Among a number of elegant biomimetic approaches that have been developed for the modification of the peptide backbone1, only a few have been applied to protein structures.2 Here, we report high resolution structural consequences of amide backbone replacements in the context of a folded peptide architecture. We show that a triazole ε2-amino acid (Figure 1a)3 can be used as a dipeptide surrogate in α-helical coiled coils and report on the effects of the substitution on the thermodynamic stability, helical secondary structure, and the four helix bundle quaternary organization.
Figure 1.
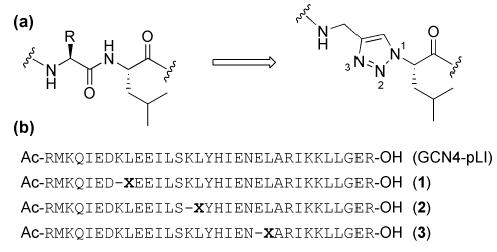
(a) A native dipeptide and the L-leucine derived triazole-ε2- amino acid incorporated as a replacement; (b) Sequences for GCN4-pLI and modified peptides 1-3; X denotes incorporation of the ε2-residue.
The reactivity and functional group tolerance of Cu(I) catalyzed 1,3-dipolar cycloaddition between azides and alkynes to give 1,2,3-triazoles4 have led to the increasing use of this reaction in bioconjugate chemistry.5 We reported recently the use of triazole ε2-amino acids as a dipeptide replacement in the design of selfassembling peptide nanotubes.6 We hypothesized that the structural and functional features of the triazole ε2-amino acids could also be potentially useful in the context peptide and protein secondary structures. The backbone of the ε-amino acid is one atom longer as compared to that of a native dipeptide (Figure 1a), leading to a calculated increase in Cα-Cαspacing of 1.1 Å.7 In addition to the amide NH and carbonyl groups flanking the residue, the triazole ring possesses two nitrogen atoms (N2 and N3) that might act as hydrogen bond acceptors. Furthermore, the triazole ring has a large dipole that could align with that of the other amides in a given peptide secondary structure.8
We selected the pLI mutant of the α-helical coiled coil GCN4 in order to test the utility of the ε2-amino acid substitution in the context of a peptide with well defined secondary and quaternary structure in solution and the solid state.9 In coiled coils, interhelical interactions between buried hydrophobic residues as well as exposed hydrophilic side chains lead to robust peptide self-assembly into α-helical bundles. We replaced dipeptides K8L9, K15L16, and E22L23 in the pLI-GCN4 sequence with an Lleucine derived triazole ε-amino acid to give sequences 1, 2, and 3, respectively (Figure 1b). In each case, the isobutyl side chain of the ε-residue was predicted to replace a leucine residue in the hydrophobic core of the native 4-helix bundle. The amino acid employed was synthesized in two steps from L-leucine and used in standard solid phase peptide synthesis conditions.
Circular dichroism (CD) spectra of peptides 1-3 (50 μM in 10 mM MOPS, pH 7.0) showed minima at 208 nm and 222 nm, characteristic of α-helical secondary structure (Figure S2). Thermal denaturation, monitoring the molar ellipticity at 222 nm, indicated broad two-state transitions for peptides 2 and 3, while peptide 1, similar to the parent pLI-GCN4 sequence, was considerably more stable and did not fully denature up to 96°C.9 Gel permeation chromatography indicated that peptides 1 and 3 adopt tetrameric oligomerization states in solution, while peptide 2 appears to exist primarily as a dimeric assembly (Table 1). These studies indicate that the modified peptides retain much of the native α-helical character, but that the position of the ε2- amino acid substitution differentially influences thermodynamic stability.
Table 1.
Biophysical Data and PDB IDs for Peptides 1-3a
| peptide | [θ]222 (deg cm2 dmol−1)b | Tm(°C) | Naggc | PDB ID |
|---|---|---|---|---|
| GCN4-pLId | −30,600 | >96 | 4 | 1GCL |
| 1 | peptide 28,000 | >96 | 4 | 1U9G |
| 2 | peptide 13,100 | 36 | 2 | 1U9F |
| 3 | peptide 21,000 | 61 | 4 | 1U9H |
see Supporting Information for experimental details and full spectra;
at 4°C;
apparent aggregation state in solution as determined by gel permeation chromatography;
values and structure from reference.9
We employed X-ray crystallography to ascertain the structural consequences of the ε2-amino acid substitutions in the context of the helical coiled coil architecture. Crystal structures were obtained for peptides 1-3 at 2.2 Å resolution. Although each peptide crystallized in a different unit cell and space group, all three exhibited the parallel tetrameric coiled coil structure of the parent pLI-GCN4 sequence with a crystallographic 2-fold symmetry axis along the center of each bundle (Figure 2, S3). In peptides 2 and 3, a complete well resolved structure similar to the parent pLI-GCN4 was obtained. However, in the case of 1, the electron density for portion of the peptide preceding the triazole ε-amino substitution (residues 1-8) was not observed, presumably due to chain disorder in the crystal (Figure S3).
Figure 2.
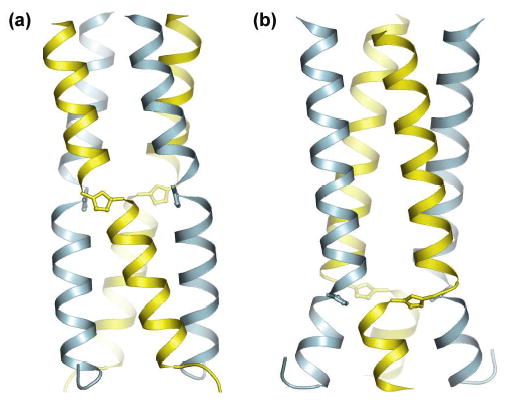
Schematic representation of the crystal structure of peptides 2 (a) and 3 (b) with atomic positions shown for the triazole residues. Each four-helix bundle superposes a crystallographic 2-fold axis and unique chains in each structure are indicated by different colors.10
The crystal structure of 2 is similar to that of the parent pLI with the hydrophobic side chain of the ε-residue projecting toward the core of the bundle (Figure S4). Notably, the ε-residue in each chain fully participates in α-helical backbone hydrogen bonding (Figure 3a). The N2 of the triazole accepts a hydrogen bond from the amide NH of Ile18 and the triazole C5-H appears to participate in a CH-O hydrogen bond with the carbonyl oxygen of Ile12.12 This observation is supported by the geometry and 2.2 Å distance between the atoms and is in agreement with the large dipole of the triazole ring.8 Hence, the normal i, i+4 hydrogen bonding that would have been provided to residues 12 and 18 in the parent pLI structure are replaced by the triazole ring. In addition, the amide NH of the ε-residue is hydrogen bonded to the carbonyl oxygen of Ile12, while the Nδof the imidazole ring of His17 is hydrogen bonded to the carbonyl of Ser14. These structural features give rise to an increase in the α-helical pitch of about 1.8 Å in the region of the ε-residue. This increase in the local pitch creates a shallow pocket adjacent to the triazole that is occupied by a water molecule providing bridging hydrogen bonds that may contribute to the stability of the helical structure (Figure S5).
Figure 3.
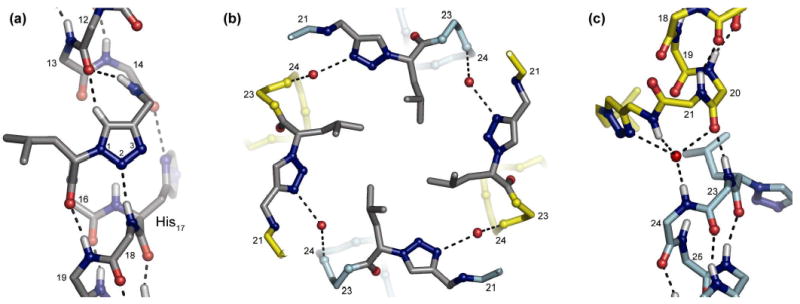
(a) Detail from the crystal structure of 2 showing participation of the triazole residue in main chain hydrogen bonding; residues and nitrogen atoms of the triazole ring are numbered. (b) A top down view of the crystal structure of 3 showing the hydrophobic plate formed by the triazole ε2 residues and inter-chain hydrogen bonds bridged by water. (c) Detail of two chains from the crystal structure of 3 showing the inter-helical crossing. Dashed lines indicate hydrogen bonds. In (a) and (c), H atoms are modeled based on N, C, and O coordinates.10
The ε2-amino acid substitution in peptide 3 creates an unusual right-handed interhelical crossover structure such that the helix formed by residues 1-22 of one chain is completed by residues 23-32 of another (Figure 3c, S6). The residues 20-24, which include the ε2-amino acid at position 22, act together as a template for the strand crossing, providing an overall helical chain register similar to that of the parent pLI fold. Inter-chain interactions consisting of a backbone hydrogen bond between the carbonyl oxygen of Glu20 and the amide NH of Ala23 and a water bridged hydrogen bond between N3 of the triazole ring and the amide NH of Arg24 further stabilize this intertwined fold (Figure 3c). The isobutyl side chains of the ε2-residue also form a core hydrophobic packing in the helix bundle similar to the parent pLI (Figure 3b).
In summary, we have shown that a non-natural 1,2,3-triazole ε2-amino acid can replace a dipeptide in an α-helical secondary structure. In light of these observations, we suggest that Cu(I) catalyzed azide-alkyne coupling could be useful in the non-native chemical synthesis of peptides and proteins.12
Supplementary Material
Acknowledgments
We thank National Institute of General Medical Sciences (GM57690) for financial support and NSF for a fellowship to W.S.H.
Footnotes
Supporting Information Available: Figures S1-S6, experimental details, crystallographic data, and CD spectra. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.For reviews on the subject, see: Spatola AF. Chemistry and Biochemistry of Amino Acids, Peptides, and Proteins. Dekker; New York: 1983. pp. 267–357.Ahn JM, Boyle NA, MacDonald MT, Janda KD. Mini-Rev Med Chem. 2002;2:463–473. doi: 10.2174/1389557023405828.
- 2.(a) Chapman E, Thorson JS, Schultz PG. J Am Chem Soc. 1997;119:7151–7152. [Google Scholar]; (b) Beligere GS, Dawson PE. J Am Chem Soc. 2000;122:12079–12082. [Google Scholar]; (c) Wales TE, Fitzgerald MC. J Am Chem Soc. 2001;123:7709–7710. doi: 10.1021/ja010775n. [DOI] [PubMed] [Google Scholar]; (d) Deechongkit S, Nguyen H, Powers ET, Dawson PE, Gruebele M, Kelly JW. Nature. 2004;430:101–105. doi: 10.1038/nature02611. [DOI] [PubMed] [Google Scholar]
- 3.For a description of the ε2 nomenclature, see Figure S1.
- 4.(a) Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. Angew Chem, Int Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]; (b) Tornoe CW, Christensen C, Meldal M. J Org Chem. 2002;67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 5.(a) Wang Q, Chan TR, Hilgraf R, Fokin VV, Sharpless KB, Finn MG. J Am Chem Soc. 2003;125:3192–3193. doi: 10.1021/ja021381e. [DOI] [PubMed] [Google Scholar]; (b) Link AJ, Tirrell DA. J Am Chem Soc. 2003;125:11164–11165. doi: 10.1021/ja036765z. [DOI] [PubMed] [Google Scholar]; (c) Deiters A, Cropp TA, Mukherji M, Chin JW, Anderson JC, Schultz PG. J Am Chem Soc. 2003;125:11782–11783. doi: 10.1021/ja0370037. [DOI] [PubMed] [Google Scholar]; (d) Fazio F, Bryan MC, Blixt O, Paulson JC, Wong CH. J Am Chem Soc. 2002;124:14397–14402. doi: 10.1021/ja020887u. [DOI] [PubMed] [Google Scholar]; (e) Speers AE, Adam GC, Cravatt BF. J Am Chem Soc. 2003;125:4686–4687. doi: 10.1021/ja034490h. [DOI] [PubMed] [Google Scholar]
- 6.Horne WS, Stout CD, Ghadiri MR. J Am Chem Soc. 2003;125:9372–9376. doi: 10.1021/ja034358h. [DOI] [PubMed] [Google Scholar]
- 7.Due to the planar aromatic ring structure of the triazole, this distance is fixed regardless of the backbone torsion angles.
- 8.Palmer MH, Findlay RH, Gaskell AJ. J Chem Soc, Perkin Trans 2. 1974;4:420–428. [Google Scholar]
- 9.Harbury PB, Zhang T, Kim PS, Alber T. Science. 1993;262:1401– 1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 10.Figure prepared using PyMOL DeLano WL. The PyMOL Molecular Graphics System. http://www.pymol.org.
- 11.For a review on CH-O hydrogen bonds, see: Derewenda ZS, Lee L, Derewenda U. J Mol Biol. 1995;252:248–262. doi: 10.1006/jmbi.1995.0492.
- 12.For a review of non-native peptide ligation strategies, see: Tam JP, Xu J, Eom KD. Biopolymers. 2001;60:194–205. doi: 10.1002/1097-0282(2001)60:3<194::AID-BIP10031>3.0.CO;2-8.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


