Abstract
Invasive cervical carcinomas frequently reveal additional copies of the long arm of chromosome 3. The detection of this genetic aberration in diagnostic samples could therefore complement the morphological interpretation. We have developed a triple-color DNA probe set for the visualization of chromosomal copy number changes directly in thin-layer cervical cytology slides by fluorescence in situ hybridization. The probe set consists of a BAC contig that contains sequences for the RNA component of the human telomerase gene (TERC) on chromosome band 3q26, and repeat sequences specific for the centromeres of chromosomes 3 and 7 as controls. In a blinded study, we analyzed 57 thin-layer slides that had been rigorously screened and classified as normal (n = 13), atypical squamous cells (ASC, n = 5), low-grade squamous intraepithelial lesions (LSIL, n = 14), and high-grade squamous intraepithelial lesions (HSIL) grade 2 (CIN2, n = 8), and grade 3 (CIN3, n = 17). The percentage of tetraploid cells (PTrend < 0.0005) and cells with multiple 3q signals increased with the severity of the cytologic interpretation (PTrend < 0.0005). While only few normal samples, ASC and LSIL lesions, revealed copy number increases of 3q, 63% of the HSIL (CIN2) lesions and 76% of the HSIL (CIN3) lesions showed extra copies of 3q. We conclude that the visualization of chromosome 3q copy numbers in routinely prepared cytological material using BAC clones specific for TERC serves as an independent screening test for HSIL and may help to determine the progressive potential of individual lesions.
Cytologic screening 1 has greatly reduced incidence and mortality of cervical cancer in industrialized nations. 2 In developing countries, however, cervical cancer remains a health problem of tremendous proportions. If detected in a timely manner, cervical cancer precursors, especially high-grade squamous intraepithelial lesions (HSILs) can be effectively treated, sparing patients the morbidity and mortality resulting from invasive cancer. Despite its success as a public health measure, a single cytologic examination is relatively insensitive, poorly reproducible and frequently yields equivocal results. Inadequate sampling, the scarcity of aberrant cells in some samples and the subjectivity of morphological interpretation are recognized limitations of cytology. 2,3 In addition, equivocal and mild cytologic abnormalities are extremely common in young women, but most of these lesions regress spontaneously, even when caused by oncogenic types of human papillomaviruses, 4,5 which play a crucial role in the pathogenesis of cervical cancer. 6,7 This has prompted efforts to discover other biomarkers and other screening techniques with the potential to supplement cytologic screening. 8-13
The sequential acquisition of specific chromosomal copy number changes and the maintenance of these aneuploidies are commonly observed in solid tumors of epithelial origin. 14,15 Previous studies using tissue microdissection followed by comparative genomic hybridization have shown that acquisition of extra copies of chromosome 3q is a recurrent genetic alteration in cervical carcinomas, which is less frequently recognized in premalignant precursor lesions. 16,17 This region is of particular significance because it contains the RNA component of the human telomerase gene. 18,19 We postulated that the acquisition of additional copies of 3q may represent an early event in malignant transformation, which could provide a useful biomarker for screening if detectable using an assay capable of identifying rare cells with this genetic aberration. Accordingly, we developed a method for detecting aneuploidies of 3q within individual cervical cells of specimens collected in a routine manner for cytological screening.
The objective visualization of chromosomal aneuploidies, such as copy number increases of chromosome 3q in the intact nuclei of cytological preparations, can be readily achieved using in situ hybridization with DNA probes that are labeled with either fluorescent or chromogenic dyes. 20 Termed interphase cytogenetics, this approach has been applied to many different tumor types, including cervical cancer, mainly using repetitive probe sequences that detect the centromeres of human chromosomes. 21-24 We have now devised a multicolor fluorescence in situ hybridization (FISH) assay using three specific DNA probes. The design of the probe panel was solely based on the pattern of genomic imbalances in cervical cancers as detected by comparative genomic hybridization (CGH). 16,17 The probe panel includes pericentromeric repetitive sequence probes for enumerating chromosomes 3 (CEP3) and 7 (CEP7) and a set of four overlapping BAC clones that contain sequences for the TERC gene. 25 This strategy allowed us to specifically assess gains and amplification of 3q, and discern these changes from those involving the entire chromosome 3 and polyploidy.
We compared the independent assessment of gain of 3q to the rigorous cytological classification of thin-layer slides prepared from the same specimens. Our goal was to determine the feasibility of applying this assay to routinely collected cytological material and to gain insights into the potential for future clinical applications.
Materials and Methods
Cytological Specimens
We obtained two to four anonymized thin-layer slides from each of 68 residual PreservCyt (Cytyc) specimens that had been used previously for clinical purposes. The appropriate review boards exempted the study because we used anonymized specimens that were no longer needed clinically. One thin-layer slide from each woman was stained by a modified Papanicolaou method at The Johns Hopkins University Cytopathology laboratory. An experienced cytotechnologist (F. H. B.) initially screened the slides, dotted possible abnormal cells and classified the specimens according to the 2001 Bethesda System. 26 Then, a cytopathologist (M. E. S.) reviewed the dotted slides independently and classified the samples in an identical manner. For the purposes of this review, specimens interpreted as HSIL were further subclassified as cervical intraepithelial neoplasia grade 2, HSIL (CIN2), or cervical intraepithelial neoplasia grade 3, HSIL (CIN3). In addition, the number of abnormal-appearing cells per slide was roughly estimated as <10, >10 to <50, >50 to 100, and >100. Discrepancies in cell counts or classification were resolved and consensus reached on all specimens by a joint review conducted using a multiheaded microscope. For the joint review, abnormal cell counts were dichotomized into <50 and >50. Before joint review, in the 57 cases evaluable by FISH, inter-observer concordance on the classification was observed in 11 of 13 (85%) of the normal cases, 0 of 5 (0%) of the ASC cases, 11 of 14 (79%) of LSIL, 4 of 8 (50%) HSIL (CIN2), and 15 of 17 (88%) of HSIL (CIN3). Abnormal cell counts <50 were found in five ASC, five LSIL, seven HSIL (CIN2), and five HSIL (CIN3) cases. The remaining thin-layer slides were used for FISH.
Cervical Cancer-Specific FISH Probe Panel
A probe panel to evaluate the gain of 3q was designed based on our previously published CGH results. 16,17 The panel consists of three probes: centromere 7 (CEP7) labeled with the fluorescent dye Spectrum Aqua (SA), centromere 3 (CEP3) with Spectrum Green (SG) and a contig consisting of four overlapping BAC clones for detection of the long arm of chromosome 3, labeled with Spectrum Orange (SO). These clones contain the sequence for the RNA component of the human telomerase (TERC) gene. All probes were provided by Vysis, Inc./Abbott Laboratories (Downers Grove, IL). Probe labeling was performed chemically as described previously. 27 Slides were pretreated with RNase, followed by pepsin digestion, and fixation in an ethanol series. Slides were denatured in 70% formamide, 2X SSC for 3.5 minutes at 80°C. After overnight hybridization at 37°C, the coverslips were removed gently, slides washed four times in 50% formamide/2X SSC at 45°C (once for 3 minutes and three times for 7 minutes), followed by washes in 2X SSC (45°C, 5 minutes) and 0.1% NP40 in 2X SSC (45°C, 5 minutes). The slides were counterstained with DAPI and embedded in an antifade solution. Images were acquired using a Leica DMRXA microscope (Leica, Wetzlar, Germany) equipped with custom optical filters for DAPI, SA, SG, and SO (Chroma Technologies, Brattleboro, VT) with a ×40 Plan Apo (NA 1.25) objective. The microscope was connected to an ORCA ER (IEEE1394 I/F) digital camera (Hamamatsu, Bridgewater, NJ).
Signal Enumeration
The signals were evaluated by screening the entire slide visually for the 3q probe (using the optical filter specific for SO). Cells with normal signal numbers for 3q were recorded as “diploid” and cells that could not be evaluated (because of insufficient hybridization or cell clumps) were recorded as “uncountable” using a manual counter and excluded from further analysis. Abnormal signal numbers for 3q were registered in relocation charts in form of patterns for the whole probe panel (eg, 2-3-3, referring to two signals for CEP7, three signals for CEP3 and three signals for the 3q probe). Multifocus images were then acquired for all probes of the probe panel using five to seven focal planes with Leica Q-FISH software. Cells with four signals for each probe (pattern 4-4-4) were considered tetraploid. Based on cell density, between 209 (case 107, normal) and 3903 (case 2, LSIL) nuclei were enumerated (Table 2) ▶ ▶ . The chromosomal instability index was calculated by dividing the number of observed hybridization patterns by the number of cases for each diagnostic group.
Table 2.
Relationship Between Cytologic Interpretations and FISH Results
| Cytology | # Nuclei | # Counted nuclei | % Counted | # Diploid cells | |||||
|---|---|---|---|---|---|---|---|---|---|
| n | Median | Range | Median | Range | Median | Range | Median | Range | |
| Normal | 13 | 792 | 592–2991 | 543 | 209–2918 | 76.1% | 31.6–97.6% | 524 | 208–2894 |
| ASC | 5 | 1106 | 668–2565 | 729 | 503–2436 | 75.3% | 48.0–95.0% | 723 | 502–2432 |
| LSIL | 14 | 2321.5 | 673–4324 | 2189.5 | 444–3903 | 93.3% | 65.4–100% | 2169.5 | 423–3895 |
| HSIL (CIN2) | 8 | 1146 | 599–2936 | 1040 | 402–2699 | 90.4% | 56.8–93.1% | 985 | 395–2614 |
| HSIL (CIN3) | 17 | 718 | 416–2334 | 674 | 308–2130 | 74.0% | 20.0–94.4% | 551 | 197–2024 |
| P† | 0.008 | 0.002 | 0.01 | 0.001 | |||||
| PTrend† | 0.9 | 0.6 | 0.3 | 0.3 | |||||
| Normal | 13 | 792 | 592–2991 | 543 | 209–2918 | 76.2% | 31.6–97.6% | 524 | 208–2894 |
| ASC or LSIL | 19 | 2020 | 668–4324 | 1827 | 444–3903 | 91.9% | 48.0–100% | 1812 | 423–3895 |
| HSIL (CIN2 or CIN3) | 25 | 1008 | 416–2936 | 694 | 308–2699 | 75.2% | 20.0–94.4% | 651 | 197–2614 |
| P† | 0.004 | 0.003 | 0.04 | 0.001 | |||||
| PTrend‡ | 0.8 | 0.6 | 0.4 | 0.4 | |||||
†Nonparametric analysis of variance (Kruskal-Wallis test);
‡Nonparametric test of trend.
The relationships of the number of nuclei per slide, number of counted nuclei, the percentage of counted nuclei, number of diploid cells (cells with two signals for each probe = pattern 2-2-2), number of tetraploid cells (cells with four signals for each probe = pattern 4-4-4), the percentage of tetraploid cells, number of cells with more than two 3q signals, the percentage of cells with more than two 3q signals and cells with a relative 3q gain (as defined by higher copy numbers of the probe for 3q compared to CEP7) with the severity of the cytologic interpretation.
Table 2A.
Continued
| # Tetraploid cells | % Tetraploid | # with Multiple 3q | % with Multiple 3q | Relative 3q gain | |||||
|---|---|---|---|---|---|---|---|---|---|
| Median | Range | Median | Range | Median | Range | Median | Range | Median | Range |
| 2 | 0–8 | 0.1% | 0.0–1.5% | 2 | 1–22 | 0.4 | 0.1–2.8 | 2 | 1–22 |
| 1 | 0–4 | 0.1% | 0.0–0.4% | 1 | 0–3 | 0.1 | 0.0–0.41 | 1 | 0–3 |
| 5.5 | 0–54 | 0.2% | 0.0–7.6% | 5.5 | 1–17 | 0.3 | 0.1–2.9 | 6 | 0–16 |
| 30 | 1–70 | 2.9% | 0.2–7.6% | 12 | 1–37 | 1.3 | 0.1–4.1 | 11 | 1–30 |
| 10 | 0–96 | 2.5% | 0.0–12.0% | 30 | 2–480 | 4.8 | 0.3–42.9 | 25 | 0–476 |
| 0.001 | 0.002 | 0.0001 | 0.0001 | 0.001 | |||||
| <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | |||||
| 2 | 0–8 | 0.1% | 0.0–1.5% | 2 | 1–22 | 0.4 | 0.1–2.8 | 2 | 1–22 |
| 3 | 0–54 | 0.2% | 0.0–7.6% | 4 | 0–17 | 0.2 | 0.0–2.9 | 4 | 0–16 |
| 25 | 0–96 | 2.6% | 0.0–12.0% | 20 | 1–480 | 4.0 | 0.1–42.9 | 15 | 0–476 |
| 0.0004 | 0.0001 | 0.0001 | 0.0001 | 0.002 | |||||
| <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | |||||
Statistical Analysis
Non-parametric tests of association (Kruskal-Wallis test) and trends were used to examine the relationships between consensus cytologic interpretations and FISH results. Parameters examined included number of nuclei per slide, number of counted nuclei, the percentage of counted nuclei, number of diploid cells, number of tetraploid cells, the percentage of tetraploid cells, number of cells with more than two 3q signals, the percentage of cells with more than two 3q signals, and the relative 3q gain (as defined by higher copy numbers of the probe for 3q compared to CEP7). For some analyses, we combined cytologic interpretations into more clinically relevant groupings of normal, ASC/LSIL, and HSIL (CIN2/3), reflecting the current clinical practice in the United States of treating confirmed HSILs (CIN2 and more severe abnormalities). We also evaluated HSIL (CIN2) and HSIL (CIN3) separately, based on data indicating that a large percentage of HSIL (CIN2) lesions regress spontaneously, whereas evidence for regression of HSIL (CIN3) is sparse. We also considered combinations of 3q parameters, percentage of cells with multiple copies of 3q and maximum number of 3q signal numbers observed for optimization of clinical performance. In addition, we compared different permutations of 3q assessment to cytology at varying thresholds including percentage of informative nuclei with 3q gain (≥1% and ≥5%) and/or identification of any nuclei with a high number of extra 3q copies (≥5 and ≥6).
Receiver Operator Characteristic (ROC) and Distance From Ideal (DFI) analyses: ROC curves were used to further establish optimal thresholds and identify FISH parameters that best distinguish HSIL from LSIL and lesser conditions (normal and ASC). ROC curves were generated by plotting the sensitivity for detecting either HSIL (CIN2 and CIN3) versus 1 minus the specificity fordetecting normal, ASC, or LSIL, obtained at percent cell thresholds ranging from 0% to 10% (0.05% increments). Curves were generated based on the percentage of tetraploid cells, the percentage of cells with 3q gain (excluding tetraploidy), and the percentage of cells with either tetraploidy or 3q gain (ie, all gains of 3q). Curves which come closest to the ideal values of 100% sensitivity and 100% specificity (top left corner of ROC graph, Figure 2A ▶ ) provide the best combination of sensitivity and specificity (assuming equal importance of each) and optimal thresholds are typically selected from points near the “breaks” in the curves (region closest to top left corner; curve slope near 45°). A better view of the dependence of sensitivity and specificity on threshold can be obtained by plotting the DFI versus threshold (Figure 2B) ▶ . DFI is defined here as the distance from the ideal point (0, 1) on the ROC plot (100% sensitivity, 100% specificity), and is calculated as [(1-sensitivity)2 + (1-specificity)2]1/2. DFI is smallest for the best combined sensitivity and specificity (giving equal weight to each) and varies from 0 for thresholds providing 100% sensitivity and 100% specificity, to 1.414 for thresholds providing 0% sensitivity and 0% specificity.
Figure 2.
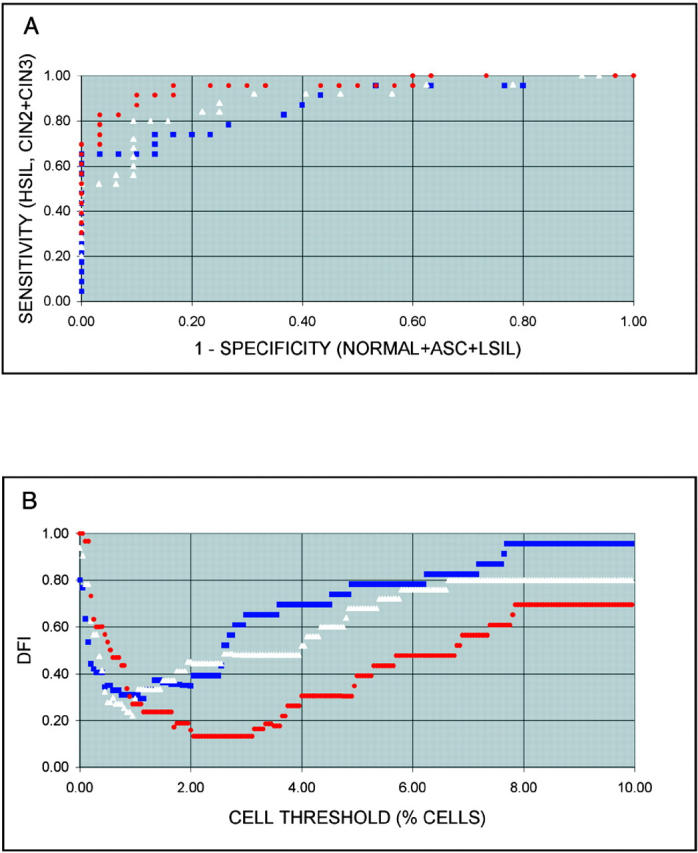
A: Plots of sensitivity versus 1-specificity at percent cell thresholds ranging from 0% to 10% at 0.05% intervals (ROCs) for 3q gain excluding tetraploidy (white triangles), tetraploidy (blue squares), and 3q gain including tetraploidy (red circles). B: DIF versus percent thresholds for 3q gain excluding tetraploidy (white triangles), tetraploidy (blue squares), and 3q gain including tetraploidy (red circles).
Results
Design of Multicolor FISH Probe Set
The pattern of chromosomal imbalances in cervical carcinomas is conserved. Numerous studies using comparative genomic hybridization as a screening test for chromosomal copy number changes showed that the gain of chromosome arm 3q is present in at least 80% of cervical cancers. 16,17,28-34 Based on this pattern of genomic imbalances we have developed a triple-color probe set that allowed us to visualize this genetic marker of early invasive disease directly in cytologic samples. The probe set consists of two repetitive DNA probes that recognize the centromeres of chromosomes 3 (CEP3) and 7 (CEP7). The probe for chromosome arm 3q comprises a set of overlapping BAC clones that contain sequences for the RNA component of TERC. This gene maps to chromosome band 3q26, which is consistently involved in the minimally overlapping region of genomic copy number increases in cervical carcinomas. 16,17 The probe set is depicted in Figure 1A ▶ , along with a display of the CGH results of 40 cervical carcinomas. CEP3 is included in the probe set to evaluate the relative copy number increases of TERC compared to the number of chromosome 3 centromeres. This is especially important in cases involving an isochromosome 3 where the 3q arm is present in three copies while the centromere is present in two copies. CEP7 serves as a control for the overall ploidy of the cells, because chromosome 7 is rarely subject to copy number variations in cervical carcinomas. 16,17,28-30,32,34 We first evaluated the performance of the probe panel on methanol/acetic acid-fixed peripheral lymphocyte cultures from karyotypically normal individuals. The results revealed that less than 1% of the cells display aberrant copy numbers with this fluorescent probe panel (data not shown).
Figure 1.
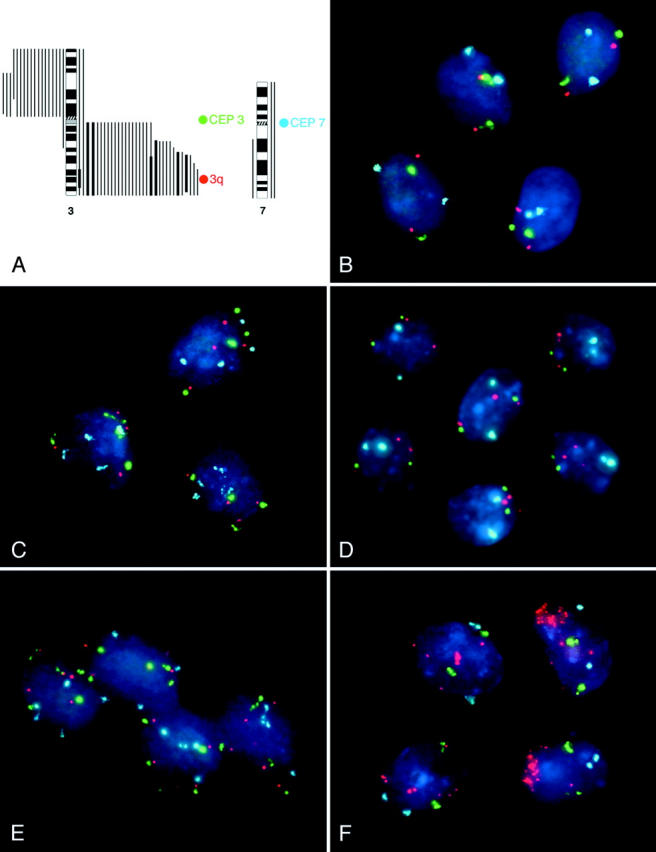
Triple-color probe set for the detection of the 3q gain in cervical dysplasia. The probe set, which was designed based on CGH results, targets the centromeres of chromosome 7 (CEP7, labeled with SA), and chromosome 3 (CEP3, labeled with SG) and the TERC gene (labeled with SO). The bars next to the ideogram show the CGH results of 40 cervical carcinomas. 16,17 . The hybridization of the 3q probe panel to normal epithelial cells of the cervix uteri reveals two signals for each probe in all of the nuclei. A tetraploid pattern (4 signals for CEP7, 4 signals for CEP3, 4 signals for TERC: 4-4-4 pattern) was frequently observed in HSIL (CIN2) and HSIL (CIN3) lesions, here imaged in case 9 (HSIL, CIN 3). Case 6 (HSIL, CIN3) exhibited a variety of aberrant clones with the most frequent one, the 2-2-3 pattern, depicted here. 2-2-3 was the most frequent 3q gain pattern observed in the lesions investigated in this study. The predominant aberrant pattern observed in case 9 was 4-5-5, indicating that a tetraploidization of the genome preceded the acquisition of extra copies of 3q. Nuclei of case 17 (HSIL, CIN3) showed high-level amplification of 3q (up to >20 copies) while the centromere probes were still diploid.
Summary of FISH Results by Cytologic Category
The probe panel was hybridized to 68 cases of thin-layer slides, 57 of which could be successfully evaluated. 11 cases were excluded because there were either too few cells on the slides [n = 7; 1 normal, 2 LSIL, and 4 HSIL (CIN3)] or the hybridization was unsuccessful [n = 4; 1 LSIL and 3 HSIL (CIN3)]. The 57 thin-layer preparations were classified in an independently performed cytologic screening as follows: normal (n = 13); ASC (n = 5); LSIL (n = 14); HSIL (CIN2) (n = 8); HSIL (CIN3) (n = 17). Examples of the hybridizations are presented in Figure 1, B to F ▶ .
Table 1, a to e ▶ ▶ ▶ ▶ ▶ summarizes the results of the enumerations. Between 209 and 3903 nuclei were counted per case. 15.4% (2 of 13) of normal, 0.0% (0 of 5) of ASC, 7.1% (1 of 15) of LSIL, 62.5% (5 of 8) of HSIL (CIN2), and 76.4% (13 of 17) of HSIL (CIN3) lesions were positive for extra copies of chromosome 3q (using 1% 3q-positive cells as the threshold based on the results of the hybridization to the normal peripheral blood lymphocytes). On average, 0.6% (normal), 0.1% (ASC), 0.3% (LSIL), 1.2% (HSIL, CIN2), and 8.8% (HSIL, CIN3) of all cells on the pap smears showed additional 3q copy numbers in the respective diagnostic groups. We also computed a chromosomal instability index for each of the different cytological groups by dividing the number of observed hybridization patterns by the number of cases for each group. These indices increased similarly to the number of 3q-positive cells: 4.0 (normal), 2.6 (ASC), 4.6 (LSIL), 6.5 (HSIL, CIN2), and 11.2 (HSIL, CIN3). This suggests that the gain of 3q can emerge in genetically stable cells, and that chromosomal instability occurs mainly in higher-grade dysplastic lesions.
Table 1A.
Normal
| Case no. | No. of nuclei counted | No. of nuclei with a diploid (2-2-2) pattern | No. of nuclei with a tetraploid (4-4-4) pattern | % of nuclei with a tetraploid (4-4-4) pattern | Hybridization patterns observed in non-tetraploid (non 4-4-4) nuclei with more than 2 copies of 3q and the number of nuclei counted for each pattern. Patterns are described in the following order: CEP7-CEP3-3q | No. of nuclei with more than 2 copies of 3q in non-tetraploid (non 4-4-4) cells | % of nuclei with more than 2 copies of 3q in non-tetraploid (non 4-4-4) cells | No. of nuclei with relative gain of 3q (3q > CEP 7) | % of nuclei with more than 2 copies of 3q including tetraploid (4-4-4) cells |
|---|---|---|---|---|---|---|---|---|---|
| 2 | 485 | 484 | nd | nd | 1 × 2-2-3? | 1 | 0.21 | 1 | 0.21 |
| 3 | 525 | 523 | nd | nd | 2 × 2-2-3? | 2 | 0.38 | 2 | 0.38 |
| 7 | 540 | 531 | 4 | 0.74 | 4 × 2-2-3?, 1 × 2-2-4? | 5 | 0.92 | 5 | 1.67 |
| 10 | 1575 | 1572 | 2 | 0.13 | 1 × 2-3-3? | 1 | 0.06 | 1 | 0.19 |
| 101 | 541 | 523 | 3 | 0.55 | 8 × 2-2-3, 4 × 2-2-4, 1 × 2-2-5, 1 × 3-3-3, 1 × 3-3-4 | 15 | 2.77 | 14 | 3.33 |
| 105 | 543 | 524 | 8 | 1.47 | 6 × 2-2-3, 1 × 2-3-3, 1 × 3-3-3, 1 × 3-4-4, 1 × 2-2-3?, 1 × 2-2-4? | 11 | 2.03 | 10 | 3.50 |
| 106 | 1838 | 1828 | 4 | 0.22 | 2 × 2-2-3, 1 × 2-2-4, 1 × 2-3-3, 2 × 2-2-3? | 6 | 0.33 | 6 | 0.54 |
| 107 | 209 | 208 | 0 | 0 | 1 × 2-2-3 | 1 | 0.48 | 1 | 0.48 |
| 112 | 451 | 447 | 2 | 0.44 | 1 × 2-2-3?, 1 × 2-3-3? | 2 | 0.44 | 2 | 0.89 |
| 119 | 2918 | 2894 | 2 | 0.07 | 12 × 2-2-3, 1 × 2-2-3?, 1 × 2-2-4, 2 × 2-2-4?, 1 × 2-4-4, 5 × 2-3-3 | 22 | 0.75 | 22 | 0.82 |
| A: 24271 | 454 | 452 | 0 | 0 | 1 × 2-2-3, 1 × 3-4-4 | 2 | 0.44 | 2 | 0.44 |
| B: 24338 | 578 | 577 | 0 | 0 | 1 × 2-2-3 | 1 | 0.17 | 1 | 0.17 |
| D: 24323 | 873 | 868 | 1 | 0.11 | 1 × 2-2-4, 2 × 2-2-3?, 1 × 2-4-4 | 4 | 0.46 | 4 | 0.57 |
n = 13.
Shown are the numbers of counted nuclei, the numbers of diploid nuclei (nuclei with two signals for each probe, pattern 2-2-2), the numbers of tetraploid nuclei (nuclei with four signals for each probe, pattern 4-4-4), the percentage of tetraploid nuclei, the hybridization patterns observed in non-tetraploid nuclei with more than two copies of 3q and the number of nuclei counted for each pattern, the number of nuclei with more than two copies of 3q in non-tetraploid nuclei, the percentage of cells with more than two copies of 3q and the relative 3q gain (as defined by higher copy numbers of 3q compared to the control probe CEP7), and the percentage of cells with more than two copies of 3q, including tetraploid cells, for every case, sorted by the cytological diagnosis. Bold numbers denote cases with more than 1% of 3q-positive cells.
*? following a pattern (eg, 2-2-3?) means that the possibility that one of the signals was an artifact cannot be excluded.
Table 1B.
ASC Lesions
| Case no. | No. of nuclei counted | No. of nuclei with a diploid (2-2-2) pattern | No. of nuclei with a tetraploid (4-4-4) pattern | % of nuclei with a tetraploid (4-4-4) pattern | Hybridization patterns observed in nuclei with more than two copies of 3q in non-tetraploid (non 4-4-4) cells and the number of nuclei counted for each pattern. Patterns are described in the following order: CEP7-CEP3-3q | No. ofnuclei with more than 2 copies of 3q in non-tetraploid (non 4-4-4) cells | % of nuclei with more than 3 copies of 3q in non-tetraploid (non 4-4-4) cells | No. of nuclei with relative gain of 3q (3q > CEP 7) | % of nuclei with more than 2 copies of 3q including tetraploid (4-4-4) cells |
|---|---|---|---|---|---|---|---|---|---|
| 14 | 508 | 507 | 1 | 0.2 | 0 | 0 | 0 | 0 | 0.20 |
| 20 | 729 | 723 | 3 | 0.41 | 2 × 2-2-3, 1 × 2-2-3? | 3 | 0.41 | 3 | 0.82 |
| 121 | 2436 | 2432 | 4 | 0.16 | 0 | 0 | 0 | 0 | 0.16 |
| C: 24255 | 1054 | 1052 | 1 | 0.09 | 1 × 2-2-3? | 1 | 0.09 | 1 | 0.19 |
| E: 14174 | 503 | 502 | 0 | 0 | 1 × 2-2-3? | 1 | 0.2 | 1 | 0.20 |
n = 5.
Table 1C.
LSIL
| Case no. | No. of nuclei counted | No. of nuclei with a diploid (2-2-2) pattern | No. of nuclei with a tetraploid (4-4-4) pattern | % of nuclei with a tetraploid (4-4-4) pattern | Hybridization patterns observed in nuclei with more than two copies of 3q in non-tetraploid (non 4-4-4) cells and the number of nuclei counted for each pattern. Patterns are described in the following order: CEP7-CEP3-3q | No. of nuclei with more than 2 copies of 3q in non-tetraploid (non 4-4-4) cells | % of nuclei with more than 2 copies of 3q in non-tetraploid (non 4-4-4) cells | No. of nuclei with relative gain of 3q (3q > CEP 7) | % of nuclei with more than 2 copies of 3q including tetraploid (4-4-4) cells |
|---|---|---|---|---|---|---|---|---|---|
| 102 | 1827 | 1812 | 9 | 0.49 | 2 × 2-3-3 (cluster), 1 × 2-2-4, 1 × 2-3-3?, 1 × 2-4-3?, 1 × 3-3-4 | 6 | 0.33 | 6 | 0.82 |
| 103 | 3903 | 3895 | 5 | 0.13 | 1 × 2-2-3, 1 × 2-3-3, 1 × 3-3-3 | 3 | 0.08 | 2 | 0.20 |
| 104 | 444 | 423 | 8 | 1.8 | 4 × 2-2-3, 3 × 2-2-3?, 4 × 2-3-3, 2 × 2-2-4 | 13 | 2.93 | 13 | 4.73 |
| 110 | 3439 | 3437 | 0 | 0 | 1 × 3-3-3, 1 × 2-2-3? | 2 | 0.06 | 1 | 0.06 |
| 111 | 1332 | 1320 | 2 | 0.15 | 6 × 2-2-3, 3 × 2-2-3?, 1 × 2-3-3 | 10 | 0.75 | 10 | 0.90 |
| 114 | 1784 | 1748 | 29 | 1.63 | 1 × 2-2-3, 1 × 2-2-3?, 1 × 3-3-3, 4 × 3-4-4 | 7 | 0.39 | 6 | 2.02 |
| 115 | 2069 | 2046 | 6 | 0.29 | 11 × 2-2-4, 2 × 2-2-3?, 2 × 3-4-4, 1 × 4-4-3, 1 × 4-4-8 | 17 | 0.82 | 16 | 1.11 |
| 118 | 2401 | 2395 | 2 | 0.08 | 2 × 2-2-3?, 2 × 2-3-3? | 4 | 0.17 | 4 | 0.25 |
| 120 | 2762 | 2707 | 54 | 1.96 | 1 × 2-3-3? | 1 | 0.06 | 1 | 1.99 |
| 123 | 1720 | 1715 | 1 | 0.06 | 3 × 2-2-3?, 1 × 2-3-3? | 4 | 0.23 | 4 | 0.29 |
| 124 | 2310 | 2293 | 10 | 0.43 | 2 × 2-2-3, 3 × 2-2-3?, 1 × 2-2-4, 1 × 5-4-8 | 7 | 0.3 | 7 | 0.74 |
| 125 | 3421 | 3414 | 1 | 0.03 | 4 × 2-2-3?, 1 × 2-2-4, 1 × 2-4-4 | 6 | 0.18 | 6 | 0.20 |
| 130 | 2917 | 2912 | 0 | 0 | 3 × 2-2-3?, 1 × 2-2-4?, 1 × 2-4-4 | 5 | 0.17 | 5 | 0.17 |
| J: 11373 | 654 | 643 | 7 | 1.07 | 1 × 2-2-3?, 1 × 4-2-4, 2 × 4-4-5 | 4 | 0.61 | 3 | 1.68 |
n = 14.
Table 1D.
HSIL (CIN 2)
| Case no. | No. of nuclei counted | No. of nuclei with a diploid (2-2-2) pattern | No. of nuclei with a tetraploid (4-4-4) pattern | % of nuclei with a tetraploid (4-4-4) pattern | Hybridization patterns observed in nuclei with more than two copies of 3q in non-tetraploid (non 4-4-4) cells and the number of nuclei counted for each pattern. Patterns are described in the following order: CEP7-CEP3-3q | No. of nuclei with more than 2 copies of 3q in non-tetraploid (non 4-4-4) cells | % of nuclei with more than 2 copies of 3q in non-tetraploid (non 4-4-4) cells | No. of nuclei with relative gain of 3q (3q > CEP 7) | % of nuclei with more than 2 copies of 3q including tetraploid (4-4-4) cells |
|---|---|---|---|---|---|---|---|---|---|
| 16 | 402 | 395 | 1 | 0.25 | 1 × 2-2-3, 3 × 2-2-3?, 1 × 2-2-4, 1 × 2-4-4 | 6 | 1.49 | 6 | 1.74 |
| 19 | 447 | 412 | 34 | 7.61 | 1 × 3-4-4 | 1 | 0.22 | 1 | 7.83 |
| 2;8 | 903 | 841 | 25 | 2.77 | 12 × 2-2-3, 4 × 2-3-3, 2 × 2-2-4, 7 × 3-3-3, 3 × 3-4-4, 7 × 2-2-3 split | 37 | 4.1 | 30 | 6.87 |
| 109 | 700 | 663 | 25 | 3.57 | 3 × 2-2-3, 1 × 2-2-3?, 1 × 2-2-9?, 1 × 2-4-4, 2 × 3-3-3, 2 × 3-4-4, 1 × 4-4-6, 1 × 4-5-5 | 12 | 1.71 | 10 | 5.29 |
| 116 | 1176 | 1129 | 35 | 2.98 | 3 × 2-2-3, 1 × 3-3-3, 1 × 3-3-4, 1 × 3-3-4?, 3 × 3-4-4, 2 × 3-5-5, 1 × 4-4-6 | 12 | 1.02 | 11 | 4.00 |
| 126 | 2699 | 2614 | 70 | 2.59 | 2 × 2-2-3, 2 × 2-2-3?, 1 × 2-4-4, 2 × 3-4-4, 2 × 4-3-4, 1 × 4-5-5, 1 × 5-5-5, 1 × 6-6-6, 2 × 8-7-8, 1 × 8-8-8 | 15 | 0.56 | 12 | 3.15 |
| 127 | 1667 | 1661 | 5 | 0.3 | 1 × 2-2-3? | 1 | 0.06 | 1 | 0.36 |
| 129 | 1358 | 1266 | 66 | 4.86 | 1 × 2-2-3?, 24 × 3-4-4, 1 × 5-6-7? | 26 | 1.91 | 26 | 6.77 |
n = 8.
Table 1E.
HSIL (CIN 3)
| Case no. | No. of nuclei counted | No. of nuclei with a diploid (2-2-2) pattern | No. of nuclei with a tetraploid (4-4-4) pattern | % of nuclei with a tetraploid (4-4-4) pattern | Hybridization patterns observed in nuclei with more than two copies of 3q in non-tetraploid (non 4-4-4) cells and the number of nuclei counted for each pattern. Patterns are described in the following order: CEP7-CEP3-3q | No. of nuclei with more than 2 copies of 3q in non-tetraploid (non 4-4-4) cells | % of nuclei with more than 2 copies of 3q in non-tetraploid (non 4-4-4) cells | No. of nuclei with relative gain of 3q (3q > CEP 7) | % of nuclei with more than 2 copies of 3q including tetraploid (4-4-4) cells |
|---|---|---|---|---|---|---|---|---|---|
| 6 | 1118 | 558 | 80 | 7.16 | 289 × 2-2-3, 81 × 3-4-4, 29 × 2-3-5, 27 × 3-5-5, 22 × 2-2-4, 3 × 2-4-5, 3 × 2-5-5, 2 × 3-6-6, 2 × 4-8-8, 2 × 2-3-3, 2 × 2-3-4, 1 × 2-4-4, 1 × 3-3-3, 1 × 3-5-4, 1 × 3-5-9, 1 × 3-4-6, 1 × 4-5-4, 1 × 4-6-5, 1 × 4-7-7, 1 × 5-7-7 | 480 | 42.93 | 476 | 50.09 |
| 8 | 347 | 320 | 7 | 2.02 | 5 × 2-2-3, 2 × 2-2-3?, 3 × 2-3-3, 2 × 3-3-3, 4 × 2-3-4, 2 × 3-3-4, 1 × 3-3-5, 1 × 4-5-6 | 20 | 5.76 | 18 | 7.78 |
| 9 | 547 | 441 | 34 | 6.21 | 32 × 4-5-5, 13 × 3-4-4, 7 × 4-6-6, 3 × 3-5-5, 2 × 5-5-5, 2 × 6-4-4, 2 × 7-7-8, 2 × 7-8-8, 1 × 2-2-3, 1 × 2-3-3, 1 × 4-4-7, 1 × 4-5-3, 1 × 4-5-4, 1 × 4-5-6, 1 × 4-6-7, 1 × 5-5-5, 1 × 5-6-5, 1 × 6-5-5, 1 × 6-6-6, 1 × 7-11-11, 1 × 8-11-11 | 72 | 13.16 | 66 | 19.38 |
| 11 | 694 | 668 | 8 | 1.15 | 7 × 2-2-3, 6 × 2-2-3?, 2 × 3-3-3, 1 × 3-4-4, 1 × 3-5-4, 1 × 5-5-4 | 18 | 2.59 | 15 | 3.75 |
| 12 | 1030 | 962 | n.d. | n.d. | 54 × 2-2-4, 8 × 2-2-3, 2 × 2-4-4, 1 × 2-1-3, 1 × 2-1-4, 1 × 2-2-6, 1 × 3-4-7 | 68 | 6.6 | 68 | 6.60 |
| 13 | 467 | 442 | n.d. | n.d. | 17 × 4-2-4, 3 × 3-2-3, 1 × 2-2-3, 1 × 2-2-4, 1 × 3-2-4, 1 × 4-2-3, 1 × 4-3-3 | 25 | 5.35 | 3 | 5.35 |
| 15 | 517 | 450 | 62 | 11.99 | 2 × 2-2-3?, 1 × 2-3-3?, 1 × 2-2-4?, 1 × 4-4-5? | 5 | 0.97 | 5 | 12.96 |
| 17 | 308 | 196 | 0 | 0 | 15 × 2-2-11, 14 × 2-2-6, 12 × 2-2-9, 12 × 2-2-10, 9 × 2-2-13, 9 × 2-2-15, 8 × 2-2-8, 6 × 2-2-12, 6 × 2-2->20, 5 × 2-2-7, 3 × 2-1-9, 3 × 2-2-8, 2 × 2-2-5, 2 × 2-4-12, 2 × 2-5-15, 2 × 2-2-14, 1 × 2-1-4, 1 × 2-2-12, 1 × 2-2-17, 1 × 2-2-18, 1 × 4-4-20 | 112 | 36.36 | 112 | 36.36 |
| 18 | 393 | 364 | 10 | 2.54 | 1 × 4-3-3, 1 × 5-4-3, 4 × 5-5-5, 11 × 5-4-4, 1 × 4-5-5or8, 1 × 6-6-6? | 19 | 4.83 | 0 | 7.38 |
| 2;5 | 758 | 670 | 58 | 7.65 | 7 × 2-2-3, 2 × 2-2-3?, 1 × 2-3-3, 1 × 2-5-5, 5 × 3-3-3, 1 × 3-5-5, 2 × 3-5-6, 1 × 3-7-7, 6 × 3-4-4, 1 × 4-4-6, 3 × 4-6-6 | 30 | 3.96 | 25 | 11.61 |
| 2;14 | 413 | 398 | 11 | 2.66 | 1 × 7-7-7, 1 × 5-5-5?, 1 × 4-5-4, 1 × 5-4-4 | 4 | 0.97 | 0 | 3.63 |
| 113 | 1323 | 1258 | 2 | 0.15 | 45 × 2-2-3, 1 × 2-3-3, 1 × 2-4-4, 1 × 3-4-3, 11 × 3-4-5, 1 × 4-4-6, 1 × 4-5-6, 1 × 5-5-6, 1 × 8-8-8 | 63 | 4.76 | 61 | 4.91 |
| 117 | 2130 | 2024 | 96 | 4.51 | 1 × 3-3-3, 1 × 3-3-4, 2 × 3-4-4, 1 × 3-5-5, 2 × 4-4-5, 1 × 4-4-6, 1 × 4-4-6?, 1 × 4-5-5 | 10 | 0.47 | 9 | 4.98 |
| 128 | 1291 | 1248 | 33 | 2.56 | 3 × 2-2-4, 1 × 2-4-4, 1 × 3-4-4, 1 × 4-2-4, 1 × 4-4-5, 1 × 7-8-8, 1 × 8-8-8?, 1 × 10-10-10 | 10 | 0.77 | 8 | 3.33 |
| F: 12781 | 404 | 330 | 2 | 0.5 | 8 × 2-2-3, 1 × 2-2-3?, 56 × 2-3-3, 1 × 2-2-4, 1 × 4-4-5, 1 × 4-4-6, 2 × 4-5-6, 1 × 4-6-5, 1 × 8-10-12 | 72 | 17.82 | 72 | 18.32 |
| H: 08045 | 674 | 551 | 3 | 0.45 | 117 × 2-2-3, 1 × 2-2-3?, 1 × 2-1-3, 1 × 4-5-5? | 120 | 17.8 | 120 | 18.25 |
| I: 11322 | 690 | 651 | 9 | 1.3 | 11 × 2-2-3, 2 × 2-3-3, 3 × 2-2-4, 1 × 3-3-5, 1 × 3-4-5, 1 × 3-4-7, 6 × 4-4-5, 2 × 4-4-6, 1 × 4-5-5, 1 × 4-5-6, 1 × 4-5-7 | 30 | 4.35 | 30 | 5.65 |
n = 17.
The inclusion of probes for the centromeres of chromosomes 3 and 7 also allowed us to score the percentage of cells that had become tetraploid (defined here as four copies for all signals). In thin-layer specimens diagnosed as normal, only one case showed more than 1% tetraploid cells, none of the ASC cases revealed tetraploid cells, while the percentage increased from 29% in case of LSIL, to 75% (HSIL, CIN2), and 73% (HSIL, CIN3). In some instances tetraploidy was observed in cases in which copy number increases for 3q were absent. In particular the hybridization patterns observed for the HSIL (CIN3) cases suggest that 3q amplification can occur either in diploid cells or after genome tetraploidization. For instance, most clones in cases 9 and 18 (HSIL, CIN3) showed copy numbers of four or more for both centromere probes and numbers greater than four for the 3q probe. In contrast, in cases 12 and 17 (HSIL, CIN3), the copy number of centromeres for chromosomes 3 and 7 is two, and various copy number increases for the 3q probe were observed (Table 1) ▶ .
We hypothesized that the acquisition of extra copies of chromosome 3q, and hence the TERC gene, would render a growth advantage to cervical epithelial cells. One might therefore expect to see some aberrant cells clustering despite the physical disaggregation during the cytological sampling. In many instances cells with aberrant copy numbers for chromosome arm 3q were indeed located next to each other in the thin-layer specimens, and this clustering increased with advanced dysplasia (Figure 1, C to F) ▶ . We suggest that this feature reflects a clonal expansion event, in which cells that carry extra copies of 3q have acquired a distinct growth advantage.
Performance of Different Classification Algorithms Based on Number and/or Percentage of Cells Showing 3q Gain
There were significant differences in the number of cells per slide (P < 0.008), number of cells counted (P < 0.002), percentages of cells counted (P < 0.01), and number of diploid cells (normal, ASC, LSIL and HSIL) but these differences were not related to the degree of cytologic severity (PTrend ≥ 0.3 for all three parameters, Table 1 and 2 ▶ ). The number of tetraploid cells, percentage of tetraploid cells, number of cells with more than two 3q signals, percentage of cells with more than two 3q signals, and relative 3q gain increased significantly with the severity of cytologic interpretation (PTrend < 0.005). These trends did not diminish when cytologic interpretations were categorized as normal, ASC/LSIL, and HSIL (Table 1 and 2) ▶ .
Cytologic severity was compared with 3q gains stratified by percentage of cells and/or identification of cells with high copy numbers of 3q to discriminate cases of normal and mild cytologic abnormalities from those with severe cytologic abnormalities (Table 3) ▶ . Using a threshold of ≥5% of cells with multiple 3q signals and/or at least one observed cell with six or more 3q signals, 0.0% normals, 0.0% ASC, 14.3% LSIL, 50% HSIL (CIN2), and 88.2% HSIL (CIN3) were positive. Changing the threshold to ≥1% of cells with multiple 3q signals and/or at least one observed cell with five or more 3q signals, 15.4% normals, 0.0% ASC, 28.6% LSIL, 75% HSIL (CIN2), and 100.0% HSIL (CIN3) were positive. Finally, using a threshold of ≥5% of cells with multiple 3q signals and/or at least one observed cell with five or more 3q signals, 7.7% normals, 0.0% ASC, 21.4% LSIL, 50% HSIL (CIN2), and 94.1% HSIL (CIN3) were positive (Table 3) ▶ .
Table 3.
Comparison of Cytologic Severity with TERC Gain
| % Cell threshold for 3q gain Max # of 3q signals in ≥1 cell | % Multiple 3q (excluding tetraploidy) | % Multiple 3q (including tetraploidy) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≥5% | ≥1% | ≥5% | ≥1% | ≥5% | ≥1% | ≥5% | ≥2.5% | |||||||||||
| ≥6 | ≥5 | ≥6 | ≥6 | ≥5 | ≥5 | |||||||||||||
| Neg | Pos | Neg | Pos | Neg | Pos | Neg | Pos | Neg | Pos | Neg | Pos | Neg | Pos | Neg | Pos | Neg | Pos | |
| Normal | 13 | 0 | 12 | 1 | 11 | 2 | 13 | 0 | 11 | 2 | 13 | 0 | 11 | 2 | 12 | 1 | 11 | 2 |
| 100.0 | 0.0 | 92.3 | 7.7 | 84.6 | 15.4 | 100.0 | 0.0 | 84.6 | 15.4 | 100.0 | 0.0 | 84.6 | 15.4 | 92.3 | 7.7 | 84.6 | 15.4 | |
| ASC | 5 | 0 | 5 | 0 | 5 | 0 | 5 | 0 | 5 | 0 | 5 | 0 | 5 | 0 | 5 | 0 | 5 | 0 |
| 100.0 | 0.0 | 100.0 | 0.0 | 100.0 | 0.0 | 100.0 | 0.0 | 100.0 | 0.0 | 100.0 | 0.0 | 100.0 | 0.0 | 100.0 | 0.0 | 100.0 | 0.0 | |
| LSIL | 12 | 2 | 11 | 3 | 13 | 1 | 14 | 0 | 11 | 3 | 12 | 2 | 10 | 4 | 11 | 3 | 13 | 1 |
| 85.7 | 14.3 | 78.6 | 21.4 | 92.9 | 7.1 | 100.0 | 0.0 | 78.6 | 21.4 | 85.7 | 14.3 | 71.4 | 28.6 | 78.6 | 21.4 | 92.9 | 7.1 | |
| HSIL (CIN2) | 4 | 4 | 4 | 4 | 3 | 5 | 8 | 0 | 2 | 6 | 4 | 4 | 2 | 6 | 4 | 4 | 2 | 6 |
| 50.0 | 50.0 | 50.0 | 50.0 | 37.5 | 62.5 | 100.0 | 0.0 | 25.0 | 75.0 | 50.0 | 50.0 | 25.0 | 75.0 | 50.0 | 50.0 | 25.0 | 75.0 | |
| HSIL (CIN3) | 4 | 13 | 2 | 15 | 4 | 13 | 9 | 8 | 1 | 16 | 2 | 15 | 0 | 17 | 1 | 16 | 0 | 17 |
| 23.5 | 76.5 | 11.8 | 88.2 | 23.5 | 76.5 | 52.9 | 47.1 | 5.9 | 94.1 | 11.8 | 88.2 | 0.0 | 100.0 | 5.9 | 94.1 | 0.0 | 100.0 | |
| Normal or ASC or LSIL | 30 | 2 | 28 | 4 | 29 | 3 | 32 | 0 | 27 | 5 | 30 | 2 | 26 | 6 | 28 | 4 | 29 | 3 |
| 93.8 | 6.3 | 87.5 | 12.5 | 90.6 | 9.4 | 100.0 | 0.0 | 84.4 | 15.6 | 93.8 | 6.3 | 81.3 | 18.8 | 87.5 | 12.5 | 90.6 | 9.4 | |
| ASC or LSIL | 17 | 2 | 16 | 3 | 18 | 1 | 19 | 0 | 16 | 3 | 17 | 2 | 15 | 4 | 16 | 3 | 18 | 1 |
| 89.5 | 10.5 | 84.2 | 15.8 | 94.7 | 5.3 | 100.0 | 0.0 | 84.2 | 15.8 | 89.5 | 10.5 | 79.0 | 21.1 | 84.2 | 15.8 | 94.7 | 5.3 | |
| HSIL (CIN2 or CIN3) | 8 | 17 | 6 | 19 | 7 | 18 | 17 | 8 | 3 | 22 | 6 | 19 | 2 | 23 | 5 | 20 | 2 | 23 |
| 32.0 | 68.0 | 24.0 | 76.0 | 28.0 | 72.0 | 68.0 | 32.0 | 12.0 | 88.0 | 24.0 | 76.0 | 8.0 | 92.0 | 20.0 | 80.0 | 8.0 | 92.0 | |
A comparison of cytologic severity with 3q gains stratified by percentage of cells (≥1% or ≥5%) and/or identification of cells with high copy number (≥5 copies or ≥6 copies) and cells with 3q gains and/or tetraploidy (threshold of ≥2.5%) to discriminate cases of normal and mild cytologic abnormalities from those with severe cytologic abnormalities (HSIL).
Neg, negative; Pos, positive.
Classification algorithms were further developed using ROC curves, which plot sensitivity versus 1-specificity over a range of threshold values. Figure 2A ▶ shows ROC curves for discriminating HSIL (CIN2 and CIN3) from LSIL, ASC, and normal cases. Curves are plotted for discrimination based on the percentage of tetraploid cells, the percentage of cells with 3q gain excluding tetraploid cells, and the percentage of cells with 3q gain including tetraploidy. Based on the curve lying closest to the top left corner of the graph, 3q gain including tetraploidy provides the best method for distinguishing HSIL. Figure 2B ▶ shows DFI curves, which reveal the threshold values used to calculate the points plotted in the ROC curves. DFI, the distance from the top left corner of the ROC graph to any plotted point, combines sensitivity and specificity into a single value, equaling 0 for 100% sensitivity and 100% specificity, and increasing as either sensitivity or specificity decrease from 100%. As expected from the ROC curves, 3q gain including tetraploidy shows the lowest DFI. The broad minimum of this curve also indicates the most robust behavior (least sensitivity to variation of the threshold value), with optimal discrimination obtained at thresholds between 2.05% and 3.10% cells. Over this range of thresholds, the sensitivity for detecting HSIL (CIN2 and CIN3) was 92% and the specificity relative to lower conditions (normal, ASC and LSIL) was 91%. For comparison to the other classification algorithms, the last column in Table 3 ▶ lists the number of negative and positive specimens, for each cytologic class, based on the percentage of cells with 3q gain, including tetraploidy, and a threshold value of 2.5%.
Discussion
Based on extensive screening of cervical carcinomas using molecular cytogenetic techniques, namely comparative genomic hybridization, it has been firmly established that tumorigenesis in the epithelial cells of the uterine cervix requires specific genomic alterations, which commonly include the acquisition of additional copies of the long arm of chromosome 3. 16,17,28-34 The observation that genetic aberrations, such as chromosomal aneuploidies are essential for tumorigenesis, applies to other human carcinomas as well. 14,15 It is therefore very likely that the commitment of cells to malignant transformation is associated, if not caused, by the acquisition of these carcinoma specific chromosomal imbalances. Such imbalances do not occur in normal epithelial cells. We have previously demonstrated that the translational application of visualizing genomic imbalances identified by CGH via interphase FISH allows the diagnosis of breast carcinomas in fine needle aspirates with high sensitivity and specificity. 35 We have now developed a three-color fluorescent probe set for the visualization of specific chromosomal aneuploidies and genomic amplification of TERC directly in thin-layer cytological specimens. This study demonstrated that it is possible to simultaneously assay routinely collected liquid-based cytology samples for 3q amplification and for copy numbers of chromosome 3 and 7, using a specifically designed probe cocktail. Based on intensive screening of thin-layer slides, we identified extra copies of 3q (≥ 1% of cells) in 2 of 13 (15%) women with normal cytology as opposed to 13 of 17 (76%) women with HSIL (CIN3), suggesting that increased 3q copy numbers are associated with the severity of cytologic findings. Furthermore, among cases containing cells showing 3q amplification, the percentage of cells carrying extra copies of 3q was higher in cases classified as HSIL (CIN3) as compared to cases with milder cytologic interpretations. A post hoc analysis of our data indicated that the performance of the FISH assay varied with the criteria used for detecting a positive result. For example, classification of slides showing ≥5% of cells with 3q amplification and at least one cell with 6 copies of 3q detected 88% of HSIL (CIN3), whereas none of the cases classified as normal or ASC were positive. Applying ROC we established that discernment of normal and ASC/LSIL from HSIL lesions can be achieved with the highest combined sensitivity and specificity when tetraploid cell counts were combined with cells with 3q gains at threshold values of 2.05% to 3.1%. Other criteria for scoring the 3q amplification assay as positive increased sensitivity at the cost of reduced specificity, whereas stringent thresholds were associated with the reverse. A priori testing using the FISH assay at one or more designated thresholds will be needed to assess its clinical performance.
It was intriguing to see that the percentages of cells that showed hybridization patterns that were consistent with a tetraploidization of the genome increased as well with severity of dysplasia (even though we did not measure the DNA content quantitatively, but deduced tetraploidization based on a 4-4-4 hybridization pattern). This genomic doubling was consistent with the impairment of TP53 and RB1-function mediated through the E6 and E7 proteins of human papillomavirus (HPV). 36 Uncoupling of DNA synthesis and cell division would increase the population of tetraploid cells. It is probable that these cells had an increased propensity for chromosome segregation errors and hence, developed genomic imbalances. Selection for those cells containing the complement of chromosomes necessary for dysregulated cervical epithelial cell growth resulted in the gain of 3q, which we frequently observed. However, such genomic evolution via tetraploidization is certainly not present in all cases and is therefore not mandatory, as we detected HSIL lesions that contain almost exclusively diploid cells with a relative increase of 3q. This suggests that detection of 3q gain could be more sensitive than measurement of the nuclear DNA content alone. 10,37,38 The interpretation of the hybridization patterns provides evidence that chromosomal instability, measured in this study as the variability of hybridization patterns among cells in a given case, increases with the grade of dysplasia as well. This suggests that early chromosomal aneuploidies can develop in an otherwise genomically stable nucleus, and that the acquisition of specific chromosomal imbalances (in cervical carcinomas the gain of 3q) coincides with the HPV-mediated compromise of TP53 and RB1 function to promote tumorigenesis and eventually the acquisition of crude nuclear aneuploidy that is present in most invasive cervical carcinomas. 17
Although cytologic screening has had an enormous beneficial impact in developed nations, cytology has significant limitations, notably limited single-test sensitivity, poor reproducibility, and relatively frequent equivocal results. 2,3 Recently, testing for oncogenic types of HPV DNA has been endorsed as a technique for identifying women with equivocal cytology who require immediate follow-up and the potential effectiveness of primary screening using HPV testing has received increased attention. The strength of sensitive HPV testing is that it provides extremely high negative predictive value; women who test negative are at low risk for cervical cancer. However, the positive predictive value of HPV testing is limited, especially among young sexually active women, among whom transient innocuous infections are very common. 4,5 Accordingly, the development of assays with high-specificity for detecting cancer precursors as well as excellent sensitivity would represent attractive alternatives as primary screening tests or as tests to complement cytology, HPV typing, or other assays. In our study, only one of 14 LSILs revealed a 3q gain, whereas recent studies have found that ∼80% of LSILs test positive for oncogenic HPV types. 4,5 The value of FISH assays in identifying women with LSIL who harbor undetected HSIL or are destined to progress has therefore great potential; however, future studies are needed because our analysis was based on relatively low case numbers.
The fact that only few cells in the premalignant lesions contained additional copies of 3q also clearly shows that a cell-based evaluation technique such as interphase FISH has great advantages over techniques that would require DNA extraction from all cells of the sample, eg, polymerase chain reaction or microarray-based analyses. Even in HSIL (CIN3) lesions, only a few cases revealed additional copy numbers for 3q in more than 10% of the cells. Accordingly, DNA extraction from the mixture of cells from the entire specimen would result in unavoidable and significant dilution of the results by the more than 90% of normal cells in a routine diagnostic sample.
Our results support the use of gene-specific probes for diagnostic interphase cytogenetics. The use of centromere-specific probes alone would have resulted in the enumeration of normal diploid copy numbers, or in some instances tetraploid cells, in cases that actually carried extra copies of 3q and the TERC gene. These genetic alterations would have therefore remained undetected. 39 The relevance of this conclusion is corroborated by our results that clearly show that initial copy number increases of the long arm of chromosome 3q can occur in diploid, chromosomally stable cells.
It is at present not proven that the TERC gene is the only or most important target for the frequent gain of chromosome arm 3q in cervical carcinomas. However, the fact that such a high percentage of dysplastic lesions carry extra copies of this gene increases the probability of a causative involvement; the use of another candidate gene on chromosome 3q, PIK3CA, showed copy number increases in only 43% of carcinomas. 23 Immortalization via increased telomerase activity is a plausible mechanism, since telomerase genes are transcriptionally activated by the E6 protein of HPV in genital keratinocytes. 40 Genomic amplification of TERC might further enhance this pathway.
Unfortunately, in this proof-of-principle study, patient follow-up was not available to us, nor was the HPV status. It would have been of great interest to explore the fate of the two women whose Pap smears were cytologically diagnosed as normal, yet in whose samples 3q-positive cells were detected at a level of 2.03% and 2.77%. One could speculate that such cases represent examples of false-negative cytologic interpretations in cervical cytology. This could be due to the fact that the few cells that carried carcinoma-specific chromosomal aneuploidies were overlooked in the morphological evaluation; alternatively, one could speculate that despite the presence of extra copies of 3q morphological changes had not yet been manifested, such that no other diagnosis would be possible, but that these women might be at future risk of developing precancerous or cancerous lesions.
In summary, we have developed a genetic test for the diagnosis of dysplastic lesions directly in routinely collected thin-layer specimens. The test is based on the visualization of extra copies of chromosome 3q, which is achieved using interphase FISH with a triple-color probe panel that includes TERC. Our results clearly suggest that, independent from any other marker, the visualization of aneuploidy of chromosome 3q and amplification of TERC can detect dysplastic cells. These results, and the availability of computer-assisted or automated image acquisition and analysis hardware and software now provide both the rationale as well as the tools to explore the diagnostic and predictive power of this genetic marker in individual cases.
Acknowledgments
We thank Buddy Chen and Joseph Cheng for editorial assistance and IT support, Dr. Scott Hall (Department of Pathology, York Hospital, York, PA) for donating specimens and Dr. Leopold Koss (Albert Einstein College of Medicine, Bronx, NY) and Turid Knutsen for critically reading the manuscript.
Footnotes
Address reprint requests to Thomas Ried, M.D., Genetics Branch, CCR/NCI/NIH, Building 50, Room 1306, 50 South Drive, Bethesda, MD 20892-8010. E-mail: riedt@mail.nih.gov.
Supported in parts through a cooperative research and development agreement (CRADA) with Vysis, Inc./Abbott Laboratories.
References
- 1.Papanicolaou GN, Traut HF: The diagnostic value of vaginal smears in carcinomas of the uterus. Am J Obstet Gynecol 1941, 42:193-206 [Google Scholar]
- 2.Koss LG: The Papanicolaou test for cervical cancer detection: a triumph and a tragedy. JAMA 1989, 261:737-743 [PubMed] [Google Scholar]
- 3.Shingleton HM, Patrick RL, Johnston WW, Smith RA: The current status of the Papanicolaou smear. CA Cancer J Clin 1995, 45:305-320 [DOI] [PubMed] [Google Scholar]
- 4.Castle PE, Wacholder S, Lorincz AT, Scott DR, Sherman ME, Glass AG, Rush BB, Schussler JE, Schiffman M: A prospective study of high-grade cervical neoplasia risk among human papillomavirus-infected women. J Natl Cancer Inst 2002, 94:1406-1414 [DOI] [PubMed] [Google Scholar]
- 5.Castle PE, Wacholder S, Sherman ME, Lorincz AT, Glass AG, Scott DR, Rush BB, Demuth F, Schiffman M: Absolute risk of a subsequent abnormal pap among oncogenic human papillomavirus DNA-positive, cytologically negative women. Cancer 2002, 95:2145-2151 [DOI] [PubMed] [Google Scholar]
- 6.Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ, Munoz N: Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol 1999, 189:12-19 [DOI] [PubMed] [Google Scholar]
- 7.zur Hausen H: Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer 2002, 2:342-350 [DOI] [PubMed] [Google Scholar]
- 8.von Knebel Doeberitz M: New molecular tools for efficient screening of cervical cancer. Dis Markers 2001, 17:123-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jarboe EA, Liaw KL, Thompson LC, Heinz DE, Baker PL, McGregor JA, Dunn T, Woods JE, Shroyer KR: Analysis of telomerase as a diagnostic biomarker of cervical dysplasia and carcinoma. Oncogene 2002, 21:664-673 [DOI] [PubMed] [Google Scholar]
- 10.Bollmann R, Mehes G, Torka R, Speich N, Schmitt C, Bollmann M: Human papillomavirus typing and DNA ploidy determination of squamous intraepithelial lesions in liquid-based cytologic samples. Cancer 2003, 99:57-62 [DOI] [PubMed] [Google Scholar]
- 11.Reesink-Peters N, Helder MN, Wisman GB, Knol AJ, Koopmans S, Boezen HM, Schuuring E, Hollema H, de Vries EG, de Jong S, van der Zee AG: Detection of telomerase, its components, and human papillomavirus in cervical scrapings as a tool for triage in women with cervical dysplasia. J Clin Pathol 2003, 56:31-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams GH, Romanowski P, Morris L, Madine M, Mills AD, Stoeber K, Marr J, Laskey RA, Coleman N: Improved cervical smear assessment using antibodies against proteins that regulate DNA replication. Proc Natl Acad Sci USA 1998, 95:14932-14937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baldwin P, Laskey R, Coleman N: Translational approaches to improving cervical screening. Nat Rev Cancer 2003, 3:217-226 [DOI] [PubMed] [Google Scholar]
- 14.Heim S, Mitelman F: Cancer Cytogenetics 1995. Wiley-Liss New York
- 15.Ried T, Heselmeyer-Haddad K, Blegen H, Schrock E, Auer G: Genomic changes defining the genesis, progression, and malignancy potential in solid human tumors: a phenotype/genotype correlation. Genes Chromosomes Cancer 1999, 25:195-204 [DOI] [PubMed] [Google Scholar]
- 16.Heselmeyer K, Schrock E, du Manoir S, Blegen H, Shah K, Steinbeck R, Auer G, Ried T: Gain of chromosome 3q defines the transition from severe dysplasia to invasive carcinoma of the uterine cervix. Proc Natl Acad Sci USA 1996, 93:479-484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heselmeyer K, Macville M, Schrock E, Blegen H, Hellstrom AC, Shah K, Auer G, Ried T: Advanced-stage cervical carcinomas are defined by a recurrent pattern of chromosomal aberrations revealing high genetic instability and a consistent gain of chromosome arm 3q. Genes Chromosomes Cancer 1997, 19:233-240 [PubMed] [Google Scholar]
- 18.Hackett JA, Greider CW: Balancing instability: dual roles for telomerase and telomere dysfunction in tumorigenesis. Oncogene 2002, 21:619-626 [DOI] [PubMed] [Google Scholar]
- 19.Maser RS, DePinho RA: Connecting chromosomes, crisis, and cancer. Science 2002, 297:565-569 [DOI] [PubMed] [Google Scholar]
- 20.Cremer T, Landegent JE, Bruckner A, Scholl HP, Schardin M, Hager H-D, Devilee P, Pearson PL, van der Ploeg M: Detection of chromosome aberrations in the human interphase nucleus by visualization of specific target DNAs with radioactive and nonradioactive in situ hybridization techniques: diagnosis of trisomy 18 with probe L1.84. Hum Genet 1986, 74:346-352 [DOI] [PubMed] [Google Scholar]
- 21.Van Dekken H, Rosenberg C, Krijtenburg P, Alers J: Interphase cytogenetics and comparative genomic hybridization of human epithelial cancers and precursor lesions. Histochem Cell Biol 1997, 108:419-430 [DOI] [PubMed] [Google Scholar]
- 22.Kurtycz D, Nunez M, Arts T, Bauman C, Harris C, Inhorn S, Meisner L: Use of fluorescent in situ hybridization to detect aneuploidy in cervical dysplasia. Diagn Cytopathol 1996, 15:46-51 [DOI] [PubMed] [Google Scholar]
- 23.Zhang A, Maner S, Betz R, Angstrom T, Stendahl U, Bergman F, Zetterberg A, Wallin KL: Genetic alterations in cervical carcinomas: frequent low-level amplifications of oncogenes are associated with human papillomavirus infection. Int J Cancer 2002, 101:427-433 [DOI] [PubMed] [Google Scholar]
- 24.Mian C, Bancher D, Kohlberger P, Kainz C, Haitel A, Czerwenka K, Stani J, Breitenecker G, Wiener H: Fluorescence in situ hybridization in cervical smears: detection of numerical aberrations of chromosomes 7, 3, and X and relationship to HPV infection. Gynecol Oncol 1999, 75:41-46 [DOI] [PubMed] [Google Scholar]
- 25.Greider CW: Telomerase and telomere-length regulation: lessons from small eukaryotes to mammals. Cold Spring Harb Symp Quant Biol 1993, 58:719-723 [DOI] [PubMed] [Google Scholar]
- 26.Solomon D, Davey D, Kurman R, Moriarty A, O’Connor D, Prey M, Raab S, Sherman M, Wilbur D, Wright T, Jr, Young N: The 2001 Bethesda System: terminology for reporting results of cervical cytology. JAMA 2002, 287:2114-2119 [DOI] [PubMed] [Google Scholar]
- 27.Bittner ML, Morrison LE, Legator MS: Direct label transaminated DNA probe compositions for chromosome identification and methods for their manufacture. United States Patent 5 1996, 491:224 [Google Scholar]
- 28.Dellas A, Torhorst J, Jiang F, Proffitt J, Schultheiss E, Holzgreve W, Sauter G, Mihatsch MJ, Moch H: Prognostic value of genomic alterations in invasive cervical squamous cell carcinoma of clinical stage IB detected by comparative genomic hybridization. Cancer Res 1999, 59:3475-3479 [PubMed] [Google Scholar]
- 29.Kirchhoff M, Rose H, Petersen BL, Maahr J, Gerdes T, Lundsteen C, Bryndorf T, Kryger-Baggesen N, Christensen L, Engelholm SA, Philip J: Comparative genomic hybridization reveals a recurrent pattern of chromosomal aberrations in severe dysplasia/carcinoma in situ of the cervix and in advanced-stage cervical carcinoma. Genes Chromosomes Cancer 1999, 24:144-150 [DOI] [PubMed] [Google Scholar]
- 30.Allen DG, White DJ, Hutchins AM, Scurry JP, Tabrizi SN, Garland SM, Armes JE: Progressive genetic aberrations detected by comparative genomic hybridization in squamous cell cervical cancer. Br J Cancer 2000, 83:1659-1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang YC, Shyong WY, Chang MS, Chen YJ, Lin CH, Huang ZD, Wang, Hsu MT, Chen ML: Frequent gain of copy number on the long arm of chromosome 3 in human cervical adenocarcinoma. Cancer Genet Cytogenet 2001, 131:48-53 [DOI] [PubMed] [Google Scholar]
- 32.Umayahara K, Numa F, Suehiro Y, Sakata A, Nawata S, Ogata H, Suminami Y, Sakamoto M, Sasaki K, Kato H: Comparative genomic hybridization detects genetic alterations during early stages of cervical cancer progression. Genes Chromosomes Cancer 2002, 33:98-102 [DOI] [PubMed] [Google Scholar]
- 33.Hidalgo A, Schewe C, Petersen S, Salcedo M, Gariglio P, Schluns K, Dietel M, Petersen I: Human papilloma virus status and chromosomal imbalances in primary cervical carcinomas and tumour cell lines. Eur J Cancer 2000, 36:542-548 [DOI] [PubMed] [Google Scholar]
- 34.Matthews CP, Shera KA, McDougall JK: Genomic changes and HPV type in cervical carcinoma. Proc Soc Exp Biol Med 2000, 223:316-321 [DOI] [PubMed] [Google Scholar]
- 35.Heselmeyer-Haddad K, Chaudhri N, Stoltzfus P, Cheng JC, Wilber K, Morrison L, Auer G, Ried T: Detection of chromosomal aneuploidies and gene copy number changes in fine needle aspirates is a specific, sensitive, and objective genetic test for the diagnosis of breast cancer. Cancer Res 2002, 62:2365-2369 [PubMed] [Google Scholar]
- 36.Ramel S, Sanchez CA, Schimke MK, Neshat K, Cross SM, Raskind WH, Reid BJ: Inactivation of p53 and the development of tetraploidy in the elastase-SV40 T antigen transgenic mouse pancreas. Pancreas 1995, 11:213-222 [DOI] [PubMed] [Google Scholar]
- 37.Kashyap V, Das BC: DNA aneuploidy and infection of human papillomavirus type 16 in preneoplastic lesions of the uterine cervix: correlation with progression to malignancy. Cancer Lett 1998, 123:47-52 [DOI] [PubMed] [Google Scholar]
- 38.Lorenzato M, Clavel C, Masure M, Nou JM, Bouttens D, Evrard G, Bory JP, Maugard B, Quereux C, Birembaut P: DNA image cytometry and human papillomavirus (HPV) detection help to select smears at high risk of high-grade cervical lesions. J Pathol 2001, 194:171-176 [DOI] [PubMed] [Google Scholar]
- 39.Ried T: Interphase cytogenetics and its role in molecular diagnostics of solid tumors. Am J Pathol 1998, 152:325-327 [PMC free article] [PubMed] [Google Scholar]
- 40.Veldman T, Horikawa I, Barrett JC, Schlegel R: Transcriptional activation of the telomerase hTERT gene by human papillomavirus type 16 E6 oncoprotein. J Virol 2001, 75:4467-4472 [DOI] [PMC free article] [PubMed] [Google Scholar]


