Abstract
Recent molecular genetic studies have revealed that two major types of genomic instabilities, chromosomal instability and microsatellite instability, exist in the colorectal carcinomas. To clarify the relationship between chromosomal abnormalities and mismatch repair gene defects in colorectal carcinomas, we performed a chromosomal analysis on 39 colorectal carcinomas with high-microsatellite instability (MSI-H) and compared the results obtained with those of 20 right-sided microsatellite-stable (MSS) colorectal carcinomas. Chromosomal imbalances (CIs) in MSS colorectal carcinomas were more frequent than in MSI-H colon carcinomas by comparative genomic hybridization analysis (70% versus 31%, P = 0.004). The CI patterns of MSI-H and MSS carcinomas were different. Frequent CIs in MSI-H colon carcinomas were gains of 4q (15%) and 8q (8%), and losses of 9q (21%), 1p (18%), and 11q (18%). In contrast, frequent CIs in right-sided MSS colon carcinomas were gains of 8q (50%), 13q (35%), and 20q (25%), and losses of 18q (55%), 15q (35%), and 17p (30%). We compared the mutation status of 45 target genes and CIs in our MSI-H tumors. Among these 45 target genes, mutation of hRAD50, a member of the DNA repair genes, and FLJ11383 were significantly related to MSI-H colorectal carcinomas with CIs (P = 0.01 and P = 0.02, respectively). Our findings indicate that unique CIs were present in a subset of MSI-H colorectal carcinomas and that these CIs are related to the mutation of several target genes, especially of hRAD50.
It is widely accepted that the molecular genetics of human cancers can be used to categorize colorectal carcinomas into two major types of genomic instabilities, chromosomal instability (CIN) and microsatellite instability (MSI). 1 The majority of colorectal carcinomas are categorized into the CIN pathway, which is characterized by a high frequency of allelic losses, deletions, and/or mutations of tumor suppressor genes such as APC and p53, and abnormal tumor DNA. 2 Aneuploidy in CIN phenotype tumors had been demonstrated in colorectal cancer cell lines 3 and tumor tissues. 4 Although CIN is a common finding in colorectal carcinomas, the mechanism of CIN has not been clearly elucidated. Defects in DNA replication check point genes and many other genes increase the rate of genome rearrangement and it is suggested to be associated with CIN. 5-8
The other pathway, namely the MSI pathway, begins with the inactivation of one of a group of genes responsible for DNA nucleotide mismatch repair, which leads to extensive mutations in both repetitive and nonrepetitive DNA sequences with low frequencies of allelic losses and rare alterations of tumor DNA content. 9,10 The mechanism of tumorigenesis in high-microsatellite instability (MSI-H) tumors is thought to involve frameshift mutations of microsatellite repeats within coding regions of the affected target genes, and the inactivation of these target genes is believed to directly contribute to tumor development and progression.
Although these two distinct major genetic pathways of genetic instabilities are widely accepted, some tumors reveal different genetic pathways; ie, some tumors show both types of genomic instabilities and some tumors do not show any of these two instabilities. 11-13 To clarify the relationship between these two genetic instabilities in colorectal carcinomas, we analyzed chromosomal imbalances (CIs) in 39 MSI-H colorectal carcinomas and compared the results with changes in 20 microsatellite-stable (MSS) colorectal carcinomas.
Materials and Methods
Patient Selection
Thirty-nine cases confirmed as MSI-H colorectal carcinomas, and 20 cases of right-sided MSS colorectal carcinomas were included in this study. In each case, grossly normal mucosa remote from the tumor was included as a control. All cases were identified consecutively at the Gastrointestinal Tumor Working Group Tissue Bank at Yonsei University Medical Center (Seoul, Korea) between December 1996 and November 1999. Cases included in this study were sporadic tumors without relevant family history or clinical evidence of familial adenomatous polyposis or hereditary nonpolyposis colorectal cancer. DNAs were extracted from fresh-frozen tissues. Tumor specimens were microdissected on a cryostat and fractionated to enrich the tumor cell population as described previously. 14,15
Screening of Microsatellite Instability
DNAs from tumors and normal mucosae were polymerase chain reaction (PCR) amplified at six microsatellite loci to evaluate the MSI. These markers included the recommended panel of five markers proposed at the National Cancer Institute Collaboratory Meeting and MSI in colorectal cancer 16 plus BAT40. PCR reactions were performed in a mixture of 20 μl containing 1.5 mmol/L MgCl2; 20 pmol of primer; 0.2 mmol/L each of dATP, dGTP, and dTTP; 5 μmol/L dCTP; 1 μCi of [α-32P]-dCTP (3000 Ci/mmol; DuPont New England Nuclear, Boston, MA); 50 ng of sample DNA; 1× PCR buffer; and 1.25 U of Taq polymerase (Life Technologies, Inc., Grand Island, NY). After denaturation at 95°C for 5 minutes, DNA amplification was performed for 25 to 30 cycles consisting of denaturation at 95°C for 30 seconds, primer annealing at 55 to 60°C for 30 seconds, and elongation at 72°C for 15 seconds. PCR products were separated in 6% polyacrylamide gels containing 5.6 mol/L of urea, followed by autoradiography. MSI was determined by the mobility shift of products from PCR. In tumors with MSI, additional bands were found in the normal allele regions. MSI in three or more markers of which more than two mononucleotide repeat markers were included was classified as MSI-H; and those showing no instability were classified as MSS (Figure 1A) ▶ . Methylation analysis of hMLH1 and expression analysis of hMLH1 and hMSH2 were performed as described previously. 17,18
Figure 1.

Examples of MSI, CGH, and PCR-LOH analysis in MSI-H (case 17) and MSS (case 60) colorectal carcinoma with CIs. A: MSI analysis. Additional alleles were found at six microsatellite markers in a MSI-H colorectal carcinoma and no changes were found in a MSS colorectal carcinoma. B: CGH analysis. CIs were found in 9q, 16p, 17p, and 17q in MSI-H colorectal carcinoma (left) and 8q, 16p, 17p, 17q, and 18q in MSS colorectal carcinoma (right). C: Comparison of CGH analysis and PCR-LOH. PCR-LOH analysis showed same allelic imbalance (arrow) in 17p and 18q in MSS colorectal carcinoma (right), whereas comparison was not possible in MSI-H carcinoma because of shifted bands in 9q, 16p, 17p, 17q, and 18q (left).
Comparative Genomic Hybridization (CGH) and Digital Image Analysis
Genomic DNA samples from tumors were labeled with Spectrum Green dUTP (Vysis Inc., Downers Grove, IL), and normal reference genomic DNA was labeled with Spectrum Red dUTP (Vysis) using the nick translation technique. Labeled tumor and reference DNA (200 to 400 ng), and 10 μg of unlabeled human Cot-1 DNA (Vysis) were dissolved in 10 μl of hybridization buffer (50% formamide, 10% dextran sulfate, and 2× standard saline citrate) and denatured at 72°C for 2 minutes. Hybridization was performed at 37°C on denatured normal metaphase spreads. After hybridization for 3 days, the slides were washed and counterstained with 4′,6-diamidino-2-phenylindole dihydrochloride in anti-fade solution. CGHs were analyzed using an Olympus fluorescent microscope and the Cytovision image analysis system (Applied Imaging; Times Square, Newcastle upon Tyne, UK). Three digital images (4′,6-diamidino-2-phenylindole dihydrochloride, Spectrum Green, Spectrum Red) were acquired from 10 to 20 metaphases in each hybridization. Normal male DNA and DNA from tumor cell lines with known aberrations were used as DNA test controls. Green-to-red intensity ratio profiles were calculated for each chromosome and threshold values defining gains and losses were set at 1.25 and 0.75, respectively (Figure 1, B and C) ▶ . Only the chromosomal losses and gains were analyzed because reciprocal translocation cannot be differentiated by CGH analysis. We regarded tumors as CIs when any chromosomal losses or gains were present in the tumors. The CIs of five chromosomal arms were validated by PCR-loss of heterozygosity (LOH) analysis using seven microsatellite markers (D9S195, D16S521, D17S578, D17S250, D18S58, D18S57, DCC).
Detection of Frameshift Mutation
We chose the 45 target genes of MSI-H carcinomas based on the reported frequency of frameshift mutations and their functions. Genes with mutation frequencies of more than 30% (24 genes), genes containing more than 10 mononucleotide repeats (16 genes), and genes that are reported to be involved in the suppression of genome instability (5 genes) 14,15,19-22 were selected. The incidence of frameshift mutations had been previously reported in 38 of 45 genes in our 39 MSI-H colorectal carcinomas. 14,15,19 DNA preparation, the MSI status of all cases, and frameshift mutations of the 38 target genes have been previously reported. 14,15,19 The analyzed target genes were ABCF1, ACVRII, AIM2, ATR, BAX, BLM, BRCA1, BRCA2, Caspase 5, CHD2, DKFZp564C2478, FLJ11186, FLJ11222, FLJ11383, FLJ11712, FLJ13615, FLJ20139, FLJ20333, GART, GRB-14, hMSH3, hMSH6, hRAD50, KIAA1052, KIAA1096, KIAA1268, KIAA1470, MAC30, MARCKS, MBD4, NADH-UOB, OGT, PRKDC, PRKWNK1, RFC3, SEC63, SLC24AI, SPINK5, SYCP1, TAF1B, TCF4, TCF6L1, TGF-β RII, UVRAG, and WISP3. All PCR products of these genes showed one band from the normal DNA, whereas monoallelic or biallelic mutations were present in some MSI-H tumors (Figure 2) ▶ .
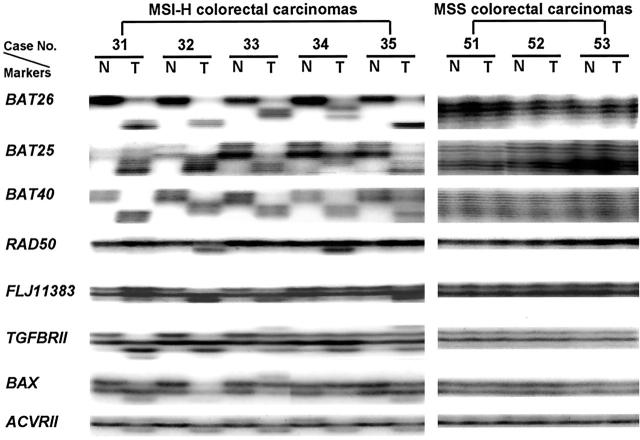
Figure 2.
Frameshift mutations of target genes in MSI-H and MSS colorectal carcinomas. All of five MSI-H colorectal carcinomas showed shifted bands in all three mononucleotide repeat microsatellite markers (BAT26, BAT25, and BAT40). Fragment shifted bands of five target genes (hRad50, FLJ11383, TGFβRII, BAX, and ACVRII) are shown in five MSI-H colorectal carcinomas, whereas no shifted bands were found in the three MSS colorectal carcinomas.
Statistical Analysis
Clinicopathological variables and the presence of frameshift mutations of the target genes were cross-tabulated with the source (MSI-H colorectal carcinomas with CIs and those without CIs), and the significance of association was determined by using the Pearson’s chi-square test or Fisher’s exact test. The mean number and mean incidence of frameshift mutations of target genes in MSI-H colorectal carcinomas with and without CIs were determined using Student’s t-test. All calculations were performed using the SPSS 10.0 for Windows statistical software package (SPSS Inc., Chicago, IL).
Results
CIs in MSI-H Colorectal Carcinomas
We found CIs in 12 (31%) of 39 MSI-H colorectal carcinomas by CGH analysis and the average number of CIs in MSI-H carcinomas was 2.1. CIs showed a rather random distribution over the chromosomes, although there were more losses than gains (ratio of losses to gains, 5.9:1). Chromosomal arm imbalances did not exceed 25% in MSI-H tumors (Figure 3A) ▶ . Most frequent CIs in MSI-H tumors were gains of 4q (15%) and losses of 9q (21%), 1p (18%), and 11q (18%).
Figure 3.

The rate of chromosomal losses and gains observed in 39 designated nonacrocentric chromosomal arms of MSI-H colorectal carcinomas (A) and 20 MSS colorectal carcinomas (B). Each bar represents the percentage loss (lower) or gain (upper) of a chromosomal arm.
Comparison of CIs in MSI-H and MSS Colorectal Carcinomas
We compared the results of CIs of the 39 MSI-H colorectal carcinomas to those of the 20 MSS colon carcinomas. We found CIs in 14 (70%) of 20 MSS colon carcinomas (Figure 3B) ▶ . In addition to more frequent CIs in the MSS tumors (31% versus 70%, P = 0.004), the average number of CIs was significantly higher in the MSS carcinomas (2.1 versus 4.4, P = 0.04). Moreover, the MSS carcinomas showed distinct and different CIs from the MSI-H carcinomas. Frequent changes in MSS carcinomas were gains of 8q (50%), 13q (35%), and 20q (25%), and losses of 18q (55%), 15q (35%), and 17p (30%). None of these changes were included in the chromosomal arms showing frequent changes in our 39 MSI-H colorectal carcinomas (Figure 3) ▶ . CIs in all of the five chromosomal arms were validated by seven microsatellite markers. The PCR-LOH results were matched with all 20 MSS tumors, however it could not be evaluated in most of the MSI-H tumors because of frequent microsatellite instability in these seven markers (Figure 1C) ▶ .
Clinicopathological Characteristics Related to CIs in MSI-H Colorectal Carcinomas
We analyzed the relationship between the incidence of CIs and clinicopathological variables in our 39 MSI-H colorectal carcinomas. No clinicopathological feature was significantly associated with CIs in MSI-H carcinomas. Tumor stage, differentiation, mucin production, and peritumoral lymphoid reaction were not related to the chromosomal changes in MSI-H colorectal carcinomas. Expression of mismatch repair proteins and methylation of hMLH1 were not related to the chromosomal changes in MSI-H colorectal carcinomas (Table 1) ▶ .
Table 1.
Comparison of Clinical and Pathological Features of MSI-H Colorectal Carcinomas with and without CIs
| Variables | Categories | % of MSI-H colorectal carcinomas | P value | |
|---|---|---|---|---|
| With CIs (no.) | Without CIs (no.) | |||
| Differentiation | Well and Moderate | 50 (6) | 41 (11) | 0.9 |
| Poor | 50 (6) | 59 (16) | ||
| Site | Right | 67 (8) | 85 (23) | 0.2 |
| Left | 33 (4) | 15 (4) | ||
| Age | ≤50 | 33 (4) | 37 (10) | 1.0 |
| >50 | 67 (8) | 63 (17) | ||
| Sex | Male | 50 (6) | 56 (15) | 1.0 |
| Female | 50 (6) | 44 (12) | ||
| Peritumoral lymphoid reaction | Absent | 17 (2) | 30 (8) | 0.5 |
| Present | 83 (10) | 70 (19) | ||
| Extracellular mucin formation | Absent | 25 (3) | 48 (13) | 0.3 |
| Present | 75 (9) | 52 (14) | ||
| Perineural invasion | Absent | 92 (11) | 96 (26) | 0.5 |
| Present | 8 (1) | 4 (1) | ||
| Extramural vessel invasion | Absent | 67 (8) | 89 (24) | 0.2 |
| Present | 33 (4) | 11 (3) | ||
| TNM stage | I & II | 75 (9) | 74 (20) | 1.0 |
| III & IV | 25 (3) | 26 (7) | ||
| hMLH1 expression | Absent | 75 (9) | 81 (22) | 0.7 |
| Present | 25 (3) | 19 (5) | ||
| hMSH2 expression | Absent | 0 (0) | 0 (0) | |
| Present | 100 (12) | 100 (27) | ||
| hMLH1 methylation | Absent | 2 (17) | 3 (11) | 0.6 |
| Present | 10 (83) | 24 (89) | ||
Relationship between CIs and Target Gene Mutations in MSI-H Colorectal Carcinomas
We analyzed the relationship between the incidence of CIs and the incidence of frameshift mutations of the 45 target genes we analyzed previously. 14,15,19 The mean number of frameshift mutations of the 45 target genes was 18.3 in the MSI-H carcinomas with CIs and 15.4 in the MSI-H carcinomas without CIs (P = 0.20). The mean incidence of frameshift mutations of the 45 target genes in MSI-H colorectal carcinomas with CIs was 41%, and in MSI-H colorectal carcinomas without CIs was 34% (P = 0.20). There was no difference in the incidence of frameshift mutations of the 45 target genes in MSI-H carcinomas with and without CIs, except for the hRAD50 and FLJ11383 genes. Mutation of hRAD50, a member of the DNA repair genes, was significantly related to MSI-H colorectal carcinomas with CIs (P = 0.01). Mutation of FLJ11383, the function of which is unknown, was also frequent in MSI-H tumors with CIs (P = 0.02). There was also a tendency toward more frequent frameshift mutations of ATR and MBD4, both of which are DNA repair genes, in MSI-H colorectal carcinomas with CIs (Table 2) ▶ .
Table 2.
Comparison of Frameshift Mutations of Target Genes in MSI-H Colorectal Carcinomas with and without CIs
| Genes | % of Frameshift mutations in MSI-H colorectal carcinomas | P value* | P value† | Type of repeat | Reported function of wild type gene product | |||
|---|---|---|---|---|---|---|---|---|
| With CIs (no.) | Without CIs (no.) | |||||||
| Total mutation* | Biallelic mutation† | Total mutation* | Biallelic mutation† | |||||
| hRAD50 | 67 (8) | 0 (0) | 19 (5) | 0 (0) | 0.01 | A (9) | DNA double strand breaks repair | |
| FLJ11383 | 100 (12) | 58 (7) | 63 (17) | 29 (5) | 0.02 | 0.12 | A (10) | Unknown |
| ATR | 50 (6) | 0 (0) | 22 (6) | 0 (0) | 0.13 | A (10) | Checkpoint kinase | |
| MBD4 | 50 (6) | 33 (2) | 22 (6) | 0 (0) | 0.13 | 0.46 | A (10) | DNA glycosylase, Methyl-CpG binding protein |
| RFC3 | 33 (4) | 0 (0) | 15 (4) | 0 (0) | 0.22 | A (10) | DNA replication factor | |
| FLJ13615 | 42 (5) | 0 (0) | 19 (5) | 0 (0) | 0.23 | A (11) | Unknown | |
| TGFβRII | 100 (12) | 83 (10) | 81 (22) | 68 (15) | 0.30 | 0.68 | A (10) | Tumor suppressor |
| BRCA2 | 8 (1) | 0 (0) | 0 (0) | 0 (0) | 0.32 | A (8) | Tumor suppressor | |
| FLJ11712 | 8 (1) | 8 (1) | 22 (6) | 0 (0) | 0.4 | 0.31 | A (10) | Unknown |
| PRKDC | 8 (1) | 0 (0) | 22 (6) | 0 (0) | 0.4 | A (10) | DNA-dependent protein kinase | |
| ABCF1 | 33 (4) | 0 (0) | 19 (5) | 0 (0) | 0.42 | A (10) | Unknown | |
| MARCKS | 83 (10) | 20 (2) | 67 (18) | 28 (5) | 0.45 | 0.68 | A (11) | Cell proliferation and differentiation |
| FLJ11186 | 75 (9) | 0 (0) | 59 (16) | 4 (1) | 0.48 | 1 | A (11) | Unknown |
| GRB-14 | 42 (5) | 0 (0) | 30 (8) | 0 (0) | 0.49 | A (9) | Growth factor bound protein | |
| NADH-UOB | 67 (8) | 0 (0) | 52 (14) | 0 (0) | 0.49 | T (9) | NADH ubiquinone oxydoreductase | |
| Caspase 5 | 42 (5) | 17 (1) | 59 (16) | 40 (6) | 0.50 | 0.61 | A (10) | Apoptosis |
| MAC30 | 8 (1) | 0 (0) | 4 (1) | 0 (0) | 0.53 | A (10) | Unknown | |
| BLM | 25 (3) | 0 (0) | 15 (4) | 0 (0) | 0.65 | A (9) | DNA helicase | |
| DKFZp564C2478 | 25 (3) | 8 (1) | 15 (4) | 4 (1) | 0.65 | 0.53 | A (10) | Unknown |
| TAF1B | 92 (11) | 0 (0) | 81 (22) | 11 (3) | 0.65 | 0.54 | A (11) | Transcription factor |
| TCF6L1 | 58 (7) | 0 (0) | 44 (12) | 0 (0) | 0.65 | A (10) | Transcription factor | |
| GART | 25 (3) | 0 (0) | 19 (5) | 0 (0) | 0.68 | A (10) | Unknown | |
| ACVRII | 83 (10) | 50 (5) | 70 (19) | 47 (9) | 0.69 | 1 | A (8) | Growth factor receptor |
| KIAA1052 | 25 (3) | 0 (0) | 37 (10) | 0 (0) | 0.71 | A (11) | Unknown | |
| SPINK5 | 25 (3) | 0 (0) | 37 (10) | 0 (0) | 0.71 | A (10) | Serine protease inhibitor | |
| TCF4 | 75 (9) | 33 (3) | 63 (17) | 24 (4) | 0.71 | 0.66 | A (9) | Transcription factor |
| BAX | 42 (5) | 20 (1) | 33 (9) | 22 (2) | 0.72 | 1 | G (8) | Apoptosis |
| UVRAG | 42 (5) | 0 (0) | 33 (9) | 0 (0) | 0.72 | A (10) | Unknown | |
| hMSH3 | 50 (6) | 17 (1) | 59 (16) | 13 (2) | 0.85 | 1 | A (8) | DNA mismatch repair |
| KIAA1470 | 50 (6) | 8 (1) | 41 (11) | 11 (3) | 0.85 | 1 | A (10) | Unknown |
| SEC63 | 58 (7) | 17 (2) | 56 (15) | 15 (4) | 0.85 | 1 | A (10) | ER membrane protein |
| AIM2 | 67 (8) | 8 (1) | 67 (18) | 19 (5) | 1 | 0.65 | A (10) | IFN inducible |
| BRCA1 | 0 (0) | 0 (0) | 4 (1) | 0 (0) | 1 | A (8) | Tumor suppresor | |
| CHD2 | 8 (1) | 8 (1) | 15 (4) | 0 (0) | 1 | 0.31 | A (10) | Sequence-selective DNA binding protein |
| FLJ11222 | 25 (3) | 0 (0) | 26 (7) | 0 (0) | 1 | A (10) | Unknown | |
| FLJ20139 | 25 (3) | 8 (1) | 30 (8) | 19 (5) | 1 | 0.65 | A (10) | Unknown |
| FLJ20333 | 17 (2) | 0 (0) | 22 (6) | 0 (0) | 1 | A (10) | Unknown | |
| hMSH6 | 17 (2) | 0 (0) | 26 (7) | 29 (2) | 1 | 1 | C (8) | DNA mismatch repair |
| KIAA1096 | 8 (1) | 0 (0) | 15 (4) | 0 (0) | 1 | A (10) | Unknown | |
| KIAA1268 | 25 (3) | 8 (1) | 22 (6) | 0 (0) | 1 | 0.31 | A (10) | Unknown |
| OGT | 42 (5) | 8 (1) | 41 (11) | 11 (3) | 1 | 1 | T (10) | O-linked GlcNAc transferase |
| PRKWNK1 | 25 (3) | 0 (0) | 26 (7) | 4 (1) | 1 | 1 | A (10) | Unknown |
| SLC24AI | 42 (5) | 0 (0) | 37 (10) | 0 (0) | 1 | C (9) | Nucleobase transporter | |
| SYCP1 | 25 (3) | 0 (0) | 22 (6) | 0 (0) | 1 | A (10) | Synaptonemal complex | |
| WISP3 | 8 (1) | 0 (0) | 15 (4) | 25 (1) | 1 | 1 | A (9) | Growth factor (Wnt pathway) |
*P value of total allelic mutations with and without CIs.
† P value of biallelic mutations with and without CIs.
Discussion
CIs are common phenomena in human cancers, but their causes and consequences are not well defined. Of the two main forms of genomic instability, CIN and MSI, CIN is known to be closely associated with CIs, and the CIN phenotype is characterized by gross rearrangement of chromosomes. Common CIs include the loss or gain of whole chromosomes or chromosomal fragments, and amplification of chromosome. 23 Chromosomal loss can lead to the inactivation of tumor suppressor genes, whereas chromosomal gains can lead to the activation of proto-oncogenes. The other frequent CIs involves translocation, which can lead to the deregulation of gene expression or aberrant gene expression related to tumorigenesis by causing two genes to fuse. 24 In contrast, the MSI phenotype is characterized by small point mutations or small deletions because of defects in the mismatch repair genes. Recent studies have found that most of the sporadic MSI-H carcinomas result from the inactivation of the hMLH1 gene, principally by transcriptional silencing. 25,26 In this study, we found CIs in a subset of MSI-H colorectal carcinomas. Although it is widely accepted that CIs are rare in MSI-H tumors, a few studies on CIs in MSI-H carcinomas are available. The majority of previous studies about CIs in MSI-H tumors have included a small number of cases, and the results obtained have been conflicting. 11,12,27-29 The co-existence of two kinds of genomic instability in several colorectal cancer cell lines has been reported. 13 In this study, we examined CIs in a relatively large number of cases (n = 39) and found CIs in 12 (31%) cases. These findings indicate that some colorectal carcinomas have both types of genomic instability, and therefore, it might show biological behaviors that differ from those showing only one of the two typical genetic instabilities.
The tumor development stage and the genetic events that initiate CIs in MSI-H colorectal carcinomas are important for the understanding of the molecular pathogenesis of colorectal carcinomas. The MSI pathway begins with the inactivation of one of a group of genes responsible for DNA nucleotide mismatch repair, which leads to extensive point mutations and frameshift mutations in repetitive DNA sequences with low frequencies of allelic losses and rare alterations of tumor DNA content. 30 Frequent somatic frameshift mutations in the genes containing nucleotide repeats in their coding sequences have been reported, 14,15,31-33 and many of these genes are regarded as target genes in MSI-H tumors. At present, many genes have been reported to be candidate target genes. 34 We previously proposed a role for frameshift mutations during carcinoma transformation. We found that the incidence and type of inactivation patterns of the mismatch-repair genes were the same in MSI-H gastric adenomas and carcinomas, but that frameshift mutations were much more frequent in MSI-H gastric carcinomas. 35 This suggests that the accumulated frameshift mutations of the target genes are related to the malignant transformation of MSI-H tumors. The incidence of frameshift mutations of the target genes in our MSI-H tumors with CIs was slightly higher than that of the MSI-H tumors without CIs, suggesting that the progression period of the MSI pathway might be longer in MSI-H tumors with CIs. We also found that chromosomal changes in MSI-H colon carcinomas are much less frequent than changes of MSS carcinomas and that the types of chromosomes involved are quite different. These findings suggest that some tumors following the MSI pathway can have mutations in the genes responsible for CIN and these defects might be selected 13 during tumor progression. However, the genetic pathways responsible for inducing CIN might differ from that of MSS tumors.
The reported target genes of MSI-H tumors can be functionally categorized as tumor suppressors, and genes involved in apoptosis and DNA repair. Many genes involved in DNA damage signaling and DNA repair pathways play critical roles in the suppression of genome instabilities. Inactivating mutations of ATM, BRCA1, BRCA2, NBS1, and BLM genes cause defects in DNA damage signaling and DNA repair, give rise to some forms of CIN, and contribute to carcinogenesis. 36 We and others have reported that several genes involved in DNA repair have mononucleotide repeats in their coding sequence and are frequently mutated in MSI-H colorectal carcinomas. 14,15,33,37 We analyzed the relationship between target gene mutation and CIs in MSI-H tumors, and found that frameshift mutation of hRAD50 is significantly related to CIs in MSI-H tumors. hRAD50 forms a complex with hMRE11 and NBS1. The role of this heterotrimer in DNA damage signaling and chromosome instability has been reported. 38,39 Based on the reported functional characteristics of the hRAD50/MRE11/NBS1 complex, our findings indicate that CIN in MSI-H colorectal carcinomas might be induced from the functional alterations of hRAD50.
We cannot explain the significance of the high frequency of mutations of FLJ11383 in MSI-H tumors with CIs, because the functionality of FLJ11383 is unknown. In addition to hRAD50 and FLJ11383, mutations of the other genes involved in DNA repair, ATR and MBD4, also showed trends toward more frequent mutations in MSI-H tumors with CIs. Future analysis of a larger series of cases and the determination of the functional mechanism of the CIN of these genes should clarify the associations between mutations of these genes and genetic progression via the CIN pathway in MSI-H carcinomas.
Acknowledgments
We thank Dr. Haeyoun Kang and Haeyoung Kim for assistance with English and Miss Se Kyung Kim for technical assistance.
Footnotes
Address reprint requests to Hoguen Kim, M.D., Department of Pathology, Yonsei University College of Medicine, CPO Box 8044, Seoul, Korea. E-mail: hkyonsei@yumc.yonsei.ac.kr.
Supported by the International Mobile Telecommunications 2000 R&D Project (grant 01-PJ11-PG9-01BT00B-0006), Ministry of Information and Communication, Republic of Korea.
References
- 1.Lengauer C, Kinzler KW, Vogelstein B: Genetic instabilities in human cancers. Nature 1998, 396:643-649 [DOI] [PubMed] [Google Scholar]
- 2.Kinzler KW, Vogelstein B: Lessons from hereditary colorectal cancer. Cell 1996, 87:159-170 [DOI] [PubMed] [Google Scholar]
- 3.Lengauer C, Kinzler KW, Vogelstein B: Genetic instability in colorectal cancers. Nature 1997, 386:623-627 [DOI] [PubMed] [Google Scholar]
- 4.Miyazaki M, Furuya T, Shiraki A, Sato T, Oga A, Sasaki K: The relationship of DNA ploidy to chromosomal instability in primary human colorectal cancers. Cancer Res 1999, 59:5283-5285 [PubMed] [Google Scholar]
- 5.Kolodner RD, Putnam CD, Myung K: Maintenance of genome stability in Saccharomyces cerevisiae. Science 2002, 297:552-557 [DOI] [PubMed] [Google Scholar]
- 6.Chen C, Kolodner RD: Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat Genet 1999, 23:81-85 [DOI] [PubMed] [Google Scholar]
- 7.Myung K, Datta A, Kolodner RD: Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell 2001, 104:397-408 [DOI] [PubMed] [Google Scholar]
- 8.Myung K, Chen C, Kolodner RD: Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 2001, 411:1073-1076 [DOI] [PubMed] [Google Scholar]
- 9.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M: Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature 1993, 363:558-561 [DOI] [PubMed] [Google Scholar]
- 10.Thibodeau SN, Bren G, Schaid D: Microsatellite instability in cancer of the proximal colon. Science 1993, 260:816-819 [DOI] [PubMed] [Google Scholar]
- 11.Curtis LJ, Georgiades IB, White S, Bird CC, Harrison DJ, Wyllie AH: Specific patterns of chromosomal abnormalities are associated with RER status in sporadic colorectal cancer. J Pathol 2000, 192:440-445 [DOI] [PubMed] [Google Scholar]
- 12.Eshleman JR, Casey G, Kochera ME, Sedwick WD, Swinler SE, Veigl ML, Willson JK, Schwartz S, Markowitz SD: Chromosome number and structure both are markedly stable in RER colorectal cancers and are not destabilized by mutation of p53. Oncogene 1998, 17:719-725 [DOI] [PubMed] [Google Scholar]
- 13.Abdel-Rahman WM, Katsura K, Rens W, Gorman PA, Sheer D, Bicknell D, Bodmer WF, Arends MJ, Wyllie AH, Edwards PA: Spectral karyotyping suggests additional subsets of colorectal cancers characterized by pattern of chromosome rearrangement. Proc Natl Acad Sci USA 2001, 98:2538-2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim NG, Choi YR, Baek MJ, Kim YH, Kang H, Kim NK, Min JS, Kim H: Frameshift mutations at coding mononucleotide repeats of the hRAD50 gene in gastrointestinal carcinomas with microsatellite instability. Cancer Res 2001, 61:36-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim NG, Rhee H, Li LS, Kim H, Lee JS, Kim JH, Kim NK, Kim H: Identification of MARCKS, FLJ11383 and TAF1B as putative novel target genes in colorectal carcinomas with microsatellite instability. Oncogene 2002, 21:5081-5087 [DOI] [PubMed] [Google Scholar]
- 16.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S: A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 1998, 58:5248-5257 [PubMed] [Google Scholar]
- 17.Kim H, Kim YH, Kim SE, Kim NG, Noh SH, Kim H: Concerted promoter hypermethylation of hMLH1, p16INK4A, and E-cadherin in gastric carcinomas with microsatellite instability. J Pathol 2003, 200:23-31 [DOI] [PubMed] [Google Scholar]
- 18.Baek MJ, Kang H, Kim SE, Park JH, Lee JS, Paik YK, Kim H: Expression of hMLH1 is inactivated in the gastric adenomas with enhanced microsatellite instability. Br J Cancer 2001, 85:1147-1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung KY, Kim NG, Li LS, Kim H, Kim H, Nam CM, Kim H, Shin DH: Clinicopathologic characteristics related to the high variability of coding mononucleotide repeat sequences in tumors with high-microsatellite instability. Oncol Rep 2003, 10:439-444 [PubMed] [Google Scholar]
- 20.Hempen PM, Zhang L, Bansal RK, Iacobuzio-Donahue CA, Murphy KM, Maitra A, Vogelstein B, Whitehead RH, Markowitz SD, Willson JK, Yeo CJ, Hruban RH, Kern SE: Evidence of selection for clones having genetic inactivation of the activin A type II receptor (ACVR2) gene in gastrointestinal cancers. Cancer Res 2003, 63:994-999 [PubMed] [Google Scholar]
- 21.Duval A, Rolland S, Compoint A, Tubacher E, Iacopetta B, Thomas G, Hamelin R: Evolution of instability at coding and non-coding repeat sequences in human MSI-H colorectal cancers. Hum Mol Genet 2001, 10:513-518 [DOI] [PubMed] [Google Scholar]
- 22.Thorstensen L, Diep CB, Meling GI, Aagesen TH, Ahrens CH, Rognum TO, Lothe RA: WNT1 inducible signaling pathway protein 3, WISP-3, a novel target gene in colorectal carcinomas with microsatellite instability. Gastroenterology 2001, 121:1275-1280 [DOI] [PubMed] [Google Scholar]
- 23.Mertens F, Johansson B, Hoglund M, Mitelman F: Chromosomal imbalance maps of malignant solid tumors: a cytogenetic survey of 3185 neoplasms. Cancer Res 1997, 57:2765-2780 [PubMed] [Google Scholar]
- 24.Nowell PC: Genetic alterations in leukemias and lymphomas: impressive progress and continuing complexity. Cancer Genet Cytogenet 1997, 94:13-19 [DOI] [PubMed] [Google Scholar]
- 25.Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R: Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res 1997, 57:808-811 [PubMed] [Google Scholar]
- 26.Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, Kane MF, Kolodner RD, Vogelstein B, Kunkel TA, Baylin SB: Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA 1998, 95:6870-6875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schlegel J, Stumm G, Scherthan H, Bocker T, Zirngibl H, Ruschoff J, Hofstadter F: Comparative genomic in situ hybridization of colon carcinomas with replication error. Cancer Res 1995, 55:6002-6005 [PubMed] [Google Scholar]
- 28.Willenbucher RF, Aust DE, Chang CG, Zelman SJ, Ferrell LD, Moore DH, Waldman FM: Genomic instability is an early event during the progression pathway of ulcerative-colitis-related neoplasia. Am J Pathol 1999, 154:1825-1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goel A, Arnold CN, Niedzwiecki D, Chang DK, Ricciardiello L, Carethers JM, Dowell JM, Wasserman L, Compton C, Mayer RJ, Bertagnolli MM, Boland CR: Characterization of sporadic colon cancer by patterns of genomic instability. Cancer Res 2003, 63:1608-1614 [PubMed] [Google Scholar]
- 30.Loeb LA: Microsatellite instability: marker of a mutator phenotype in cancer. Cancer Res 1994, 54:5059-5063 [PubMed] [Google Scholar]
- 31.Fujiwara T, Stolker JM, Watanabe T, Rashid A, Longo P, Eshleman JR, Booker S, Lynch HT, Jass JR, Green JS, Kim H, Jen J, Vogelstein B, Hamilton SR: Accumulated clonal genetic alterations in familial and sporadic colorectal carcinomas with widespread instability in microsatellite sequences. Am J Pathol 1998, 153:1063-1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malkhosyan S, Rampino N, Yamamoto H, Perucho M: Frameshift mutator mutations. Nature 1996, 382:499-500 [DOI] [PubMed] [Google Scholar]
- 33.Mori Y, Yin J, Rashid A, Leggett BA, Young J, Simms L, Kuehl PM, Langenberg P, Meltzer SJ, Stine OC: Instabilotyping: comprehensive identification of frameshift mutations caused by coding region microsatellite instability. Cancer Res 2001, 61:6046-6049 [PubMed] [Google Scholar]
- 34.Duval A, Hamelin R: Mutations at coding repeat sequences in mismatch repair-deficient human cancers: toward a new concept of target genes for instability. Cancer Res 2002, 62:2447-2454 [PubMed] [Google Scholar]
- 35.Kim JJ, Baek MJ, Kim L, Kim NG, Lee YC, Song SY, Noh SH, Kim H: Accumulated frameshift mutations at coding nucleotide repeats during the progression of gastric carcinoma with microsatellite instability. Lab Invest 1999, 79:1113-1120 [PubMed] [Google Scholar]
- 36.Schar P: Spontaneous DNA damage, genome instability, and cancer—when DNA replication escapes control. Cell 2001, 104:329-332 [DOI] [PubMed] [Google Scholar]
- 37.Woerner SM, Gebert J, Yuan YP, Sutter C, Ridder R, Bork P, von Knebel Doeberitz M: Systematic identification of genes with coding microsatellites mutated in DNA mismatch repair-deficient cancer cells. Int J Cancer 2001, 93:12-19 [DOI] [PubMed] [Google Scholar]
- 38.Chen C, Kolodner RD: Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat Genet 1999, 23:81-85 [DOI] [PubMed] [Google Scholar]
- 39.Giannini G, Ristori E, Cerignoli F, Rinaldi C, Zani M, Viel A, Ottini L, Crescenzi M, Martinotti S, Bignami M, Frati L, Screpanti I, Gulino A: Human MRE11 is inactivated in mismatch repair-deficient cancers. EMBO Rep 2002, 3:248-254 [DOI] [PMC free article] [PubMed] [Google Scholar]