas cyclin D3 expression results only in epidermal hyperplasia Am J Pathol 2000, 157:1039-1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodriguez-Puebla ML, LaCava M, Conti CJ: Cyclin D1 overexpression in mouse epidermis increases cyclin-dependent kinase activity and cell proliferation in vivo but does not affect skin tumor development. Cell Growth Differ 1999, 10:467-472 [PubMed] [Google Scholar]
- 50.Fantl V, Stamp G, Andrews A, Rosewell I, Dickson C: Mice lacking cyclin D1 are small and show defects in eye and mammary gland development. Genes Dev 1995, 9:2364-2372 [DOI] [PubMed] [Google Scholar]
- 51.Ledda-Columbano GM, Pibiri M, Concas D, Cossu C, Tripodi M, Columbano A: Loss of cyclin D1 does not inhibit the proliferative response of mouse liver to mitogenic stimuli. Hepatology 2002, 36:1098-1105 [DOI] [PubMed] [Google Scholar]
- 52.Rodriguez-Puebla ML, de Marval PL, LaCava M, Moons DS, Kiyokawa H, Conti CJ: Cdk4 deficiency inhibits skin tumor development but does not affect normal keratinocyte proliferation. Am J Pathol 2002, 161:405-411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Galvez-Aranda MV, Herrera-Ceballos E, Sanchez-Sanchez P, Bosch-Garcia RJ, Matilla-Vicente A: Pilomatrix carcinoma with lymph node and pulmonary metastasis: report of a case arising on the knee. Am J Dermatopathol 2002, 24:139-143 [DOI] [PubMed] [Google Scholar]
- 54.Kurtin PJ: Mantle cell lymphoma. Adv Anat Pathol 1998, 5:376-398 [DOI] [PubMed] [Google Scholar]