Abstract
A constitutively active form of mitogen-activated protein kinase kinase (MEK1) was synthesized under control of a zinc-inducible promoter in NIH 3T3 fibroblasts. Zinc treatment of serum-starved cells activated extracellular signal-regulated protein kinases (ERKs) and induced expression of cyclin D1. Newly synthesized cyclin D1 assembled with cyclin-dependent kinase-4 (CDK4) to form holoenzyme complexes that phosphorylated the retinoblastoma protein inefficiently. Activation of the MEK1/ERK pathway neither triggered degradation of the CDK inhibitor kinase inhibitory protein-1 (p27Kip1) nor led to activation of cyclin E- and A-dependent CDK2, and such cells did not enter the DNA synthetic (S) phase of the cell division cycle. In contrast, zinc induction of active MEK1 in cells also engineered to ectopically overexpress cyclin D1 and CDK4 subunits generated levels of cyclin D-dependent retinoblastoma protein kinase activity approximating those achieved in cells stimulated by serum. In this setting, p27Kip1 was mobilized into complexes containing cyclin D1; cyclin E- and A-dependent CDK2 complexes were activated; and serum-starved cells entered S phase. Thus, although the activity of p27Kip1 normally is canceled through a serum-dependent degradative process, overexpressed cyclin D1-CDK complexes sequestered p27Kip1 and reduced the effective inhibitory threshold through a stoichiometric mechanism. A fraction of these cells completed S phase and divided, but they were unable to continuously proliferate, indicating that other serum-responsive factors ultimately became rate limiting for cell cycle progression. Therefore, the MEK/ERK pathway not only acts transcriptionally to induce the cyclin D1 gene but functions posttranslationally to regulate cyclin D1 assembly with CDK4 and to thereby help cancel p27Kip1-mediated inhibition.
Regulation of mammalian cell proliferation by extracellular signals occurs during the first gap (G1) phase of the cell division cycle. During this interval, growth stimulatory and growth inhibitory signals transduced from the extracellular environment converge on the cell cycle control machinery, the engine of which is driven by cyclins and cyclin-dependent kinases (CDKs) and opposed by CDK inhibitors (1). Enzymes that regulate G1 phase progression include CDK4 and CDK6, which can be activated through their association with any one of three D-type cyclins, and CDK2, which forms active holoenzyme complexes with cyclins E and A (2, 3).
Mitogens stimulate synthesis of D-type cyclins and their assembly with CDK4 or CDK6 (4–6). Cyclin D-CDK complexes phosphorylate the retinoblastoma protein (RB) (4, 5, 7–9), helping to cancel its growth suppressive function by eliminating its ability to function as a transcriptional corepressor (10). In addition, they “titrate” CDK inhibitors, such as kinase inhibitory protein-1 (p27Kip1), into ternary complexes, thereby freeing cyclin E-CDK2 complexes from such constraint (1, 11–16). The cyclin E-CDK2 holoenzyme contributes to RB phosphorylation (17–19), phosphorylates p27Kip1 to trigger its ubiquitin-mediated degradation (20–22), and likely modifies components of preinitiation complexes to trigger DNA replication per se (23, 24). Gene products that coordinate S phase entry include cyclin A, which is induced in late G1 and is essential for DNA synthesis (25–27). The irreversible decision to enter S phase, which is made at the so-called restriction point late in G1 (28), therefore is marked by several molecular events, including (i) RB phosphorylation, (ii) p27Kip1 degradation, (iii) initiation of cyclin A synthesis, and (iv) CDK2 activation (1, 10, 29).
Growth factor receptor tyrosine kinases activate a class of intracellular serine/threonine protein kinases termed mitogen-activated protein kinases (MAPKs) or extracellular signal-regulated kinases (ERKs) (30, 31). A pathway from receptor kinases to ERKs sequentially involves the small GTP-binding protein Ras and the Raf-1 protein kinase (32–36). Raf-1 phosphorylates MAPK/ERK kinases (MEKs), which in turn activate ERKs and facilitate their nuclear translocation (37). Several observations implicate the Ras/ERK signaling pathway in cyclin D1 regulation. Microinjection of activated, but not wild-type, forms of Ras can initiate DNA synthesis in quiescent fibroblasts (38), whereas neutralizing Ras antibodies block DNA synthesis induced by mitogens (39–41). The mitogen-dependent induction of the cyclin D1 gene depends on Ras, Raf-1, and ERK activities, with their induced expression being necessary and sufficient for cyclin D1 transcription (40, 42–46). Sustained activation of ERKs is required for fibroblasts to pass the G1 restriction point (47), and in Ras- or Raf-transformed fibroblasts, cyclin D1 levels are constitutively elevated (48, 49). Specific inhibitors of cyclin D-dependent kinases (INK4 proteins) block Ras-mediated cell proliferation and transformation in an RB-dependent manner (40, 41, 50, 51), arguing that cyclin D-dependent kinases are key physiologic targets in this pathway. Here, we report posttranslational effects of MEK/ERK signaling on the assembly and activation of cyclin D-dependent kinases.
MATERIALS AND METHODS
Special Reagents.
Rabbit polyclonal antibodies against ERK1 (K-23), ERK2 (C-14), cyclin E (M-20), cyclin A (C-19), and p21Cip1 (C-19) were purchased from Santa Cruz Biotechnology. Hybridoma cells producing mAb 9E10 to a myc-epitope (ATCC CRL-1729) were purchased from the American Type Culture Collection, and culture medium containing antibody was produced as described (52). mAb against mouse cyclin D1 (72–13G-11) (53), and rabbit antisera to CDK4 (RY to full-length CDK4 and RZ to a CDK4 C-terminal peptide) (4), to full-length p27Kip1 (RLL) (14), and to a CDK2 C-terminal peptide (TKPVPHLRL) (sera RCC and RDD) (54) were produced in our laboratory. Myelin basic protein was from Sigma, histone H1 from Boehringer Mannheim, and G418 and puromycin from GIBCO/BRL.
Cells and Culture Conditions.
NIH 3T3 cells were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine and 100 units/ml each of penicillin and streptomycin. To make them quiescent, cells were washed twice with PBS and cultured for 24–48 hr in serum-depleted medium (DMEM with 0.1% FBS, 0.4 mg/ml BSA, glutamine, penicillin and streptomycin). Quiescent cells were treated with 25 μM ZnSO4 in serum-depleted medium or with complete medium containing 10% FBS, and entry into S phase was monitored by estimating their DNA content by fluorescence-activated flow cytometry (55).
Expression Vectors.
A constitutively active MEK1 mutant (56) was provided by Natalie G. Ann (University of Colorado, Boulder). MEK1 cDNA was amplified by using a PCR performed with a 5′ oligonucleotide (CGGGGTACCACTAGTATGGAACAAAAGCTTATTTCTGAAGAAGACTTGCTGCCAAGAAGAAGCCG) encoding recognition sequences for KpnI (underlined), a myc epitope tag, and the first 18 bases of the MEK1 sense strand; and a 3′ oligonucleotide (GCTCTAGATCAGATGCTGGCAGCGTGGGTTGGTGTGCTGGG) containing an XbaI site (underlined) and the last 20 bases (antisense strand) of MEK1 coding sequences. The PCR product was subcloned into the KpnI and XbaI sites of the expression vector pMTCB6, which contains a zinc-inducible metallothionein promoter and a linked neomycin-resistance gene (57). Coding regions of the pMTCB6-MEK1 plasmid were validated by nucleotide sequencing analysis. MEK1* denotes the constitutively active, myc-tagged mutant.
Selection of Stable Cell Lines.
pMTCB6-MEK1* DNA (25 μg) was transfected into NIH 3T3 cells (58), selected for 3 weeks in 800 μg/ml of G418, and maintained in medium containing 400 μg/ml of drug. pMTCB6-MEK1* (20 μg) was cotransfected with pJ6Ω-puro (5 μg) encoding puromycin resistance (59) into NIH 3T3 cells engineered as described (60) to ectopically synthesize cyclin D1 and CDK4. Cells were selected for 3 weeks in complete medium containing 7.5 μg/ml puromycin, and MEK1*-positive subclones were obtained by limiting dilution of single cells in 96-well microtiter plates.
Immunoblotting.
Cells were disrupted in lysis buffer containing 50 mM Hepes (pH 7.4), 1% Nonidet P-40, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 1 mM DTT, 4 μg/ml aprotinin, 4 μg/ml pepstatin (both from Sigma), 0.5 mM sodium orthovanadate, 1 mM sodium fluoride, and 10 mM β-glycerophosphate. Proteins (100 μg) were electrophoretically resolved on polyacrylamide gels containing SDS and transferred to nitrocellulose membranes (Millipore). Nonspecific binding sites were blocked by incubation for 1 hr at room temperature with Tris-buffered saline (TBS) (25 mM Tris, pH 8.0/150 mM NaCl/2 mM KCl) containing 0.05% Triton X-100 (TBS-T) and 3% nonfat dry milk. Filters were exposed to antibodies in TBST-milk for 1 hr at room temperature, and sites of binding were visualized by using horseradish peroxidase-conjugated goat anti-rabbit or anti-mouse Ig G (IgG) (Cappel) followed by enhanced chemiluminescent detection (ECL kit; Amersham). For measuring assembly of cyclin D1 with CDK4, anti-CDK4 (RZ plus RY) precipitates electrophoretically resolved on denaturing polyacrylamide gels were transferred to nitrocellulose and probed with mAb to cyclin D1.
Protein Kinase Assays.
Kinase assays using immune complexes containing ERK1 or ERK2 were carried out as described by using 400 μg of cell lysate protein per precipitation, 2 μg of antibody, and myelin basic protein as substrate (61). RB immune complex kinase assays were performed after precipitation of 500 μg of lysate protein with anti-CDK4 (RZ) and with 1 μg of fresh glutathione S-transferase-RB fusion protein as substrate (4). CDK2 activity was assayed by using histone H1 as substrate (62) with 200 μg of protein per precipitation and antisera to cyclin E (M-20) or CDK2 (RCC plus RDD). Reactions were stopped with 1/3 volume 3× gel sample buffer and heating at 85°C for 5 min. Labeled proteins were resolved on denaturing polyacrylamide gels, which were dried and subjected to autoradiography.
RESULTS AND DISCUSSION
Induced Expression of Constitutively Active MEK1 Stimulates ERK1 and ERK2.
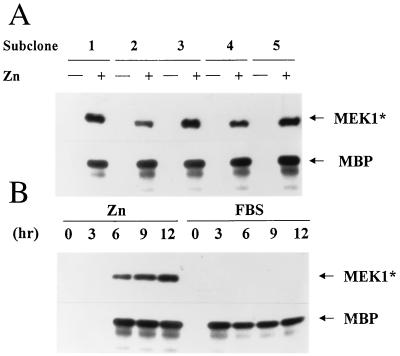
We introduced an expression plasmid encoding active myc-tagged MEK1 under the control of a zinc-inducible metallothionein promoter into NIH 3T3 fibroblasts and derived subclones that conditionally expressed the protein. Ectopically expressed, tagged MEK1 protein (designated MEK1*) was detected by immunoblotting with a mAb (9E10) directed to the myc epitope. With continued passage, uncloned transfected cells began to acquire transformed properties, losing contact inhibition and forming colonies in semisolid medium even when grown in the absence of heavy metals (data not shown). We therefore selected subclones by limiting dilution of single cells in microtiter dishes, obtaining several in which basal MEK1* levels remained low but were induced after zinc treatment.
In quiescent, serum-starved subclones, no basal expression of MEK1* was detected, but cells transferred to medium containing zinc expressed the protein, and ERK1 precipitated from the same lysates became enzymatically active (Fig. 1A); similar results were obtained with antibodies to ERK2 (data not shown). MEK1* was detected and endogenous ERK1 was activated between 3 and 6 hr after addition of zinc to the culture medium (Fig. 1B). Cells stimulated with 10% serum did not express exogenous MEK1*, but endogenous ERK activity was induced within 3 hr and was sustained for at least 12 hr. Zinc treatment of cells containing the inducible MEK1* gene generated roughly physiologic levels of ERK activity, although the kinetics of induction were slower than those observed with serum (Fig. 1B). Comparable data were obtained with each of five subclones (data not shown).
Figure 1.
Inducible expression of MEK1* in NIH 3T3 fibroblasts leads to activation of ERK1. (A) Five subclones expressing inducible MEK1* were serum-starved and restimulated with ZnS04 for 12 hr. MEK1* was detected by immunoblotting with antibody 9E10 to the myc tag, and activation of ERK1 myelin basic protein kinase activity was evaluated in immune complexes. (B) Starved cells from subclone 3 were restimulated with ZnSO4 or 10% FBS for the indicated times and assayed for MEK1* expression and ERK1 kinase activity as in A.
MEK1* Induces Cyclin D1 Synthesis and CDK4 Assembly.
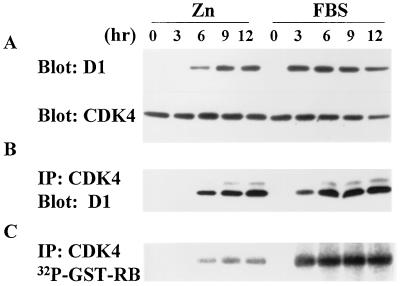
On expression of MEK1* and activation of ERK1, cyclin D1 synthesis was observed (Fig. 2A). Similar levels of cyclin D1 appeared after serum stimulation but with faster kinetics (Fig. 2A), again correlating with ERK activation (Fig. 1B). In both cases, the onset of cyclin D1 synthesis lagged behind MEK1 induction and ERK1 activation by 60–90 min (data not shown). Overall, the levels of cyclin D1 induced by zinc in five individual subclones were 60–75% of those induced by serum. CDK4 synthesis was constitutive (63), and once synthesized, cyclin D1 coprecipitated with it, indicating that MEK1* could induce cyclin D1-CDK4 assembly (Fig. 2B, Left). Amounts of cyclin D1 bound to CDK4 after zinc treatment of the cells were comparable to those obtained after serum stimulation (Fig. 2B, Right).
Figure 2.
MEK1* induces cyclin D1 synthesis and assembly with CDK4. Serum-starved cells (subclone 3) harboring inducible MEK1* were stimulated with ZnSO4 or FBS and assayed at the indicated times thereafter (hr) for cyclin D1 and CDK4 expression (A), assembly (B), and RB kinase activity (C).
Assembly of cyclin D-CDK4 complexes facilitates their nuclear transport and phosphorylation by the CDK-activating kinase (CAK) (54, 64). The latter step stabilizes the enzyme in an active conformation and is essential for its activity as a RB kinase (3). Because CAK is active throughout the cell cycle (54, 65), assembled cyclin D-CDK4 complexes promptly undergo CAK phosphorylation unless they are blocked by excess CDK inhibitors, such as p27Kip1 (14, 62). Although cyclin D1-CDK4 complexes formed in response to serum stimulation exhibited robust RB kinase activity, those formed after zinc induction were much less active (Fig. 2C). The amounts of cyclin D1-CDK4 complexes assembled after zinc or serum treatment were roughly equivalent (Fig. 2B), so serum must induce a MEK1*-independent function that is required for full activation of the assembled holoenzyme.
MEK1* Activity Is Insufficient for p27Kip1 Degradation and S Phase Entry.
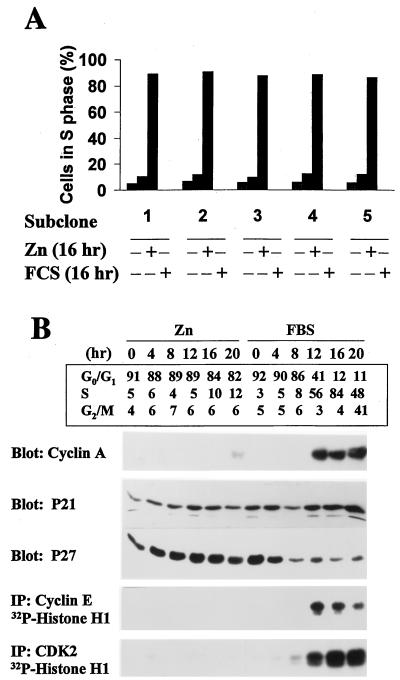
Eighty-five to 90% of quiescent cells were in S phase by 16 hr after serum treatment, but only a small fraction (5–10%) entered S phase in response to zinc (Fig. 3A). Serum-treated cells progressed into G2/M by 20 hr (Fig. 3B, Upper), and by 24 hr, >90% passed through mitosis into the next G1 interval (data not shown). Because expression of MEK1* failed to induce NIH 3T3 cells to enter S phase, we studied additional molecular correlates of late G1 phase progression to try to determine what additional events were serum dependent. Specifically, we quantitated the relative amounts of two CDK inhibitors, p21Cip1 and p27Kip1, studied cyclin A induction, and assayed cyclin E-dependent and total CDK2 kinase activity. In serum-treated cells, the levels of p21Cip1 remained relatively constant throughout G1 phase, whereas much of the p27Kip1 was degraded (Fig. 3B, Right). Levels of p27Kip1 were highest in quiescent cells but fell by late G1 phase (≈8 hr after stimulation) and remained low thereafter (14, 66). Throughout the remainder of the cycle and in continuously proliferating cells, virtually all residual p27Kip1 remained in cyclin D-CDK complexes, preventing it from interfering with the activities of cyclin E- and A-dependent kinases (data not shown). Cyclin A was induced as serum-stimulated cells entered S phase, and CDK2 activity was detected just before the G1/S transition (Fig. 3B, Right).
Figure 3.
Molecular correlates of S phase entry. (A) Cells from five individual subclones expressing inducible MEK1* were serum-starved and restimulated with ZnSO4 or FBS for 16 hr, and the S phase fraction was determined by using flow cytometry to estimate the DNA content of propidium iodide-stained nuclei. (B) Serum-starved cells (subclone 3) stimulated with ZnSO4 or FBS were assayed at 4-hr intervals for DNA content (Upper). Cyclin A, p21Cip1, and p27Kip1 levels were determined by immunoblotting. Precipitates recovered with antibodies to cyclin E and CDK2 were assayed for histone H1 kinase activity.
In contrast, no degradation of p27Kip1 was observed after zinc treatment, cyclin A was not induced, and no cyclin E- or A-dependent CDK2 activity was detected (Fig. 3B, Left). The inability of cyclin D1-CDK4 complexes to undergo full activation in response to MEK1* induction (Fig. 2C) together with the absence of CDK2 activity (Fig. 3B) suggest that sustained, high levels of p27Kip1 prevent the full activation of both CDKs. In turn, a failure to phosphorylate and cancel the transcriptional corepressor activities of RB-family proteins may prevent induction of cyclin A (67–70). Degradation of p27Kip1 must require serum-induced signals in addition to, or apart from, those mediated via the MEK/ERK pathway (22, 40, 46, 71).
Overexpression of Cyclin D1 and CDK4 with MEK1* Induces S Phase Entry.
When cyclin D1 and CDK4 both are ectopically expressed in fibroblasts, they do not form holoenzyme complexes unless cells are stimulated with serum (4). However, MEK1* enabled newly induced cyclin D1 to assemble with CDK4 to form a partially active RB kinase (Fig. 2). Because the failure of the assembled holoenzyme to become fully active appeared to be caused by sustained levels of p27Kip1 in zinc-induced cells (Fig. 3B), we reasoned that overexpression of cyclin D1 and CDK4 in cells engineered to synthesize MEK1* might stoichiometrically titrate more p27Kip1 into cyclin D1-CDK4 complexes, thereby lowering the effective CDK inhibitory threshold. In principle, this overexpression might facilitate the activation of CDK2 and so enable these cells to enter S phase, even in the absence of serum.
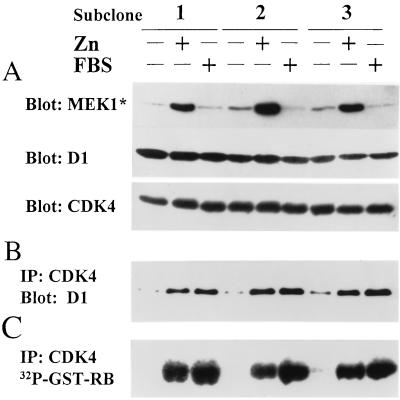
The expression plasmid containing inducible MEK1* therefore was introduced into NIH 3T3 cells that were engineered to ectopically express both cyclin D1 and CDK4. Low basal levels of MEK1* detected in serum-starved cells were much increased after zinc treatment (Fig. 4A). Although ectopically expressed D1 and CDK4 subunits were detected in starved cells (Fig. 4A), few complexes formed (Fig. 4B). Yet, their assembly was augmented to a comparable extent after serum or zinc treatment (Fig. 4B). After 12-hr stimulation, the cyclin D1-CDK4 complexes induced by MEK1* were highly active and yielded as much RB kinase activity as those induced by serum (Fig. 4C). Therefore, under circumstances in which cyclin D1 and CDK4 subunits were overexpressed, equivalent amounts of active cyclin D-dependent kinases were formed in response to serum or zinc induction.
Figure 4.
Expression of MEK1* in cells overexpressing cyclin D1 and CDK4. Serum-starved cells from three subclones coexpressing cyclin D1, CDK4, and inducible MEK1* were stimulated with ZnSO4 or FBS for 12 hr. Cell lysates were assayed for expression of MEK1*, cyclin D1, and CDK4 by immunoblotting (A), cyclin D1-CDK4 complex formation by immunoprecipitation with CDK4 antibody followed by immunoblotting with cyclin D1 antibody (B), and CDK4-associated RB kinase activity (C).
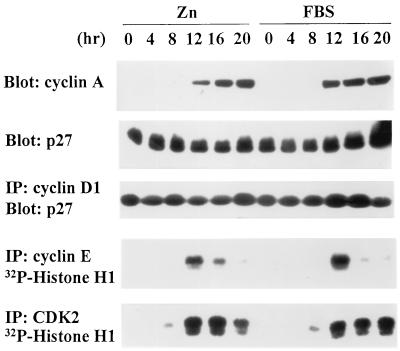
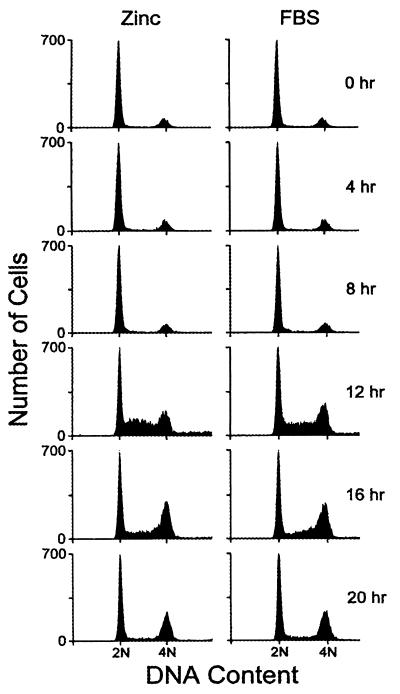
In cells overexpressing cyclin D1 and CDK4, p27Kip1 levels did not significantly decrease when serum-depleted cells were stimulated to enter the cycle (Fig. 5). Indeed, most of the p27Kip1 (>85% in several such experiments) coprecipitated with overexpressed cyclin D1, regardless of whether the cells were deprived of serum or restimulated with serum or zinc. Under these conditions, restimulation with zinc alone led to the induction of cyclin A and generated CDK2 activity as cells neared the G1/S boundary (Fig. 5). A substantial fraction of these cells were able to enter S phase (Fig. 6). Yet, zinc-stimulated cells overexpressing MEK1*, cyclin D1, and CDK4 did not proliferate continuously in medium lacking serum, undergoing only one population doubling on average (data not shown). Therefore, although certain gene products required for DNA synthesis initially may not have been rate limiting, cells that completed one S phase required serum-induced factors to remain in cycle.
Figure 5.
Overexpression of cyclin D1, CDK4, and MEK1* induces cyclin A synthesis and CDK2 activation, but not p27Kip1 degradation. Cells (subclone 2) ectopically expressing cyclin D1, CDK4, and inducible MEK1* were starved and restimulated with ZnSO4 or FBS. Cells harvested at intervals thereafter (hr) were assayed for cyclin A and p27Kip1 expression by immunoblotting (top two panels), for cyclin D1-associated p27Kip1 by immunoprecipitation with cyclin D1 followed by immunoblotting with anti-p27Kip1 (middle panel), and for cyclin E- and CDK2-associated histone H1 kinase activity (bottom two panels). More than 85% of p27Kip1 coprecipitated with cyclin D1.
Figure 6.
Ectopic expression of cyclin D1, CDK4, and MEK1* induces S phase entry. Serum-starved cells (subclone 2) ectopically expressing cyclin D1, CDK4, and inducible MEK1* were stimulated with ZnSO4 or FBS, harvested at indicated times thereafter, and assayed for DNA content by flow cytometry.
Implications for Restriction Point Control.
The MEK1*/ERK signaling pathway not only acts to induce the cyclin D1 gene but also facilitates assembly of cyclin D1 into catalytically active complexes with CDK4. This action might somehow be mediated by phosphorylation of cyclin D1 or CDK4 on as-yet-unmapped phosphoserine residues (14, 72). Alternatively, ERKs might phosphorylate molecular chaperones such as cdc37 (73) or “assembly factors” (4) such as p21Cip1 or other molecules that have the potential to enter into higher-order complexes with the cyclin-CDKs (6). Whatever the mechanism(s), formation of cyclin D-dependent kinases depends on growth factor signals that act at both transcriptional and posttranslational levels.
Activation of the cyclin D1-CDK4 complex is opposed by p27Kip1, which normally builds to high levels in the absence of serum-dependent signals that otherwise lead to its destruction. Cyclin D1-CDK complexes accumulating during the G0 to S interval lower the effective p27Kip1 inhibitory threshold, facilitating the initial activation of cyclin E-dependent CDK2 as cells approach the restriction point. In this regard, it may be significant that cyclin D-dependent kinases are less susceptible than cyclin E- or A-dependent kinases to inhibition by p27Kip1 (12, 13). Once activated, phosphorylation by cyclin E-CDK2 may help trigger p27Kip1 ubiquitination and degradation (20–22), accounting in part for the irreversibility of the transition. Cyclin D- and E-dependent kinases also collaborate to phosphorylate RB-family proteins, leading to the induction of E2F responsive genes necessary for DNA synthesis. Hence, both catalytic and stoichiometric activities of cyclin D-dependent kinases contribute to restriction point control.
In quiescent cells induced by zinc to express MEK1* alone, cyclin D1-CDK4 complexes were effectively assembled, but p27Kip1 was not degraded and the activities of both CDK4 and CDK2 remained low. In serum-stimulated cells expressing a dominant-negative Ras mutant, p27Kip1 also continued to be expressed at high levels (46). Together, these data argue that degradation of p27Kip1 depends both on Ras-dependent and Ras-independent signals. However, MEK1*-induced assembly of ectopically overexpressed cyclin D1 and CDK4 subunits was sufficient to neutralize p27Kip1 inhibitory activity, so that CDK4 and CDK2 were activated in the absence of serum stimulation. In this setting, p27Kip1 was not degraded, and the sequestration of the CDK inhibitor by supra-physiologic levels of cyclin D1-CDK4 complexes triggered entry of cells into S phase. Cells that entered S phase in response to induced MEK1*, D1, and CDK4 expression were unable to proliferate continuously, underscoring the fact that additional proteins synthesized in response to mitogens must contribute to the G1/S transition in later cycles.
Acknowledgments
We thank Natalie G. Ann for providing active MEK1 cDNA and Frank Rauscher, Jr. for the pMTCB6 plasmid; Shawn Hawkins, Carol Bockhold, and Joseph Watson for excellent technical assistance; Richard Ashmun for flow cytometric analysis of DNA content; Alan Diehl and Jason Weber for comments on the manuscript; and other members of our laboratory for helpful criticisms and encouragement. This work was supported in part by National Institutes of Health Grant CA-56819 (M.F.R.), Cancer Center Core Grant CA-21765, Howard Hughes Medical Institute (C.J.S.), and the American Lebanese Syrian Associated Charities of St. Jude Children’s Research Hospital. V.S. is a Fellow of the Austrian Fond zur Foerderung der Wissenschaflichen Forschung (FWF).
ABBREVIATIONS
- CDK
cyclin-dependent kinase
- RB
retinoblastoma protein
- Kip
kinase inhibitory protein
- MEK
mitogen-activated protein kinase kinase
- ERK
extracellular signal-regulated kinase
- FBS
fetal bovine serum
References
- 1.Sherr C J, Roberts J M. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 2.Sherr C J. Cell. 1994;79:551–555. doi: 10.1016/0092-8674(94)90540-1. [DOI] [PubMed] [Google Scholar]
- 3.Morgan D O. Nature (London) 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 4.Matsushime H, Quelle D E, Shurtleff S A, Shibuya M, Sherr C J, Kato J-Y. Mol Cell Biol. 1994;14:2066–2076. doi: 10.1128/mcb.14.3.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyerson M, Harlow E. Mol Cell Biol. 1994;14:2077–2086. doi: 10.1128/mcb.14.3.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.LaBaer J, Garrett M D, Stevenson L F, Slingerland J M, Sandhu C, Chou H S, Fattaey A, Harlow E. Genes Dev. 1997;11:847–862. doi: 10.1101/gad.11.7.847. [DOI] [PubMed] [Google Scholar]
- 7.Ewen M E, Sluss H K, Sherr C J, Matsushime H, Kato J-Y, Livingston D M. Cell. 1993;73:487–497. doi: 10.1016/0092-8674(93)90136-e. [DOI] [PubMed] [Google Scholar]
- 8.Kato J-Y, Matsushime H, Hiebert S W, Ewen M E, Sherr C J. Genes Dev. 1993;7:331–342. doi: 10.1101/gad.7.3.331. [DOI] [PubMed] [Google Scholar]
- 9.Connell-Crowley L, Harper J W, Goodrich D W. Mol Biol Cell. 1997;8:287–301. doi: 10.1091/mbc.8.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weinberg R A. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 11.Polyak K, Kato J-Y, Solomon M J, Sherr C J, Massague J, Roberts J M, Koff A. Genes Dev. 1994;8:9–22. doi: 10.1101/gad.8.1.9. [DOI] [PubMed] [Google Scholar]
- 12.Polyak K, Lee M-H, Erdjument-Bromage H, Koff A, Roberts J M, Tempst P, Massagué J. Cell. 1994;78:59–66. doi: 10.1016/0092-8674(94)90572-x. [DOI] [PubMed] [Google Scholar]
- 13.Toyoshima H, Hunter T. Cell. 1994;78:67–74. doi: 10.1016/0092-8674(94)90573-8. [DOI] [PubMed] [Google Scholar]
- 14.Kato J, Matsuoka M, Polyak K, Massagué J, Sherr C J. Cell. 1994;79:487–496. doi: 10.1016/0092-8674(94)90257-7. [DOI] [PubMed] [Google Scholar]
- 15.Reynisdóttir I, Polyak K, Iavarone A, Massagué J. Genes Dev. 1995;9:1831–1845. doi: 10.1101/gad.9.15.1831. [DOI] [PubMed] [Google Scholar]
- 16.Reynisdóttir I, Massagué J. Genes Dev. 1997;11:492–503. doi: 10.1101/gad.11.4.492. [DOI] [PubMed] [Google Scholar]
- 17.Hinds P W, Mittnacht S, Dulic V, Arnold A, Reed S I, Weinberg R A. Cell. 1992;70:993–1006. doi: 10.1016/0092-8674(92)90249-c. [DOI] [PubMed] [Google Scholar]
- 18.Hatakeyama M, Brill J A, Fink G R, Weinberg R A. Genes Dev. 1994;8:1759–1771. doi: 10.1101/gad.8.15.1759. [DOI] [PubMed] [Google Scholar]
- 19.Mittnacht S, Lees J A, Desai D, Harlow E, Morgan D O, Weinberg R A. EMBO J. 1994;13:118–127. doi: 10.1002/j.1460-2075.1994.tb06241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pagano M, Tam S W, Theodoras A M, Beer-Romano P, Dal Sal G, Chau V, Yew P R, Draetta G F, Rolfe M. Science. 1995;269:682–685. doi: 10.1126/science.7624798. [DOI] [PubMed] [Google Scholar]
- 21.Sheaff R, Groudine M, Gordon M, Roberts J, Clurman B. Genes Dev. 1997;11:1464–1478. doi: 10.1101/gad.11.11.1464. [DOI] [PubMed] [Google Scholar]
- 22.Vlach J, Hennecke S, Amati B. EMBO J. 1997;16:5334–5344. doi: 10.1093/emboj/16.17.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stillman B. Science. 1996;274:1659–1664. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- 24.Krude T, Jackman M, Pines J, Laskey R A. Cell. 1997;88:109–119. doi: 10.1016/s0092-8674(00)81863-2. [DOI] [PubMed] [Google Scholar]
- 25.Girard F, Strausfeld U, Fernandez A, Lamb N J C. Cell. 1991;67:1169–1179. doi: 10.1016/0092-8674(91)90293-8. [DOI] [PubMed] [Google Scholar]
- 26.Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G. EMBO J. 1992;11:961–971. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zindy F, Lamas E, Chenivesse X, Sobczak J, Wang J, Fesquet D, Henglein B, Brechot C. Biochem Biophys Res Commun. 1992;182:1144–1154. doi: 10.1016/0006-291x(92)91851-g. [DOI] [PubMed] [Google Scholar]
- 28.Pardee A B. Science. 1989;246:603–608. doi: 10.1126/science.2683075. [DOI] [PubMed] [Google Scholar]
- 29.Sherr C J. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 30.Marshall C J. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 31.Cobb M H, Goldsmith E J. J Biol Chem. 1995;270:14843–14846. doi: 10.1074/jbc.270.25.14843. [DOI] [PubMed] [Google Scholar]
- 32.Wood K W, Sarnecki C, Roberts T M, Blenis J. Cell. 1992;68:1041–1050. doi: 10.1016/0092-8674(92)90076-o. [DOI] [PubMed] [Google Scholar]
- 33.Thomas S M, DeMarco M, D’Arcangelo G, Halegoua S, Brugge J S. Cell. 1992;68:1031–1040. doi: 10.1016/0092-8674(92)90075-n. [DOI] [PubMed] [Google Scholar]
- 34.Robbins D J, Cheng M, Zhen E, Vanderbilt C A, Feig L A, Cobb M H. Proc Natl Acad Sci USA. 1992;89:6924–6928. doi: 10.1073/pnas.89.15.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warne P H, Viciana P R, Downward J. Nature (London) 1993;364:352–355. doi: 10.1038/364352a0. [DOI] [PubMed] [Google Scholar]
- 36.Zhang X-F, Settleman J, Kyriakis J M, Takeuchi-Suzuki E, Elledge S J, Marshall M S, Bruder J T, Rapp U R, Avruch J. Nature (London) 1993;364:308–313. doi: 10.1038/364308a0. [DOI] [PubMed] [Google Scholar]
- 37.Chen R-H, Sarnecki C, Blenis J. Mol Cell Biol. 1992;12:915–927. doi: 10.1128/mcb.12.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feramisco J R, Gross M, Kamata T, Rosenberg M, Sweet R W. Cell. 1984;38:109–117. doi: 10.1016/0092-8674(84)90531-2. [DOI] [PubMed] [Google Scholar]
- 39.Mulcahy L S, Smith M R, Stacey D W. Nature (London) 1985;313:241–243. doi: 10.1038/313241a0. [DOI] [PubMed] [Google Scholar]
- 40.Peeper D S, Upton T M, Ladha M H, Neuman E, Zalvide J, Bernards R, DeCaprio J A, Ewen M E. Nature (London) 1997;386:177–181. doi: 10.1038/386177a0. [DOI] [PubMed] [Google Scholar]
- 41.Mittnacht S, Paterson H, Olson M F, Marshall C J. Curr Biol. 1997;7:219–221. doi: 10.1016/s0960-9822(97)70094-0. [DOI] [PubMed] [Google Scholar]
- 42.Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, Pestell R G. J Biol Chem. 1995;270:23589–23597. doi: 10.1074/jbc.270.40.23589. [DOI] [PubMed] [Google Scholar]
- 43.Winston J T, Coats S R, Wang Y-Z, Pledger W J. Oncogene. 1996;12:127–134. [PubMed] [Google Scholar]
- 44.Lavoie J N, L’Allemain G, Brunet A, Müller R, Pouysségur J. J Biol Chem. 1996;271:20608–20616. doi: 10.1074/jbc.271.34.20608. [DOI] [PubMed] [Google Scholar]
- 45.Kerkhoff E, Rapp U R. Mol Cell Biol. 1997;17:2576–2586. doi: 10.1128/mcb.17.5.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aktas H, Cai H, Cooper G M. Mol Cell Biol. 1997;17:3850–3857. doi: 10.1128/mcb.17.7.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pages G, Lenormand P, L’Allemain G, Chambard J-C, Meloche S, Pouysségur J. Proc Natl Acad Sci USA. 1993;90:8319–8323. doi: 10.1073/pnas.90.18.8319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Filmus J, Robles A I, Shi W, Wong M J, Colombo L L, Conti C J. Oncogene. 1994;9:3627–3633. [PubMed] [Google Scholar]
- 49.Liu J-J, Chao J-R, Jiang M-C, Ng S-Y, Yen J J Y, Yang-Yen H-F. Mol Cell Biol. 1995;15:3654–3663. doi: 10.1128/mcb.15.7.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Serrano M, Gomez-Lahoz E, DePinho R A, Beach D, Bar-Sagi D. Science. 1995;267:249–252. doi: 10.1126/science.7809631. [DOI] [PubMed] [Google Scholar]
- 51.Lukas J, Bartkova J, Rohde M, Strauss M, Bartek J. Mol Cell Biol. 1995;15:2600–2611. doi: 10.1128/mcb.15.5.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Evan G I, Lewis G K, Ramsay G, Bishop J M. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vallance S J, Lee H-M, Roussel M F, Shurtleff S A, Kato J-Y, Strom D K, Sherr C J. Hybridoma. 1994;13:37–44. doi: 10.1089/hyb.1994.13.37. [DOI] [PubMed] [Google Scholar]
- 54.Matsuoka M, Kato J, Fisher R P, Morgan D O, Sherr C J. Mol Cell Biol. 1994;14:7265–7275. doi: 10.1128/mcb.14.11.7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matsushime H, Roussel M F, Ashmun R A, Sherr C J. Cell. 1991;65:701–713. doi: 10.1016/0092-8674(91)90101-4. [DOI] [PubMed] [Google Scholar]
- 56.Mansour S J, Matten W T, Hermann A S, Candia J M, Rong S, Fukusawa K, Vande Woude G F, Ahn N G. Science. 1994;265:966–969. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- 57.Inaba T, Inukai T, Yoshihara T, Seyschab H, Ashmun R A, Canman C E, Laken S J, Kastan M B, Look A T. Nature (London) 1996;382:541–544. doi: 10.1038/382541a0. [DOI] [PubMed] [Google Scholar]
- 58.Chen C, Okayama H. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morgenstern J P, Land H. Nucleic Acids Res. 1990;18:1068–1077. doi: 10.1093/nar/18.4.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quelle D E, Ashmun R A, Shurtleff S E, Kato J-Y, Bar-Sagi D, Roussel M, Sherr C J. Genes Dev. 1993;7:1559–1571. doi: 10.1101/gad.7.8.1559. [DOI] [PubMed] [Google Scholar]
- 61.Reuter C W M, Catling A D, Weber M J. Methods Enzymol. 1995;255:245–256. doi: 10.1016/s0076-6879(95)55027-5. [DOI] [PubMed] [Google Scholar]
- 62.Koff A, Ohtsuki M, Polyak K, Roberts J M, Massagué J. Science. 1993;260:536–539. doi: 10.1126/science.8475385. [DOI] [PubMed] [Google Scholar]
- 63.Matsushime H, Ewen M E, Strom D K, Kato J-Y, Hanks S K, Roussel M F, Sherr C J. Cell. 1992;71:323–334. doi: 10.1016/0092-8674(92)90360-o. [DOI] [PubMed] [Google Scholar]
- 64.Kato J-Y, Matsuoka M, Strom D K, Sherr C J. Mol Cell Biol. 1994;14:2713–2721. doi: 10.1128/mcb.14.4.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tassan J P, Schultz S J, Bartek J, Nigg E A. J Cell Biol. 1994;127:467–478. doi: 10.1083/jcb.127.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nourse J, Firpo E, Flanagan W M, Coats S, Polyak K, Lee M H, Massagué J, Crabtree G R, Roberts J M. Nature (London) 1994;372:570–573. doi: 10.1038/372570a0. [DOI] [PubMed] [Google Scholar]
- 67.Schultz A, Zerfass K, Spitkovsky D, Middendorp S, Berges J, Helin K, Jansen-Durr P, Henglein B. Proc Natl Acad Sci USA. 1995;92:11264–11268. doi: 10.1073/pnas.92.24.11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zwicker J, Lucibello F C, Wolfraim L A, Gross C, Truss M, Engeland K, Muller R. EMBO J. 1995;14:4514–4522. doi: 10.1002/j.1460-2075.1995.tb00130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huet X, Rech J, Plet A, Vie A, Blanchard J M. Mol Cell Biol. 1996;16:3789–3798. doi: 10.1128/mcb.16.7.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hurford R K, Cobrinik D, Lee M-H, Dyson N. Genes Dev. 1997;11:1447–1463. doi: 10.1101/gad.11.11.1447. [DOI] [PubMed] [Google Scholar]
- 71.Hirai A, Nakamura S, Noguchi Y, Yasuda T, Kitagawa M, Tatsuno I, Oeda T, Tahara K, Terano T, Narumiya S, Kohn L D, Saito Y. J Biol Chem. 1997;272:13–16. [PubMed] [Google Scholar]
- 72.Diehl J A, Zindy F, Sherr C J. Genes Dev. 1997;11:957–972. doi: 10.1101/gad.11.8.957. [DOI] [PubMed] [Google Scholar]
- 73.Stepanova L, Leng X, Parker S B, Harper J W. Genes Dev. 1996;10:1491–1502. doi: 10.1101/gad.10.12.1491. [DOI] [PubMed] [Google Scholar]