Abstract
In the present study, we cloned and characterized a novel actin-binding molecule, designated skeletrophin, from aggregated neuroblastoma cells. The putative amino acid sequence of human skeletrophin cDNA contained a cysteine-rich zinc-finger motif which was also found in dystrophin and five ankyrin repeats. Northern blot analysis revealed that the 3.2-kb skeletrophin mRNA was expressed in normal skeletal muscle, and to a lesser extent in heart, brain, and kidney. Specific antibody was prepared to human skeletrophin peptide, and a single protein band with an approximate molecular weight of 70 kd was detected in tissue extracts by immunoblotting using the antibody. To better understand the biological properties of skeletrophin, we used a yeast two-hybrid system to screen for molecules interacting with skeletrophin and found that skeletrophin bound to actin monomer. Co-immunoprecipitation experiments also demonstrated that skeletrophin was able to bind to actin monomer. Fluorescence in situ hybridization mapped the skeletrophin gene on human chromosome 1p36.2–36.3, in which putative tumor suppressor genes for malignant melanoma have been postulated to exist. We therefore immunohistochemically stained benign nevi and malignant melanoma tissues. Notably, 23 of 25 benign nevi expressed skeletrophin in cytoplasm, but 18 of 38 cases of primary skin melanoma appeared to lack skeletrophin expression. Treatment with a demethylating agent, 5′-aza-2-deoxycytidine, restored skeletrophin expression in cultured Mewo melanoma cells. The present findings suggest that skeletrophin may be a novel actin-binding cytoskeleton-related molecule, expression of which is silenced in a considerable number of melanoma specimens.
We previously showed that neuroblastoma cells transfected with truncated human SWI1 which contain AT-rich interactive domain (ARID) but lack one of two putative glucocorticoid-receptor binding domains, aggregated tightly and exhibited increased cell-cell adhesion. 1 While exploring for molecules differently expressed in original and aggregated neuroblastoma cells, we isolated several candidate cDNA fragments by differential display of mRNA reverse transcription-polymerase chain reaction (DDRT-PCR). We partially sequenced and reported three representative cDNAs in a previous report 1 ; however, subsequent study to isolate and sequence the entire cDNAs revealed that none of them encode a cytoskeleton-related protein. Therefore, we further isolated other genes, whose expression was overexpressed in B120-transfected neuroblastoma cells, from bands of DDRT-PCR analysis. In the present study, we cloned and characterized a putative cytoskeleton-related molecule which was strongly expressed in truncated human SWI1-transfected neuroblastoma cells but less expressed in original untransfected cells.
Materials and Methods
Cells, Cell Culture, Expression Plasmids, and Transfection
SCCH-26, a neuroblastoma cell line, and Mewo, a melanoma cell line, were obtained from the Japan Health Science Research Resources Bank (Osaka, Japan) and cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco Life Technologies, Grand Island, NY) containing 10% heat-inactivated fetal bovine serum (FBS) and 50 μg/ml gentamicin (Gibco Life Technologies). COS-7 cells were also cultured in DMEM containing 10% fetal calf serum. The construction and stable transfection of a pCI-neo expression vector (Promega, Madison, WI) containing a truncated form of human SWI1 (SMARCF1; SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily f, member 1) was previously reported. 1 Notably, this truncated form, designated B120, includes a novel DNA-binding region, termed ARID, but lacks one of the two putative glucocorticoid receptor binding domains. Skeletrophin cDNA isolated from a human brain library described below was also subcloned to pCI-neo vector and transfected to SCCH-26 cells as previously reported. 1 As a control, the pCI-neo vector alone transfected cell line was also established.
Isolation and Characterization of Human cDNA Encoding Skeletrophin
Full-length cDNA was isolated from a human brain λTriplEx library (Clontech, Palo Alto, CA) using a previously isolated partial cDNA as a hybridization probe. This cDNA fragment was isolated in a previous study because it is strongly expressed in truncated human SWI-transfected neuroblastoma cells but less expressed in non-transfected cells. After cre-lox alteration of obtained λTriplEx phage to pTriplEx plasmid according to the manufacturer’s protocol, sets of nested deletions were prepared according to the method of Henikoff. 2 cDNA insert was sequenced using an Automated Laser Fluorescent ALF sequencer (Amersham Bioscience, Piscataway, NJ). The rapid amplification of cDNA end (RACE) technique was also performed using the SMRAT RACE cDNA amplification kit (Clontech).
Northern Blot Analysis
Multiple-tissue Northern blots (Clontech) were screened with an approximately 600-b cDNA fragment spanning the N terminus of the coding sequence for skeletrophin as previously described. 3 Notably, this cDNA region did not encode ankyrin-repeats. The probe was radiolabeled with [α-32P]dCTP using random-primed labeling kits (Roche GmbH, Mannheim, Germany). Membranes were washed in 2X standard saline citrate (SSC)/0.1% sodium dodecyl sulfate (SDS) at 55°C, and then with 0.1X SSC/0.1% SDS at 55°C. Radioactivities were visualized using a Fujix BAS 2000 Image Analyzer (Fujifilm, Tokyo, Japan).
Antibodies and Western Immunoblotting
Details of the procedures of preparation of conventional rabbit antibody to synthesized peptides were described previously. 4 In brief, rabbits were immunized with human skeletrophin peptide, LDLLRRRPEQVDTKNQGR, conjugated with keyhole limpet hemocyanin. After the fifth immunization, antibody was purified by affinity-chromatography using the same peptide. Control normal rabbit IgG was also prepared in our laboratory. COS-7 cells were transfected with pCI-neo expression vector containing the human skeletrophin cDNA (pCI-neo-skeletrophin) or vector alone by the DEAE-dextran method as previously described. 3 Seventy-two hours later, the transfected COS-7 cells were harvested and solubulized with a lysis buffer (20mmol/L Tris-HCL, 0.01% SDS, 0.5% Nonidet P-40, 1mmol/L ethylenediaminetetraacetate, 1 mmol/L phenylmethyl sulfonyl fluoride (PMSF), 5 μg/ml aprotinin). Extracted protein mixtures from liver, skeletal muscle, thymus, lung, heart, and brain were purchased from Clontech. Western immunoblotting was carried out according to the methods of Towbin and colleagues. 5 Briefly, equal amounts of proteins were electrophoresed on SDS polyacrylamide gel electrophoresis gels and electroblotted to a nitrocellulose membrane. After blocking with FBS, membranes were incubated with affinity-purified rabbit antibody to human skeletrophin peptide or normal rabbit IgG. Membranes were then incubated with a second goat antibody to rabbit IgG labeled with horseradish peroxidase and developed with diaminobenzidine. Notably, the second antibody was preadsorbed with human IgG.
Yeast Two-Hybrid Library Screening
A yeast two-hybrid cDNA library derived from human skeletal muscle was screened for interacting proteins using the pretransformed Matchmaker two-hybrid system as described by the distributor (Clontech). Briefly, skeletrophin cDNA (nucleotides 754-3250) was inserted in-frame downstream of the Gal-4 DNA binding domain in the pGBKT7 bait vector (Clontech). Subsequently, AH 109 cells, MATa strain, were transformed with the pGBKT7-skeletrophin plasmid. A pretransformed library constructed in Y187 yeast cells, MATα strain, with pACT2 Gal-4 activation domain vector was mated with the transformed AH 109 cells. Mating yeast mixtures were then spread on synthetic dropout/-Ade/-His/-Leu/-Trp/+3-amino-1,2,4-triazole/+X-α-GAL plates and incubated at 30°C until colonies appeared. Colonies able to grow on the minimal plates and exhibit α-galactosidase activity were isolated after 8 days of incubation. The inserted cDNAs were amplified by PCR and sequenced using an Automated Laser Fluorescent ALF sequencer (Amersham Bioscience)
Direct Binding of Skeletrophin and α-Actin in Vitro
To test whether the interaction between skeletrophin and α-actin molecule is direct, and to confirm the results of the yeast two-hybrid assay, in vitro transcription/translation followed by co-immunoprecipitation assay was performed. A cDNA which encode α-actin labeled with hemagglutinin epitope (HA) was obtained from yeast two-hybrid screening and subcloned to pGEM-T Easy Vector (Promega). A skeletrophin cDNA was also subcloned to pGEM-T Easy Vector. Biotinylated α-actin labeled with HA and skeletrophin proteins were produced with TNT Coupled Reticulocyte Lysate System and Transcend tRNA as described by the distributor (Promega). Biotinylated proteins were mixed with a lysis buffer (20 mmol/L Hepes-KOH, pH 7.6, 100 mmol/L KCL, 0.2 mmol/L ATP, 0.5 mmol/L dithiothreitol (DTT), 0.2 mmol/L CaCl2, 0.2 mmol/L MgCl2, 1 mmol/L PMSF, 0.2% Nonidet P-40) and incubated for 2 hours at 4°C. Subsequently, immunoprecipitation with rabbit antibodies was performed as previously described. 6 Briefly, either 5 μg of rabbit anti-HA tag polyclonal antibody (Clontech) or control rabbit IgG was added to protein mixtures and reincubated for 3 hours at 4°C. After this step, suspended Protein A Sepharose (Amersham) was added and shaken at 4°C overnight. Protein A Sepharose was preincubated with 2% bovine serum albumin to prevent non-specific protein binding. The next day, the pellet was washed three times with a lysis buffer and resuspended in 2X Tris-glycine sample buffer with 50 mmol/L DTT. The samples were heated to 95°C for 3 minutes, subjected to SDS-PAGE, transferred to polyvinylidene difluoride membranes. Then, membranes were incubated with Streptavidin-alkaline phosphatase (Promega) according to the manufacturer’s protocol. Finally, proteins were visualized with Western Blue substrate (Promega).
Fluorescence in Situ Hybridization (FISH)
Lymphocytes isolated from human blood were cultured in α-minimal essential medium (MEM) supplemented with 10% FBS and phytohemagglutinin (PHA) at 37°C for 68 to 72 hours. The lymphocyte cultures were treated with BrdU (0.18 mg/ml, ς-Aldrich, St Louis, MO) to synchronize the cell population. The synchronized cells were washed three times with serum-free medium to release the block and recultured at 37°C for 6 hours in a α-MEM with thymidine (2.5 mg/ml; ς-Aldrich). Cells were harvested, fixed, and air-dried, and slides were prepared using standard procedures including hypotonic treatment,. We isolated approximately 6 kb genomic DNA which included exon 2 and exon 3 using Clontech GenomeWalker Kits. Briefly, human genomic DNAs were digested with DraI restriction enzyme and ligated with adaptor. The fragment was amplified by two round PCR (nested PCR) with adaptor-specific primers and human skeletrophin primers. The primers 5′-agcagtcacagatgatgttggggtgcc-3′ and 5′-ttgtcgtacagcagcaggtcgtgcgcg-3′ were used for initial and second-round PCR, respectively. We confirmed the sequence of amplified PCR products and found that it contains exon 2 and exon 3. This DNA probe was biotinylated with dATP using the Gibco BRL BioNick labeling kit (15°C, 1 hour). 7 The procedure for FISH detection was performed as described by Heng et al 7 and Heng and Tsui. 8 Briefly, slides were baked at 55°C for 1 hour. After RNase A treatment, the slides were denatured in 70% formamide in 2X SSC for 2 minutes at 70°C followed by dehydration with ethanol. Probes were denatured at 75°C for 5 minutes. in a hybridization mix consisting of 50% formamide and 10% dextran sulfate. Probes were loaded on the denatured chromosomal slides. After overnight hybridization, slides were washed and detected, as well as amplified using a published method. 8 FISH signals and the DAPI banding pattern were recorded separately. Images were captured and combined by a CCD camera, and assignment of the FISH mapping data to chromosomal bands was performed by superimposing FISH signals with DAPI-banded chromosomes. 8
Immunohistochemical Staining
Details of the procedures used for immunohistochemical staining were described previously. 9 In the present study, we examined archival skin tissue specimens comprising a spectrum of various melanomas and benign nevi. Normal skeletal muscle in surgically-resected margin were also examined. All specimens were surgically resected, fixed in 10% buffered formalin, and paraffin-embedded. Deparaffinized sections were placed first in plastic Coplin jars filled with preheated citrate buffer (pH 6.0) and microwave-irradiated for 5 minutes at 650 W. The slides were incubated for 30 minutes in FBS. The tissues were then immunostained with affinity-purified anti-human skeletrophin peptide or control antibody using a streptavidin-biotin complex peroxidase kit (DAKO LSAB Kit; Dakopatts, Kyoto, Japan). The procedures were performed according to the manufacturer’s protocol. In several experiments, anti-skeletrophin antibody was preadsorbed with the immunized peptide. Finally, reaction was developed with 3-amino-9-ethylcarbazole (ς-Aldrich). The tissue sections were couterstained with hematoxylin and semiquantitatively scored as weakly positive (+; 5> to 20%), moderately positive (++; >20 to 80%), or markedly positive (+++; >80%).
Aza-CdR Treatment and RT-PCR
Mewo melanoma cells were incubated in culture medium with and without 5-Aza-2′-deoxycytidine (Aza-CdR) (ς-Aldrich) at a concentration of 0, 1, or 2 μg/ml. Cells were harvested at the end of the fourth day for extraction of total RNA using RNA-zol B (Biotex Laboratory, Houston, TX). Total RNA was also extracted from SCCH-26 cells which were transfected with pCI-neo-truncated human SWI1 expression vector. We used total RNA as substrate for cDNA synthesis with an RT-PCR kit (Life Technologies, Gaithersburg, MD) as previously described. 9 The cDNA synthesis and subsequent PCR were performed according to the manufacturer’s instructions. The primer sets used in this study were: sense 5′-GCAGCCCCGCCCAACATGGACC-3′ and antisense 5′-AGCAGTCACAGATGATGTTGGGGTGCC-3′ for skeletrophin, and sense 5′-TCCACCACCCTGTTGCTGTA-3′ and antisense 5′-ACCACAGTCCATGCCATCAC-3′ for G3PDH.
Results
Cloning and Sequencing of Skeletrophin cDNA
An approximately 100-bp partial cDNA fragment, which was overexpressed in truncated human SWI1 (B120)-transfected IMR-32 neuroblastoma cells, was previously isolated by DDRT-PCR. In the present study, we isolated a clone with the longest insert of 3242 bp from a human adult brain λ phage library using the partial cDNA fragment. It was entirely sequenced and appeared to encode 1013 amino acids, and designated skeletrophin (Figure 1) ▶ . 5′-RACE exhibited an additional 8-bp fragment of 5′ nocoding region without any prolongation of the open reading frame. The deduced amino acid sequence of skeletrophin cDNA contained a Zinc binding domain seen in dystrophin and five ankyrin repeats, as shown in Figure 1 ▶ . The entire nucleotide sequence of skeletrophin can be seen in GenBank nucleotide sequence databases under the accession number AB074480.
Figure 1.

Deduced amino acid sequence of human skeletrophin cDNA. The cysteine-rich zinc-binding ZZ domain and five ankyrin repeats are boxed and underlined, respectively. The immunizing peptide used is shown by dots on amino acids.
Northern Blot Analysis, RT-PCR, and Transfection of Skeletrophin to Neuroblastoma Cells
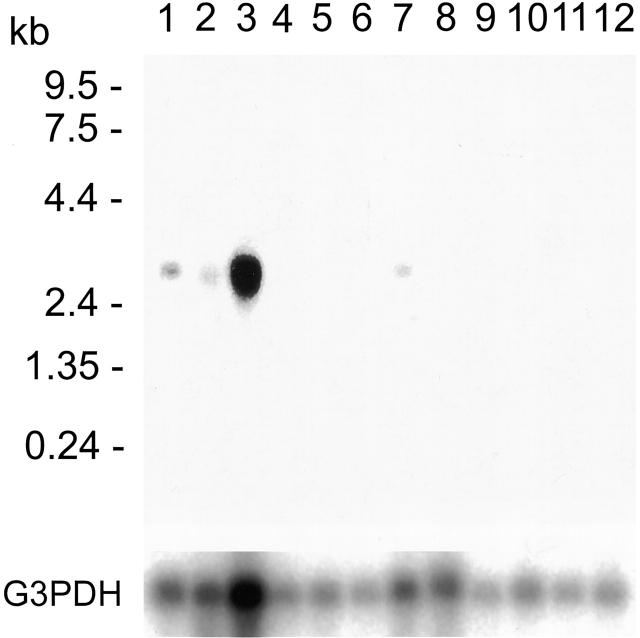
As demonstrated in Figure 2 ▶ , an approximately 3.2-kb band was detected in polyA+ RNA from the brain, heart, skeletal muscle, and kidney. RT-PCR confirmed that exogenous expression of truncated form of human SWI1 (B120) induced skeletrophin expression in SCCH-26 neuroblastoma cells (Figure 3A) ▶ . The transfection of both skeletrophin and B120 cDNAs with expression vector into a neuroblastoma cell line, SCCH-26, caused cell aggregation. (Figure 3, B–D) ▶
Figure 2.
mRNA tissue distribution of skeletrophin. A Northern blot of mRNA isolated from human tissues was probed with cDNA containing the coding region of human skeletrophin. RNA was obtained from brain (lane 1), heart (lane 2), skeletal muscle (lane 3), colon (no mucosa) (lane 4), thymus (lane 5), spleen (lane 6), kidney (lane 7), liver (lane 8), small intestine (lane 9), placenta (lane 10), lung (lane 11), and peripheral blood leukocytes (lane 12). The blot was rehybridized with G3PDH cDNA as a control after being stripped with the probe (lower panel).
Figure 3.
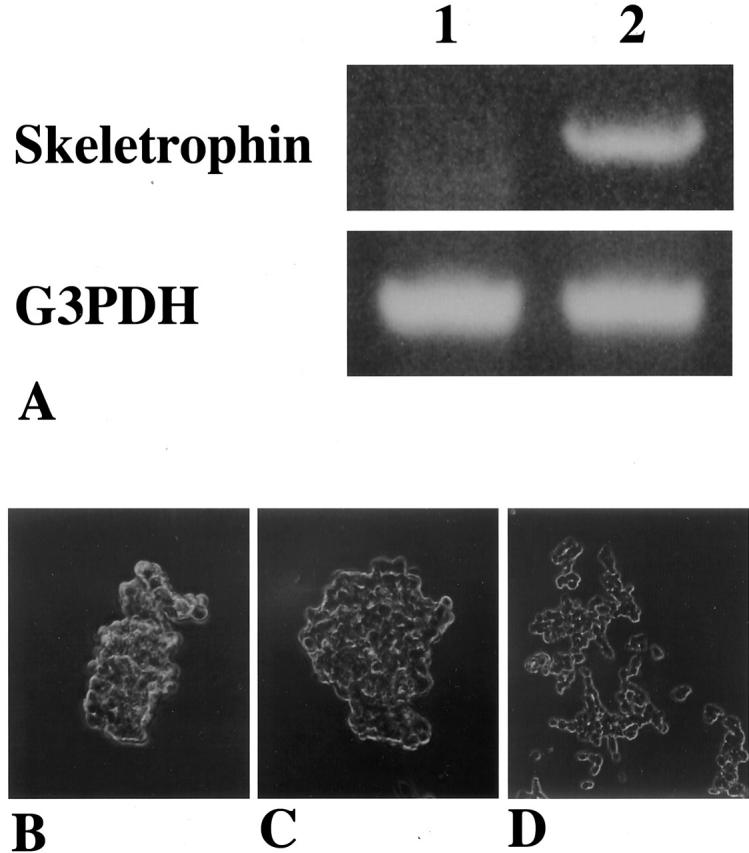
Truncated form of human SWI1-induced skeletrophin in SCCH-26 neuroblastoma cells and transfection of skeletrophin and B120 cDNA with expression vector into neuroblastoma cells RT-PCR using total RNA isolated from SCCH-26 cells which had been transfected with pCI-neo vector containing truncated human SWI1 (B120) or vector alone. Skeletrophin mRNA was not observed in SCCH-26 cells transfected with pCI-neo vector alone (lane 1), but was observed in SCCH-26 cells transfected with vector containing truncated human SWI1 cDNA (lane 2). Note the even intensity of internal control bands of G3PDH. Both B-120 and skeletrophin-transfected SCCH-26 neuroblastoma cells adhered to each other and formed small clusters (B and C, respectively). In contrast, no significant change was observed in control plasmid alone transfected SCCH-26 cells (D).
Western Immunoblotting
In the present study, we prepared affinity-purified rabbit anti-skeletrophin peptide for Western blotting. As demonstrated in Figure 4A, a ▶ major band with an approximately molecular weight of 70 kd was detected in extracted protein mixtures from human skeletal muscle, heart, and brain but not those from liver, thymus, or lung with anti-skeletrophin peptide antibody. A single band with approximate molecular weight of 70 kd was also detected in lysates of COS-7 cells, which was transfected with pCI-neo-skeletrophin but not in vector alone (Figure 4B) ▶ . No significant band obtained with control rabbit antibody.
Figure 4.
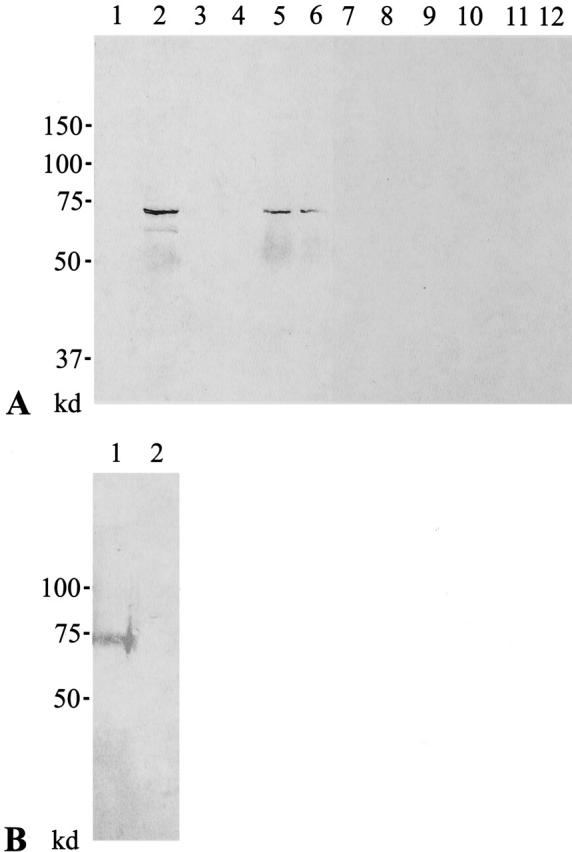
Western immunoblotting. A: Extracts of human liver (lanes 1 and 7), skeletal muscle (lanes 2 and 8), thymus (lanes 3 and 9), lung (lanes 4 and 10), heart (lanes 5 and 11), and brain (lanes 6 and 12) were subjected to SDS-PAGE and transferred to membrane, and reacted with affinity-purified rabbit antibody to human skeletrophin peptide (lanes 1 to 6) or control antibody (lanes 7 to 11). A band with an approximate molecular weight of 70 kd was detected in human skeletal muscle, heart, and brain. In contrast, any significant band was observed with control antibody. B: Lysates from COS-7 cells, which were transfected with pCI-neo-skeletrophin (lane 1) or vector alone (lane 2), were subjected to SDS-PAGE, transferred to membrane, and reacted with anti-skeletrophin antibody. A major band with approximate molecular weight of 70 kd was detected in lysates of COS-7 cells transfected with pCI-neo-skeletrophin but not in those of COS-7 cells transfected with vector alone.
Yeast Two-Hybrid Screening and Co-Immunoprecipitation Assay
To screen for proteins that interact with skeletrophin, yeast two-hybrid screens were performed with a bait plasmid containing the entire human skeletrophin coding sequence. Approximately 3.5 × 10 6 independent transformant clones were screened from the human skeletal muscle cDNA libraries. This screen resulted in the isolation of 3 clones, which differ in length of the inserted cDNA. Sequencing of these clones revealed that all three were derived from the same gene, α-actin; however, they are in-frame ligated to Gal-4 activation domain at different 5′-sites (as a result, about 2.5, 2.4, and 1.6 kd truncated α-actin molecules appeared to bind skeletrophin). To verify the results of the two-hybrid screen, in vitro co-immunoprecipitation experiments were performed with HA-tagged 2.5-kd α-actin and untagged full-length skeletrophin proteins.
Proteins were expressed and biotinylated with the TNT quick-coupled transcription/translation system and Transcend tRNA (Promega). The anti-HA tag antibody co-immunoprecipitated skeletrophin protein in the mixture of HA-tagged α-actin and skeletrophin (Figure 5 ▶ , lane 1). In contrast, control antibody immunoprecipitated neither α-actin nor skeletrophin (lane 2).
Figure 5.
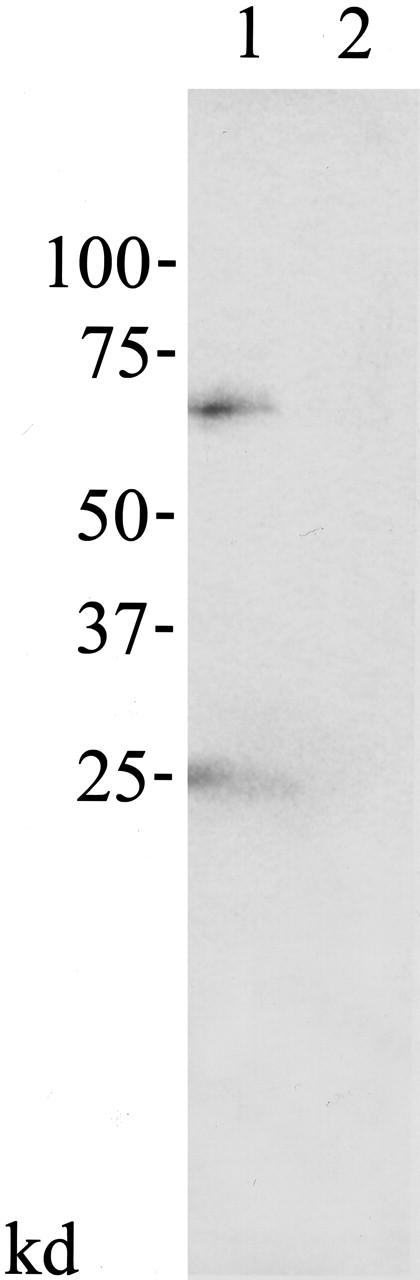
Co-immunoprecipitation of skeletrophin and actin monomer. The in vitro direct binding between skeletrophin and α-actin was shown by co-immunoprecipitation assay. The biotinylated HA-tagged α-actin and untagged skeletrophin were expressed by in vitro transcription/translation and mixed together. α-actin complexes were isolated by immunopreciptation with rabbit antibody to HA epitope (lane 1), subsequently separated in SDS/PAGE. Immunoprecipitation using control rabbit antibody was subjected in parallel (lane 2). After immunoblotting, biotinylated proteins which was co-immunoprecipitated with α-actin molecule was visualized with streptoavidin-alkaline phosphatase and substrate buffer. Note the 70-kd skeletrophin protein band in lane 1. A 25-kd HA-tagged α-actin protein band was also observed in lane 1.
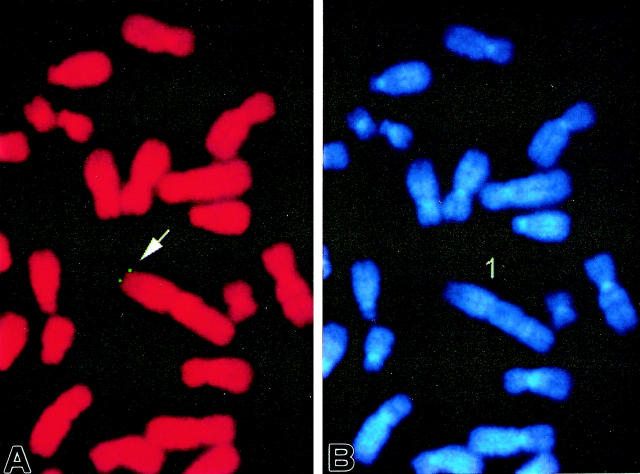
FISH Mapping
Under the conditions used, the FISH detection efficiency was approximately 48% for this probe at this locus (among 100 checked mitotic figures, 48 exhibited positive hybridization signals). Since DAPI banding was used to identify the specific chromosome, assignment of signals from the probe and the short arm of chromosome 1 was possible. Detailed positions were further determined based on a summary from 10 photos (Figure 6) ▶ . This probe was thus mapped to human chromosome 1, region p36.2–36.3.
Figure 6.
Example of FISH mapping of probe. A: FISH signals on the chromosome; B shows the same mitotic figure stained with DAPI to identify chromosome 1.
Immunohistochemical Staining
Representative staining results are demonstrated in Figure 7 ▶ . Normal skeletal muscle was immunohistochemically stained with anti-skeletrophin antibody. As demonstrated in Figure 7, A and B ▶ , perisurface cytoplasm of skeletal muscle was focally stained with antibody to skeletrophin. Preadsorption of antibody with immunized skeletrophin peptide eliminated this staining (Figure 7C) ▶ . Twenty-three of 25 benign nevi specimens were immunohistochemically stained with anti-skeletrophin antibody (Figure 7, D and E) ▶ . Preadsorption of antibody with the peptide again diminished staining in benign nevi (Figure 7F) ▶ . Skeletrophin expression was also observed in 20 of 38 malignant melanoma specimens (Figure 7G) ▶ , whereas skeletrophin was not expressed in 18 of 38 malignant melanoma tissues (Figure 7, H and I) ▶ . The results of immunohistochemical staining are summarized in Table 1 ▶ .
Figure 7.
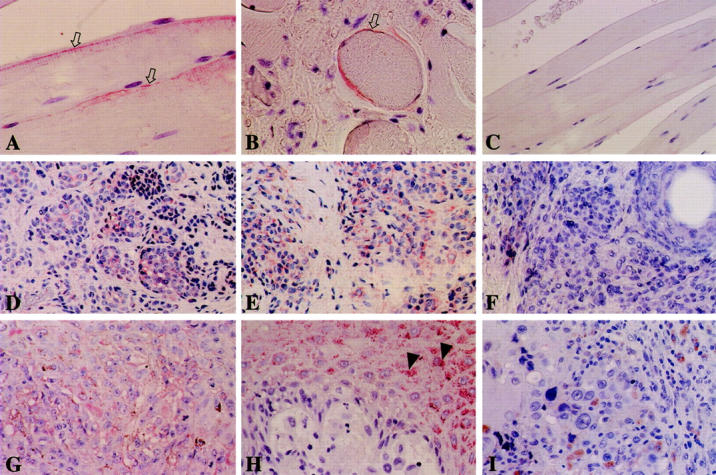
Immunohistochemical staining of benign nevi and malignant melanoma. Representative skeletrophin expression in skeletal muscle (A and B), benign nevi (D and E), and malignant melanoma (G, H, and I) is demonstrated. Perisurface membrane of skeletal muscle was focally stained with anti-skeletrophin antibody (A and B, white arrow). Most tumor cells in 23 of 25 benign nevi were stained with anti-skeletrophin antibody (D and E). Preadsorption of antibody with immunized peptide diminished staining in skeletal muscle (C) and benign nevi (F). Skeletrophin expression was observed in only 20 of 38 malignant melanoma specimens (G). Eighteen of 38 malignant melanoma specimens did not express skeletrophin (H and I). Note the skeletrophin expression in non-cancerous supraepithelial epidermal cells (H, arrowhead) in malignant melanoma tissue specimens. Original magnification: ×640 (A, B), ×200 (C, D, E, and F), ×400 (G, H, and I).
Table 1.
Immunohistochemical Staining of Benign Nevi and Malignant Melanoma Specimens with Anti-Skeletrophin Antibody
| Lesion | Skeletrophin staining intensity | ||||
|---|---|---|---|---|---|
| n | − | + | ++ | +++ | |
| Benign nevi | 25 | 2 | 0 | 8 | 15 |
| Primary melanoma | |||||
| (superficial spreading melanoma) | 8 | 5 | 0 | 3 | 0 |
| (other than superficial spreading melanoma) | 30 | 13 | 8 | 5 | 4 |
The relative intensity of staining of tissues with anti-skeletrophin antibody was semiquantitatively scored as negative (−; <5%), weakly positive (+; 5> to 20%), moderately positive (++; >20 to 80%), or markedly positive (+++; >80%). n, number of specimens tested. Data are shown as the number of specimens negative or positive for skeletrophin staining. Benign nevi include both congenital and acquired types.
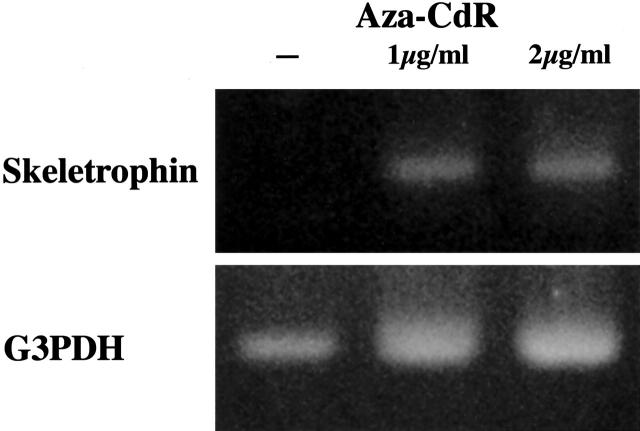
Aza-CdR Restored Skeletrophin Expression in Melanoma Cells
A melanoma cell line, Mewo, was cultured with or without the demethylating agent Aza-CdR. Incubation of the cells with 1 and 2 μg/ml Aza-CdR for 4 days restored skeletrophin mRNA expression in Mewo cells (Figure 8) ▶ .
Figure 8.
Restoration of skeletrophin expression after Aza-CdR treatment. Cultured Mewo human malignant melanoma cells were incubated with or without Aza-CdR (final concentration 1 or 2 μg/ml) for 4 days. Restoration of skeletrophin expression was observed in Mewo cells by RT-PCR. Note the even intensity of internal control bands of G3PDH.
Discussion
Previous studies revealed that neuroblastoma cells, in which exogenous truncated human SWI1 molecule (B120) was overexpressed, aggregated tightly and exhibited increased cell-cell adhesion. 1 The DNA binding domain ARID is conserved in the truncated human SWI1; however, truncated human SWI1 lacked the C-terminal LXXLL motif, which has been shown to be critical for the binding of a variety of nuclear proteins to liganded nuclear hormone receptors. 10 Recent studies reveal that mutation of other SWI/SNF complexes subunits act as dominant negatives and have been shown to inhibit gene activation events that normally require SWI/SNF function. 11 These recent reports suggested to us that overexpression of the truncated human SWI1 might also act as a dominant negative, cause dysfunction of SWI/SNF complexes, and alter cell-cell adhesion of neuroblastoma cells.
In the present study we characterized a putative cytoskeleton-related molecule, termed skeletrophin. IMR-32 neuroblastoma cells, which was used for DDRT-PCR, weakly expressed skeletrophin. To facilitate the subsequent analysis, we screened several neuroblastoma cells, in which we could not find any detectable skeletrophin expression even by the sensitive RT-PCR. SCCH-26 neuroblastoma cells did not express skeletrophin and subsequent RT-PCR confirmed that skeletrophin expression was induced in pCI-neo-B120 transfected-SCCH-26 cells (Figure 3A) ▶ . We confirmed skeletrophin expression in pCI-neo-B120-transfected SCCH-26 cells but not in control SCCH-26 cells by Western immunoblotting (data not shown). Notably, both B120-transfected and skeletrophin-transfected SCCH-26 neuroblastoma cells also adhered to each other and aggregated as previously found in B120-transfected neuroblastoma cells (Figure 3, B–D) ▶ .
Insufficient function of SWI/SNF complexes plays a role in the tumorigenecity in various malignant tumors. For example, synovial sarcomas are typified by a unique chromosomal translocation t(X;18)(p11.2;q11.2) that results in fusion of the SYT gene on chromosome 18 with the SSX1 or SSX2 gene in Xp11.2 and, consequently, production of chimeric SYT-SSX proteins. 12,13 SYT is a ubiquitously expressed protein with a QPGY domain, whereas the SSX proteins carry KRAB-like domains and are almost exclusively expressed in the testis. 14,15 Human SWI1 also contained many QPGY domains and thus may interact with SYT and SYT-SSX proteins. 16 Since the SYT-SSX and SYT proteins are confined to, in particular, nuclear speckles in which hBRM protein is present, altered localization of the SYT-SSX proteins and associated proteins may account for the pathogenesis of synovial sarcoma. Very interestingly, recent studies also revealed that SSX1, 2, 4, and 5 proteins were heterogeneously expressed in human melanoma lesions. 17 Moreover, SSX2 protein may act as a tumor-associated antigen (also designated HOM-MEL-40), eliciting humoral immune responses in a subset of melanoma patients. 18
The present FISH examination mapped the skeletrophin gene to human chromosome 1p36.2–36.3 (Figure 6) ▶ . Recently, human genome projects have also identified human skeletrophin gene on chromosome 1p36.2–36.3. Interestingly, murine skeletrophin was also mapped to murine chromosome 4 by murine genome projects. Comparative genome sequence analysis has demonstrated conserved synteny between the murine genes on chromosome 4 and human genes on 1p36. 19 These findings clearly showed that the human skeletrophin gene is located in chromosome 1p36.2–36.3, where putative tumor suppressor genes for malignant melanoma have long been postulated to exist. 20 Together with the finding of aberrant expression of SSX in melanomas, these findings encouraged us to examine skeletrophin expression in malignant melanoma specimens. An affinity-purified rabbit antibody to human skeletrophin peptide was generated to examine skeletrophin expression in archival tissue sections. Before immunization, we isolated a murine skeletrophin cDNA and determined the amino acid homology between human and mouse skeletrophin (the murine homologue of skeletrophin can be found in GenBank under the accession number AB072336). There is a high degree of amino acid homology for skeletrophin between human and mouse (deduced amino acid homology was approximately 80%). We synthesized a peptide, which differed somewhat between human and mouse. Prepared antibody to human skeletrophin peptide located on amino acids 506–524 (Figure 1) ▶ detected a 70-kd protein band in proteins extracted from human skeletal muscle, heart, and brain (Figure 4A) ▶ . We transfected COS-7 cells with pCI-neo-skeletrophin expression vector and performed immunoblotting. The major 70-kd protein band was also detected in COS-7 cells which were transfected with pCI-neo-skeletrophin vector for 72 hours but not those transfected with pCI-neo vector alone (Figure 4B) ▶ . As demonstrated in Figure 4 ▶ , lane 2, another minor bands with an approximate molecular weight of 65-kd was detected in skeletal muscle lysates. To now, we isolated three alternative splicing forms of skeletrophin and thought that this 65-kd band represented a splicing form in skeletal muscle (manuscript in preparation).
We carried out immunohistochemical staining using archival tissue sections of benign nevi and melanomas. Very interestingly, 20 of 38 malignant melanoma expressed skeletrophin, whereas 23 of 25 benign nevi expressed it. Lack of skeletrophin expression was observed in 18 of 38 (47%) cutaneous malignant melanoma specimens. Skeletrophin expression was not related to pathological subclassification, radial growth pattern, vertical growth pattern, or depth of invasion. To elucidate the differences between skeletrophin-positive and -negative melanoma cells, we are examining the biological behavior of malignant melanoma cells, in which expression of skeletrophin was reconstructed, especially in terms of cell-cell attachment. Interestingly, none of the two Spitz nevus expressed skeletrophin in the present study. Spitz nevus (also known as the spindle and epithelioid nevus) usually grows progressively for several months from previously normal skin but almost always has a benign clinical course. Spitz nevus is rare and it needs a further study to conclude whether none of the Spitz nevus expresses skeletrophin. Cytogenetic studies of human cutaneous melanocytic tumors have demonstrated a variety of nonrandom abnormalities on chromosomes 1p36, 6q27, 7, and 9p21. 20-22 These chromosome analyses indicate that human cutaneous malignant melanomas are very heterogeneous and suggested that several molecules may participate in tumorigenesis and progression of human melanomas. In contrast, clear cell sarcoma, a malignant melanoma of soft parts, almost always exhibits chromosome translocation involving chromosome 22q12. 23
We further examined whether skeletrophin expression was silenced in cultured cutaneous melanoma cells. One malignant melanoma cell line, Mewo, of four malignant melanoma cell lines tested appeared to lack skeletrophin expression on both immunohistochemical staining and RT-PCR (data not shown). Recent studies revealed that aberrant methylation of 5′ gene promoter regions associated with gene silencing is an epigenetic phenomenon observed in many types of cancer. 24 We found that skeletrophin expression was restored by Aza-CdR demethylating agent treatment in skeletrophin-negative Mewo cells (Figure 8) ▶ . This result indicated that skeletrophin expression was silenced by aberrant methylation in Mewo cells. Taken together with the results of the present immunohistochemical study and chromosome localization of the skeletrophin gene, it suggests that skeletrophin could be a candidate tumor suppressor factor for a considerable number of cutaneous malignant melanomas. Since skeletrophin expression was not observed in 5 of 8 superficial spreading melanoma specimens tested, we speculate that silencing of skeletrophin expression may occur in an early step of carcinogenesis of skeletrophin-negative melanoma cells.
What is the physiological role of skeletrophin and how might lack of skeletrophin expression be related to tumorigenicity of melanoma cells? Skeletrophin cDNAs encoded proteins which had a very interesting molecular structure (Figure 1) ▶ , since the deduced amino acid structure of skeletrophin lacked a hydrophobic region including a signal peptide or transmembrane region and was entirely hydrophilic. It also lacks a nuclear transport motif. Skeletrophin is thus most likely a cytoplasmic protein, as we confirmed by immunohistochemical staining. A novel zinc finger domain found in dystrophin is present in the first half of skeletrophin and five ankyrin repeats are found in relatively the C-terminal region of skeletrophin. Ponting et al 25 named this novel zinc finger motif the ZZ domain, partly by reference to the WW domain 26 and partly for its potential role in binding two Zn2+ ions. A ZZ motif was also found in utrophin, a close homologue of dystrophin. 27 Dystrophin is the gene responsible for Duchenne muscular dystrophy; 28,29 however, in-frame deletions within the non-functional region may cause Becker muscular dystrophy, the phenotype of which is much milder than that of Duchenne muscular dystrophy. 30 In contrast, mutations within the ZZ domain almost invariably result in severe Duchenne muscular dystrophy and appear to bind calmodulin. 31 Therefore, the ZZ domain found in skeletrophin may bind calmodulin, as reported for dystrophin. Notably, this ZZ domain was also well-conserved in murine skeletrophin. Dystrophin is a cytoskeletal protein distributed throughout the sarcolemma in muscle, and the dystrophin-glycoprotein complex has been found to physically stabilize the sarcolemmal membrane from shear stresses imposed during eccentric muscle contraction. 32,33 Notably, skeletrophin as characterized in the present study has five ankyrin repeats in the C-terminal region (Figure 1) ▶ . Ankyrin repeats are a conspicuous feature of ankyrin, and the motifs in erythrocyte ankyrin are the binding sites for several integral membrane proteins to form bridges between the cytoskeleton and membrane components. 34
Two-hybrid yeast assay followed by co-immunoprecipitation experiments demonstrated that skeletrophin could bind α-actin monomer (Figure 5) ▶ . The actin cytoskeleton is essential for diverse cellular processes such as morphogenesis, motility, endocytosis, and cell division. The structure and dynamics of actin filaments are regulated by a large number of actin-binding proteins which interact with actin filaments. 35 In addition, overexpression of skeletrophin molecule aggregated neuroblastoma cells as previously observed in B120-transfected neuroblastoma cells (Figure 3) ▶ . These facts together with the molecular structure of skeletrophin suggested that skeletrophin probably plays a role in cytoskeleton regulation and increases cell-cell attachment. Although further study is needed to elucidate whether skeletrophin can bind actin filament as well as actin monomer, lack of skeletrophin could play a role in aberrant regulation of the cytoskeleton and be related to the malignant phenotype of melanoma cells.
Acknowledgments
We thank the Japan Health Science Research Resources Bank (Osaka, Japan) for the gift of human melanoma cells. We are grateful to Drs. Nicholas J. Skelton (Department of Protein Engineering, Genentech Inc., CA), Katoh Hiroyuki (National Institute of Infectious Diseases, Japan), and Michael J. Atkinson (Institute für Pathologie, GSF-Forschungszentrum, Germany) for helpful suggestions and encouragement. We thank Yamaguchi Takuya, Nakamura Naoyo (Department of Pathology, Kochi Medical School, Japan), and Rumi Matumura (Division of Molecular Biology, Kochi Medical School, Japan) for skillful technique.
Footnotes
Address reprint requests to Tamotsu Takeuchi, M.D., Department of Pathology, Kochi Medical School, Nankoku, Japan 783-8505. E-mail: takeutit@med.kochi-ms.ac.jp.
This study was supported by grants from the Ministry of Education of Japan (12670165, 13670177), the Medical Research Fund of Kochi Medical School, and a Project grant for short study abroad from a vice chancellor of Kochi Medical School.
References
- 1.Takeuchi T, Nicole S, Misaki A, Furihata M, Iwata J, Sonobe H, Ohtsuki Y: Expression of SMARCF1, a truncated form of SWI1, in neuroblastoma. Am J Pathol 2001, 158:663-672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henikoff S: Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene 1984, 28:351-359 [DOI] [PubMed] [Google Scholar]
- 3.Takeuchi T, Kuro-o M, Miyazawa H, Ohtsuki Y, Yamamoto H: Transgenic expression of a novel thymic epithelial cell antigen stimulates aberrant development of thymocytes. J Immunol 1997, 159:726-733 [PubMed] [Google Scholar]
- 4.Takeuchi T, Misaki A, Liang S-B, Tachibana A, Hayashi N, Sonobe H, Ohtsuki Y: Expression of T-cadherin (CDH13, H-cadherin) in human brain and its characteristics as a negative growth regulator of EGF in neuroblastoma cells. J Neurochem 2000, 74:1489-1497 [DOI] [PubMed] [Google Scholar]
- 5.Towbin H, Staehelin T, Gordon J: Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 1979, 76:4350-4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takeuchi T, Barcos MP, Seon BK: Monoclonal antibody SN10 which shows a highly selective reactivity with human B leukemia-lymphoma and is effectively internalized into cells. Cancer Res 1991, 51:2985-2993 [PubMed] [Google Scholar]
- 7.Heng HHQ, Squire J, Tsui L-C: High resolution mapping of mammalian genes by in situ hybridization to free chromatin. Proc Natl Acad Sci USA 1992, 89:9509-9513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heng HHQ, Tsui L-C: Models of DAPI banding and simultaneous in situ hybridization. Chromosoma 1993, 102:325-332 [DOI] [PubMed] [Google Scholar]
- 9.Takeuchi T, Liang S-B, Matsuyoshi N, Zhou S, Miyachi T, Sonobe H, Ohtsuki Y: Loss of T-cadherin (CDH13, H-cadherin) expression in cutaneous squamous cell carcinoma. Lab Invest 2002, 82:1023-1029 [DOI] [PubMed] [Google Scholar]
- 10.Ko L, Cardona GR, Iwasaki T, Bramlett KS, Burris TP, Chin WW: Ser-884 adjacent to the LXXLL motif of coactivator TRBP defines selectivity for ERs and TRs. Mol Endocrinol 2002, 16:128-140 [DOI] [PubMed] [Google Scholar]
- 11.Reisman DN, Strobeck MW, Betz BL, Sciariotta J, Funkhouser WJ, Murchardt C, Yaniv M, Sherman LS, Knudsen ES, Weissman BE: Concomitant down-regulation of BRM and BRG1 in human tumor cell lines: differential effects on RB-mediated growth arrest vs. CD44 expression. Oncogene 2002, 21:1196-1207 [DOI] [PubMed] [Google Scholar]
- 12.Clark J, Rocques OJ, Crew AJ, Gill S, Shipley J, Chan A M-L, Gusterson BA, Cooper CS: identification of novel genes, SYT and SSX, involved in the t(X;18)(p11.2;q11.2) translocation found in human synovial sarcoma. Nat Genet 1994, 7:502-508 [DOI] [PubMed] [Google Scholar]
- 13.de Leeuw B, Balemans M, Olde Weghuis D, Seruca R, Janz M, Geraghty MT, Gilgenkrantz S, Ropers HH, Geurts van Kessel A: Molecular cloning of the synovial sarcoma-specific translocation (X;18)(p11.2;q11.2) breakpoint. Hum Mol Genet 1994, 3:745-749 [DOI] [PubMed] [Google Scholar]
- 14.Crew AJ, Clark J, Fisher C, Gill S, Grimer R, Chand A, Shipley J, Gusterson BA, Cooper CS: Fusion of SYT to two genes, SSX1 and SSX2, encoding proteins with homology to the Krüppel-associated box in human synovial sarcoma. EMBO J 1995, 14:2333-2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Güre AO, Türeci O, Sahin U, Tsang S, Scanlan MJ, Jäger E, Knuth A, Pfreundschuh M, Old LJ, Chen Y-T: SSX: a multigene family with several members transcribed in normal testis and human cancer. Int J Cancer 1997, 72:965-971 [DOI] [PubMed] [Google Scholar]
- 16.Kato H, Tjernberg A, Zhang W, Krutchinsky AN, An W, Takeuchi T, Ohtsuki Y, Sugano S, Chaii BT, Roeder RG: SYT associates with human SNF/SWI complexes and the C-terminal region of its fusion partner SSX1 targets histones. J Biol Chem 2002, 277:5498-5505 [DOI] [PubMed] [Google Scholar]
- 17.dos Santos NR, Torensma R, de Vries TJ, Schreurs MWJ, de Bruijn DRH, Kater-Baats E, Ruiter DJ, Adema GJ, van Muijen GNP, van Kessel AG: Heterogeneous expression of the SSX cancer/testis antigens in human melanoma lesions and cell lines. Cancer Res 2000, 60:1654-1662 [PubMed] [Google Scholar]
- 18.Türeci O, Sahin U, Schobert I, Koslowski M, Schmitt H, Schild H-J, Stenner F, Seitz G, Rammensee G, Pfreundschuh M: The SSX-2 gene, which is involved in the t(X;18) translocation of synovial sarcomas, codes for the human tumor antigen HOM-MEL-40. Cancer Res 1996, 56:4766-4772 [PubMed] [Google Scholar]
- 19.Shapiro DN, Sublett JE, Li B, Valentine MB, Morris SW, Noll M: The gene for PAX7, a member of the paired-box-containing genes, is localized on human chromosome arm 1p36. Genomics 1993, 17:767-769 [DOI] [PubMed] [Google Scholar]
- 20.Bale SJ, Dracopoli NC, Tucker MA, Clark WH, Jr, Fraser MC, Stanger BZ, Green P, Donis-Keller H, Housman DE, Greene MH: Mapping of the gene for hereditary cutaneous malignant melanoma-dysplastic nevus to chromosome 1p. N Engl J Med 1989, 320:1367-1372 [DOI] [PubMed] [Google Scholar]
- 21.Millikin D, Meese E, Vogelstein B, Witkowski C, Trent J: Loss of heterozygosity for loci on the long arm of chromosome 6 in human malignant melanoma. Cancer Res 1991, 51:5449-5453 [PubMed] [Google Scholar]
- 22.Cannon-Albright LA, Meyer LJ, Goldgar DE, Lewis CM, McWhorter WP, Jost M, Harrison D, Anderson DE, Zone JJ, Skolnick MH: Penetrance and expressivity of the chromosome 9p melanoma susceptibility locus (MLM). Cancer Res 1994, 54:6041-6044 [PubMed] [Google Scholar]
- 23.Zucman J, Delattre O, Desmaze C, Epstein AL, Stenman G, Speleman F, Fletchers CD, Aurias A, Thomas G: EWS and ATF-1 gene fusion induced by t(12: 22) translocation in malignant melanoma of soft parts. Nat Genet 1993, 4:341-345 [DOI] [PubMed] [Google Scholar]
- 24.Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP: Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res 1998, 72:141-196 [PubMed] [Google Scholar]
- 25.Ponting CP, Blake DJ, Davies KE, Kendrick-Jones J, Winder SJ: ZZ and TAZ: new putative zinc fingers in dystrophin and other proteins. Trends Biochem Sci 1996, 21:11-13 [PubMed] [Google Scholar]
- 26.Sudol M, Chen HI, Bougeret C, Einbond A, Bork P: Characterization of a novel protein-binding module: the WW domain. FEBS Lett 1995, 369:67-71 [DOI] [PubMed] [Google Scholar]
- 27.Tinsley JM, Blake DJ, Roche A, Byth BC, Knight AE, Kendrick-Jones J, Suthers GK, Love DR, Edwards YH, Davies KE: Primary structure of dystrophin-related protein. Nature 1992, 360:591-593 [DOI] [PubMed] [Google Scholar]
- 28.Kunkel LM, Monaco AP, Middlesworth W, Ochs HD, Latt SA: Specific cloning of DNA fragments absent from the DNA of a male patient with an X chromosome deletion. Proc Natl Acad Sci USA 1985, 82:4778-4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffman EP, Brown RH, Kunkel LM: Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell 1987, 51:919-928 [DOI] [PubMed] [Google Scholar]
- 30.Monaco AP, Bertelson CJ, Liechti-Gallati S, Moser H, Kunkel LM: An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics 1988, 2:90-95 [DOI] [PubMed] [Google Scholar]
- 31.Anderson JT, Rogers RP, Jarrett HW: Ca2+-calmodulin binds to the carboxyl-terminal domain of dystrophin. J Biol Chem 1996, 271:6605-6610 [DOI] [PubMed] [Google Scholar]
- 32.Petrof BJ, Shrager JB, Stedman HH, Kelly AM, Sweeney HL: Dystrophin protects the sarcolemma from stresses developed during muscle contraction. Proc Natl Acad Sci USA 1993, 90:3710-3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pasternak C, Wong S, Elson EL: Mechanical function of dystrophin in muscle cells. J Cell Biol 1995, 128:355-361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bennett V: Ankyrins. J Biol Chem 1992, 267:8703-8706 [PubMed] [Google Scholar]
- 35.Pollard TD, Blanchoin L, Mullins RD: Molecular mechanism controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct 2000, 29:545-576 [DOI] [PubMed] [Google Scholar]