Abstract
The induction of organ-specific autoimmune diseases, such as experimental allergic encephalomyelitis (EAE) the principal animal model of multiple sclerosis (MS), relies on the use of complete Freund’s adjuvant (CFA) emulsions. In this study we report that the physical structure of the particles comprising neuroantigen-CFA emulsions significantly influences the genetic control of the incidence and sexual dimorphism seen in EAE. Immunization of (B10.S/SgMcdJ × SJL/J) F2 mice segregating the quantitative trait loci (QTL) controlling EAE in susceptible SJL/J and resistant B10.S/SgMcdJ mice with emulsions consisting of particles where the Mycobacterium tuberculosis and neuroantigens are localized on the phase surfaces led to severe EAE in 98.8% of the mice, overriding all sex-specific and non-sex-specific genetic checkpoints. In contrast, F2 mice immunized with emulsions where the bacterial products and encephalitogens are buried inside the water/oil vesicles exhibited a significant reduction in disease incidence (7.5%) and a sexual dimorphism (5% male versus 10% female). A genome scan identified QTL on chromosomes 7 and 11 controlling the sexual dimorphism as a function of the physical structure of the emulsion. The chromosome 11 QTL co-localizes with eae6b, and with Il12b and heptatitis A virus cellular receptor 2 (Havcr2, formerly known as Timd3), both of which are candidate genes for this QTL. Sequence analysis of the SJL/J and B10.S/SgMcdJ alleles indicates that both gene products are structurally monomorphic. Expression analysis also excluded both as candidates for this sex-specific QTL. These results reinforce the importance of gene-environment interactions in initiating and propagating autoimmune disease of the central nervous system, particularly in the context of susceptibility to MS and disease heterogeneity.
The use of complete Freund’s adjuvant (CFA) to induce organ-specific autoimmune disease dates back to 1947 when Freund reported the induction of experimental allergic encephalomyelitis (EAE) in guinea pigs using a single injection of central nervous system (CNS) antigens emulsified in CFA. 1 Before this time, multiple injections without adjuvants were used. 2 Subsequently, the use of CFA resulted in eliciting a plethora of experimental models of organ-specific autoimmune disease in a wide variety of species 3 including humans. 4,5 The initial theories put forth by Freund to explain the adjuvanticity of CFA centered around three mechanisms: 1) persistence of the antigen at the site of injection, 2) transport of the antigen to the lymphatic system, and 3) development of sensitization. 6 These hypotheses served as the impetus for the studies establishing that autoantigen sensitization using CFA emulsions preferentially stimulates delayed-type hypersensitivity reactions mediated by CD4+ Th1 effector T cells. 3
Additional adjuvants, such as Bordetella pertussis 7,8 and pertussis toxin (PTX), 9,10 the major exotoxin produced by B. pertussis, 11 were also found to be useful for inducing certain diseases, particularly in rats and mice. However, B. pertussis and PTX elicit both strong DTH and anaphylactic antibody responses. 12 With respect to disease induction, it was recently demonstrated that the histamine H1 receptor underlies Bphs, a shared autoimmune disease susceptibility locus between EAE and autoimmune orchitis, associated with the adjuvanticity of PTX and controlling the development of pathogenic CD4+ Th1 effector T cells. 13-15
We recently demonstrated that PTX also overrides many of the genetic checkpoints identified in mapping studies where CFA was used as the sole adjuvant. 16 Additionally, the use of PTX as an ancillary adjuvant resulted in less severe disease in female (B10.S/DvTe × SJL/J) × B10.S/DvTe backcross mice compared to female backcross mice immunized without PTX. In this same study, comparative mapping studies using backcross mice immunized with and without PTX led to the identification of adjuvant-dependent, sex-specific QTL. 16 These results suggest that the sexual dimorphism seen in EAE may be due in part to sex-specific QTL controlling responsiveness to the adjuvants used to induce disease. We report here that immunization of (B10.S/SgMcdJ × SJL/J) F2 mice with CFA emulsions consisting of particles where the Mycobacterium tuberculosis and encephalitogens are localized on the phase surfaces do not exhibit a sexual dimorphism. In contrast, F2 mice immunized with emulsions where the bacteria and neuroantigens are buried inside the water/oil vesicles exhibit a significant sexual dimorphism (5% male versus 10% female) and that QTL on chromosomes 7 and 11 control susceptibility to sexually dimorphic EAE as a function of the particulate structure of neuroantigen-CFA emulsions.
Materials and Methods
Animals
Male and female SJL/J and B10.S/SgMcdJ mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Retired male and female SJL/J breeders were purchased from Harlan (Indianapolis, IN). Male and female (B10.S/SgMcdJ × SJL/J) F1 hybrid animals were generated and (B10.S/SgMcdJ × SJL/J) F2 mice were produced by intercrossing F1 animals. The same parental and F1 hybrid mice were used to generate all F2 mice used in this study. F1 hybrid and F2 intercross mice were kept in the same room within the vivarium at the College of Veterinary Medicine at the University of Illinois, Urbana-Champaign (Urbana, IL). Animals were maintained in accordance with the Animal Welfare Act and the Public Health Service Policy on the Humane Care and Use of Laboratory Animals.
Mouse Spinal Cord Homogenate (MSCH)
MSCH was generated using retired breeder SJL/J mice purchased from The Jackson Laboratory. Spinal cords were extracted by insufflation, mixed with an equal volume of distilled H2O, homogenized in a Pyrex tissue homogenizer, and filtered through nylon mesh. The MSCH was lyophilized overnight and resuspended at 50 mg/ml in phosphate-buffered saline (PBS) and 1-ml aliquots stored at −70°C until used. The same pool of SJL/J MSCH was used for all injections in this study.
Preparation of MSCH-CFA Emulsions
MSCH-CFA emulsions with the Mycobacterium tuberculosis and encephalitogens localized on the external surface phase (ext-ENC-CFA) or buried within the water/oil vesicles (int-ENC-CFA) were prepared by high-speed homogenization and syringe extrusion, respectively. Homogenized emulsions were prepared by adding equal volumes of CFA (IFA supplemented with 0.1 mg/ml M. tuberculosis, H37Ra, as described above) and SJL/J MSCH in PBS into a 20-ml homogenization chamber fitted to a high-speed Waring blender. Homogenization was performed at room temperature for 1 minute. Syringe extruded MSCH-CFA emulsions were prepared using disposable syringes and a 21-gauge double-hub microemulsifying needle. The oil and water phases were emulsified at a 1:1 ratio. One syringe contained incomplete Freund’s adjuvant (Sigma, St. Louis, MO) supplemented with 0.1 mg/ml M. tuberculosis, H37Ra (Difco Laboratories, Detroit, MI). The other syringe contained an equal volume of SJL/J MSCH in PBS at a concentration of 6.67 mg/ml. Both syringes were cleared of trapped air, affixed to the microemulsifying needle, and the plunger of the syringe containing the aqueous phase was first depressed to extrude the entire contents into the oil phase. Subsequently, the plungers were depressed alternately until a non-separating emulsion was achieved. Both emulsions were non-separating on a water surface.
Induction and Evaluation of EAE
Mice were immunized for the induction of EAE as previously described. 17 Briefly, they were inoculated with 0.3 ml of SJL/J MSCH-CFA emulsion via two subcutaneous injections in the posterior right and left flank (2 × 0.15 ml). One week later all mice were similarly injected at two sites on the right and left flanks anterior of the initial injection sites. In this way, each animal received a total of 2.0 mg dry weight SJL/J MSCH and 30.0 μg of M. tuberculosis H37Ra. A cohort of B10.S/SgMcdJ mice were immunized with proteolipid protein peptide 139–151 (HSLGKWLGHPDKF) (PLP139–151) emulsified in CFA by injecting 0.2 ml of emulsion made by mixing equal volumes of 1 mg/ml PLP in PBS and 4 mg/ml M. tuberculosis H37Ra. Twenty-four hours later each mouse received by i.v. injection 200 ng PTX (List Biological Laboratories, Inc., Campbell, CA).
Mice were weighed and scored for clinical signs of disease daily from day 10 post-injection through day 60. Severity scores ranging from 0 to 4 were assigned as previously described 18 : 0, no clinical signs; 1, flaccid tail without hind leg weakness; 2, flaccid tail with hind limb weakness; 3, complete hind limb paralysis and floppy tail; 4, hind leg paralysis accompanied by a floppy tail and urinary or fecal incontinence; 5, quadriplegia and/or death. The incidence of EAE was recorded as positive for any mouse exhibiting clinical signs for one or more days. Susceptibility was also analyzed as a quantitative trait, using a disease index generated by averaging the clinical score for each animal over the course of the experiment. Severity of disease among affected animals was analyzed using the single high score for each mouse exhibiting clinical signs and onset was the day clinical signs were first observed. A weight index was calculated as the average weight change from the preinjection baseline to the peak of clinical disease for the population.
Genotyping
Genomic DNA was isolated from individual liver samples as previously described. 14 Microsatellite markers were purchased from Research Genetics (Huntsville, AL), or synthesized according to published sequences (http://www-genome.wi.mit.edu/cgi-bin/mouse/index). Polymerase chain reaction (PCR) parameters for microsatellite typing were previously described. 17 Microsatellite size variants were resolved by electrophoresis on large-format denaturing polyacrylamide gels, and visualized by autoradiography.
Linkage Analysis
Thirty-seven of 494 (B10.S/SgMcdJ × SJL/J) F2 mice immunized with int-ENC-CFA emulsions exhibited clinical signs of EAE. A set of 39 randomly selected sex-matched littermates served as the unaffected population for linkage analysis. Linkage analysis was performed using information derived from a genome scan with 127 informative microsatellite markers covering all 19 autosomes and the X-chromosome. Linkage maps were generated using the Kosambi mapping function in the MAPMAKER/EXP computer package. 19,20
QTL analysis was performed using the disease index (susceptibility) and severity or high score. Composite interval mapping (CIM) was carried out using model 6 of the Zmapqtl program in QTL Cartographer v1.30 software package (http://statgen.ncsu.edu/qtlcart/cartographer.html). By combining classical interval mapping with multiple regression analysis, CIM allows for more precise QTL localization than classical interval mapping. Additionally, CIM controls for spurious ghost loci. 21-22 Significant markers are first chosen using a linear regression model with a forward/backward selection procedure in the SRmapqtl module of QTL Cartographer. Markers flanking the test interval are added to the regression model to control for the presence of linked QTL. Additional markers, unlinked to the interval, but with significant effects on the trait are added to the model to control for the genetic background. Two cM increments with a window size of 10 cM and all 9 significant background markers selected via SRmapqtl were used in our CIM analyses. Tests of significant linkage are reported as likelihood ratio test (LRT) statistics. Significance of the linkage between marker loci and putative QTL was assessed by permutation-based threshold analysis. 23,24 Significant (α = 0.05) and suggestive (α = 0.10) experiment-wise critical values were determined using the distribution of maximum LRT statistics from 1000 permutations of our data. Analysis of linkage stratified by sex was carried out using QTManager (http://mapmgr.roswellpark.org/mapmgr.html).
Havcr2/Timd3 Allotyping
DNA samples from a number of inbred strains were purchased from The Jackson Laboratory or were prepared from tissues on hand, as described above. We designed two sets of oligonucleotide primers that amplify two portions of the Havcr2/Timd3 genomic sequence: one set for a positive control amplification (Havcr2/Timd3 control), and one set that flanked a microsatellite region (Havcr2/Timd3 MS) between nucleotides 1686 and 1862 in the 3′-UTR of the AKR/J mouse Havcr2/Timd3 sequence (AF450241): Havcr2/Timd3 MS forward primer: 5′-AGCCTAGG TGCTGAGTTCCA-3′, Havcr2/Timd3 MS reverse: 5′-GAAGGTTCTCTCTGTGCCTGC-3′. Havcr2/Timd3 control forward primer: 5′-CTACCTACATCTGGGACACTTG-3′, reverse: 5′-GACTCTGGATGACC ATGGGAC-3′. Fragments amplified by radiolabeled primer pairs were run on polyacrylamide gels as above, and Havcr2/Timd3 alleles read directly from the autoradiographic image. The absence of an AflIII restriction site in the PCR product amplified by the control primers distinguished DBA/2J (d) or AKR/J (a) Havcr2/Timd3 alleles. 25 The results were combined into unique haplotypes representing the combination of alleles at each of these Havcr2/Timd3 sequence characteristics.
Nucleic Acid Sequencing
Total RNA was isolated from the spleen of adult B10.S/SgMcdJ and SJL/J mice using TRIzol Reagent (Invitrogen, Carlsbad, CA). Reverse transcription (RT)-PCR was performed to obtain cDNA. First-strand cDNA was synthesized by reverse transcription of 1.0 μg total RNA primed with poly (dT16) oligonucleotide and SuperscriptII reverse transcriptase (Invitrogen). cDNAs were PCR-amplified using Taq polymerase and Havcr2/Timd3 specific primer pairs (Operon, Alameda, CA) flanking the mRNA coding regions (Forward: 5′-CTGCTGCTGC TGCAACTACTA-3′ and reverse: 5′-GGACTGCCACTTTTAAAGGCT-3′). The amplified fragments were cloned and PCR-screened. Insert-positive plasmid DNAs were sequenced using vector primers and the ABI PRIZM Dye Terminator Reading Reaction Cycle Sequencing Kit on a model 373A automated DNA sequencer (PerkinElmer, Applied Biosystems Division, Foster City, CA). PCR fragments were sequenced from both insert termini.
CD4+ T-Cell Preparation and Activation
CD4+ T cells were isolated from spleen and lymph nodes of adult B10.S/SgMcdJ and SJL/J mice by negative selection as described 26 using a combination of anti-MHC class II (m5/115), anti-CD8 (TIB105), anti-NK1.1, and anti-Mac1 mAbs (PharMingen, San Diego, CA). Cells were activated with plate-bound anti-CD3 (145–2C11, 5 μg/ml) and soluble anti-CD28 (1 μg/ml) mAbs (PharMingen) in the presence or absence of IFNγ (R&D Systems Inc., Minneapolis, MN) at a final concentration of 200 U/ml for 48 hours.
Real-Time PCR Analysis of Gene Expression
Total RNA was extracted from non-activated and activated CD4+ T cells using Ultraspec RNA Isolation Reagent (Biotecx Laboratory, Houston, TX) as recommended by the manufacturer. 1.0 μg of total RNA was used as a template to synthesize first-strand cDNA using random primers (Promega, Madison, WI) with SuperscriptII reverse transcriptase (Invitrogen). Real-time quantitative RT-PCR was performed using the TaqMan system (Applied Biosystems). PCRs were performed using the TaqMan Universal PCR Master Mix and the ABI PRISM 7700 Sequence Detection System. Havcr2/Timd3, Il12b and γ-actin (Actg) (internal reference) were measured by real-time PCR using probes labeled with 6-carboxyfluorescein (FAM) and Black Hole Quencher (BHQ). The Havcr2/Timd3 and Il12b probes and primer set sequences used were as previously described. 27,28 The Actg probe and primer sequences were: probe, 5′-6-FAM-d(CATTGCTCCCCCTGAGCGCAA)-BHQ-1–3′; forward primer, 5′-d(GCACCTAGCACGAT GAAGATTAA GA)-3′; reverse primer, 5′-d(AGCCACCGATCCAGACTGAGT)-3′ (Biosearch Technologies, Novato, CA). Havcr2/Timd3, IL12b, and Actg mRNA levels were analyzed separately using the standard curve method. A no-RT control was also prepared for each sample and no genomic DNA was detected. As a positive control for Havcr2/Timd3 expression, cDNA was prepared from Th1-skewed CD4+ T cells. Briefly, CD4+ T cells were stimulated with anti-CD3 and anti-CD28 mAbs in the presence of IL-12 (3.5 ng/ml; R&D Systems) and IFNγ (200 U/ml) for 4 days. For IL12β, cDNA was prepared from the spleens of mice immunized with CFA + PTX taken at 3 days post-injection. Total RNA was isolated and reverse-transcribed into first-strand cDNA. Varying amounts of the positive control cDNA were used to determine the linear range of the PCR reactions. 1 μl of the sample cDNA was used in the PCR reaction and the comparative threshold cycle (CT) value obtained was used to calculate the amount of sample cDNA required to reach the linear range established by the standard. The values obtained from separate reactions were averaged and the ratio of Havcr2/Timd3:Actg and Il12β:Actg calculated. For each sample, the Havcr2/Timd3 and Il12β CT value was expressed as an n-fold difference compared to the unstimulated B10.S/SgMcdJ sample.
Results
Cohorts of SJL/J, B10.S/SgMcdJ, (B10.S/SgMcdJ × SJL/J) F1 hybrid and (B10.S/SgMcdJ × SJL/J) F2 intercross mice were immunized under the same conditions and at the same sites with identical preparations of SJL/J MSCH-CFA emulsions with the exception of the distribution of the bacterial products and encephalitogens within the particles comprising the emulsions. F1 hybrid and F2 intercross mice were matched for parents, sex, age, and season. The incidence of EAE was significantly greater in animals immunized with ext-ENC-CFA emulsions compared to mice immunized with int-ENC-CFA emulsions (Table 1) ▶ . Similar results were obtained with B10.S/SgMcdJ mice immunized with ext-PLP139–151-CFA (9 of 10 mice sick) and int-PLP139–151-CFA (1 of 10 mice sick) emulsions.
Table 1.
Incidence of EAE in Mice Immunized with Int-ENC-CFA and Ext-ENC-CFA Emulsions
| Strain | Int-ENC-CFA | Ext-ENC-CFA | P value |
|---|---|---|---|
| SJL/J (5–6 weeks) | |||
| Female | 4/5 | 5/5 | |
| Male | 0/5 | 4/5 | |
| Total | 4/10 | 9/10 | 0.019 |
| SJL/J (>24 weeks) | |||
| Female | 4/5 | 5/5 | |
| Male | 4/5 | 5/5 | |
| Total | 8/10 | 10/10 | |
| B10.S/SgMcdJ | |||
| Female | 0/5 | 5/5 | |
| Male | 0/5 | 5/5 |
Animals were scored as unaffected or affected (any clinical signs) by D30 post-injection.
All (B10.S/SgMcdJ × SJL/J) F1 hybrid mice (24 of 24) and nearly all (B10.S/SgMcdJ × SJL/J) F2 intercross mice (248 of 251) immunized with ext-ENC-CFA developed EAE (Table 2) ▶ . In contrast, only 22% (11 of 50) of the F1 hybrid and 8% (37 of 494) of the F2 intercross mice immunized with int-ENC-CFA developed clinical disease. In addition, ext-ENC-CFA immunized F1 hybrid and F2 intercross mice developed EAE earlier (15.4 ± 1.4 days vs. 23.6 ± 12.0 days; P = 0.0021 and 15.4 ± 4.4 vs. 21.1 ± 7.7 days; P < 0.0001, respectively) and more severe disease (3.8 ± 0.4 vs. 2.3 ± 0.8; P < 0.0001 and 3.1 ± 1.8 vs. 2.3 ± 0.9, P = 0.0086, respectively) compared to int-ENC-CFA immunized mice. Additionally, ext-ENC-CFA immunized mice exhibited significantly greater weight loss compared to int-ENC-CFA immunized animals (4.6 ± 2.4 vs. 0.1 ± 2.6 grams; P < 0.0001 and 2.5 ± 2.5 vs. 0.9 ± 1.6 grams; P < 0.0001, respectively).
Table 2.
Clinical Signs Associated with EAE in (B10.S/SgMcdJ × SJL/J) F1 Hybrid and F2 Intercross Mice Injected with Ext-ENC-CFA and Int-ENC-CFA Emulsions
| Trait | F1 hybrid | P value | F2 intercross | P value |
|---|---|---|---|---|
| Incidence | ||||
| Ext-ENC-CFA | 24/24 (100)* | 248/251 (99) | ||
| Int-ENC-CFA | 11/50 (22) | <0.0001 | 37/494 (8) | <0.0001 |
| Onset | ||||
| Ext-ENC-CFA | 15.4 ± 1.4 | 15.4 ± 4.4 | ||
| Int-ENC-CFA | 23.6 ± 12.0 | 0.0021 | 21.1 ± 7.8 | <0.0001 |
| Severity | ||||
| Ext-ENC-CFA | 3.8 ± 0.4 | 3.1 ± 1.8 | ||
| Int-ENC-CFA | 2.3 ± 0.8 | <0.0001 | 2.3 ± 0.9 | 0.0086 |
| Weight loss† | ||||
| Ext-ENC-CFA | 4.6 ± 2.4 | 2.5 ± 2.5 | ||
| Int-ENC-CFA | 0.1 ± 2.6 | <0.0001 | 0.9 ± 1.6 | <0.0001 |
*Percent affected.
†The average weight loss was calculated using the total loss in grams for each animal from the baseline weight at injection to the peak of clinical disease.
Stratification of the populations by sex revealed that there was no significant difference in the clinical signs of disease between male and female mice immunized with ext-ENC-CFA with the exception of weight loss (Table 3) ▶ . Both male F1 hybrid and F2 intercross ext-ENC-CFA immunized mice exhibited significantly greater weight loss compared to female mice (5.7 ± 2.4 vs. 3.5 ± 2.0 grams; P = 0.028 and 3.2 ± 2.7 vs. 1.8 ± 2.1 grams; P < 0.0001, respectively). Among the int-ENC-CFA immunized animals, the only significant sexual dimorphism observed was for disease incidence in the F2 intercross population (24 of 233 female mice vs. 13 of 261 male mice; P = 0.025). These proportions (approximately 2:1) recapitulate our previous observations with this strain combination. 29
Table 3.
Clinical Signs Associated with EAE in Female and Male (B10.S/SgMcdJ × SJL/J) F1 Hybrid and F2 Intercross Mice Injected with Ext-ENC-CFA and Int-ENC-CFA Emulsions
| Trait | F1 hybrid | P value | F2 intercross | P value |
|---|---|---|---|---|
| Incidence | ||||
| Ext-ENC-CFA | ||||
| Female | 12/12 (100)* | 125/127 (98) | ||
| Male | 12/12 (100) | 123/124 (99) | ||
| Int-ENC-CFA | ||||
| Female | 7/26 (27) | 24/233 (10) | ||
| Male | 4/24 (17) | 13/261 (5) | 0.025 | |
| Onset | ||||
| Ext-ENC-CFA | ||||
| Female | 16.0 ± 1.5 | 15.4 ± 4.5 | ||
| Male | 14.8 ± 1.1 | 15.3 ± 4.4 | ||
| Int-ENC-CFA | ||||
| Female | 24.9 ± 14.9 | 22.1 ± 8.3 | ||
| Male | 21.0 ± 4.6 | 19.1 ± 6.7 | ||
| Severity | ||||
| Ext-ENC-CFA | ||||
| Female | 3.7 ± 0.5 | 2.9 ± 1.2 | ||
| Male | 3.9 ± 0.3 | 3.3 ± 2.3 | ||
| Int-ENC-CFA | ||||
| Female | 2.2 ± 0.7 | 2.1 ± 0.9 | ||
| Male | 2.5 ± 1.1 | 2.7 ± 0.9 | ||
| Weight loss† | ||||
| Ext-ENC-CFA | ||||
| Female | 3.5 ± 2.0 | 1.8 ± 2.1 | ||
| Male | 5.7 ± 2.4 | 0.028 | 3.2 ± 2.7 | <0.0001 |
| Int-ENC-CFA | ||||
| Female | 0.02 ± 1.9 | 0.9 ± 1.4 | ||
| Male | 0.3 ± 3.2 | 0.9 ± 1.9 |
*Percent affected.
†The average weight loss was calculated using the total change in grams for each animal from the baseline weight at injection to the peak of clinical disease.
Given these results, it was of interest to map the QTL controlling differential susceptibility to EAE as a function of the MSCH-CFA emulsion. The F2 intercross mice immunized with ext-ENC-CFA could not be used for this purpose due to complete penetrance and the lack of phenotypic variation among the mice. However, CIM based QTL analysis using disease index and severity were performed using 121 informative microsatellite markers on the 37 affected F2 intercross mice immunized with int-ENC-CFA and 39 unaffected sex-matched littermates. The use of unaffected littermates allowed for better control of prenatal, perinatal, and postnatal environmental conditions, which may alter the immunological profile. 30,31
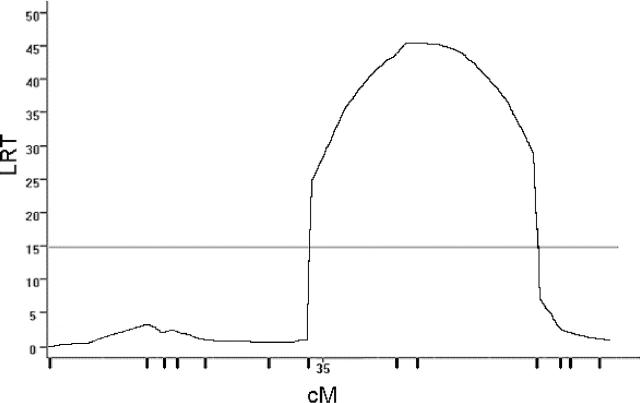
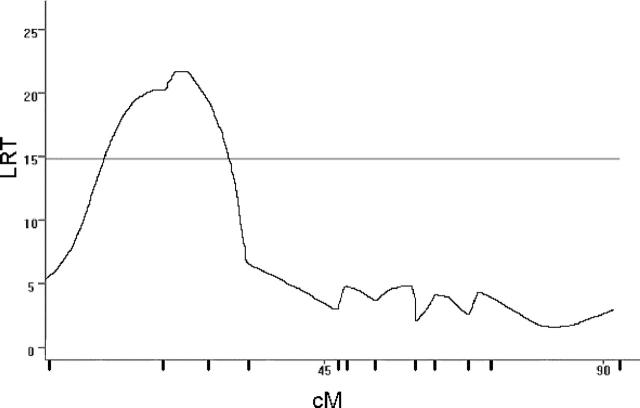
Using this population, significant linkage of susceptibility to EAE (disease index) was detected to marker loci on chromosomes 7 (Figure 1) ▶ and 11 (Figure 2) ▶ (Table 4) ▶ . Susceptibility induced with int-ENC-CFA was associated with B10.S alleles at both QTL. Suggestive linkage was also detected to D18Mit24 on proximal chromosome 18 (LRT = 12.2; P = 0.0022) in a region where linkage has not been previously detected. Similar linkage results were obtained with disease severity for all three QTL (data not shown). Stratification by sex revealed that the suggestive linkage to proximal chromosome 18 (♀; LRT = 11.3; P = 0.0036 vs. ♂; LRT = 17.4; P = 0.0002), is primarily male-specific whereas linkage to the chromosome 11 (♀; LRT = 11.7; P = 0.0029 vs. ♂; LRT <6.0) is female specific. Previous analysis of the eae6b QTL, which co-localizes with this female-specific QTL in the present study, did not show female bias, and the SJL/J-derived allele was associated with higher severity. 17,29,32
Figure 1.
Genetic linkage on chromosome 7. CIM results for linkage of susceptibility to EAE (disease index) to chromosome 7 using the combined male and female F2 intercross mice immunized with int-ENC-CFA emulsion. Permutation-derived significance cut-offs were based on 1000 permutations at α = 0.05. Significance cutoffs for this trait are 14.8.
Figure 2.
Genetic linkage on chromosome 11. CIM results for linkage of susceptibility to EAE (disease index) to chromosome 11 using the combined male and female F2 intercross mice immunized with int-ENC-CFA emulsion. Permutation-derived significance cut-offs were based on 1000 permutations at α = 0.05. Significance cutoffs for this trait are 14.8.
Table 4.
Susceptibility to EAE Induced Using Int-ENC-CFA Emulsion is Linked to Marker Loci on Chromosomes 7 and 11*
| Marker | Affected | Unaffected | χ2 | P value | ||||
|---|---|---|---|---|---|---|---|---|
| S | H | B | S | H | B | |||
| D7Mit281 | ||||||||
| Total | 2 | 18 | 18 | 15 | 22 | 1 | 25.6 | 0.0001 |
| Female | 2 | 13 | 10 | 11 | 12 | 1 | 13.6 | 0.0011 |
| Male | 0 | 5 | 8 | 4 | 10 | 0 | 13.7 | 0.0011 |
| D11Mit307 | ||||||||
| Total | 2 | 11 | 25 | 15 | 14 | 9 | 13.2 | 0.0013 |
| Female | 0 | 20 | 5 | 10 | 9 | 5 | 14.2 | 0.0008 |
| Male | 2 | 5 | 6 | 5 | 5 | 4 | 1.7 | 0.43 |
*Mice were classified as susceptible if they exhibited clinical signs for two or more consecutive days. D7Mit281 and D11Mit307 are the chromosome 7 and 11 markers exhibiting maximal linkage.
These linkage results indicate that the QTL on chromosomes 7 and 11 controlling susceptibility to EAE as a function of the particulate structure of the MSCH-CFA emulsions colocalized with eae4 and eae6b. 17,32 Two obvious candidate genes for the QTL on chromosome 11 and eae6b are IL12β and the T-cell immunoglobulin and mucin domain containing gene-3 (Timd3 now designated Havcr2 for heptatitis A virus cellular receptor 2) located at ∼20 cM on chromosome 11. 25 Havcr2/Timd3 is a cell surface protein, found on Th1-like T cells, that plays a role in regulating macrophage activation and has been implicated in EAE. 27 Therefore, we searched for Havcr2/Timd3 locus-specific polymorphisms as well as structural polymorphisms in the gene product that may segregate with susceptibility and resistance to EAE. Two Havcr2/Timd3 allele-specific genomic markers were developed and their segregation with susceptibility and resistance to EAE among inbred strains of mice examined (Table 5) ▶ . No clear correlation was detected. Second, we sequenced the SJL/J and B10.S/SgMcdJ Havcr2/Timd3 alleles to look for potential structural polymorphisms. Although there is one mutation between the nucleotide sequences of B10.S/SgMcdJ and SJL/J Havcr2/Timd3 alleles, at the third base position of codon 76 (Thr); this change, A->G, is silent (Figure 3) ▶ . Thus, there does not appear to be a structural polymorphism in the SJL/J and B10.S/SgMcdJ Havcr2/Timd3 alleles that underlies linkage to eae6b or susceptibility to EAE, despite the fact that the Havcr2/Timd3 locus itself is quite polymorphic. However, we did not do an exhaustive analysis to rule out the possible existence of polymorphic splice variants. With respect to IL12β, we previously sequenced the SJL/J and B10.S/SgMcdJ alleles and found no differences in the predicted amino acid sequences. 32
Table 5.
Havcr2/Timd3 Allotypes in EAE Susceptible and Resistant Strains of Mice
| Strain | ASO* | SSLP† | RFLP‡ | Haplotype§ | eae6b¶ |
|---|---|---|---|---|---|
| NOD/LtJ | d | 197 | + | a | Yes |
| Biozzi ABH | c | 201 | − | c | |
| B10.S/DvTe | c | 199 | − | b | Yes |
| SJL/J | c | 201 | − | c | |
| RIII.S/J | c | 201 | − | c | Yes |
| (B10.S/SgMcdJ, B10.A/SgSnJ) | c | 199 | − | b | |
| BALB/cByJ | c | 201 | − | c | Unknown |
| BALB/cJ | c | 201 | − | c | |
| PL/J | c | 201 | − | c | |
| SWR/J | c | 191 | − | e | |
| CBA/J | c | 201 | − | c | |
| C3H/HeJ | c | 201 | − | c | |
| DBA/2/J | d | 205 | + | d | |
| AKR/J | d | 197 | + | a | |
| C57BL/6J | d | 205 | + | d |
*Indicates the presence of either the DBA/2J (d) or BALB/c (c) Havcr2 (formerly Timd3) allele as determined by using allele-specific oligonucleotide (ASO) primers. The presence of a signature codon in this fragment was confirmed by the detection of a BstNI site (at position 132 relative to the ATG) in the PCR product from each d strain.
†The fragment sizes of a polymorphic microsatellite marker within the Havcr2/Timd3 locus is given in nucleotides, including the primer and flanking sequence.
‡Indicates the presence of a polymorphic AflIII restriction site at position 277 relative to the ATG.
§Havcr2/Timd3 haplotypes based on the combined genotypes at the three polymorphic regions are indicated by a letter (a–e). The AKR/J subline sample (DNA from Jackson Laboratory) showed a different haplotype than the sequence in NCBI (AF450241).
¶Indicates if either significant or suggestive linkage of EAE to the eae6b region of chromosome 11 was observed in previous publications of the progeny of pairs indicated. For the RIII.S/J × B10.RIII/SnJ cross, the haplotype of B10.RIII/SnJ is inferred from the haplotype found in B10.S/DvTe and B10.A/SgSnJ.
Figure 3.

cDNA sequences for Havcr2/Timd3 alleles. SJL/J and B10.S/SgMcdJ were obtained using total RNA isolated from adult spleen. cDNAs were PCR-amplified using Taq polymerase and specific primer pairs flanking the mRNA coding region of the gene. The amplified fragments were TA-cloned and PCR-screened for inserts using gene-specific primers. At least three clones for each PCR fragment were sequenced from both insert termini. Although there is one mutation between the nucleotide sequences of B10.S/SgMcdJ and SJL/J Havcr2/Timd3 alleles, at the third base position of codon 76 (Thr); this change, A->G, is silent.
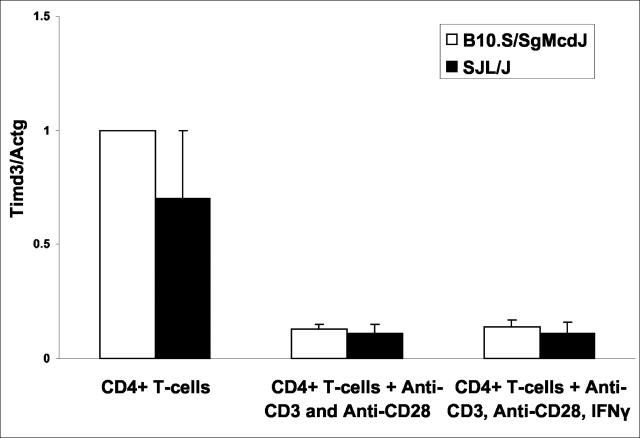
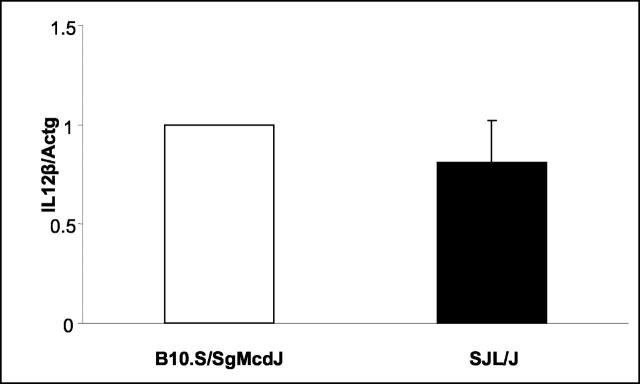
To more fully explore Havcr2/Timd3 and IL12β as candidates for eae6b, we examined the levels of expression of these two genes in SJL/J and B10.S/SgMcdJ mice. Havcr2/Timd3 mRNA expression levels were assessed by real-time PCR using RNA from freshly isolated CD4+ T cells, CD4+ T cells stimulated in vitro with anti-CD3 and anti-CD28, or anti-CD3 and anti-CD28 plus IFNγ. There was no significant difference in the basal levels of expression of Havcr2/Timd3 by freshly isolated CD4+ T cells between the two strains (P = 0.10) (Figure 4) ▶ . Interestingly, stimulation in vitro resulted in a significant reduction in the levels of expression of Havcr2/Timd3 in both strains. With unstimulated cells, there was, however, no significant difference in the level of Havcr2/Timd3 expression between the two strains. IL12β expression levels were determined using unstimulated splenic RNA. Again, no significant difference in the level of expression of IL12β was seen between the two strains (P = 0.24) (Figure 5) ▶ . These results are in agreement with earlier reports on IL12β secretion by SJL/J and B10.S/SgMcdJ macrophages 33 ; however, a subsequent report shows that SJL/J mouse macrophages make more IL12 p40 when they are stimulated with lipopolysaccharide (LPS) compared with B10.S mouse macrophages. 34
Figure 4.
Havcr2/Timd3 expression by CD4+ T cells. CD4+ T cells were isolated from B10.S/SgMcdJ and SJL/J mice and Havcr2/Timd3 expression levels assessed by TaqMan PCR using cDNA generated by reverse transcription of mRNA isolated from naïve cells and CD4+ T cells stimulated with either anti-CD3 and anti-CD28 or anti-CD3 and anti-CD28 and IFNγ. Expression levels were normalized with respect to B10.S/SgMcdJ levels. Data are presented as the normalized means ± SD. No significant differences in Havcr2/Timd3 expression were observed between B10.S/SgMcdJ and SJL/J under any of the conditions studies (P > 0.05).
Figure 5.
Il12β expression by SJL/J and B10.S/SgMcdJ splenic cells. IL12β expression was assessed by TaqMan PCR using cDNA generated by reverse transcription of mRNA isolated from individual, freshly excised spleens. Expression levels were normalized with respect to B10.S/SgMcdJ levels. Data are presented as the normalized means ± SD. Statistical significance was assessed using Student’s t-test (P = 0.24).
Discussion
In this study we used SJL/J, B10.S/SgMcdJ, (B10.S/SgMcdJ × SJL/J) F1 hybrid, and (B10.S/SgMcdJ × SJL/J) F2 intercross mice to demonstrate that MSCH-CFA emulsions consisting of particles where the M. tuberculosis and neuroantigens are localized on the phase external surfaces (ext-ENC-CFA) are significantly more encephalitogenic than MSCH-CFA emulsions in which the bacteria and neuroantigens are buried inside the water/oil vesicles (int-ENC-CFA). These results are consistent with those seen in inbred and congenic strains considered to be resistance to EAE immunized with ext-ENC-CFA emulsions generated by sonication. 35,36 Importantly, we see the same effects with or without the use of pertussis toxin as an ancillary adjuvant and when encephalitogenic peptides are used as the immunogen. Clearly, emulsification techniques and the resultant physical structure of the particles comprising the emulsions play a major role in their encephalitogenic activity and the genetic control of susceptibility and resistance to disease. In fact, the use of either ext-ENC-CFA and int-ENC-CFA emulsions accounts for greater phenotypic variation than the underlying genetic differences among susceptible and resistant inbred strains of mice. Additionally, our results support the hypothesis that the sexual dimorphism seen in EAE may be due in part to sex-specific QTL controlling responsiveness to the adjuvants used to induce disease 16 and that the QTL identified on chromosomes 7 and 11 may function in this capacity.
Historically, significant species differences were observed in the adjuvanticity of oil-in-water and water-in-oil emulsions, and changes in both the structure and size of the particles comprising the emulsions have been shown to influence the type of immune response elicited. 35,37-44 In this regard, syringe-extrusion, high-speed homogenization and sonication lead to emulsions with markedly different physical structures. The particles generated by the later two techniques are much smaller than those generated by syringe-extrusion, and the M. tuberculosis and antigen are localized on the surface of the particles as compared to being within the particles.
Since dendritic cells (DCs) constitutively sample the local microenvironment via macropinocytosis and cell signaling using different receptors, 45 it is reasonable to assume that in emulsion where the M. tuberculosis and neuroantigens are localized on the phase surfaces effectively increases the antigen specific immunostimulatory activity of DCs compared to emulsions in which the bacteria and neuroantigens are buried inside the water/oil vesicles. DCs encountering the latter must first ingest the larger particles and then release the bacterial products and encephalitogens from within these particles before their activation and maturation, and the presentation of disease inducing epitopes to T cells 46,47 ; dendritic cells that encounter ext-ENC-CFA emulsions do not.
Thus, the QTL controlling susceptibility to EAE induced with the less encephalitogenic int-ENC-CFA emulsions are likely to encode for genes that modify the ability of DCs to regulate or carry out one or more of these processes or activation states associated with these processes. IL12β and Havcr2/Timd3, located at ∼19 and 20 cM on chromosome 11, respectively, are both associated with macrophage/T-cell activation and EAE severity 27,33 and are therefore attractive candidate genes the QTL identified on chromosome 11 in this study and eae6b. However, the lack of both structural and expression-level polymorphisms for Havcr2/Timd3 between SJL/J and B10.S/SgMcdJ mice, combined with the discordance in Havcr2/Timd3 allotypes among EAE-susceptible and -resistant inbred strains, suggests that it is excluded as a candidate. However, we did not do an exhaustive analysis to rule out the possible existence of polymorphic splice variants. Similarly, IL12β can be excluded as a candidate based on the same criteria. 32,33 Importantly, any candidate gene for the chromosome 11 QTL must take into account the sex specificity of the linkage observed.
The mechanisms underlying sex-specific QTL are unknown but may arise as a result of sex hormone regulation of the polymorphic genes underlying these QTL or interactions between mitochondrially or Y-chromosome-linked genes. The role of sex hormones in the sexual dimorphism observed in immune responsiveness as well as in immunopathologically based diseases has been well documented 48,49 as has the regulation of immunologically relevant genes such as cytokines. 50 With respect to potential interactions with mitochondrially linked genes, there is evidence to suggest that mutations in mitochondrially encoded genes contribute to an MS-like syndrome, Leber’s hereditary optic neuropathology, which occurs primarily in women. 51 Such mutations, however, do not appear to be a risk factor for MS per se. 52 Additionally, a number of immunologically relevant genes are known to be on the X-chromosome (http://www.informatics.jax.org/searches/linkmap.cgi).
Although somewhat speculative, it is worth noting several candidate genes residing within the chromosome 7 QTL interval. Pgia3 (proteoglycan induced arthritis-3) and Cia7 (collagen-induced arthritis QTL-7) at 42.6 cM and 50.0 cM, respectively, are of interest in light of the recent validation of the shared autoimmune disease susceptibility gene hypothesis. 13-15 Additionally, Tria4, a QTL controlling T-cell receptor-induced activation, resides within the interval as do a number of immunologically relevant candidate genes. These include Art2a and Art2b (ADP-ribosyltransferase 2a and 2b), Art1 and Art4 (ADP-ribosyltransferase 1 and 2), Cd19, Ly14, Itgal, Mph1 (macrophage antigen-1), and Spn (sialophorin or CD43) (www.informatics.jax.org/searches/linkmap.cgi). Two additional candidate genes of particular relevance to our hypothesis that the QTL identified in this study may play a role in the efficiency of processing and presenting antigens to T cells as a function of the emulsions particulate microstructure are Aqp8 (aquaphorin 8) at 61.0 cM and Psma1 (proteosome subunit, α type 1 or macropain) at 53.0 cM especially given their respective roles in antigen uptake, concentration, processing, and presentation to T cells. 53-56 Clearly, a structural or expression-level polymorphism that affects the ability of aquaporins to facilitate this process, or degrade the concentrated macrosolutes (Psma1), could lead to significant quantitative differences in disease. Interestingly, Psma1 maps to 11p15.1 in the human, which is in an MS susceptibility region at 11p15 identified by the Multiple Sclerosis Genetics Group in 1996. 57
Several EAE genetics studies in the mouse have been completed during the past few years. 16,17,32,58-64 These studies were carried out with little regard for the importance that the microstructure of the particles comprising the CFA emulsions play in disease susceptibility. In light of our results and those reported previously, 35,36 this variable may be singularly responsible for failing to identify a more uniform, robust set of susceptibility loci across these studies. However, the results of this study and those of Baker et al 59 do exhibit a significant degree of concordance in that the most significant linkages observed in both studies were to chromosomes 7 (eae4), 11 (eae6b), and 18 and int-ENC-CFA emulsions were used to immunize the mapping populations.
Our results raise the intriguing possibility that similar genetically controlled mechanisms may play a role in the gene-environment interactions underlying the disease heterogeneity seen in MS. 65 It is highly likely that some of the genes controlling susceptibility to MS control the host’s response to the microstructure-based adjuvant effects of the infectious or environmental agents associated with MS rather than their infectivity, latency, or the ability of the anticipatory immune system to mount a pathogenic T- or B-cell immune response via molecular mimicry. This appears to be the case with respect to particulate pollution. 66 For example, the IgE and IgG3a antigen-independent, adjuvant-enhancing effects of physically and chemically defined particles is also genetically controlled. Clearly, a better understanding of the genes, their products, and the pathways controlling responsiveness to immunogens and infectious agents solely as a function of the physicality of their microstructure may lead to an improved understanding of how specific environmental and genetic factors, and their interactions, are involved in initiating and propagating immunopathologically based diseases.
Footnotes
Address reprint requests to Dr. Cory Teuscher, C317 Given Medical Building, University of Vermont, Burlington, VT 05405. E-mail: cteusche@zoo.uvm.edu.
Supported by National Institutes of Health grants NS36526, AI41747, and AI45105 (to C.T.), NS255195 (to E. P. B.), and National Multiple Sclerosis Society Grant RG3129 (to C.T.).
References
- 1.Freund J, Stern ER, Pisami TM: Isoallergic encephalomyelitis and radiculitis in guinea pigs after one injection of brain and mycobacteria in water-in-oil emulsion. J Immunol 1947, 57:179-194 [PubMed] [Google Scholar]
- 2.Rivers TM, Sprunt DH, Berry GP: Observations on attempts to produce acute disseminated encephalomyelitis in monkeys. J Exp Med 1933, 58:39-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Billiau A, Matthys P: Modes of action of Freund’s adjuvants in experimental models of autoimmune diseases. J Leuk Biol 2001, 70:849-860 [PubMed] [Google Scholar]
- 4.Mancini RE, Andrada JA, Saraceni A, Bachmann AE, Lavieri JC, Nemirovsky M: Immunological and testicular response in man sensitized with human testicular homogenate. J Clin Endocrinol Metab 1965, 25:859-875 [DOI] [PubMed] [Google Scholar]
- 5.Drachman DA, Paterson PY, Bornstein MB: Experimental allergic encephalomyelitis in man: a laboratory accident. Neurology 1974, 24:364 [Google Scholar]
- 6.Freund J: The mode of action of immunologic adjuvants. Adv Tuberc Res 1956, 7:130-148 [PubMed] [Google Scholar]
- 7.Lee JM, Olitzky PK: Simple methods of enhancing development of acute disseminated encephalomyelitis in mice. Proc Soc Exp Biol Med 1955, 89:263-266 [DOI] [PubMed] [Google Scholar]
- 8.Levine S, Wenk EJ, Devlin HB, Pieroni RE, Levine L: Hyperacute allergic encephalomyelitis: adjuvant effect of pertussis vaccine and extracts. J Immunol 1966, 97:363-368 [PubMed] [Google Scholar]
- 9.Munoz JJ, Arai H, Bergman RK, Sadowski PL: Biological activities of crystalline pertussigen from Bordetella pertussis. Infect Immun 1981, 33:820-826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergman RK, Munoz JJ, Portis JL: Vascular permeability changes in the central nervous system of rats with hyperacute experimental allergic encephalomyelitis induced with the aid of a substance from Bordetella pertussis. Infect Immun 1978, 21:627-637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rappouli R, Pizza M: Structure and evolutionary aspects of ADP-ribosylating toxins. Abuf J Freer J eds. Source Book of Bacterial Protein Toxins. 1991:pp 1-20 Academic Press London
- 12.Munoz JJ: Biological activities of pertussigen (pertussis toxin). Sekura RD Moss J Vaughan M eds. Pertussis Toxin. 1985:pp 1-18 Academic Press Orlando, FL
- 13.Teuscher C: Experimental allergic orchitis in mice: II. association of disease susceptibility with the locus controlling Bordetella pertussis-induced sensitivity to histamine. Immunogenetics 1985, 22:417-425 [DOI] [PubMed] [Google Scholar]
- 14.Sudweeks JD, Todd JA, Blankenhorn EP, Wardell BB, Woodward SR, Meeker ND, Estes SS, Teuscher C: Locus controlling Bordetella pertussis-induced histamine sensitization (Bphs), an autoimmune disease-susceptibility gene, maps distal to T-cell receptor β-chain gene on mouse chromosome 6. Proc Natl Acad Sci USA 1993, 90:3700-3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma RZ, Gao J, Meeker ND, Fillmore PD, Tung KSK, Watanabe T, Zachary JF, Offner H, Blankenhorn EP, Teuscher C: Identification of Bphs, an autoimmune disease locus, as histamine receptor H1. Science 2002, 297:620-623 [DOI] [PubMed] [Google Scholar]
- 16.Blankenhorn EP, Butterfield RJ, Rigby R, Cort L, Giambrone D, McDermott P, McEntee K, Solowski N, Meeker ND, Zachary JF, Doerge RW, Teuscher C: Genetic analysis of the influence of pertussis toxin on experimental allergic encephalomyelitis susceptibility: an environmental agent can override genetic checkpoints. J Immunol 2000, 164:3420-3425 [DOI] [PubMed] [Google Scholar]
- 17.Butterfield RJ, Sudweeks JD, Blankenhorn EP, Korngold R, Marini JC, Todd JA, Roper RJ, Teuscher C: New genetic loci that control susceptibility and symptoms of experimental allergic encephalomyelitis in inbred mice. J Immunol 1998, 161:1860-1867 [PubMed] [Google Scholar]
- 18.Teuscher C, Hickey WG, Korngold R: An analysis of the role of TNF in the phenotypic expression of actively induced experimental allergic orchitis and experimental allergic encephalomyelitis. Clin Immunol Immunopathol 1990, 54:442-453 [DOI] [PubMed] [Google Scholar]
- 19.Lander E, Green P, Abrahamson J, Barlow A, Daly M, Lincoln S, Newburg L: MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1987, 1:174-181 [DOI] [PubMed] [Google Scholar]
- 20.Lincoln S, Daly M, Lander E: Constructing genetic maps withMAPMAKER/EXP 3.0. Whitehead Institute Technical Report. 3rd ed. 1992. Whitehead Institute Cambridge MA
- 21.Zeng ZB: Theoretical basis for separation of multiple linked gene effects in mapping quantitative trait loci. Proc Natl Acad Sci USA 1993, 90:10972-10976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng ZB: Precision mapping of quantitative trait loci. Genetics 1994, 136:1457-1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doerge RW, Zeng ZB, Weir BS: Statistical issues in the search for genes affecting quantitative traits in experimental populations. Stat Sci 1997, 3:195-219 [Google Scholar]
- 24.Churchill GA, Doerge RW: Empirical threshold values for quantitative trait mapping. Genetics 1994, 138:963-971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McIntire JJ, Umetsu SE, Akbari O, Potter M, Kuchroo VK, Barsh GS, Freeman GJ, Umetsu DT, DeKruyff RH: Identification of Tapr (an airway hyperreactivity regulatory locus) and the linked Tim gene family. Nat Immunol 2001, 2:1109-1116 [DOI] [PubMed] [Google Scholar]
- 26.Rincón M, Flavell RA: AP-1 transcriptional activity requires both T-cell receptor-mediated and co-stimulatory signals in primary T lymphocytes. EMBO J 1994, 13:4370-4381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monney L, Sabatos CA, Gaglia JL, Ryu A, Waldner H, Chernova T, Manning S, Greenfield SA, Coyle AJ, Sobel RA, Freeman GJ, Kuchroo VK: Th1-specific cell surface protein Tim-3 regulates macrophage activation and severity of an autoimmune disease. Nature 2002, 415:536-541 [DOI] [PubMed] [Google Scholar]
- 28.Overbergh L, Valckx D, Waer M, Mathieu C: Quantification of murine cytokine mRNAs using real-time quantitative reverse-transcriptase PCR. Cytokine 1999, 11:305-312 [DOI] [PubMed] [Google Scholar]
- 29.Butterfield RJ, Blankenhorn EP, Roper RJ, Zachary JF, Doerge RW, Sudweeks JD, Rose J, Teuscher C: Genetic analysis of disease subtypes and sexual dimorphisms in mouse experimental allergic encephalomyelitis (EAE): relapsing/remitting and monophasic remitting/nonrelapsing EAE are immunologically distinct. J Immunol 1999, 162:3096-3102 [PubMed] [Google Scholar]
- 30.Neveu PJ, Deleplanque B, Puglisi-Allegra S, D’Amato FR, Cabib S: Influence of early life events on immune reactivity in adult mice. Dev Psychobiol 1994, 27:205-213 [DOI] [PubMed] [Google Scholar]
- 31.Laban O, Markovic BM, Dimitrijevic M, Jankovic BD: Maternal deprivation and early weaning modulate experimental allergic encephalomyelitis in the rat. Brain Behav Immun 1995, 9:9-19 [DOI] [PubMed] [Google Scholar]
- 32.Teuscher C, Butterfield RJ, Ma RZ, Zachary JF, Doerge RW, Blankenhorn EP: Sequence polymorphisms in the chemokines Scya1 (TCA-3), Scya2 (monocyte chemoattractant protein (MCP)-1), and Scya12 (MCP-5) are candidates for eae7, a locus controlling susceptibility to monophasic remitting/nonrelapsing experimental allergic encephalomyelitis. J Immunol 1999, 163:2262-2266 [PubMed] [Google Scholar]
- 33.Chang JT, Shevach EM, Segal BM: Regulation of interleukin (IL)-12 receptor β2 subunit expression by endogenous IL-12: a critical step in the differentiation of pathogenic autoreactive T cells. J Exp Med 1999, 189:969-978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alleva DG, Johnson EB, Wilson J, Beller DI, Conlon PJ: SJL and NOD macrophages are uniquely characterized by genetically programmed, elevated expression of the IL-12(p40) gene, suggesting a conserved pathway for the induction of organ-specific autoimmunity. J Leukoc Biol 2001, 69:440-448 [PubMed] [Google Scholar]
- 35.Määttä JA, Erälinna JP, Röyttä M, Salmi AA, Hinkkanen AE: Physical state of the neuroantigen in adjuvant emulsions determines encephalitogenic status in the BALB/c mouse. J Immunol Meth 1996, 190:133-141 [DOI] [PubMed] [Google Scholar]
- 36.Määttä JA, Nygårdas PT, Hinkkanen AE: Enhancement of experimental autoimmune encephalomyelitis severity by ultrasound emulsification of antigen/adjuvant in distinct strains of mice. Scand J Immunol 2000, 51:87-90 [DOI] [PubMed] [Google Scholar]
- 37.Hilleman MR: Critical appraisal of emulsified oil adjuvants applied to viral vaccines. Prog Med Virol 1966, 8:131-182 [PubMed] [Google Scholar]
- 38.Steinberg P, Norman PS: Mechanism of action of mineral oil adjuvants: I. conditions influencing the in vitro stability of water-in-oil emulsions J Allergy Clin Immunol 1973, 51:238-244 [DOI] [PubMed] [Google Scholar]
- 39.Goto N: Comparative studies on effects of incomplete oil adjuvants with different physical properties. Jpn J Med Sci Biol 1978, 31:53-79 [DOI] [PubMed] [Google Scholar]
- 40.Edelman R: Vaccine adjuvants. Rev Infect Dis 1980, 2:370-383 [DOI] [PubMed] [Google Scholar]
- 41.Edelman R, Tacket CO: Adjuvants. Int Rev Immuno 1990, 7:51-66 [DOI] [PubMed] [Google Scholar]
- 42.Yamanaka M, Hiramatsu K, Hirahara T, Okabe T, Nakai M, Sasaki N, Goto N: Pathological studies on local tissue reactions in guinea pigs and rats caused by four different adjuvants. J Vet Med Sci 1992, 54:685-692 [DOI] [PubMed] [Google Scholar]
- 43.Vulliet R: Improved technique for the preparation of water-in-oil emulsions containing protein antigens. Biotechniques 1996, 20:797-800 [DOI] [PubMed] [Google Scholar]
- 44.Aucouturier J, Dupuis L, Ganne V: Adjuvants designed for veterinary and human vaccine. Vaccine 2001, 19:2666-2672 [DOI] [PubMed] [Google Scholar]
- 45.Thery C, Amigorena S: The cell biology of antigen presentation in dendritic cells. Cur Opin Immun 2001, 13:45-51 [DOI] [PubMed] [Google Scholar]
- 46.Collins DS, Findlay K, Harding CV: Processing of exogenous liposome-encapsulated antigens in vivo generates class I MHC-restricted T-cell responses. J Immunol 1992, 148:3336-3341 [PubMed] [Google Scholar]
- 47.Harding CV, Collins DS, Slot JW, Geuze HJ, Unanue ER: Liposome-encapsulated antigens are processed in lysosomes, recycled, and presented to T cells. Cell 1991, 64:393-401 [DOI] [PubMed] [Google Scholar]
- 48.Da Silva JA: Sex hormones and glucocorticoids: interactions with the immune system. Ann NY Acad Sci 1999, 879:102-117 [DOI] [PubMed] [Google Scholar]
- 49.Grossman CJ, Roselle GA, Mendenhall CL: Sex steroid regulation of autoimmunity. J Steroid Biochem Mol Biol 1991, 40:649-659 [DOI] [PubMed] [Google Scholar]
- 50.Cutolo M, 0Seriolo B, Villaggio B, Pizzorni C, Craviotto C, Sulli A: Androgens and estrogens modulate the immune and inflammatory responses in rheumatoid arthritis. Ann NY Acad Sci 2002, 966:131-142 [DOI] [PubMed] [Google Scholar]
- 51.Wissinger B, Besch D, Baumann B, Fauser S, Christ-Adler M, Jurklies B, Zrenner E, Leo-Kottler B: Mutation analysis of the ND6 gene in patients with Lebers hereditary optic neuropathy. Biochem Biophys Res Commun 1997, 234:511-515 [DOI] [PubMed] [Google Scholar]
- 52.Reynier P, Penisson-Besnier I, Moreau C, Savanger F, Vielle B, Emile J, Dubas F, Malthiery Y: mtDNA haplogroup J: a contributing factor of optic neuritis. Eur J Hum Genet 1999, 7:404-406 [DOI] [PubMed] [Google Scholar]
- 53.Sallusto F, Lanzavecchia A: Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and down-regulated by tumor necrosis factor-α. J Exp Med 1994, 179:1109-1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sallusto F, Cella M, Danieli C, Lanzavecchia A: Dendritic cells use macropinocytosis and the mannose receptor to concentrate macromolecules in the major histocompatibility complex class II compartment: down-regulation by cytokines and bacterial products. J Exp Med 1995, 182:389-400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Norbury CC, Chambers BJ, Prescott AR, Ljunggren HG, Watts C: Constitutive macropinocytosis allows TAP-dependent major histocompatibility complex class I presentation of exogenous soluble antigen by bone marrow-derived dendritic cells. Eur J Immunol 1997, 27:280-288 [DOI] [PubMed] [Google Scholar]
- 56.de Baey A, Lanzavecchia A: The role of aquaporins in dendritic cell macropinocytosis. J Exp Med 2000, 191:743-748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haines JL, Ter-Minassian M, Bazyk A, Gusella JF, Kim DJ, Terwedow H, Pericak-Vance MA, Rimmler JB, Haynes CS, Roses AD, Lee A, Shaner B, Menold M, Seboun E, Fitoussi RP, Gartioux C, Reyes C, Ribierre F, Gyapay G, Weissenbach J, Hauser SL, Goodkin DE, Lincoln R, Usuku K, Oksenberg JR: A complete genomic screen for multiple sclerosis underscores a role for the major histocompatibility complex. The Multiple Sclerosis Group. Nat Genet 1996, 13:469-471 [DOI] [PubMed] [Google Scholar]
- 58.Sundvall M, Jirholt J, Yang HT, Jansson L, Engström Å, Pettersson U, Holmdahl R: Identification of murine loci associated with susceptibility to chronic experimental autoimmune encephalomyelitis. Nat Genet 1995, 10:313-317 [DOI] [PubMed] [Google Scholar]
- 59.Baker D, Rosenwasser OA, O’Neill JK, Turk JL: Genetic analysis of experimental allergic encephalomyelitis in mice. J Immunol 1995, 155:4046-4051 [PubMed] [Google Scholar]
- 60.Encinas JA, Lees MB, Sobel RA, Symonowicz CJ, Greer M, Shovlin CL, Weiner HL, Seidman CE, Seidman JG, Kuchroo JK: Genetic analysis of susceptibility to experimental autoimmune encephalomyelitis in a cross between SJL/J and B10.S mice. J Immunol 1996, 157:2186-2192 [PubMed] [Google Scholar]
- 61.Butterfield RJ, Blankenhorn EP, Roper RJ, Zachary JF, Doerge RW, Teuscher C: Identification of genetic loci controlling the characteristics and severity of brain and spinal cord lesions in experimental allergic encephalomyelitis. Am J Pathol 2000, 157:637-645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Encinas JA, Lees MB, Sobel RA, Symonowicz C, Weiner HL, Seidman CE, Seidman JG, Kuchroo VK: Identification of genetic loci associated with paralysis, inflammation and weight loss in mouse experimental autoimmune encephalomyelitis. Int Immunol 2001, 13:257-264 [DOI] [PubMed] [Google Scholar]
- 63.Jirholt J, Lindqvist AK, Karlsson J, Andersson A, Holmdahl R: Identification of susceptibility genes for experimental autoimmune encephalomyelitis that overcome the effect of protective alleles at the eae2 locus. Int Immunol 2002, 14:79-85 [DOI] [PubMed] [Google Scholar]
- 64.Croxford JL, O’Neill JK, Baker D: Polygenic control of experimental allergic encephalomyelitis in Biozzi ABH and BALB/c mice. J Neuroimmunol 1997, 74:205-211 [DOI] [PubMed] [Google Scholar]
- 65.Compston A, Coles A: Multiple sclerosis. Lancet 2002, 359:1221-1231 [DOI] [PubMed] [Google Scholar]
- 66.Granum B, Gaarder PI, Eikeset A, Stensby BA, Lovik M: The adjuvant effect of particles: importance of genetic background and pre-sensitization. Int Arch Allergy Immunol 2000, 122:167-173 [DOI] [PubMed] [Google Scholar]