Abstract
Aberrant hypermethylation of promoter CpG islands is an important mechanism for the inactivation of tumor suppressor genes. CpG island hypermethylation occurs in relation to tumorigenesis or aging. Gastric cancer is one of the tumors with a high level of aberrant CpG island methylation. However, the data on the methylation status of normal gastric mucosa has been very limited. The present study attempted to compare the methylation status of nonneoplastic gastric mucosa, using clinicopathological parameters, including age, gender, Helicobacter pylori (H. pylori), acute and chronic inflammation, and intestinal metaplasia. Two hundred sixty-eight nonneoplastic gastric mucosa samples were studied for the methylation status of 11 genes (COX-2, DAP-kinase, E-cadherin, GSTP1, MGMT, hMLH1, p14, p16, THBS1, TIMP3, and RASSF1A), using methylation-specific PCR. CpG island hypermethylation was found in 53.7, 41, 37.7, 23.1, 18.7, 10.9, 10, 4.1, 3.4, 1.7, 0.4% for DAP-kinase, E-cadherin, THBS1, TIMP3, p14, MGMT, p16, COX-2, GSTP1, hMLH1 and RASSF1A, respectively. Five genes (DAP-kinase, E-cadherin, p14, THBS1, and TIMP-3) showed a general progressive increase in the methylation frequency as a function of aging, whereas the other genes (COX-2, GSTP1, MGMT, hMLH1, p16, and RASSF1A) were rarely methylated. Male patients showed higher numbers of methylated genes than females (3.2 vs. 2.1, respectively, P = 0.002). Gastritis samples with marked intestinal metaplasia, showed higher numbers of genes methylated than those without (3.7 vs. 2.6, respectively, P = 0.021). Gastritis samples with marked infiltration of mononuclear cells displayed higher numbers of genes methylated than those with mild or moderate infiltration of mononuclear cells (3.4 vs. 2.5 or 2.5, respectively, P < 0.05). Our results demonstrated that many genes are methylated in the stomach as a function of age, and suggested that male gender, intestinal metaplasia, and chronic inflammation are closely associated with increased methylation in nonneoplastic gastric mucosa samples.
Aberrant DNA methylation is a feature of human cancers, characterized by generalized hypomethylation and regional hypermethylation. 1-4 The regional hypermethylation involves CpG islands located in the promoter and upstream exons. Hypermethylation of CpG islands recruits methyl DNA binding proteins, and subsequently histone deacetylases. 5,6 Deacetylation of the histone backbones makes the DNA structure of the promoter into a closed chromatin structure, inaccessible to transcription factors, resulting in gene inactivation. 7,8 Aberrant hypermethylation of CpG islands located in the promoter, and/or upstream exons, acts as an alternative to genetic changes for the inactivation of tumor suppressor genes. 3,4
CpG islands are normally protected from DNA methylation, 9 but in relation to cancer or aging, they are aberrantly methylated. 3,10 The stomach is one of the organs that shows frequent methylation of CpG islands of genes in nonneoplastic epithelial cells. 11-13 Many genes have been demonstrated to be methylated in nonneoplastic gastric mucosae, whether in association with gastric cancer or not. 11-13 Our previous study has demonstrated that these genes are not methylated in pediatric gastric mucosae, or at a frequency significantly lower than that in gastric mucosae from adults. 11 If these methylations are an age-related event, they would be expected to be progressively prominent with age. However, no study has ever displayed plots correlating the methylation frequency of genes in the normal stomach with age.
In our previous study, aberrant promoter CpG island methylation of some genes was a frequent event in nonneoplastic gastric mucosae. 11 The methylation status of these genes, in normal gastric mucosae, have not been analyzed in relation to the histological parameters of chronic gastritis. The present study attempted to determine whether the histological parameters of chronic gastritis, as well as aging and gender, affect DNA methylation in nonneoplastic gastric mucosae. The aim of the present study was not to investigate the functional consequences of CpG island hypermethylation events, but to investigate the occurrence of CpG island hypermethylation in chronic gastritis in relation to aging or clinicopathologic variables. We examined 11 genes, whose expressions were frequently silenced by aberrant CpG island methylation in gastric, or other, cancers. The tested genes included those involved in cell-cycle regulation (COX2, p14, p16), signal transduction (RASSF1A), DNA repair or protection (hMLH1, MGMT, and GSTP1), apoptosis (DAP-kinase), and angiogenesis (THBS1), or those related to metastasis and invasion (E-cadherin and TIMP-3). MSP method was used for the analysis of the methylation frequency for the 11 genes, and the findings were correlated with the clinicopathological parameters.
Materials and Methods
A total of 268 archival samples from gastroscopic biopsies were collected from the Department of Pathology, Seoul National University Hospital, Seoul, Korea, and the Department of Diagnostic Pathology, Asan Medical Center, Seoul, Korea. The patients underwent endoscopic examination for the complaint of abdominal discomfort and diagnosed as having chronic gastritis. They did not have gastric cancers. The sex ratio was 160:108, and the ages ranged from 2 to 79 years. Through microscopic examination, hematoxylin and eosin-stained histological slides were scored for their histological parameters; H. pylori, chronic inflammation, acute inflammation, and intestinal metaplasia. The scoring was performed according to the modified Sydney system. 14 Giemsa-stained histological slides were microscopically examined for the densities of H. pylori in the biopsy sample.
DNA Extraction and Bisulfite Modification
Ten histological sections of 20-μm thickness were dewaxed in xylene, with subsequent ethanol treatment. After digestion with proteinase K (Invitrogen, Carlsbad, CA), the DNA was extracted with phenol/chloroform/isoamyl alcohol, and precipitated in ethanol. The purified DNA was subjected to bisulfite modification, as described by Herman et al 15 In brief, 2 μg of genomic DNA and 1 μg of salmon sperm DNA (as a carrier) (Invitrogen) were heated at 97°C for 6 minutes, and then cooled on ice. Fifteen μl of 1 mol/L NaOH was added to 35 μl of the denatured DNA solution, and this mixture was stored at room temperature for 15 minutes. Five hundred fifty μl of bisulfite (0.22 g) (Sigma-Aldrich, St. Louis, MO) and hydroquinone (0.08 mg) (pH 5.0) (Sigma-Aldrich) was added to the denatured DNA solution, and the mixture incubated at 55°C for 16 hours. After extraction with a JET-SORB gel extraction kit (Genomed, Bad Oeynhausen, Germany), the purified DNA (35 μl) was desulphonated with 15 μl of 1 mol/L NaOH. The modified DNA was precipitated in ethanol, and then resuspended in 50 μl TE buffer [10 mmol/L Tris and 1 mmol/L EDTA (pH 8.0)].
MSP (Methylation-Specific PCR)
The bisulfite-modified DNA (2 μl) was amplified with primers specific for either the methylated, or unmethylated, sequences. The methylated primers were selected because the primer sequences are located in upstream sequences of the genes (around transcription start sites or in the first exon) and the inverse relation between methylation and expression of the gene has been demonstrated in cancer cell or tissues by other studies using these primers. In a preliminary study, nine SNU gastric cancer cell lines were analyzed for the relationship between CpG island methylation and gene expression of the specific gene, using the same primer sets in the present study. The cell lines showed a loss or decrease of mRNA for the genes methylated, and re-expression of the genes after treatment of demethylating agent. The primer sequences of all genes for the methylated and unmethylated forms, and the annealing temperatures, cycle numbers and references, are summarized in Table 1 ▶ . PCR was performed in 25-μl reaction volumes, containing 1X PCR buffer [16.6 mmol/L (NH4)2SO4, 67 mmol/L Tris (pH 8.8), 6.7 mmol/L MgCl2, 10 mmol/L β-mercaptoethanol], deoxynucleotide triphosphates (each at 0.2 mmol/L), primers (10 pmol each) and 1 unit of Taq polymerase. Seven μl of PCR products underwent electrophoresis on 2.5% agarose gel, then visualized under UV illumination using an ethidium bromide stain. Samples showing signals approximately equivalent to that of the size marker (7 ng/μl) were scored as methylated. Samples giving faint positive signals were repeated three times and only those samples with consistent positive signals were scored as methylated. Normal peripheral blood lymphocytes, obtained from patients with no evidence of cancer (n = 20), were used as a control group. Twelve men and 8 women, with a median age of 46 years (range, 23 to 68), were included in this group. The 11 genes were not methylated in the normal peripheral blood lymphocytes.
Table 1.
Primer Sequences and PCR Conditions for MSP Analysis
| Primer name* | Primer sequence (5′-3′) forward | Primer sequence (5′-3′) reverse | Product size (bp) | AT† (°C) | No. of cycles | References | |
|---|---|---|---|---|---|---|---|
| COX-2 | m | TTAGATACGGCGGCGGCGGC | TCTTTACCCGAACGCTTCCG | 161 | 61 | 3516 | |
| u | ATAGATTAGATATGGTGGTGGTGGT | CACAATCTTTACCCAAACACTTCCA | 171 | 61 | 35 | ||
| DAP- | m | GGATAGTCGGATCGAGTTAACGTC | CCCTCCCAAACGCCGA | 98 | 60 | 35 | 17 |
| kinase | u | GGAGGATAGTTGGATTGAGTTAATGTT | CAAATCCCTCCCAAACACCAA | 98 | 60 | 35 | |
| E- | m | TTAGGTTAGAGGGTTATCGCGT | TAACTAAAAATTCACCTACCGAC | 115 | 57 | 33 | 15 |
| cadherin | u | TAATTTTAGGTTAGAGGGTTATTGT | CACAACCAATCAACAACACA | 97 | 53 | 33 | |
| GSTP1 | m | TTCGGGGTGTAGCGGTCGTC | GCCCCAATACTAAATCACGACG | 91 | 59 | 35 | 18 |
| u | GATGTTTGGGGTGTAGTGGTTGTT | CCACCCCAATACTAAATCACAACA | 97 | 59 | 35 | ||
| MGMT | m | TTTCGACGTTCGTAGGTTTTCGC | GCACTCTTCCGAAAACGAAACG | 81 | 59 | 35 | 19 |
| u | TTTGTGTTTTGATGTTTGTAGGTTTTTGT | AACTCCACACTCTTCCAAAAACAAAACA | 93 | 59 | 35 | ||
| hMLH1 | m | TATATCGTTCGTAGTATTCGTGT | TCCGACCCGAATAAACCCAA | 153 | 60 | 33 | 20 |
| u | TTTTGATGTAGATGTTTTATTAGGGTTGT | ACCACCTCATCATAACTACCCACA | 124 | 60 | 33 | ||
| p14 | m | GTGTTAAAGGGCGGCGTAGC | AAAACCCTCACTCGCGACGA | 122 | 60 | 33 | 21 |
| u | TTTTTGGTGTTAAAGGGTGGTGTAGT | CACAAAAACCCTCACTCACAACAA | 132 | 60 | 33 | ||
| p16 | m | TTATTAGAGGGTGGGGCGGATCGC | GACCCCGAACCGCGACCGTAA | 150 | 65 | 33 | 15 |
| u | TTATTAGAGGGTGGGGTGGATTGT | CAACCCCAAACCACAACCATAA | 151 | 60 | 33 | ||
| RASSF1A | m | GTGTTAACGCGTTGCGTATC | AACCCCGCGAACTAAAAACGA | 93 | 60 | 35 | 22 |
| u | TTTGGTTGGAGTGTGTTAATGTG | CAAACCCCACAAACTAAAAACAA | 105 | 60 | 35 | ||
| THBS1 | m | TGCGAGCGTTTTTTTAAATGC | TAAACTCGCAAACCAACTCG | 74 | 62 | 35 | 23 |
| u | GTTTGGTTGTTGTTTATTGGTTG | CCTAAACTCACAAACCAACTCA | 115 | 62 | 35 | ||
| TIMP3 | m | CGTTTCGTTATTTTTTGTTTTCGGTTTTC | CCGAAAACCCCGCCTCG | 116 | 59 | 35 | 24 |
| u | TTTTGTTTTGTTATTTTTTGTTTTTGGTTTT | CCCCCCAAAAACCCCACCTCA | 122 | 59 | 35 |
*m, methylated sequence; u, unmethylated sequence.
†AT, annealing temperature.
Statistical Analysis
All statistical calculations were made using the SPSS software (version 10.0 SPSS, Chicago, IL). Student’s t-test or analysis of variance test and χ2 test were used to compare the numbers of methylated genes, and the frequency of CpG island hypermethylation for the tested genes, respectively, in chronic gastritis by anatomical site, gender, age, and scores of histological parameters of the modified Sydney system (H. pylori, acute inflammation, chronic inflammation, and intestinal metaplasia). Of the collected samples, those from the patients younger than 40 years showed a skewed distribution of intestinal metaplasia, H. pylori, and gender. Pediatric patients showed no intestinal metaplasia at all, and very low frequencies of H. pylori-positivity, whereas young patients (third and fourth decades) showed high frequencies of Helicobacter-positivity and low frequencies of intestinal metaplasia. When the distribution of the scores for each histological parameter was analyzed by decades, for the patients over 40 years of age, the distribution was not skewed for any of the histological parameters and thus, the statistical analysis of the methylation frequency, with respect to histological parameters, was confined to the patients over 40 years of age (n = 169).
Results
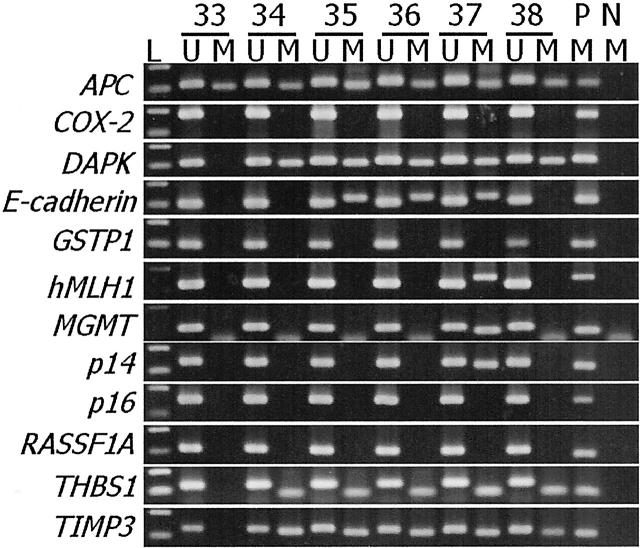
Two hundred sixty eight nonneoplastic gastric mucosa samples were studied for the methylation status of 11 genes (COX-2, DAP-kinase, E-cadherin, GSTP1, MGMT, hMLH1, p14, p16, RASSF1A, THBS1, and TIMP3) using methylation-specific PCR (Figure 1) ▶ . CpG island methylation was found in 53.7% for DAP-kinase, 41% for E-cadherin, 37.7% for THBS1, 23.1% for TIMP3, 18.7% for p14, 10.9% for MGMT, 10% for p16, 4.1% for COX-2, 3.4% for GSTP1, 1.7% for hMLH1, and 0.4% for RASSF1A. The frequencies of CpG island hypermethylation for the genes tested in each age group are displayed in Figure 2 ▶ . The numbers of genes methylated for the 11 genes tested ranged from 0.4 to 3.3, exhibiting a general progressive increase with aging (Figure 2A) ▶ . Five genes (DAP-kinase, E-cadherin, p14, THBS1, and TIMP-3) showed a general increase in the methylation frequency as a function of aging (Figure 2B) ▶ , whereas the other genes (COX-2, GSTP1, MGMT, hMLH1, p16, and RASSF1A) were rarely methylated (Figure 2C) ▶ .
Figure 1.
Electrophoresis of PCR products of representative samples. The PCR products in the lanes marked U show the presence of unmethylated templates of each gene, whereas the products in the lanes marked M indicate the presence of methylated templates. L, size marker (100-bp DNA ladder); P, positive control; N, negative control. Positive control is normal lymphocyte DNA treated with Sss1 methyl transferase before bisulfite modification. Negative control is distilled water without template DNA.
Figure 2.
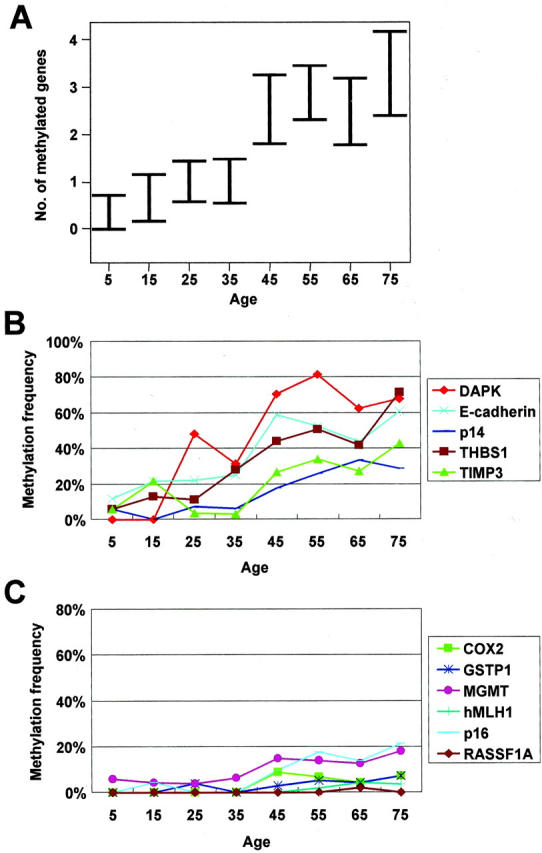
The number of methylated genes in 10-year age groups. Results are given as mean ± SE (A). The methylation frequency of DAP-kinase, E-cadherin, p14, THBS1, and TIMP-3 increased with aging (B) but that of COX2, GSTP1, MGMT, hMLH1, p16, and RASSF1A did not (C).
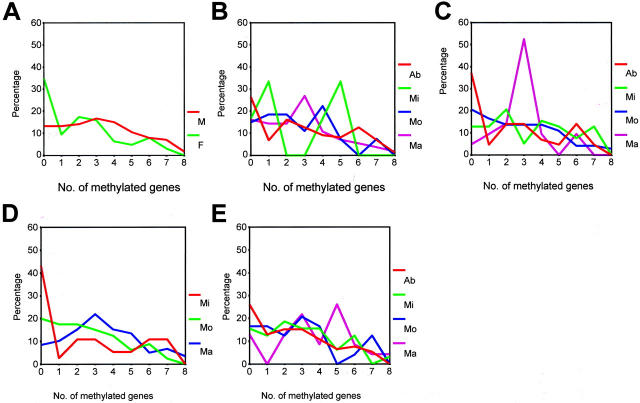
When the number of genes methylated, or the frequency of CpG island methylation, of each gene tested was compared in chronic gastritis patients (aged over 40 years, n = 169), by gender and histological parameters (Table 2 ▶ and Figure 3 ▶ ), the male patients (mean age, 58 years; n = 108) showed higher numbers of methylated genes than the females (mean age, 57 years; n = 61) (3.2 vs. 2.1, respectively, Student’s t-test, P = 0.002). DAP-kinase, E-cadherin, THBS1, and TIMP-3 were more frequently methylated in the male patients than in females, with the difference in the methylation frequency reaching statistical significance (P < 0.05). No significant difference was found in the numbers of the genes methylated between subjects either with, or without, H. pylori. The subjects with marked intestinal metaplasia showed higher numbers of genes methylated than the subjects without (3.7 vs. 2.6, P = 0.021, Student’s t-test). Samples with a marked degree of mononuclear cell infiltration displayed higher numbers of genes methylated than samples with mild or moderate degrees of chronic inflammation (3.4 vs. 2.5 or 2.5, P = 0.022, analysis of variance test). DAP-kinase, E-cadherin, and THBS1 were more frequently methylated in samples with a marked degree of mononuclear cell infiltration than in samples with mild or moderate degrees of chronic inflammation, with the difference in the methylation frequency of the respective genes reaching statistical significance. However, no statistical difference in the numbers of genes methylated was noted in the samples in relation to the degree of neutrophilic infiltration. The numbers of genes methylated were 2.6, 3.2, and 2.9 for samples from the antrum, body, and cardia, respectively, but they showed no significant difference.
Table 2.
The Number of Genes Methylated in Chronic Gastritis According to Grades of Histologic Parameters
| Absent | Mild | Moderate | Marked | |
|---|---|---|---|---|
| Helicobacter pylori | 2.9 (n = 84) | 2.7 (n = 5) | 2.7 (n = 26) | 2.7 (n = 54) |
| Intestinal metaplasia | 2.5* (n = 91) | 2.9† (n = 31) | 2.8‡ (n = 24) | 3.7*,†,‡ (n = 23) |
| Acute inflammation | 2.4 (n = 41) | 3.3 (n = 37) | 2.7 (n = 71) | 2.9 (n = 20) |
| Chronic inflammation | 2.5 (n = 35) | 2.5§ (n = 78) | 3.4§ (n = 57) |
*, absent vs. marked, P = 0.015 by Student’s t-test.
†, mild vs. marked, P = 0.161 by Student’s t-test.
‡, moderate vs. marked, P = 0.165 by Student’s t-test.
§, moderate vs. marked, P = 0.01 by Student’s t-test.
Figure 3.
The number of genes methylated was analyzed in relation to clinicopathologic parameters: gender (A), H. pylori density (B), grade of acute inflammatory cell infiltration (C), grade of chronic inflammatory cell infiltration (D), and grade of intestinal metaplasia (E). Results are given as percentage of methylation-positive samples. Ab, absent; Mi, mild; Mo, moderate; Ma, marked.
Discussion
Aberrant CpG island methylation is a frequent finding in human cancers, and an important molecular mechanism of inactivation of tumor suppressor genes in the development of human cancers, regardless of the tissue type. 3,4,25 Against the original findings of the lack of CpG island methylation in normal tissues, CpG island methylation is often found in histologically normal tissues, in relation to aging. 26 Aging-related methylation has been demonstrated for the ER gene by Issa et al 27 and for multiple genes by Ahuja et al. 28 The susceptibility to aging-related CpG island methylation differs among genes and tissues. 28 The present study has demonstrated that DAP-kinase, E-cadherin, and THBS1 were methylated at frequencies higher than 35%, whereas COX-2, GSTP1, hMLH1, and RASSF1A were methylated at frequencies less than 5% in nonneoplastic gastric mucosae. When the genes frequently methylated in nonneoplastic gastric mucosae were analyzed for their methylation status in normal tissues of the colon, liver, or prostate, they were found to be rarely methylated (data not shown). This finding exhibits a tissue-, or gene-difference, for aging-related methylation. The finding that the stomach is one of the normal tissues with a high frequency of aging-related methylation, is consistent with the finding that gastric cancer is one of the tumors with a high frequency of CpG island methylation. 29 The causes for such a difference among tissue types remain unclear, but it may be related to the accessibility of the tissue to exogenous agents; the stomach is directly exposed to exogenous agents. Dietary factors or heavy metals may be possible candidates. Issa 30 has suggested that diets high in fiber might be associated with reduced levels of ER methylation in colonic mucosa. In vitro studies have demonstrated that exposure to nickel causes CpG island methylation. 31,32
In addition to exogenous agents, endogenously-produced materials, including reactive oxygen species, are suspected as possible causes for aging-related methylation in the stomach. 10,33 Epithelial cell injury, caused by H. pylori, implicates several pathways, with oxygen-radical injury being one. 34,35 However, in this study, no difference was noted in the frequencies of CpG island methylation for the genes tested between chronic gastritis with, and without, H. pylori. The present study has a limitation in the evaluation of H. pylori, as the absence of H. pylori in the biopsy materials did not exclude the possibility of the presence of H. pylori in the adjacent sites that were not biopsied, or the previous residence of H. pylori in the same biopsied sites. However, the urea breath test, which was performed on a small proportion of the patients, showed correlation with the results of histological assessment of H. pylori.
The present study has demonstrated a significant difference in the number of genes methylated between male and female patients. The average number of genes methylated was 3.2 and 2.1 in male and female patients, respectively. However, when the numbers of genes methylated was compared in gastric cancers by gender, no difference was noted (data not shown). It is plausible that the gender-difference of the extent of methylation in chronic gastritis may be related to the gender-difference in the incidences of gastric cancers. It is currently unclear why such a gender difference for the extent of methylation exists in chronic gastritis. Larger studies are needed to validate the gender difference in aging-related methylation in nonneoplastic gastric mucosae.
In the present study, a general progressive increase in the numbers of the genes methylated, in relation to aging, was displayed. The fact that gastric cancer occurs in older people and has a high frequency of CpG island methylation, suggests the link of aging and cancer via increase in CpG island methylation. In the present study, a patient-to-patient variation in the extent of DNA methylation existed in the same age group. Among the patients aged over 40 years, 15% showed CpG island methylation at more than 50% of the genes tested. Considering that nonneoplastic gastric mucosae, from patients with gastric cancers, harbor higher levels of CpG island methylation than nonneoplastic gastric mucosae from patients without gastric cancer, 12,13 these individuals with higher levels of CpG island methylation are expected to be at increased risk of tumor formation. A prospective study is required to confirm the risk.
The present study has demonstrated that many genes are frequently methylated in chronic gastritis. The methylation detected by MSP might not affect gene expression although MSP analysis using the same primer sets displayed an inverse relationship between methylation and expression of mRNA of the corresponding gene in gastric cancer cell lines and bisulfite-sequencing of MSP products of each of the genes from chronic gastritis samples showed that all of the CpG sites, in between both primers, were also methylated (data not shown). This is because the methylation of the CpG sites along CpG islands might be patchy and incomplete or the methylation detected by MSP might affect only a few cells over 1000 cells in each sample 15 and the vast majority of the cells might not harbor the CpG island methylation of the specific gene. The present study just demonstrated the presence of the methylated allele but did not say about the quantitative prevalence of the methylated allele. It is apparent that the occurrence of the methylated alleles of some genes increased with aging or was associated with some clinicopathologic variables (gender, intestinal metaplasia, and chronic inflammation), but it is unknown whether the proportion of the methylated alleles of the genes does so. For the quantitative determination of the methylated allele, microdissection of the epithelial cells and quantitative assay, including Ms-SNuPE 36 or MethyLight, 37 should be performed.
In conclusion, we have studied chronic gastritis samples, from pediatric to geriatric patients, for the methylation status of the 11 genes using MSP assay, and determined the methylation frequency of the genes as a function of age, and correlated them with the clinicopathologic parameters. We found that aberrant CpG island methylation occurred frequently in chronic gastritis, which showed aging relationship, and that aging-related methylation was gene type-specific; DAP-kinase, E-cadherin, p14, THBS1, and TIMP3 all showed a progressive increase of methylation frequency with aging, whereas COX-2, GSTP1, MGMT, hMLH1, p16, and RASSF1A were rarely methylated, with no aging relationship. Increased numbers of genes methylated were closely associated with male gender, marked chronic inflammation, marked intestinal metaplasia, as well as aging.
Footnotes
Address reprint requests to Gyeong Hoon Kang, M.D., Department of Pathology, Seoul National University Hospital, 28 Yongon-dong, Chongno-gu, Seoul, 110–744, Korea. E-mail: ghkang@snu.ac.kr.
Supported by grant no. R02–2002-00040–0 from the Basic Research Program of the Korea Science and Engineering Foundation and by the 2002 BK21 project for Medicine, Dentistry, and Pharmacy, Seoul, Korea.
References
- 1.Feinberg AP, Vogelstein B: Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 1983, 301:89-92 [DOI] [PubMed] [Google Scholar]
- 2.Rideout WM, Eversole-Cire P, Spruck CH, III, Hustad CM, Coetzee GA, Gonzales FA, Jones PA: Progressive increases in the methylation status and heterochromatinization of the myoD CpG island during oncogenic transformation. Mol Cell Biol 1994, 14:6143-6152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP: Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res 1998, 71:141-146 [PubMed] [Google Scholar]
- 4.Jones PA, Laird PW: Cancer epigenetics comes of age. Nat Genet 1999, 21:163-167 [DOI] [PubMed] [Google Scholar]
- 5.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A: Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 1998, 393:386-389 [DOI] [PubMed] [Google Scholar]
- 6.Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP: Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet 1998, 19:187-191 [DOI] [PubMed] [Google Scholar]
- 7.Bird AP, Wolffe AP: Methylation-induced repression-belts, braces, and chromatin. Cell 1999, 99:451-454 [DOI] [PubMed] [Google Scholar]
- 8.Jones PA, Takai D: The role of DNA methylation in mammalian epigenetics. Science 2001, 293:1068-1070 [DOI] [PubMed] [Google Scholar]
- 9.Bird AP: CpG-rich islands and the function of DNA methylation. Nature 1986, 321:209-213 [DOI] [PubMed] [Google Scholar]
- 10.Issa JP: CpG-island methylation in aging and cancer. Curr Top Microbiol Immunol 2000, 249:101-118 [DOI] [PubMed] [Google Scholar]
- 11.Kang GH, Shim YH, Jung HY, Kim WH, Ro JY, Rhyu MG: CpG island methylation in premalignant stages of gastric carcinoma. Cancer Res 2001, 61:2847-2851 [PubMed] [Google Scholar]
- 12.To KF, Leung WK, Lee TL, Yu J, Tong JHM, Chan MWY, Ng EKW, Sydney Chung SC, Sung JJY: Promoter hypermethylation of tumor-related genes in gastric intestinal metaplasia of patients with and without gastric cancer. Int J Cancer 2002, 102:623-628 [DOI] [PubMed] [Google Scholar]
- 13.Waki T, Tamura G, Tsuchiya T, Sato K, Nishizuka S, Motoyama T: Promoter methylation status of E-cadherin, hMLH1, and p16 genes in nonneoplastic gastric epithelia. Am J Pathol 2002, 161:399-403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dixon MF, Genta RM, Yardley JH, Correa P: Classification and grading of gastritis: the updated Sydney system. Am J Surg Pathol 1996, 20:1161-1181 [DOI] [PubMed] [Google Scholar]
- 15.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB: Methylation-specific PCR. A novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA 1996, 93:9821-9826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akhtar M, Cheng Y, Magno RM, Ashktorab H, Smoot DT, Meltzer SJ, Wilson KT: Promoter methylation regulates Helicobacter pylori-stimulated cyclooxygenase-2 expression in gastric epithelial cells. Cancer Res 2001, 61:2399-2403 [PubMed] [Google Scholar]
- 17.Katzenellenbogen RA, Baylin SB, Herman JG: Hypermethylation of the DAP-kinase CpG island is a common alteration in B-cell malignancies. Blood 1999, 93:4347-4353 [PubMed] [Google Scholar]
- 18.Esteller M, Corn PG, Urena JM, Gabrielson E, Baylin SB, Herman JG: Inactivation of glutathione S-transferase P1 gene by promoter hypermethylation in human neoplasia. Cancer Res 1998, 58:4515-4518 [PubMed] [Google Scholar]
- 19.Esteller M, Gaidano G, Goodman SN, Zagonel V, Capello D, Botto B, Rossi D, Gloghini A, Vitolo U, Carbone A, Baylin SB, Herman JG: Hypermethylation of the DNA repair gene O6-methylguanine DNA methyltransferase and survival of patients with diffuse large B-cell lymphoma. J Natl Cancer Inst 2002, 94:26-32 [DOI] [PubMed] [Google Scholar]
- 20.Kang GH, Shim YH, Ro JY: Correlation of methylation of the hMLH1 promoter with lack of expression of hMLH1 in sporadic gastric carcinomas with replication error. Lab Invest 1999, 79:903-909 [PubMed] [Google Scholar]
- 21.Esteller M, Tortola S, Toyota M, Capella G, Peinado MA, Baylin SB, Herman JG: Hypermethylation-associated inactivation of p14(ARF) is independent of p16(INK4a) methylation and p53 mutational status. Cancer Res 2000, 60:129-133 [PubMed] [Google Scholar]
- 22.Lo KW, Kwong J, Hui ABU, Chan SYY, To KF, Chan ASC, Chow LSN, Teo PML, Johnson PJ, Huang DP: High frequency of promoter hypermethylation of RASSF1A in nasopharyngeal carcinoma. Cancer Res 2001, 61:3877-3881 [PubMed] [Google Scholar]
- 23.Ueki T, Toyota M, Sohn T, Yeo CJ, Issa JPJ, Hruban RH, Goggins M: Hypermethylation of multiple genes in pancreatic adenocarcinoma. Cancer Res 2000, 60:1835-1839 [PubMed] [Google Scholar]
- 24.Bachman KE, Herman JG, Corn PG, Merlo A, Costello JF, Cavenee WK, Baylin SB, Graff JR: Methylation-associated silencing of the tissue inhibitor of metalloproteinase-3 gene suggests a suppressor role in kidney, brain, and other human cancers. Cancer Res 1999, 59:798-802 [PubMed] [Google Scholar]
- 25.Esteller M, Corn PG, Baylin SB, Herman JG: A gene hypermethylation profile of human cancer. Cancer Res 2001, 61:3225-3229 [PubMed] [Google Scholar]
- 26.Issa JP: The epigenetics of colorectal cancer. Ann NY Acad Sci 2000, 910:140-153 [DOI] [PubMed] [Google Scholar]
- 27.Issa JP, Ottaviano YL, Celano P, Hamilton SR, Davidson NE, Baylin SB: Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet 1994, 7:536-540 [DOI] [PubMed] [Google Scholar]
- 28.Ahuja N, Li Q, Mohan AL, Baylin SB, Issa JP: Aging and DNA methylation in colorectal mucosa and cancer. Cancer Res 1998, 58:5489-5494 [PubMed] [Google Scholar]
- 29.Esteller M: CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene 2002, 21:5427-5440 [DOI] [PubMed] [Google Scholar]
- 30.Issa JP: Epigenetic variation and human disease. J Nutr 2002, 132:2388S-2392S [DOI] [PubMed] [Google Scholar]
- 31.Lee YW, Klein CB, Kargacin B, Salnikow K, Kitahara J, Dowjat K, Zhitkovich A, Christie NT, Costa M: Carcinogenic nickel silences gene expression by chromatin condensation and DNA methylation: a new model for epigenetic carcinogens. Mol Cell Biol 1995, 15:2547-2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sutherland JE, Peng W, Zhang QW, Costa M: The histone deacetylase inhibitor A reduces nickel-induced gene silencing in yeast and mammalian cells. Mutat Res 2001, 479:225-233 [DOI] [PubMed] [Google Scholar]
- 33.Shen L, Ahuja N, Shen Y, Habib NA, Toyota M, Rashid A, Issa JP: DNA methylation and environmental exposures in human hepatocellular carcinoma. J Natl Cancer Inst 2002, 94:755-761 [DOI] [PubMed] [Google Scholar]
- 34.Bagchi D, Bhattacharya G, Stohs SJ: Production of reactive oxygen species by gastric cells in association with Helicobacter pylori. Free Radic Res 1996, 24:439-450 [DOI] [PubMed] [Google Scholar]
- 35.Obst B, Wagner S, Sewing KF, Beil W: Helicobacter pylori causes DNA damage in gastric epithelial cells. Carcinogenesis (Lond) 2000, 21:1111-1115 [PubMed] [Google Scholar]
- 36.Gonzalgo ML, Jones PA: Rapid quantitation of methylation differences at specific sites using methylation-sensitive single nucleotide primer extension (Ms-SNuPE). Nucleic Acids Res 1997, 25:2529-2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, Danenberg PV, Laird PW: MethylLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res 2000, 28:e32. [DOI] [PMC free article] [PubMed] [Google Scholar]