Abstract
X inactivation is a chromosome-specific form of genetic regulation in which thousands of genes on one homologue become silenced early in female embryogenesis. Although many aspects of X inactivation are now understood, the spread of the X inactivation signal along the entire length of the chromosome remains enigmatic. Extending the Gartler–Riggs model [Gartler, S. M. & Riggs, A. D. (1983) Annu. Rev. Genet. 17, 155–190], Lyon recently proposed [Lyon, M. F. (1998) Cytogenet. Cell Genet. 80, 133–137] that a nonrandom organization of long interspersed element (LINE) repetitive sequences on the X chromosome might be responsible for its facultative heterochromatization. In this paper, we present data indicating that the LINE-1 (L1) composition of the human X chromosome is fundamentally distinct from that of human autosomes. The X chromosome is enriched 2-fold for L1 repetitive elements, with the greatest enrichment observed for a restricted subset of LINE-1 elements that were active <100 million years ago. Regional analysis of the X chromosome reveals that the most significant clustering of these elements is in Xq13–Xq21 (the center of X inactivation). Genomic segments harboring genes that escape inactivation are significantly reduced in L1 content compared with X chromosome segments containing genes subject to X inactivation, providing further support for the association between X inactivation and L1 content. These nonrandom properties of L1 distribution on the X chromosome provide strong evidence that L1 elements may serve as DNA signals to propagate X inactivation along the chromosome.
Among placental mammals, the basic features of X chromosome inactivation are well established (1, 2). X inactivation is a chromosome-wide mechanism of gene regulation, transcriptionally silencing the majority of genes on the X chromosome during mammalian female embryogenesis. This process serves to maintain the correct dosage relationship of genes between females (XX) and males (XY). X inactivation is believed to involve three distinct steps: initiation of inactivation early in development, spreading of inactivation in cis along the length of the chromosome, and subsequent maintenance throughout all successive somatic cell divisions. XIST, a functional non-protein-encoding RNA, maps to the X inactivation center in human and mouse (3–5). Xist has been shown by transgenic and knockout experiments in mice to play a pivotal role in initiating X inactivation (6–9). Once inactivated, the chromosome acquires a number of features associated with transcriptionally inactive chromatin: the X becomes late-replicating, hypoacetylated, and hypermethylated at cytosine residues in CpG islands of housekeeping genes (1, 2). More recent studies have shown that the inactive X becomes coated with XIST RNA and that the chromatin structure incorporates at least one unique histone variant, MacroH2A (10–14). This has led to speculation that an XIST RNA–protein–DNA complex may be an important component in delineating the heterochromatic structure. All of these features indicate that a complex system has evolved to create and maintain the inactive state.
Although genetic and molecular aspects of initiation and maintenance of X inactivation have been characterized, the propagation of the signal in cis remains unknown. In particular, it is unknown whether cis-acting DNA sequences participate in this process. Early studies of mice carrying X:autosome translocations and later autosomally integrated Xist yeast artificial chromosome transgenes indicate that inactivation can spread into and effectively silence autosomal genes (8, 9, 15, 16). However, these and other studies of human X:autosome translocations (17) suggest that X inactivation is spread less efficiently into autosomal DNA. Therefore, if a DNA signal exists for spreading of X inactivation, it is likely not restricted to the X chromosome but may be organized or enriched on this chromosome in such a manner as to favor its nearly complete heterochromatization. Additionally, the differential organization of cis-acting sequences might explain why some X-linked genes are inactivated, whereas others are expressed on both the active and inactive chromosomes (18).
In 1983, Gartler and Riggs (19) put forward the concept of “booster” elements or “way stations,” which were concentrated at the X inactivation center and at other positions throughout the X chromosome. In their model, expanded upon by Riggs (20), the unique organization of these elements served to amplify and spread the X inactivation signal along the entire length of the chromosome. In 1998, Lyon (21) proposed long interspersed repeat element (LINE)-1 (L1) as a candidate for these “booster” elements. L1 elements are mammal-specific (22) retrotransposons with currently active members (23) in the human genome (for recent reviews see refs. 24 and 25). Lyon's “repeat hypothesis” was based largely on two observations. First, fluorescent in situ hybridization (FISH) studies in human (26) and mouse (27) using L1 repeat elements as probes had shown that the X chromosome of each species hybridized more intensely than autosomes and was therefore presumably enriched for these elements. Second, her reexamination of mouse X:autosome translocation data found that failure of the X inactivation signal to spread often correlated with cytogenetic bands that were deficient in L1 elements. Using data collected from the Human Genome Project, we sought to investigate more precisely the pattern of L1 distribution along the X chromosome, to compare this pattern to its distribution in other human autosomes, and to determine whether its nonrandom organization was consistent with the known biology of X inactivation.
Methods
Data.
Nonoverlapping human genomic sequences were obtained from GenBank (NT division) (28). The data set (2,046 segments; average length 198 kb) represented an estimated 12.6% (405/3213 Mb) of the human haploid genomic equivalent. Both cytogenetic and radiation hybrid mapping data were used to assign chromosomal location (http://www.ncbi.nlm.nih.gov/genome/seq/). At the time of this analysis, Nov. 19, 1999, the X chromosome sequence was 34.5% (56.7/164 Mb) complete. Five other chromosomes had been completed to similar levels: 19.0% (34.8/183 Mb) for chromosome 6, 43.0% (73.6/171 Mb) for chromosome 7, 21.8% (15.7/72 Mb) for chromosome 20, 39.2% (15.3/39 Mb) for chromosome 21, and 68.9% (29.6/43 Mb) for chromosome 22. These chromosomes were used as controls for comparison with the X chromosome data. Additionally, more detailed analysis for chromosome 7 was done (Figs. 2b and 3b).
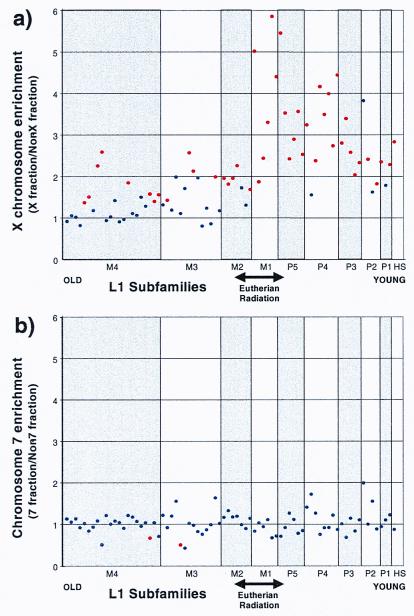
Figure 2.
L1 subfamily enrichment for chromosomes X and 7. L1 elements were grouped into 75 different subclasses based on the original ORF2 and 3′ untranslated region classification (29). In a, an enrichment factor for the X chromosome was determined for each element (bp fraction of X chromosome sequence/bp fraction of sequence not from the X chromosome) and is depicted (y axis). The various L1 elements are grouped into 10 major subfamilies (x axis) and are arranged from the evolutionarily most ancient [L1M4 (mammalian group 4)] to the currently active element in the human lineage (L1Hs). Within each of the 10 major L1 classes, individual subclasses were arranged on the basis of increasing average similarity to the consensus. L1 subclasses that differed significantly from random (P < 0.006 after a Bonferroni correction for 75 subclasses) are colored red (n = 10,000 replicates; 56 Mb). The alignment number, total bp, and fraction of bp for each element were calculated for both the X and non-X groups (see Table 2 in supplemental material at www.pnas.org). In b, a similar analysis was performed for chromosome 7. Two subclasses (red) were significantly depleted.
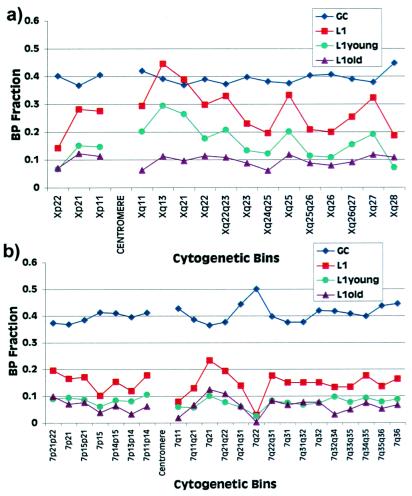
Figure 3.
Percent L1 vs. percent G + C composition. Nonoverlapping genomic sequences from the X chromosome (a) and chromosome 7 (b) were binned into cytogenetic band intervals based on cytogenetic and radiation hybrid mapping data (http://www.ncbi.nlm.nih.gov/genome/seq/). The percentage L1 content and G + C content for each bin of sequence is depicted graphically. Since the precise boundaries of cytogenetic bands are not known at the sequence level, many of the bins bracket two or more cytogenetic band intervals. L1s have been separated into old (L1M4 to L1M2) and young (L1M1, L1P5 to L1P1, L1Hs). The clustering of younger L1s at Xq13 and Xq21 suggests both a temporal and a positional bias in the accumulation of L1s on the X chromosome. The linear correlation (r2) between G + C and L1 content was 0.24 for the X chromosome and 0.50 for chromosome 7.
Sequence Analysis.
The identity of interspersed repeat elements was determined by using the program RepeatMasker v.3.0 (A. F. A. Smit & P. Green, http://ftp.genome.washington.edu/RM/RepeatMasker.html). The program implements a modified Smith–Waterman algorithm to match sequences against a curated Repbase library (http://www.girinst.org). L1 elements were classified into various subclasses on the basis of a series of diagnostic changes in either the 3′ untranslated region or ORF2 (29). In our data set, 95% (56,039/58,995 Mb) of all L1 sequences could be assigned to one of ten major L1 subfamilies (L1M4 to L1M1, L1P5 to L1P1, and L1Hs), and a subset of these could be further subdivided into 65 minor L1 subfamily variants. RepeatMasker analysis was performed at the sensitive/slow setting, and matches were identified at prescribed Smith–Waterman scores: 300 for low-complexity LINE-1 search and 180 for very old MIR, LINE-2, and MER-2 sequences. A series of perl scripts were designed to analyze the RepeatMasker output. The percent composition of every repeat analyzed in this study was based on the actual number of bases masked per segment.
Using cDNA sequences of known X inactivation status as queries, we performed blastn sequence similarity searches (30) to identify X chromosomal segments that contain genes that are either subject to or escape X inactivation. Thirty-nine genes that escape X inactivation and 228 genes that are subject to X inactivation were considered (18). Ten segments (1.78 Mb, average length = 178 kb) and 42 segments (total = 9.78 Mb, average length = 232 kb) contained genes that respectively escape and are subjected to X inactivation were considered. Because the domains of X inactivation regions are not precisely known, the entire genomic segment was categorized as either escaping or subject based on the status of known genes (18). We consider this treatment of the data conservative, because only 10–15% of X-linked genes escape inactivation (18). Untrimmed data, therefore, would be more likely to include genes that were subject to X inactivation. One segment (NT_001190) that had a mixed X inactivation status was omitted from this analysis. Each of the segments identified was subsequently aligned with the cDNA by using the program sim4 (31), and the data were inspected to confirm the intron/exon structure of each gene by using the program viewGene (C. Kashuk and A.C., unpublished work).
Statistical Analyses.
The mean percentage of interspersed repetitive elements was calculated for the X chromosome (51.5%) and non-X chromosome (40.0%) as the quotient of interspersed repeat and total number of base pairs (29.2/56.7 Mb and 139.4/348.8 Mb, respectively). To determine whether the X chromosomal sequence repeat content was significantly different from the non-X, the difference between the mean repeat contents of X and non-X was used as a test statistic. The observed repeat content difference (51.5% − 40.0% = 11.5%) was compared with the distribution of mean differences under the null hypothesis that there is no difference in the mean percentage of chromosomal repeat content. To generate this distribution, a Monte Carlo simulation program (MonteCarlo_table.pl) was developed, whereby, for each replicate, 56.7 Mb of genomic segments from the total sample of 405.5 Mb were randomly designated as “X” chromosome and the remaining sequences were designated as “non-X” chromosome. For each replicate, the difference in mean percentage of interspersed repetitive elements was calculated and compared with the observed value (11.5%). A P value was estimated by counting the number of replicates whose difference in means met or exceeded the observed mean difference, and dividing this number by the total number of replicates. For total interspersed repetitive elements, no simulated replicates met or exceeded the observed difference (P < 0.0001). Analogous Monte Carlo simulations were done to test other means: the major classes of interspersed repeats and L1 families and subfamilies.
The X-inactivation status of genomic segments was determined on the basis of content of genes that escape and are subject to X inactivation. To determine whether the mean fraction of L1 elements differed significantly, a permutation test was performed with the perl program (permute_table.pl). The X-inactivation status was randomly reassigned to the genomic segments and the mean L1 fraction for the subject and the escape categories was calculated for each of 10,000 replicates. The difference between the means was determined for each replicate. The probability that the observed difference between L1 escape and subject domains (10.1%) occurred randomly was simply the fraction of replicates that exceeded or equaled this difference (P = 40/10,000).
Results
X Chromosome L1 Bias.
We examined the repeat structure of available nonoverlapping human genomic sequence segments by using RepeatMasker software (Methods). A total of 42.6% (168/405.5 Mb) of the DNA was identified as interspersed repetitive sequence, primarily LINE, SINE (short interspersed element), LTR (long terminal repeat), and DNA repeat elements. The X chromosome had the highest mean interspersed repeat content (23/56.7 Mb = 51.5%); this composition was significantly different from that of non-X chromosome sequences (40.0%) (P < 10−6; Methods). L1 repeat elements accounted for 87% of this increase (Table 1). A similar study by Smit (32), who analyzed 327 Mb of sequence in terms of G + C content, found a comparable level of enrichment for L1 elements on the X chromosome. The 2-fold difference in L1 repeat composition (X: 26.5% vs. non-X: 13.4%) was determined to be highly significant (P < 0.0001; Fig. 1). Because only a portion of the total human genome was available for analysis and the level of completion differed among various chromosomes, one concern was that the observed differences in L1 content might reflect ascertainment biases. To address this question, L1 content was independently calculated for six human autosomes that showed the greatest levels of sequencing completion (>20%) (data not shown). The autosomes either show no significant difference (chromosomes 6 and 7) or a significant decrease (chromosomes 17, 20, 21, and 22) in mean L1 content compared with randomly sampled genomic segments. These comparisons confirm that the X chromosome has been a preferential target of L1 retrotransposition compared with human autosomes.
Table 1.
Sequence repeat content for major interspersed repeat families
| Family | X chromosome
|
Non-X chromosomes
|
X vs. non-X Δ, % | P value | ||||
|---|---|---|---|---|---|---|---|---|
| bp | No. | % | bp | No. | % | |||
| SINE | 5,420,458 | 23,405 | 9.56 | 50,020,310 | 220,539 | 14.34 | −4.78 | <0.0001 |
| Alu | 4,254,319 | 15,859 | 7.51 | 41,809,635 | 162,001 | 11.99 | −4.48 | <0.0001 |
| MIR | 1,166,139 | 7,546 | 2.06 | 8,597,603 | 58,538 | 2.46 | −0.40 | 0.0008 |
| LINE | 17,090,789 | 19,682 | 30.16 | 59,450,470 | 101,989 | 17.04 | 13.12 | <0.0001 |
| LINE-1 | 15,015,013 | 13,018 | 26.50 | 46,839,144 | 55,565 | 13.43 | 13.07 | <0.0001 |
| LINE-2 | 1,963,431 | 6,394 | 3.56 | 12,761,310 | 44,233 | 3.66 | −0.10 | 0.3172 |
| LTR | 5,090,853 | 8,941 | 8.98 | 23,688,836 | 48,984 | 6.79 | 2.19 | <0.0001 |
| MaLRs | 2,247,808 | 4,315 | 3.97 | 10,684,587 | 25,161 | 3.06 | 0.91 | <0.0001 |
| Retrovirus | 1,689,316 | 2,590 | 2.98 | 8,269,778 | 13,815 | 2.37 | 0.61 | 0.0010 |
| MER4 | 961,207 | 1,692 | 1.69 | 3,930,795 | 8,181 | 1.13 | 0.56 | <0.0001 |
| DNA | 1,411,008 | 5,483 | 2.49 | 8,515,500 | 33,176 | 2.44 | 0.05 | 0.2685 |
| MER1 | 757,640 | 3,689 | 1.34 | 4,463,695 | 21,663 | 1.28 | 0.06 | 0.1140 |
| MER2 | 499,003 | 995 | 0.88 | 3,217,455 | 6,510 | 0.92 | −0.04 | 0.2170 |
| Mariner | 49,394 | 190 | 0.09 | 210,674 | 1,057 | 0.06 | 0.03 | 0.01590 |
| Total | 29,184,062 | NA | 51.50 | 139,432,284 | NA | 39.97 | 11.53 | <0.0001 |
| Total bp | 56,666,472 | 348,844,905 | ||||||
Major classes for interspersed repeat content are shown in terms of the number (No.), total base pairs (bp), and percent bp (%) of repeats. LINE and LINE-1 content are dramatically increased on the X chromosome. The absolute difference (Δ) between mean X and non-X percentages was used to determine significance (Monte Carlo simulation; 10,000 replicates). A similar analysis has been performed addressing enrichment on the X chromosome in terms of G + C content (32). SINE, short interspersed element; MIR, mammalian-wide interspersed repeat; MaLR, mammalian LTR retrovirus; MER, medium reiteration frequency; NA, not applicable.
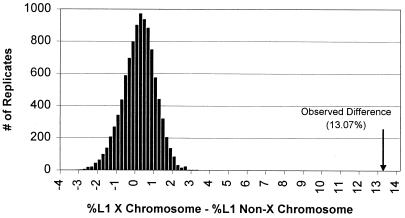
Figure 1.
Nonrandom distribution of L1 elements on the X chromosome. Assuming no chromosomal bias in L1 content, the figure shows a simulated distribution (10,000 replicates) of the difference in the mean L1 percentage between the X chromosome sequence (56.7 Mb randomly assigned) and non-X sequence (the remaining 348 Mb). Under this model variation mostly represents the random sampling of regional differences that occur between sequences. The observed difference (13.07%) between X and non-X sequence did not occur in 1,000,000 replicates (P < 10−6).
A Eutherian Burst of L1 Retransposition.
Current models for L1 transposition indicate that all mammalian L1 elements share a monophyletic origin in which specific subclasses have been active at different times during mammalian evolution (29, 33). L1 elements, based on subfamily stratification, may therefore serve as molecular fossils to date any evolutionary event. To investigate the evolution of the L1 retrotransposition on the X chromosome, we categorized all X chromosome and non-X chromosome L1 elements into various subclasses based on a previously published phylogeny (29). We then compared the representation of each L1 subclass between the X and non-X portions of the sequence (Fig. 2 and Methods).
Our analysis indicates that all L1 subclasses are not uniformly over-represented. When L1 subfamilies are arranged in descending order of evolutionary age (Fig. 2; Table 2 in supplemental material at www.pnas.org), two different groups emerge. The older L1 elements (L1M4 to L1M2) show only a slight enrichment on the X chromosome. In contrast, most younger L1 elements (L1M1, L1P5 to L1P1) are much more significantly over-represented on the X chromosome. To confirm that this effect was X-specific, a similar analysis was performed on chromosome 7 (Fig. 2b and Methods). No significant enrichments were observed for any L1 subclass. If the rate of mutation or deletion on the X chromosome is increased relative to the autosomes, then an enrichment of older L1 elements will be obscured by their rapid removal from the X chromosome. However, LINE-2 and MIR elements allow us to exclude this possibility. Both of these ancient retroposons are equally abundant on the X chromosome and autosomes, suggesting that in general elements are not removed more rapidly from the X chromosome. The most pronounced effect on the X chromosome (3- to 5-fold increase) was observed for L1 repeats belonging largely to the L1M1 and L1P4 subclasses. These families are hypothesized to have been active during the eutherian to prosimian transition, 60–100 million years ago (29). The temporal specificity indicated by this subfamily analysis corresponds with the evolution of the more complex, multistep eutherian X-inactivation pathway (34).
Regional L1 Clustering on the X Chromosome Independent of G + C Content.
What might account for the observed preference for L1 elements to accumulate on the X chromosome? One explanation might be an unusually reduced G + C content on the X compared with other human chromosomes. Early in situ experiments and studies of G + C isochores in mouse and human demonstrated an inverse relationship between G + C content and L1 chromosomal distribution (35, 36). With few exceptions, Giemsa-staining dark bands, which are generally A+T-rich, hybridized most intensely with LINE fluorescent in situ hybridization probes (26, 27, 37). These data have been interpreted as evidence for an L1 preference to integrate within A+T-rich regions (26, 36). Our analysis indicates that the X chromosome is significantly reduced in average G + C content compared with non-X chromosome sequence (39.6% vs. 43.1%; P < 0.0001). However, Smit (32) has determined that the X chromosome was 1.5- to 2.0-fold enriched over autosomal regions when the same % G + C isochore was compared.
To investigate this relationship of L1s, G + C content, and Giemsa banding patterns, we partitioned the X chromosome and a representative human autosome (chromosome 7) sequences into bins corresponding to Giemsa-staining light and dark patterns (Fig. 3). The percent G + C and L1 composition for each bin were estimated (Methods). The band assignments were based on available radiation hybrid and cytogenetic mapping data. While there is good correlation between L1 and G + C content on chromosome 7, the L1 content for the X chromosome fluctuates much more dramatically as a function of G + C content and band location (Fig. 3). There is clearly not a good correlation between L1 and G + C content at the level of the X chromosomal cytogenetic bands. However, Smit (32) has shown that some correlation can be found if smaller windows of 50 kb in length are used. Combined, the data indicate significant fluctuation in both L1 and G + C content within bands, which is clearly different from autosomal patterns. We also examined chromosomal band gene density and found no obvious correlation with L1 content (data not shown; http://www.ncbi.nlm. nih.gov/genemap/).
One prediction of the Gartler–Riggs model was that the site of X inactivation nucleation should be particularly rich in elements that promote the spread of the X-inactivation signal (19). In the absence of such “stabilizing sequences” it was hypothesized that the signal might effectively deteriorate, resulting in the incomplete inactivation of the X chromosome. An unusually strong clustering effect of L1 elements is observed for cytogenetic bands Xq21 and Xq13, where 39% and 45% of available sequence is composed of L1 repeats (Fig. 3a; Table 3 in supplemental material at www.pnas.org). The distribution within this region of the X chromosome is significantly enriched relative to other X chromosome segments (P = 0.013). In humans, the X-inactivation center (XIC) and the XIST locus have been mapped to Xq13 (4, 5), the cytogenetic band which is most enriched for L1 elements. This clustering on Xq13 and Xq21 does not correlate simply with a compensatory reduction in G + C content, but involves selection of L1 elements in excess of that observed for other regions of the X chromosome with similar G + C content. On the X chromosome, this effect is due largely to younger (classes L1M1 and L1Hs) primate L1 elements (Fig. 3a). Among other human autosomes (Fig. 3b; data not shown), no such differences in regional distribution are observed between older and younger L1 elements. Additional clusters of L1 elements within Xq22, Xq24–q25, and Xq27 (Fig. 3a) may represent “way stations” (19) that would serve to efficiently spread the X-inactivation signal along the remaining portions of the X chromosome. It is interesting that one region of the X chromosome appears uncharacteristically reduced in L1 content (Xp22). Indeed, the L1 content of Xp22 sequences (15.3%) is very similar to the genomic average (15.6%). This region of the X chromosome corresponds partially to what has been considered a recent autosomal translocation or strata IV (38, 39). The reduced L1 composition in this region would be consistent with its recent autosomal origin. It will be important to compare the properties of genes that escape X inactivation from Xp22 to other regions of the X chromosome that harbor X-inactivation escape loci.
L1 Composition: Escape vs. Subject to X Inactivation.
For at least two regions of the X chromosome, genes that escape inactivation cluster (1). This observation suggests regulatory control of X-inactivation status at the level of chromosomal domains. One prediction is that genes that escape inactivation lie within genomic environments deficient in “booster” elements. To test this association between L1 composition and X-inactivation status, we examined the L1 content of genomic segments known to contain genes that escape X inactivation. A total of 39 genes that were recently shown to escape X inactivation (18) were searched by blastn sequence similarity searches (30), against the human genomic data set (Methods). Thirteen genes from Xp11 and Xp22 corresponding to 10 genomic segments were identified (Table 4 in the supplemental material at www.pnas.org). The L1 content of genomic segments that carry genes capable of escaping X inactivation is significantly lower (P = 4.8 × 10−5; Methods) than the X chromosome average (10.3% vs. 26.5%, respectively; see Methods). Because the presence of genic sequence necessarily reduces the proportion of repetitive sequence, we wished to compare these estimates of L1 content with genic regions that are subject to X inactivation. blastn sequence similarity searches identified 42 genomic segments that contained 64 genes exhibiting complete or nearly complete X inactivation (Table 4) (18). A random permutation test between these two sets of data for L1 content found a significant difference (P = 0.004) between genomic segments that escape (10.3%) and those that are subject to X inactivation (20.5%). This finding suggests that regions that escape X inactivation have been protected from L1 retrotransposition. It should be noted, however, that the amount of X chromosome sequence containing genes that escape X inactivation is still relatively small compared with X chromosome sequence subject to X inactivation (1.7 vs. 9.8 Mb). Furthermore, although the difference in distribution is significant, the L1 content between escape and subject groups clearly overlaps (Table 4).
Discussion
Our analysis has shown that the organization and distribution of L1 elements on the X chromosome differs significantly from a random genome model. A nearly 2-fold enrichment of L1 elements is detected on the X chromosome compared with other human autosomes. The most significant increase has been observed for sequences in Xq13, which contains the X-inactivation center. Genomic loci that escape X inactivation are significantly reduced in L1 content compared with loci that were subject to inactivation. These three nonrandom properties of L1 elements on the X chromosome are remarkably consistent with Gartler, Riggs, and Lyon's predictions of cis-acting elements or way stations that propagate the inactivation signal along the X chromosome. Although the data at this point are still largely indirect, and L1 enrichment may simply be a consequence of the heterochromatic nature of the inactive X, we believe the most prosaic explanation for the L1 distribution pattern includes a functional role of L1 elements in X inactivation. One possible scenario is that clusters of L1 elements serve indirectly through a protein complex, as X chromosome binding sites for XIST RNA. In situ studies in both humans and rodents have shown that XIST RNA coats or paints the condensed inactive X chromosome in a nonuniform fashion (10, 12, 14). It has been suggested that XIST RNA is a structural component of the inactive X involved in high-level chromatin packaging associated with Barr body heterochromatization. We propose that XIST RNA/protein complex interacts with clusters of L1 elements to mediate this repackaging. The overabundance of L1 elements within Xq13–q21 ensures appropriate initiation at the X-inactivation center, whereas other clusters, spread throughout the X chromosome, facilitate the nearly complete inactivation of the entire chromosome through a synergistic condensation. Regions deficient in L1 binding sites may be excluded from heterochromatization and therefore escape X inactivation. Alternatively, such sites may be initially subject to inactivation early in embryogenesis and subsequently escape because of lack of maintenance (40). Since our data suggest that reduced L1 content (<15%) is necessary but not sufficient to define regions that escape X inactivation, we conclude that additional factors must be important determinants in establishing chromosomal domains that escape X inactivation.
One of the surprising findings of our study has been that the bulk of the L1 enrichment has been due to younger subfamilies. The temporal specificity indicated by our subfamily analysis suggests that a major L1 invasion of the ancestral human X chromosome occurred near the time of the eutherian radiation. Our examination of chromosome 7 and other autosomes indicate that this effect was not genome-wide but was specific to the X chromosome. Our estimates predict that the initial burst of activity occurred after the divergence of the metatherian (marsupial) and eutherian (placental) lineages and has continued beyond the emergence of the primates. In contrast to X inactivation among eutherians, that among marsupials is much less stable and incomplete with no hypermethylation of CpG dinucleotides (34). The evolution of the more complex, multistep eutherian X inactivation pathway may coincide with the timing of the L1 invasion. We propose that L1 accumulation has contributed to a more-efficient locking mechanism of X inactivation observed among eutherians. The continued accumulation of L1 repeat elements after the emergence of the primates may simply be a result of creating a more competent inactive X among these particular eutherians. In this regard, it will be interesting to compare the pattern of L1 distribution on the murine X chromosome and its relationship to X inactivation.
In addition to our observations, several other properties of L1 elements make them a particularly attractive candidate as a signal for the spreading of X inactivation. First, L1 elements have been identified in all mammalian species, including the genomes of both metatherians and eutherians (22, 41). The fact that X inactivation status can be faithfully maintained in mouse–human cell hybrids suggests that a way-station signal may be conserved among different orders of eutherians (19). Other repeat elements such as endogenous retroviruses and Alu elements do not share such an ancient monophyletic origin and are therefore unlikely candidates. Second, tandem reiterations of L1 repeat are capable of forming heterochromatin-like structures in other species. Among whales and dolphins, for example, the core repeat of the α-heterochromatin satellite DNA consists of a 450-bp DNA fragment with 63% similarity to L1 retrotransposons. Similarly, in both the short-tailed field vole (Microtus agrestis) and the Syrian hamster (Mesocricetus auratus), β-heterochromatin structures are highly enriched for L1 elements (42, 43). These data suggest that clusters of L1 elements, perhaps vis-à-vis mechanisms involving repeat induced silencing, are competent to repackage and condense chromatin. Third, L1 elements are common throughout the genome. This pangenomic distribution would explain how inactivation spreads into autosomal material among X:autosome translocations (17). Depending on the site of translocation, the extent of spreading of inactivation may vary or even skip a region, particularly in areas of low L1 abundance (21). As more of both the human and mouse genomes become sequenced, it should be possible to test this hypothesis directly. Although final verification of the precise relationship between L1s and X inactivation will require experimental validation, it is an intriguing possibility that repetitive material, often dismissed as “junk,” may have evolved to play such a fundamental role in the genetic regulation of our genome.
Supplementary Material
Acknowledgments
We thank Dr. David Cutler and Carl Kashuk for computational assistance, and Dr. Huntington Willard and Dr. Rob Nicholls for helpful comments in the preparation of this manuscript. This work was supported, in part, by National Institutes of Health Grants GM58815-01 to E.E.E. and HG01847-01 to E.E.E. and A.C. The financial support of the W. M. Keck Foundation and a Howard Hughes Medical Institute grant to Case Western Reserve University School of Medicine are also gratefully acknowledged.
Abbreviations
- LINE
long interspersed element
- L1
LINE-1
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 6248.
References
- 1.Willard H F. In: The Metabolic and Molecular Basis of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, Childs B, Vogelstein B, editors. New York: McGraw-Hill; 2000. , in press. [Google Scholar]
- 2.Heard E, Clerc P, Avner P. Annu Rev Genet. 1997;31:571–610. doi: 10.1146/annurev.genet.31.1.571. [DOI] [PubMed] [Google Scholar]
- 3.Borsani G, Tonlorenzi R, Simmler M C, Dandolo L, Arnaud D, Capra V, Grompe M, Pizzuti A, Muzny D, Lawrence C, et al. Nature (London) 1991;351:325–329. doi: 10.1038/351325a0. [DOI] [PubMed] [Google Scholar]
- 4.Brown C J, Ballabio A, Rupert J L, Lafreniere R G, Grompe M, Tonlorenzi R, Willard H F. Nature (London) 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 5.Brockdorff N, Ashworth A, Kay G F, Cooper P, Smith S, McCabe V M, Norris D P, Penny G D, Patel D, Rastan S. Nature (London) 1991;351:329–331. doi: 10.1038/351329a0. [DOI] [PubMed] [Google Scholar]
- 6.Penny G D, Kay G F, Sheardown S A, Rastan S, Brockdorff N. Nature (London) 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 7.Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- 8.Lee J T, Strauss W M, Dausman J A, Jaenisch R. Cell. 1996;86:83–94. doi: 10.1016/s0092-8674(00)80079-3. [DOI] [PubMed] [Google Scholar]
- 9.Lee J T, Jaenisch R. Nature (London) 1997;386:275–279. doi: 10.1038/386275a0. [DOI] [PubMed] [Google Scholar]
- 10.Clemson C M, McNeil J A, Willard H F, Lawrence J B. J Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Panning B, Dausman J, Jaenisch R. Cell. 1997;90:907–916. doi: 10.1016/s0092-8674(00)80355-4. [DOI] [PubMed] [Google Scholar]
- 12.Duthie S M, Nesterova T B, Formstone E J, Keohane A M, Turner B M, Zakian S M, Brockdorff N. Hum Mol Genet. 1999;8:195–204. doi: 10.1093/hmg/8.2.195. [DOI] [PubMed] [Google Scholar]
- 13.Costanzi C, Pehrson J R. Nature (London) 1998;393:599–601. doi: 10.1038/31275. [DOI] [PubMed] [Google Scholar]
- 14.Brown C J, Hendrich B D, Rupert J L, Lafreniere R G, Xing Y, Lawrence J, Willard H F. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 15.Russell L B, Montgomery C S. Genetics. 1970;64:281–312. doi: 10.1093/genetics/64.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rastan S. J Embryol Exp Morphol. 1983;78:1–22. [PubMed] [Google Scholar]
- 17.White W M, Willard H F, Van Dyke D L, Wolff D J. Am J Hum Genet. 1998;63:20–28. doi: 10.1086/301922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carrel L, Cottle A A, Goglin K C, Willard H F. Proc Natl Acad Sci USA. 1999;96:14440–14444. doi: 10.1073/pnas.96.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gartler S M, Riggs A D. Annu Rev Genet. 1983;17:155–190. doi: 10.1146/annurev.ge.17.120183.001103. [DOI] [PubMed] [Google Scholar]
- 20.Riggs A D. Aust J Zool. 1990;37:419–441. [Google Scholar]
- 21.Lyon M F. Cytogenet Cell Genet. 1998;80:133–137. doi: 10.1159/000014969. [DOI] [PubMed] [Google Scholar]
- 22.Burton F H, Loeb D D, Voliva C F, Martin S L, Edgell M H, Hutchison C A., 3rd J Mol Biol. 1986;187:291–304. doi: 10.1016/0022-2836(86)90235-4. [DOI] [PubMed] [Google Scholar]
- 23.Dombroski B A, Mathias S L, Nanthakumar E, Scott A F, Kazazian H H., Jr Science. 1991;254:1805–1808. doi: 10.1126/science.1662412. [DOI] [PubMed] [Google Scholar]
- 24.Kazazian H H, Jr, Moran J V. Nat Genet. 1998;19:19–24. doi: 10.1038/ng0598-19. [DOI] [PubMed] [Google Scholar]
- 25.Smit A F. Curr Opin Genet Dev. 1996;6:743–748. doi: 10.1016/s0959-437x(96)80030-x. [DOI] [PubMed] [Google Scholar]
- 26.Korenberg J R, Rykowski M C. Cell. 1988;53:391–400. doi: 10.1016/0092-8674(88)90159-6. [DOI] [PubMed] [Google Scholar]
- 27.Boyle A L, Ballard S G, Ward D C. Proc Natl Acad Sci USA. 1990;87:7757–7761. doi: 10.1073/pnas.87.19.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jang W, Chen H C, Sicotte H, Schuler G D. Trends Genet. 1999;15:284–286. doi: 10.1016/s0168-9525(99)01762-x. [DOI] [PubMed] [Google Scholar]
- 29.Smit A F, Toth G, Riggs A D, Jurka J. J Mol Biol. 1995;246:401–417. doi: 10.1006/jmbi.1994.0095. [DOI] [PubMed] [Google Scholar]
- 30.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 31.Florea L, Hartzell G, Zhang Z, Rubin G M, Miller W. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smit A F. Curr Opin Genet Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- 33.Scott A F, Schmeckpeper B J, Abdelrazik M, Comey C T, O'Hara B, Rossiter J P, Cooley T, Heath P, Smith K D, Margolet L. Genomics. 1987;1:113–125. doi: 10.1016/0888-7543(87)90003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wakefield M J, Keohane A M, Turner B M, Graves J A. Proc Natl Acad Sci USA. 1997;94:9665–9668. doi: 10.1073/pnas.94.18.9665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soriano P, Meunier-Rotival M, Bernardi G. Proc Natl Acad Sci USA. 1983;80:1816–1820. doi: 10.1073/pnas.80.7.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernardi G. Mol Biol Evol. 1993;10:186–204. doi: 10.1093/oxfordjournals.molbev.a039994. [DOI] [PubMed] [Google Scholar]
- 37.Saccone S, De Sario A, Wiegant J, Raap A K, Della Valle G, Bernardi G. Proc Natl Acad Sci USA. 1993;90:11929–11933. doi: 10.1073/pnas.90.24.11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lahn B T, Page D C. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- 39.Graves J A, Disteche C M, Toder R. Cytogenet Cell Genet. 1998;80:94–103. doi: 10.1159/000014963. [DOI] [PubMed] [Google Scholar]
- 40.Lingenfelter P A, Adler D A, Poslinski D, Thomas S, Elliott R W, Chapman V M, Disteche C M. Nat Genet. 1998;18:212–213. doi: 10.1038/ng0398-212. [DOI] [PubMed] [Google Scholar]
- 41.Dorner M, Pääbo S. Mol Biol Evol. 1995;12:944–948. doi: 10.1093/oxfordjournals.molbev.a040272. [DOI] [PubMed] [Google Scholar]
- 42.Kapitonov V V, Holmquist G P, Jurka J. Mol Biol Evol. 1998;15:611–612. doi: 10.1093/oxfordjournals.molbev.a025963. [DOI] [PubMed] [Google Scholar]
- 43.Neitzel H, Kalscheuer V, Henschel S, Digweed M, Sperling K. Cytogenet Cell Genet. 1998;80:165–172. doi: 10.1159/000014974. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.