Summary
Differentiation of embryonic stem (ES) cells from a pluripotent to a committed state involves global changes in genome expression patterns. Gene activity is critically determined by chromatin structure and interactions of chromatin binding proteins. Here, we show that major architectural chromatin proteins are hyperdynamic and bind loosely to chromatin in ES cells. Upon differentiation, the hyperdynamic proteins become immobilized on chromatin. Hyperdynamic binding is a property of pluripotent cells, but not of undifferentiated cells that are already lineage committed. ES cells lacking the nucleosome assembly factor HirA exhibit elevated levels of unbound histones, and formation of embryoid bodies is accelerated. In contrast, ES cells, in which the dynamic exchange of H1 is restricted, display differentiation arrest. We suggest that hyperdynamic binding of structural chromatin proteins is a functionally important hallmark of pluripotent ES cells that contributes to the maintenance of plasticity in undifferentiated ES cells and to establishing higher-order chromatin structure.
Introduction
Embryonic stem (ES) cells possess an unlimited potential to self-renew and the capacity to differentiate into multiple lineages. During differentiation, ES cells lose their pluripotency and undergo dramatic morphological and molecular changes. One of the key events during this process is the selective silencing and activation of specific subsets of genes (Eiges and Benvenisty, 2002;Keller, 1995; Loebel et al., 2003; Weiss and Orkin, 1996). In addition, the genome of differentiating ES cells undergoes global chromatin reorganizations via chromatin remodeling events and epigenetic modulations (Hajkova et al., 2002; Jaenisch and Bird, 2003; Rasmussen, 2003; Surani, 2001). These changes are brought about by the interaction of a multitude of proteins with chromatin and changes in chromatin structure.
Chromatin is generally composed of transcriptionally permissive, less condensed euchromatin and the highly condensed and often repressed heterochromatin (Patterton and Wolffe, 1996). The core subunit of chromatin, the nucleosome, consists of a histone octamer of four core histones, H2A, H2B, H3, and H4, wrapped inside 146 bp of double-stranded DNA. Adjacent nucleosomes are connected via linker DNA bound by the linker histone H1. Higher-order organization of this basic subunit defines the nature of the chromatin structure and its accessibility. In addition to the four core histones, several additional histone variants are present in mammalian cells (Sarma and Reinberg, 2005). Although some of their functions are yet to be determined, some appear to play active cellular roles, such as H2Ax in DNA damage responses (Rogakou et al., 1999) or H3.3, which accumulates in actively transcribed regions of the genome (Ahmad and Henikoff, 2002).
The interaction of proteins with chromatin is highly dynamic in vivo. Fluorescent recovery after photobleaching (FRAP) approaches have demonstrated that most chromatin proteins are highly mobile within the mammalian cell nucleus and transiently interact with chromatin in vivo (Phair et al., 2004). An exception to this rule is the core histones that bind relatively stably to chromatin (Kimura and Cook, 2001; Phair et al., 2004). Other structural chromatin proteins, such as linker histones and the heterochromatin protein HP1, have residence times on chromatin on the order of a few seconds to minutes (Cheutin et al., 2003; Festenstein et al., 2003; Lever et al., 2000; Misteli et al., 2000). These observations suggest that establishment, maintenance, and functional modulation of chromatin domains involve the dynamic interaction of proteins with chromatin.
While it seems likely that chromatin proteins play key roles in bringing about the changes in genome expression patterns associated with ES cell differentiation and possibly also have a role in maintenance of pluripotency, little is known about how proteins interact with chromatin in pluripotent ES cells and whether their interactions are modulated as cells begin their differentiation process. In order to compare the interaction of proteins with chromatin in pluripotent cells to their behavior in differentiated cells, we analyze chromatin structure and chromatin protein dynamics in pluripotent mouse ES cells during early neuronal differentiation.
Results
Morphological Changes in Chromatin Structure during ES Cell Differentiation
To compare nuclear architecture in pluripotent ES cells to that in lineage-committed cells, we investigated architectural features of the cell nucleus of murine R1 ES cells during their differentiation from pluripotent cells into neural progenitor cells (NPCs) (Lee et al., 2000). We triggered the differentiation of R1 cells by depletion of the leukemia inhibitory factor (LIF) from the medium, leading to formation of embryoid bodies (EBs) after 4 days and eventually NPCs within 7 days (see Experimental Procedures for details). We limited our analysis to undifferentiated ES cells, an early time point of 24 hr after withdrawal of LIF, and a later time point of 7 days to avoid the heterogeneity associated with EBs. The three selected time points consisted of largely homogenous populations suitable for imaging. As expected, more than 90% of pluripotent ES cells expressed the stem cells marker Oct4. After 24 hr, Oct4 expression was still highly abundant, but it was limited to roughly 80% of the cells, and after 7 days Oct4 was no longer detectable (Figures S1A and S1D; see the Supplemental Data available with this article online). The neural progenitor marker nestin was detected in ∼2.5% of undifferentiated ES cells, in ∼20% of cells after 24 hr, and in over 80% of 7-day-old NPCs (Figures S1B and S1D). By day 7, almost all of the progenitors are already committed to the neural lineage, as evidenced by immunostaining with TUJ1 antibody against the neuronal marker β-tubulin III, which was undetectable in both undifferentiated ES cells and cells after 24 hr (Figures S1C and S1D).
To compare the global organization of the genome in pluripotent ES cells to early stages of differentiation, heterochromatin was visualized by double immunostaining of heterochromatin protein 1 (HP1α) and Oct4 (Figure 1A, top) or nestin (Figure 1A, bottom). In addition, we employed double immunostaining of HP1α with histone H3 trimethylated on lysine 9 (H3-triMeK9), a modification generally associated with heterochromatin, in undifferentiated ES cells, 24 hr after LIF removal or in 7-day-old NPCs (Figure 1B). HP1 staining (Figures 1A and Figures 1B, green) appeared as large, poorly defined regions in undifferentiated ES cells, but it was confined to small, discrete foci with well-defined borders in NPCs. The H3-triMeK9 patterns (Figure 1B, red) were similar to those observed by DAPI (Figures 1A and Figures 1B, blue) or HP1 staining, with diffusely labeled regions in undifferentiated ES cells and the appearance of discrete foci in NPCs. These morphological changes occurred during the later stages of differentiation, since, 24 hr after removal of LIF, heterochromatin appeared similar to that in ES cells (data not shown). In order to quantify the differentiation-dependent chromatin rearrangement, we counted the number of H3-triMeK9-labeled foci in undifferentiated ES cells and NPCs, and we measured both the total H3-triMeK9-stained area and the labeling intensity. The number of heterochromatin foci per nucleus increased (Figure 1D, p < 0.0001), while the average focus size decreased (Figure 1E, p < 0.05) as cells differentiated. Both H3-triMeK9 intensity (Figure 1F) and the total heterochromatin area per nucleus (Figure 1G) increased in ES cells and NPCs (p < 0.0001, p < 0.05, respectively). This increase occurred in spite of the unchanged nuclear area (2D) in ES cells and NPCs (Figure S2B, p > 0.5) and an overall decrease in nuclear volume (Figure S2B), ruling out the possibility that an increase in nuclear volume was the cause for the observed increase in heterochromatin area. Finally, when heterochromatin was visualized directly by FISH, rather than by examination of heterochromatin binding proteins, by using a specific DNA probe against the major satellite repeat, undifferentiated ES cells displayed a more diffuse heterochromatin structure (Figure 1C, left), while regions of major satellite repeats appeared as well defined foci in NPCs (Figure 1C, right). Taken together, these observations suggest that chromatin is reorganized globally as ES cells differentiate and lose their pluripotency.
Figure 1.
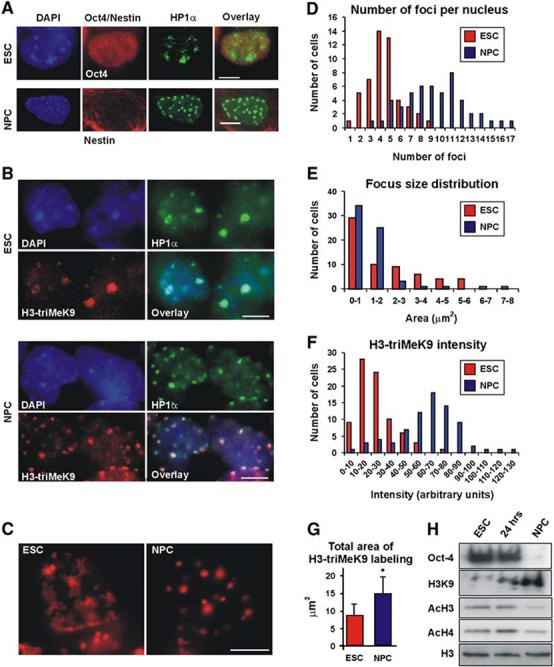
Chromatin Rearrangement in ES Cells and NPCs (A) ES cells (top) or neural progenitor cells (NPCs) (bottom) were coimmunolabeled with HP1α (green) together with the stem cell marker Oct4 (red, top) or with the NPC marker nestin (red, bottom). Left, DAPI staining; right, overlay. (B) Double immunolabeling of ES cells (top) or NPCs (bottom) with anti-HP1α antibody (green, right) and H3-triMeK9 (red, left). DAPI staining (blue), left; overlay, right. (C) DNA-FISH (red) for the mouse major satellite repeat. Left, an undifferentiated ES cell; right, NPCs. A similar length scrambled 5′-biotinylated probe yielded no signal (data not shown). (D) Distribution of heterochromatin foci number per nucleus in ES cells (red bars) or NPCs (blue bars). The average number of foci per nucleus increased from 4.4 ± 1.7 in ES cells to 9.4 ± 3.1 in NPCs (p < 0.0001). (E) Average focus size in ES cells (red) and NPCs (blue) (p < 0.05). (F) Distribution of H3-triMeK9 intensity in ES cells (red bars) or NPCs (blue bars). At least 50 nuclei from each group were analyzed. After background subtraction, the average intensity values increased from 23.8 ± 2.0 in ES cells to 62.5 ± 11.4 in NPCs (arbitrary units) (p < 0.0001). (G) Total area of H3-triMeK9 labeling per nucleus in ES cells (red) and NPCs (blue) (p < 0.05). In (D)-(G), values represent quantitation of at least 50 nuclei from each group ± SD. (H) Western blot for Oct4, trimethylated lysine 9 of histone H3 (H3K9), acetylated H3 (AcH3), and acetylated H4 (AcH4) in nuclei from undifferentiated ES cells (left), 24 hr after LIF withdrawal (middle), and NPCs (right). Anti-H3 (bottom) was used as a loading control. All scale bars are 5μm.
The observed differentiation-induced increase in H3-triMeK9 is consistent with previous reports (Keohane et al., 1996; Lee et al., 2004), in which progress in differentiation was accompanied by both an increase in H3-triMeK9 as well as a decrease in the acetylation of histones H3 and H4, a modification generally associated with euchromatin (Muller and Leutz, 2001). To study histone modifications during differentiation of R1 ES cells, we used both Western blotting and immunofluorescence microscopy with antibodies specific for H3-triMeK9, pan-acetylated histone H3, or pan-acetylated histone H4. By using Western blot analysis, an increase was observed in H3-triMeK9, but not in HP1 expression during differentiation (Figure 1H and Figure S3C), in agreement with our immunofluorescence results (Figure 1B). In contrast to the H3 methylation status, the acetylation level of both histone H3 (Figure 1H and Figure S3A) and histone H4 (Figure 1H and Figure S3B) was reduced in NPCs (Figure 1H, right and Figures S3A and S3B, bottom) compared to ES cells (Figure 1H, left and Figures S3A and S3B, top). However, 24 hr after the withdrawal of LIF (Figure 1H, middle and Figures S3A and S3B, middle), a slight increase was observed in acetylation of H4, and no significant change was observed in acetylated H3.
Dynamics of Architectural Chromatin Proteins in ES Cells
Heterochromatin domains are maintained by dynamic structural proteins including heterochromatin protein HP1 and linker histones (Cheutin et al., 2003; Festenstein et al., 2003; Lever et al., 2000; Misteli et al., 2000; Schmiedeberg et al., 2004). To ask whether proteinchromatin interactions differ in pluripotent ES cells compared to differentiated cells, we analyzed the in vivo binding properties of architectural chromatin proteins in living ES cells by using fluorescence recovery after photobleaching (FRAP). Using an established FRAP protocol (Cheutin et al., 2003), we measured the chromatin binding dynamics of HP1α-GFP expressed at low levels in undifferentiated ES cells, ES cells 24 hr after LIF withdrawal, or in NPCs (Figure 2A). No obvious difference was observed when HP1α-GFP in euchromatic regions was bleached where it is bound with short residence time (Cheutin et al., 2003) (data not shown). In contrast, HP1α-GFP exchange in heterochromatin foci was significantly faster in undifferentiated ES cells than in NPCs or in cells 24 hr after LIF withdrawal; the recovery signal reached 50% of the initial intensity within 3.5 s after bleaching in ES cells, but 7.3 s in NPCs (p < 0.01) and 8.2 s in cells 24 hr after LIF withdrawal (p < 0.01) (Figure 2B, p > 0.25 for NPCs versus 24 hr after LIF withdrawal). In addition, recovery was complete within ∼20 s in ES cells, but it took more than 60 s in NPCs. Similar results were obtained for HP1β and HP1γ (data not shown). The differences in recovery times were due to faster recovery in the earliest time points after photobleaching (Figure 2). The rate of recovery in ES cells at later time points was identical to that observed in NPCs. As previously demonstrated, the faster recovery of HP1 in ES cells in the early stages after bleaching is indicative of an increased fraction of loosely bound or soluble HP1 in ES cells compared to NPCs (Phair et al., 2004). The faster recovery kinetics of HP1-GFP cannot be accounted for by changes in expression levels during differentiation, as shown by Western blot (Figure S4), and they are not due to nonphysiological levels of the expressed proteins since HP1-GFP was only moderately overexpressed (Figure S4). In addition, HP1-GFP dynamics have previously been shown to be largely insensitive to expression levels (Cheutin et al., 2003).
Figure 2.
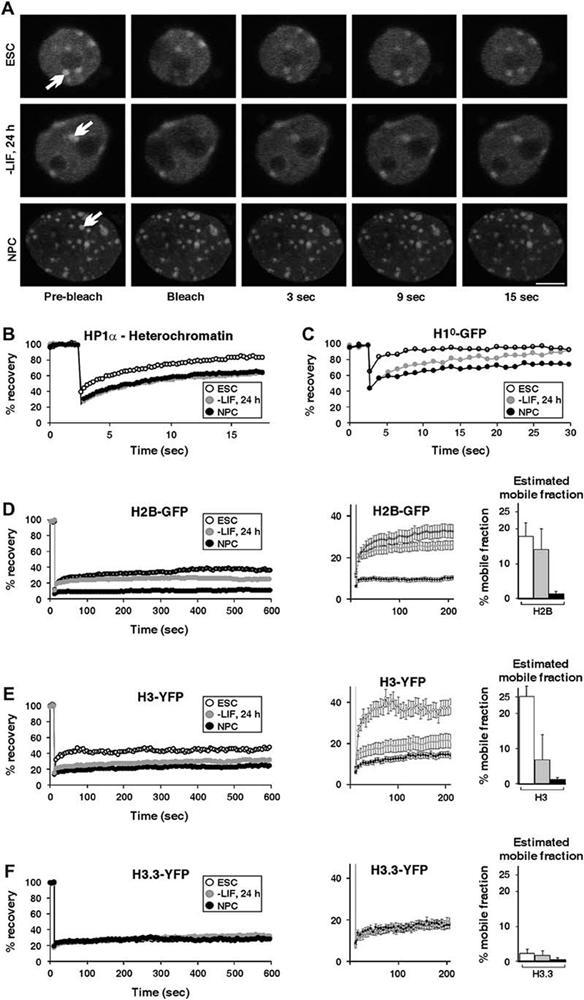
Hyperdynamic Binding of Architectural Chromatin Proteins in ES Cells (A) Fluorescence recovery after photobleaching (FRAP) to study the dynamics of chromatin-associated proteins. A heterochromatic (white arrow) region in cells expressing HP1α-GFP was bleached, and the recovery was measured. In each experiment, undifferentiated ES cells (top), cells 24 hr after LIF withdrawal (middle), and NPCs (bottom) were analyzed. The scale bar is 5μm. (B) FRAP curves of heterochromatin foci of transiently expressed HP1α-GFP. Heterochromatin recovery was significantly faster in ES cells compared to either NPCs or cells 24 hr after LIF withdrawal. (C) FRAP curves for transiently expressed H1°-GFP. (D) FRAP curves for transiently expressed H2B-GFP. Left: recovery kinetics over a period of 10 min in undifferentiated ES cells (empty circles), cells 24 hr after LIF withdrawal (gray circles), and NPCs (black circles). Middle: the first 200 s are enlarged and are shown with error bars. Right: estimated mobile fractions of undifferentiated ES cells (white), cells 24 hr after LIF withdrawal (gray), and NPCs (black). (E) FRAP curves for transiently expressed H3-YFP as in (D). (F) FRAP curves for transiently expressed H3.3-YFP as in (D). In (C)-(F), half of the nucleus was bleached, including heterochromatin and euchromatin regions. Values represent averages from at least 20 cells from 3 experiments ± SD.
To ask whether increased binding dynamics were limited to HP1 or were a general property of structural chromatin proteins, we determined the exchange dynamics of the linker histone H1° and the core histones H2B, H3, and H3.3 by using well-characterized functional YFP or GFP fusion proteins (Ahmad and Henikoff, 2002; Phair et al., 2004). In somatic cells, the recovery of H1 has previously been shown to take place over several minutes (Lever et al., 2000; Misteli et al., 2000), while the recovery time of core histones has been shown to be on the order of hours (Kimura and Cook, 2001; Phair et al., 2004). The overall recovery kinetics of transiently expressed H1°-GFP (Figure 2C), H2B-GFP (Figure 2D), and H3-YFP (Figure 2E) in undifferentiated ES cells were significantly faster than in NPCs when half of the nucleus, including heterochromatin and euchromatin regions, was bleached (p < 0.05; calculated at t = 100 s for H3 and H2B and at 15 s for H1°). In all cases, the faster overall exchange in ES cells was attributable to increased recovery during the first few seconds after bleaching; this was followed by a slower phase (after w30 s for H2B and H3, and after 5 s for H1°), which was not significantly different between ES cells and NPCs. As previously shown, the rapid initial phase is diagnostic of an increased loosely bound or soluble pool of rapidly diffusing molecules (Misteli et al., 2000; Phair et al., 2004). Although we cannot distinguish whether these molecules are soluble or loosely bound, we favor the latter since fully soluble molecules are known to recover much faster than observed here (Phair et al., 2004). Quantitative analysis of the FRAP data indicated that the size of the loosely bound pool of H2B and H3 in ES cells was 18% and 25%, respectively, whereas it was less than 3% in NPCs for both proteins (Figures 2D and Figures 2E, right). This latter value is similar to that found in somatic cells (Kimura and Cook, 2001). Interestingly, the histone variant H3.3-YFP displayed slow-exchange dynamics at all stages of differentiation (Figure 2F). Since H3.3 is considered to be a marker of transcriptionally active, open chromatin regions (Ahmad and Henikoff, 2002), this behavior is consistent with the notion that ES cells contain increased levels of transcriptionally open chromatin. Moreover, it is consistent with the formation of heterochromatin domains during differentiation (Ahmad and Henikoff, 2002). The reduced dynamicsof H1, H2B, and H3 in NPCs were not due to changes in the expression levels of these proteins during ES cell differentiation, as shown by Western blotting (Figure S3C), and they were not due to nonphysiological levels of the fusion protein since overexpressed core and linker histones typically constitute less than 5% of the total cellular histone levels (Kimura and Cook, 2001; Misteli et al., 2000)(Figure S4).
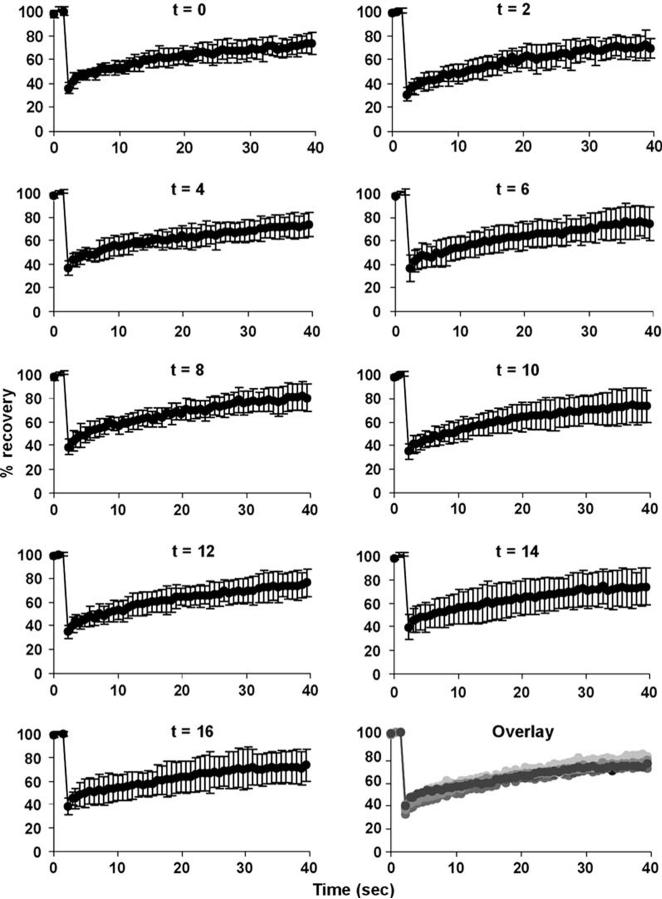
Since ES cells cycle rapidly between S phase and M phase with short G1 and G2 phases, we sought to rule out that the observed differences in binding dynamics were due to the changed cell cycle dynamics of ES cells compared to NPCs. To this end, we arrested R1 ES cells stably expressing H1°-GFP at G1/S with aphidicolin and performed FRAP analysis every 2 hr for a total of 16 hr after release from the G1/S block (Figure 3). No difference in FRAP recovery was observed at any point in the cell cycle (Figure 3, bottom right, overlay), consistent with previous observations (Cheutin et al., 2003; Kimura and Cook, 2001; Phair et al., 2004). These results rule out that changes in cell cycle dynamics were the cause for the reduced mobility of chromatin-associated proteins during ES cell differentiation.
Figure 3.
H1°-GFP Kinetics during Cell Cycle R1 cells stably expressing H1°-GFP were treated overnight with aphidicolin (5μg/ml) to arrest cells at G1/S. The following day, aphidicolin was washed out, and cells were subjected to FRAP analysis every 2 hr for two complete cell cycles. At time 0, the population of cells is enriched with cells at G1. After 2 hr, most cells are in S phase. No significant change in kinetics was observed during the different cell cycle stages. Values represent averages from at least 20 cells from 2 experiments ± SD.
Reduced Binding of Endogenous Chromatin Proteins in Undifferentiated ES Cells
To independently verify and extend the FRAP experiments to endogenous proteins, we tested the association of structural proteins with chromatin by biochemical extraction. Upon salt extraction of isolated nuclei, fractions of both endogenous H1 and HP1 were released at lower salt concentrations in ES cells than in NPCs (Figure 4A). While less than 50% of H1 was extracted at 500 mM NaCl from NPCs, more than 80% was extractable in ES cells (Figure 4A, left). Similarly, whereas less than 30% of HP1 was extractable with 250 mM NaCl in NPCs, more than 90% was released from ES cells (Figure 4A, right). All endogenous core histones were similarly more extractable (Figure 4B). Between 20%-30% of histones were removed by 200 mM NaCl from chromatin isolated from ES cells (Figure 4B, compare lanes 1 and 2), whereas no detectable core histones were released from NPC chromatin (Figure 4B, compare lanes 3 and 4). In ES cells and NPCs, extraction with up to 500 mM NaCl resulted in no further loss of core histones (data not shown), suggesting that the increased extractability in ES cells represents a distinct fraction of core histones rather than reduced binding of the entire pool of core histones. This later observation is in line with the observation of a rapid and slow phase in the FRAP recovery curves. Finally, in nuclei prepared from undifferentiated ES cells (Figure 4C, lanes 1-3), core histones were readily released upon micrococcal nuclease (MNase) digestion (Figure 4C, arrow). No core histones were detected in the soluble supernatant of whole nuclei (lane 1). Release of almost all histones was achieved after 10 min of digestion (lane 2), and no additional release was observed after 20 min of digestion (lane 3). In contrast, in nuclei prepared from NPCs (Figure 4C, lanes 4-6), release was slower, with only a partial release after 10 min (lane 5) and a ∼2-fold increase after 20 min of MNase digestion (lane 6). These observations are in agreement with our estimates from FRAP analysis and are consistent with the identification of a loosely bound fraction of chromatin proteins identified by FRAP. The in vivo imaging and biochemical extraction data on endogenous proteins together strongly support the conclusion of the existence of a hyperdynamically bound fraction of chromatin proteins in undifferentiated ES cells.
Figure 4.
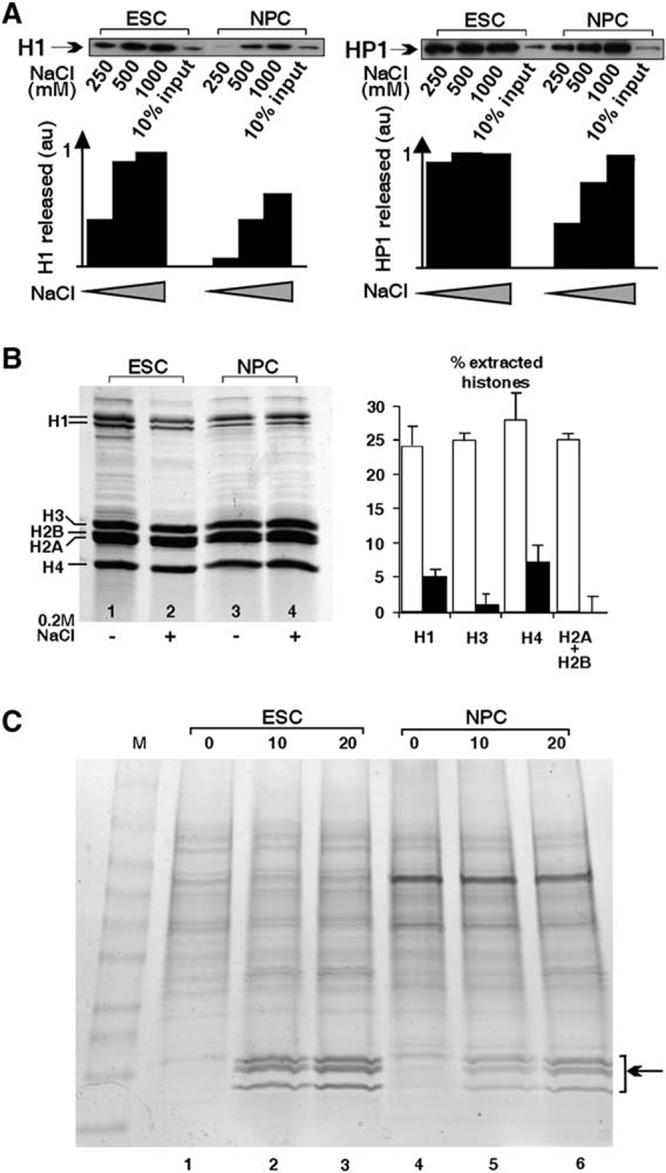
Biochemical Evidence for Looser Binding Fraction of Histones in Undifferentiated ES Cells versus NPCs.
(A) Isolated nuclei from either undifferentiated ES cells (ESC) or NPCs were extracted with increasing salt concentrations (NaCl), and the extracted fraction was detected by Western blotting. Note the increased release of both proteins from ES cells at low salt concentration. au = arbitrary units.
(B) Total histones were purified from ES cells (lanes 1 and 2) and NPCs (lanes 3 and 4) in the absence (−) or presence (+) of 0.2 M NaCl. All core histones were more readily extractable from ES cells compared to NPCs. Right: quantified densitometry of three independent experiments presented as the percentage of extracted histones ± SD. (C) Nuclei from ESCs (lanes 1-3) or NPCs (lanes 4-6) were incubated for 0, 10, or 20 min with 1 U/ml micrococcal nuclease, and supernatants were run on a 4%-20% gradient gel. Note the faster release of the histone fraction (arrow) in ESCs versus NPCs (compare lanes 2 and 5).
Available Pools of Architectural Chromatin Proteins Affect Differentiation
These results suggest that several key architectural chromatin proteins exist in undifferentiated ES cells in a hyperdynamic, loosely bound, or soluble fraction. To test whether the hyperdynamic nature of these proteins is functionally important for efficient ES cell differentiation, we asked whether perturbing the dynamic balance of core histone influences the ability of cells to differentiate.
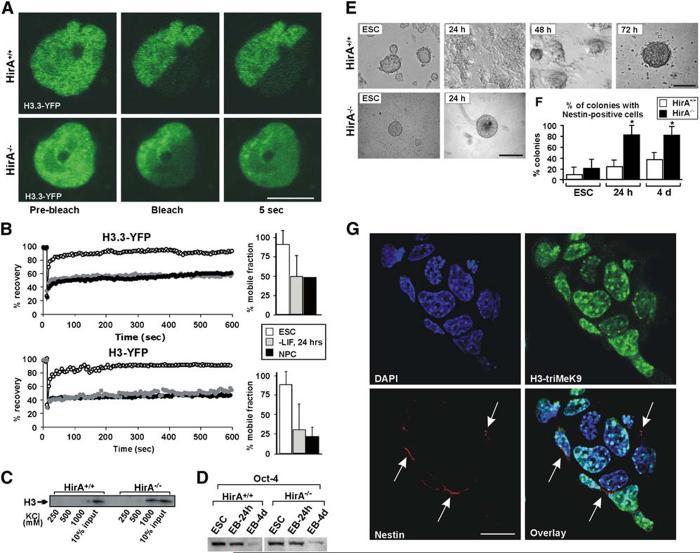
We first tested whether an increase in the hyperdynamic fraction of core histones affects differentiation. To this end, we analyzed ES cells lacking the nucleosome assembly factor HirA (Roberts et al., 2002). HirA is associated with the histone variant H3.3, promoting DNA synthesis-independent nucleosome assembly (Ray-Gallet et al., 2002; Tagami et al., 2004), and it is essential for development since HirA—/— mice die in utero (Roberts et al., 2002). We hypothesized that the absence of HirA from ES cells would result in reduced efficiency of incorporation of core histones H3 and H3.3 and, as a consequence, in an increased nucleoplasmic fraction of these core histones. FRAP analysis and biochemical extraction of endogenous core histones confirmed this prediction. FRAP analysis showed a dramatic increase in the rapidly recovering unbound and loosely bound fractiond of both H3 and H3.3 in undifferentiated ES cells (Figures 5A and Figures 5B, Movies S1 and S2). While parental HirA+/+ wild-type (wt) cells displayed similar FRAP kinetics of H3 and H3.3 to that of R1 ES cells, with a rapid recovery fraction of H3 and H3.3 of 20%-40%, this fraction was more than 80% in HirA—/— cells (Figures 5A and Figures 5B). Similarly, endogenous H3 was more soluble in HirA—/— cells, as assayed by salt extractions (Figure 5C). In HirA—/—, but not in HirA+/+ cells, H3 was extractable from isolated nuclei with 1 M KCl (Figure 5C), supporting the presence of an increased fraction of soluble core histones in HirA—/— cells.
Figure 5.
Accelerated Differentiation of HirA—/— ES Cells
(A) FRAP images of transiently expressed H3.3-YFP before bleach (left), immediately after bleach (middle), and 5 s after bleach (right) in HirA+/+ cells (top) or HirA—/— ES cells (bottom). The scale bar is 5μm. See also Movies S1 and S2.
(B) FRAP curves of H3.3-YFP (top) and H3-YFP (bottom) in HirA—/— ES cells (empty circles), 24 hr after LIF withdrawal (gray circles), and NPCs (black circles). Half nuclei were bleached in these experiments. A total of 20 cells were analyzed from 2 experiments. For clarity, error bars are omitted. Typical standard deviations were < 10% of the average value. Right: estimated mobile fractions of H3.3 (top) and H3 (bottom) in HirA—/—ES cells ± SD.
(C) Isolated nuclei from either wt parental HirA+/+ cells or HirA—/— cells were extracted with increasing salt concentrations (KCl) and were detected with anti-H3 antibodies.
(D) Western blots of Oct4 in undifferentiated ES cells and EBs in wt HirA+/+ (left) and HirA—/— (right) cells.
(E) 24 hr intervals during the course of differentiation of control or HirA—/— ES cells into embryoid bodies (EBs). In wt HirA+/+ cells, EB formation is completed within ca. 72 hr (top), whereas, in HirA—/— cells, it is completed within ca. 24 hr (bottom). The scale bars are 50μm.
(F) Immunostaining quantitation of colonies containing Nestin-positive cells in HirA+/+ (white) and HirA—/— (black) undifferentiated ES cells (left), 24 hr EBs (middle), and 4 day EBs (right).
(G) HirA—/— colony 24 hr after LIF withdrawal double labeled for H3-triMeK9 (top right) and Nestin (bottom left, arrows). Top left: DAPI staining; bottom right: overlay. The scale bar is 10μm.
When we analyzed the ability of HirA—/— ES cells to differentiate, we found accelerated progression through the early stages of differentiation (Figure 5E). In HirA—/— cells, embryoid bodies (EBs) formed within less than 24 hr upon LIF withdrawal (Figure 5E, bottom), whereas this process took ∼72 hr in control HirA+/+ cells (Figure 5E, top). The rapid differentiation into EBs is unlikely due to the presence of elevated levels of differentiation inducing factors in HirA—/— cells, since microarray analysis has not revealed misregulation of any differentiation-specific genes in these cells (P.J.S., unpublished data). Western blot analysis of Oct4 levels in undifferentiated ES cells and EBs in both wt and HirA—/— cells showed sustained levels of Oct4 protein in both HirA+/+ and HirA—/— EBs (24 hr) (Figure 5D). Most Oct4 was lost by day 4 (Figure 5D). In contrast, HirA—/— EBs had considerably more nestin-containing colonies than wt EBs after 24 hr (Figure 5F), supporting the notion of facilitated differentiation. Nestin-positive cells emerged in almost all HirA—/— colonies analyzed (Figure 5G, bottom left, arrows), while the number and morphology of chromatin foci, detected both by DAPI staining (Figure 5G, top left) and H3-triMeK9 staining (Figure 5G, top right), appeared similar to HirA+/+ cells. This accelerated differentiation suggests that increased availability of histones facilitates early differentiation.
We conversely analyzed differentiation in ES cells in which the binding dynamics of chromatin proteins is inhibited. To this end, we took advantage of an H1° mutant with increased binding capacity to chromatin. Since the C-terminal domain of H1 contributes significantly to its binding (Hendzel et al., 2004; Misteli et al., 2000), duplication of this domain is expected to confer stronger binding of H1° to chromatin. We generated ES cells stably expressing H1°cc-GFP or a control H1°-GFP under a Zn-inducible metallothionein (MT) promoter (Figure 6A). The increased binding of H1°cc-GFP in vivo was confirmed by a significantly slower FRAP exchange rate of H1°cc-GFP compared to H1°-GFP in living ES cells and cells 24 hr after LIF withdrawal (Figure 6B) and decreased sensitivity to salt extraction from isolated nuclei (Figure S5D). In addition, in vitro binding assays revealed that both GFP-tagged and untagged H1°cc proteins bound significantly more tightly than the corresponding wt H1° proteins (Figures S5A-S5C)(Gunjan et al., 2001). GFP-tagged and untagged wt H1° bound with similar affinity to purified nucleosomes as endogenous H1 (Figure S5C).
Figure 6.
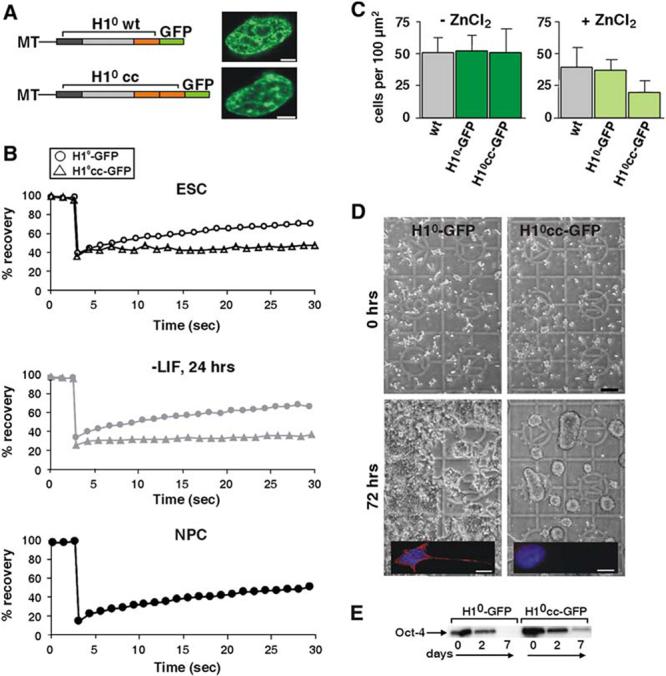
Restricted Dynamic Exchange of Linker Histone H1 Confers Differentiation Arrest
(A) Zn-inducible metallothionein (MT) promoter-driven H1°-GFP fusion constructs used for stable transfections. Wt H1° (top) consists of an N-terminal domain (dark gray), a core domain (light gray), and the chromatin binding C-terminal domain (orange). In the mutant H1°cc (bottom), the C-terminal domain was duplicated. Not drawn to scale. Right: stable ES cell clones expressing H1°-GFP (top) and H1°cc-GFP (bottom). The scale bar is 2μm.
(B) FRAP curves of stable H1°-GFP (circles) and H1°cc-GFP (triangles) in ES cells (top), 24 hr following LIF withdrawal (middle), and in NPCs (bottom, H1°-GFP only). Half nuclei were bleached. Values represent averages from at least 20 cells from 3 experiments. For clarity, error bars are omitted. Typical standard deviations were < 10% of the average value.
(C) Cell counts per 100 μm2 of wt, H1°-GFP stable cells, and H1°cc-GFP stable cells 48 hr after plating. All cells were plated at similar densities of 10 cells/100 μm2. Values represent quantitation of at least 100 cells ± SD.
(D) Normal differentiation of stable H1°-GFP ES cells (left), but not of H1°cc-GFP cells (right), which remained as round, undifferentiated colonies 4 days after LIF withdrawal. The scale bar is 100μm. Inset: immunofluorescence staining with TUJ1 antibody of a differentiating cell (7 days after LIF withdrawal), expressing the intermediate filament β-tubulin III (red), a neuroblast marker. DNA staining (DAPI) is shown in blue. The scale bar is 10μm.
(E) Western blot of Oct4 in stable H1°-GFP cells (left) and stable H1°cc-GFP cells (right) during differentiation.
To assess the effect of the dynamics of H1 binding on differentiation, we monitored the fate of H1°cc-GFP-expressing cells during the course of differentiation. When stable cell clones were grown in the absence of ZnCl2, they displayed normal growth kinetics similar to that of the wt ES cells (Figure 6C, left). When cells were grown in the presence of 50μM ZnCl2, the H1°-GFP line displayed similar behavior to that of the wt, while the H1°cc-GFP line displayed reduced growth rates (Figure 6C, right). We then differentiated these stable lines in the absence or presence of ZnCl2. In the absence of ZnCl2, both H1°-GFP and H1°cc-GFP lines differentiated normally (data not shown). However, in the presence of ZnCl2, while cells expressing H1°-GFP differentiated normally into neuroblasts (Figure 6D, left and inset), cells expressing H1°cc-GFP failed to differentiate and remained as round ES cell-like colonies (Figure 6D, bottom right and inset). These cells did not further differentiate into NPCs or neurons, as indicated by the absence of the neuronal marker b-tubulin III (Figure 6D, insets), and died upon prolonged culturing (data not shown). In these cells, Oct4 did not disappear completely as in their H1°-wt counterparts (Figure 6E), suggesting differentiation arrest rather than progression into a different lineage. These observations show that the dynamic exchange of linker histone is required for ES cell differentiation.
Hyperdynamic Binding Is a Property of Pluripotent Stem Cells
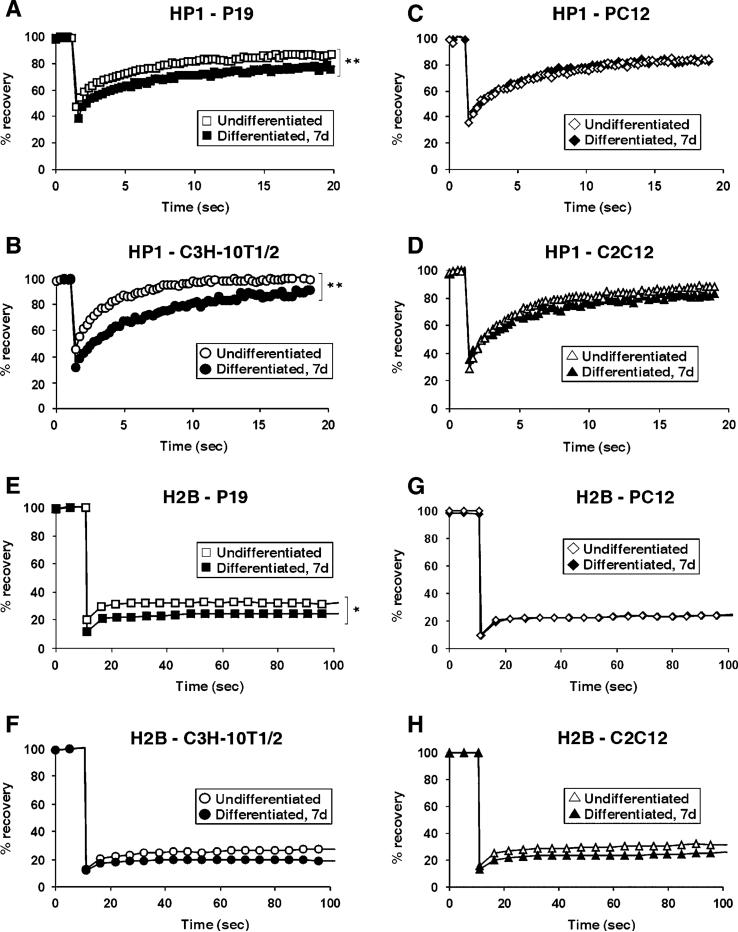
The hyperdynamic binding of chromatin-associated proteins could either be a general feature of undifferentiated cells or might be a specific property of pluripotent cells. In order to distinguish between these two possibilities, we tested the dynamic behavior of HP1 and H2B in additional multipotent cell types (P19, C3H/10T1/2) and compared it to that in undifferentiated, but already lineage-committed, cell lines (P12, C2C12). P19 mouse embryonal carcinoma (EC) cells are multipotent progenitors that can give rise to all three germ cell layers (McBurney, 1993). C3H/10T1/2 mouse mesenchymal stem cells are pluripotent mesodermal progenitors capable of differentiating into myogenic, chondrogenic, adipogenic, and osteogenic lineages (Pinney and Emerson, 1989). When heterochromatin FRAP kinetics of HP1-GFP were measured in either P19 (Figure 7A) or C3H/10T1/2 (Figure 7B), a significant decrease, similar to that seen in R1 cells, in mobility upon differentiation into neural ectoderm and mesoderm, respectively, was observed in both cases (p < 0.005). In contrast, no significant differences in FRAP recovery kinetics were evident in PC12 rat pheochromocytoma or C2C12 myoblasts upon differentiation into neurons via NGF or into myotubes by serum deprivation, respectively. In both cases, the transition from dividing to differentiated cells did not involve changes in heterochromatin FRAP kinetics for HP1-GFP (Figures 7C and Figures 7D). Similar results were observed for the core histone H2B. H2B-GFP displayed decreased FRAP mobility upon differentiation of pluripotent P19 cells (Figure 7E, p < 0.05), while a less pronounced decrease in mobility was observed in multipotent C3H/10T1/2 cells (p > 0.05). In already committed PC12 cells, no change in FRAP kinetics was recorded after differentiation (Figure 7G), and a slight, but statistically not significant, decrease was measured in committed C2C12 cells (Figure 7H, p > 0.1). These observations suggest that chromatin-associated proteins are hyperdynamic in several multipotent cell types and that decreased mobility is associated with loss of multipotency rather than differentiation per se.
Figure 7.
Dynamic Binding of HP1 and H2B in Multipotent and Unilineage Cell Types (A-D) FRAP analysis of heterochromatin foci in pluripotent (A) P19 cells and (B) C3H/10T1/2 cells, or in lineage-committed (C) PC12 cells and (D) C2C12 cells transiently transfected with HP1α-GFP. White, undifferentiated cells; black, cells differentiated for 7 days. (E-H) FRAP analysis of H2B-GFP in (E) pluripotent P19 cells or (F) multipotent C3H/10T1/2 cells, or in lineage-committed (G) PC12 cells and (H) C2C12 cells. Hyperdynamic fractions of HP1 and H2B were found in pluripotent, but not in lineage-committed, cell lines. White, undifferentiated cells; black, cells differentiated for 7 days. Values in (A)-(H) represent averages from at least 20 cells from 3 experiments. (*, p < 0.05; **, p < 0.005).
Discussion
Based on quantitative single cell in vivo imaging and biochemical analysis of endogenous proteins, we report here that several major architectural chromatin proteins exist in undifferentiated ES cells in a hyperdynamic fraction. This soluble or loosely bound fraction of chromatin proteins is a hallmark of pluripotent ES cells and is not a general feature of differentiation processes, as it does not occur in unilineage differentiating cells. We propose that this loosely bound or soluble pool of structural chromatin proteins contributes to the maintenance of the pluripotent state of ES cells and is essential in the early stages of ES cell differentiation for reshaping the global architecture of the genome, particularly for the reorganization of heterochromatin.
Genome Architecture during ES Cell Differentiation
By comparing the morphological appearance of heterochromatin regions in undifferentiated pluripotent ES cells and ES cell-derived neuronal precursor cells, we find evidence that heterochromatin undergoes substantial spatial rearrangements during the very earliest stages of ES cell differentiation. While heterochromatin showed a more dispersed pattern, heterochromatin in NPCs appeared more similar to what is typically observed in somatic cell types showing heterochromatin compaction and concentration in distinct foci. In addition, in ES cells, FISH signals of satellite repeats show a more diffuse signal not restricted to distinct foci, and these regions became more condensed in NPCs. These results resemble the differences in heterochromatin foci observed between undifferentiated F9 cells and after treatment with retinoic acid (Cammas et al., 2002). In another system, no significant differences in the extent of centromere clustering were observed between undifferentiated human ES cells and two diploid differentiated cell types, including a lymphoblastoid cell line (FATO LCL) and primary fibroblasts (Wiblin et al., 2005). However, centromeres in ES cells were mainly found within the nuclear interior, whereas, in differentiated cells, centromeres tend to localize at the nuclear periphery (Wiblin et al., 2005). If indeed pericentric heterochromatin is confined to a smaller portion of the nuclear interior in undifferentiated ES cells, this might explain the seemingly fewer, and larger, heterochromatin foci detected in these cells. These morphological changes are paralleled by an increase in H3-triMeK9, an epigenetic marker for silenced heterochromatin, and a decrease in acetylation of H3 and H4, which are both associated with transcriptionally active euchromatin. These morphological changes and the increase in epigenetic histone modifications characteristic of euchromatin suggest that chromatin in ES cells assumes a globally more open conformation than in differentiated or partially differentiated cells. These chromatin properties of ES cells might reflect a functionally important hallmark of pluripotency.
Hyperdynamic Chromatin Proteins Are a Hallmark of Pluripotent ES Cells
ES cells possess two qualities that distinguishes them from other cell types: they retain an unlimited capacity to self-renew, and, unlike immortalized cells, they are also able to generate the three embryonic germ layers and further differentiate to essentially every type of cell and tissue (O’Shea, 2004). This unlimited potential suggests that their genome has not yet been determined to fit any particular cell type and is still plastic. We suggest that hyperdynamic binding is a hallmark of pluripotency. This conclusion is supported by the presence of a hyperdynamic fraction in all pluripotent cell lines (R1, parental HirA cells, P19, and C3H/10T1/2) analyzed here. More importantly, we were unable to detect a hyperdynamic fraction in three undifferentiated, but lineage-committed, cell lines (NPC, PC12, and C2C12). The hyperdynamic nature of chromatin protein binding may contribute to maintaining chromatin in a globally relatively open, plastic state and, in this way, to the maintenance of pluripotency. Our data support a model in which ES cells preserve the potential to differentiate into multiple cell types by maintaining a loosely bound fraction of histones and other chromatin-associated proteins, which through free exchange with bound histones and chromatin, generate a state of active, “breathing” chromatin.
Hyperdynamic Binding in Genome Reorganization
The transition from undifferentiated, pluripotent ES cells to differentiated or partially differentiated cells involves dramatic changes in genome expression profiles (Ahn et al., 2004; Kelly and Rizzino, 2000; Loring et al., 2001; Sperger et al., 2003). During commitment, the parts of the genome that are not required for the newly forming lineage are presumably silenced (Eckfeldt et al., 2005). This silencing process involves epigenetic modifications and global reorganization of chromatin (Muller and Leutz, 2001), including condensation of heterochromatin into distinct foci as observed here. We suggest that the loosely bound fraction of architectural chromatin proteins is functionally important in the remodeling process during the early stages of differentiation by facilitating the structural chromatin changes that are required during this transition. As differentiation progresses and cell type-specific genome expression programs are implemented, the available structural proteins are incorporated into chromatin to establish the cell type-specific global chromatin architecture. When this occurs, the transcriptional potential of the genome is restricted. One noteworthy observation is the slight increase in the acetylation level of H4 24 hr after the onset of differentiation. The concurrent increase in both H4 acetylation and H3-triMeK9 methylation might suggest that, during the very early stages of differentiation, formation of heterochromatin parallels a transient rise in the transcriptional potential of euchromatin, allowing simultaneous activation and repression of different parts of the genome to fit the differentiation needs. Nevertheless, our observation that the reduction of dynamic binding of the major structural heterochromatin protein HP1 precedes the formation of distinct heterochromatin foci might suggest that immobilization of structural proteins is an early step in the differentiation-dependent chromatin remodeling and silencing process.
Consistent with a globally highly transcriptionally active genome in ES cells, we find that the only structural chromatin protein without an increased hyperdynamic pool is H3.3, which preferentially associates with transcriptionally active regions (Ahmad and Henikoff, 2002). This finding is also consistent with the observed accelerated differentiation of HirA—/— cells, since, in those cells, less H3.3 is incorporated into the open chromatin regions, thus facilitating the formation of heterochromatin regions and promoting differentiation.
Hyperdynamic Binding and Differentiation
We provide two lines of evidence to suggest that the dynamic properties of architectural chromatin proteins are functionally relevant for the differentiation process. Upon inhibition of HirA, the available loosely bound pool of core histones H3 and H3.3 increases, as shown by FRAP analysis and biochemical extraction. This increased pool is now available for formation of heterochromatin, facilitating the rearrangements observed during early differentiation. On the other hand, the presence of a dominant, tightly binding linker histone reduces the availability of these molecules, thus impeding the formation of heterochromatin. Thus, prevention of chromatin proteins from assembly into chromatin accelerates differentiation, while restriction of linker histone in ES cells blocks differentiation. These data imply that the dynamic nature of chromatin is functionally important for stem cell differentiation.
The observed fate of HirA—/— ES cells is in agreement with the phenotype of HirA—/— embryos. Upon loss of HirA, embryos typically die around day 10, although developmental defects occur much earlier, displaying severe early gastrulation defects at day 6 or earlier (Roberts et al., 2002). Similarly, HirA—/— ES cells exhibit efficient and rapid differentiation in the early stages, and 6 or 7 days after the onset of differentiation, they begin to deteriorate and die. While HirA—/— ES cells were able to differentiate into early NPCs they never progressed to the next level of differentiation to produce neurons (E.M., unpublished data). The fact that HirA—/— ES cells were able to undergo early differentiation also clearly demonstrates that these steps of differentiation are HirA independent, and that deposition of core histones must occur via different, possibly redundant, pathways.
On the other hand, ES cells stably expressing the strongly bound H1°cc mutant displayed both reduced growth rate and perturbed differentiation. Thus, interference with the dynamic exchange of the linker histone H1 from chromatin affects two of the classical stem cell features. Although the mode of action of the H1°cc mutant is not clear, it seems likely that, upon expression of the mutant, the tight association between H1°cc and chromatin slowly out-competes the chromatin binding of the endogenous H1°. After a while, most of the linker histone will be replaced by H1°cc, thus restricting the dynamics of the genome globally and preventing its “breathing.” The rigid genome is likely less flexible, and the activation and suppression of the desired chromatin domains may be more difficult. Differentiation is hence blocked, and the cells die upon prolonged culturing.
Taken together, our observations demonstrate the existence of a hyperdynamic fraction of architectural chromatin protein in the nucleus of pluripotent ES cells. We suggest that this soluble, more loosely bound fraction is a specific hallmark of ES cells. The presence of a hyperdynamic fraction of chromatin proteins points to altered chromatin structure in ES cells. We propose that these properties of chromatin have functional relevance by contributing to the maintenance of pluripotency of ES cells and facilitate the timely formation of higher-order chromatin domains during differentiation.
Experimental Procedures
Cells
Mouse R1 ES cells were from A. Nagy (Toronto, Canada). R1 ES cells were grown on gelatin-coated plates with DMEM, 15% ES-grade fetal calf serum (FCS), 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, 0.1 mM β-mercaptoethanol, and 1000 U/ml leukemia inhibitory factor (LIF). Nestin-positive NPCs were generated by using a protocol kindly provided by R. McKay (National Institutes of Health) (Lee et al., 2000). Briefly, ES cells were plated on bacterial culture dishes without LIF for 4 days to allow for embryoid body (EB) formation. EBs were replated on poly-L-lysin/fibronectin (Sigma)-coated plates in DMEM/F12 medium supplemented with ITS (5 μg/ml insulin, 50 mg/ml transferrin, 30 nM selenium chloride) and fibronectin (5 μg/ml). HirA—/— and HirA+/+ cells were grown on a feeder layer of γ-irradiated mouse embryonic fibroblasts (MEF). P19 embryonic carcinoma cells were grown in DMEM with 10% FBS and 1 mM sodium pyruvate. Differentiation was with 1 μM all-trans retinoic acid (ATRA). C3H/10T1/2 mesenchymal stem cells were grown in high-glucose DMEM with 10% FBS. Differentiation was with 2% horse serum. Rat PC12 pheochromocytoma cells were grown in DMEM with 8% horse serum and 8% FBS. Differentiation was induced with NGF (50 ng/ml). C2C12 myoblast cells were grown in DMEM with 10% FBS. Differentiation was with 2% horse serum. All cell culture media were supplemented with 2 mM glutamine, 100 U/ml penicillin, and 50 μg/ml streptomycin. Lipofectamine-2000 was used for all transfection experiments. All cell culture reagents were purchased from GIBCO-BRL (Invitrogen; Carlsbad, CA), and plates were purchased from Falcon (BD Biosciences, San Jose, CA), unless indicated otherwise. For photobleaching experiments, cells were grown in chambered cover glasses (Lab-Tek; Rochester, NY) or in glass-bottom culture dishes (MatTek; Ashland, MA).
Plasmids
CMV-H3-YFP and CMV-H3.3-YFP were kindly provided by K. Ahmad and S. Henikoff (Seattle, WA). CMV-HP1α-GFP, H1°-GFP under MT or CMV promoters, and CMV-H2B-GFP have previously been described (Cheutin et al., 2003; Misteli et al., 2000; Phair et al., 2004). The mutant MT-H1°cc-GFP consists of (from amino to carboxy termini) the entire coding region of H1° (amino acids 1-193), a single alanine residue, amino acids 94-193 of H1°, a single alanine residue, and the complete coding region of EGFP. The stable R1 clones were generated by introducing the expression vectors via electroporation, selection for resistance to G418, followed by identification of expressing clones by direct observation under epifluorescence.
Antibodies and Immunofluorescence
The following antibodies were used: Oct4 (goat polyclonal, Santa Cruz Biotechnologies, Santa Cruz, CA); Nestin (rabbit polyclonal), kindly provided by R. McKay (National Institutes of Health); H1, H2B, and H3 (rabbit polyclonal), kindly provided by M. Bustin (National Institutes of Health); TUJ1 (mouse monoclonal, Chemicon; Temecula, CA) against β-tubulin III; H3-triMeK9 (rabbit polyclonal, Abcam; Cambridge, MA); and HP1a (mouse monoclonal, Euromedex; Mundolsheim, France). Detection was with anti-rabbit or anti-mouse antibodies conjugated to either Texas red or FITC (Jackson Immuno-Research; West Grove, PA). IF was performed as described (Misteli et al., 2000). For EB imaging, EBs were paraffin embedded, sectioned to 7 μm intervals, and adhered to microscopic slides. Before IF, slides were treated with xylene (2 × 10 min), decreasing concentrations of EtOH (100%, 75%, 50%, and 25%), and PBT (2 3 5 min).
DNA FISH
Cells grown on glass coverslips were fixed (4% paraformaldehyde in PBS, 15 min), washed three times (PBS, 5 min each), permeabilized (0.5% Triton X-100 in PBS, 5 min), and washed again (PBS, 5 min, 2× SSC, 5 min). Probe was denatured at 90°C for 8 min and transferred to ice. Cells were denatured in 70% formamide/2× SSC solution for 7 min at 85°C. A 5′-biotinylated DNA probe (Invitrogen) recognizing the mouse major satellite repeat (GenBank accession number X06899) was applied overnight in a hybridization solution (50% formamide, 2× SSC, 10% dextran sulfate, 1 mg/ml tRNA) at 37°C. Washes (2× SSC, 5 min) were followed by blocking (3% BSA, 0.1% Tween-20, 4× SSC, 20 min) and detection with a streptavidin-Cy3 conjugate (Amersham Biosciences; Buckinghamshire, UK). A scrambled probe was used as a negative control. Probe sequences: major satellite: 5′-CTCGCCATATTTCACGTCCTAAAGT GTGTATTTCTC-3′; scrambled: 5′-TCTACGTTACCATCTCAGTGCG TATCGTTCTATTCA-3′
Salt Extractions
For H1 and HP1, cells were washed in PBS, harvested, dounced in buffer A (0.32 M sucrose, 15 mM HEPES [pH 7.9], 60 mM KCl, 2 mM EDTA, 0.5 mM EGTA, 0.5% BSA, 0.5 mM spermidine, 0.15 mM spermine, and 0.5 mM DTT), layered over a cushion of high-sucrose Buffer A (30% sucrose), and centrifuged (15 min, 3000 × g). Pelleted nuclei were resuspended in buffer B (15 mM HEPES [pH 7.9], 60 mM KCl, 15 mM NaCl, 0.34 mM sucrose, 10% glycerol) and incubated with different NaCl or KCl concentrations (250-1000 mM) at 4°C for 30 min. Supernatants were separated on 4%-20% gradient Tris-HCl SDS gels (BioRad; Hercules, CA), blotted, and incubated with the appropriate antibodies. For core histones, the pellet remaining after salt treatment was extracted with 0.2 M H2SO4. Acid-soluble maerial was precipitated with 20% TCA and separated on 18% Tris-HCl SDS gels (BioRad).
Micrococcal Nuclease Digestion
Nuclei from undifferentiated ES cells or NPCs were prepared as described above and were digested with 1 U/ml micrococcal nuclease (MNase) (Worthington; Lakewood, NJ) in 10 mM Tris-HCl buffer supplemented with 5 mM CaCl2. Reactions were then centrifuged at 14,000 × g for 10 min, and supernatants were collected and run on 4%-20% gradient Tris-HCl SDS gels (BioRad).
Microscopy and Photobleaching
A Zeiss confocal LSM 510 META was used for all photobleaching experiments and fluorescent image acquisitions. The 30 mW Argon/Neon laser at 75% power was used for bleaching. Photobleaching and quantitation was performed as described (Cheutin et al., 2003; Lever et al., 2000; Phair et al., 2004). For core histones, half of the nucleus, including both euchromatin and heterochromatin, was bleached, and images were collected every 5 s for 10 min. For H1, 30 images were collected every 1 s. For HP1, the scan time between images was reduced to zero for maximal image collection speed. A total of 60 images were collected. Image analysis was performed with MetaMorph imaging software (Molecular Devices; Downingtown, PA).
Supplementary Material
FRAP of H3.3-YFP in wt ES cell. Shown are a total of 12 frames taken at 5 sec intervals. Bleaching was after frame 3. Note immediate recovery at frame 4.
FRAP of H3.3-YFP in HirA-/- ES cell. Shown are a total of 12 frames taken at 5 sec intervals. Bleaching was after frame 3.
Acknowledgments
We thank R. McKay and D. Hoeppner for reagents, technical assistance, and for critical comments on the manuscript; S. Henikoff, K. Ahmad, and M. Bustin for reagents; and T. Karpova for technical support. Imaging was performed at the National Cancer Institute Imaging Facility. This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. P.J.S. is supported by the British Heart Foundation. D.T.B. is supported by grant MCB0235800 from the National Science Foundation. T.M. is a Fellow of the Keith R. Porter Endowment for Cell Biology.
References
- Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- Ahn JI, Lee KH, Shin DM, Shim JW, Lee JS, Chang SY, Lee YS, Brownstein MJ, Lee SH. Comprehensive transcriptome analysis of differentiation of embryonic stem cells into midbrain and hindbrain neurons. Dev. Biol. 2004;265:491–501. doi: 10.1016/j.ydbio.2003.09.041. [DOI] [PubMed] [Google Scholar]
- Cammas F, Oulad-Abdelghani M, Vonesch JL, Huss-Garcia Y, Chambon P, Losson R. Cell differentiation induces TIF1β association with centromeric heterochromatin via an HP1 interaction. J. Cell Sci. 2002;115:3439–3448. doi: 10.1242/jcs.115.17.3439. [DOI] [PubMed] [Google Scholar]
- Cheutin T, McNairn AJ, Jenuwein T, Gilbert DM, Singh PB, Misteli T. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science. 2003;299:721–725. doi: 10.1126/science.1078572. [DOI] [PubMed] [Google Scholar]
- Eckfeldt CE, Mendenhall EM, Verfaillie CM. The molecular repertoire of the ‘almighty’ stem cell. Nat. Rev. Mol. Cell Biol. 2005;6:726–737. doi: 10.1038/nrm1713. [DOI] [PubMed] [Google Scholar]
- Eiges R, Benvenisty N. A molecular view on pluripotent stem cells. FEBS Lett. 2002;529:135–141. doi: 10.1016/s0014-5793(02)03191-5. [DOI] [PubMed] [Google Scholar]
- Festenstein R, Pagakis SN, Hiragami K, Lyon D, Verreault A, Sekkali B, Kioussis D. Modulation of heterochromatin protein 1 dynamics in primary mammalian cells. Science. 2003;299:719–721. doi: 10.1126/science.1078694. [DOI] [PubMed] [Google Scholar]
- Gunjan A, Sittman DB, Brown DT. Core histone acetylation is regulated by linker histone stoichiometry in vivo. J. Biol. Chem. 2001;276:3635–3640. doi: 10.1074/jbc.M007590200. [DOI] [PubMed] [Google Scholar]
- Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA. Epigenetic reprogramming in mouse primordial germ cells. Mech. Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- Hendzel MJ, Lever MA, Crawford E, Th’ng JP. The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J. Biol. Chem. 2004;279:20028–20034. doi: 10.1074/jbc.M400070200. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Keller GM. In vitro differentiation of embryonic stem cells. Curr. Opin. Cell Biol. 1995;7:862–869. doi: 10.1016/0955-0674(95)80071-9. [DOI] [PubMed] [Google Scholar]
- Kelly DL, Rizzino A. DNA microarray analyses of genes regulated during the differentiation of embryonic stem cells. Mol. Reprod. Dev. 2000;56:113–123. doi: 10.1002/(SICI)1098-2795(200006)56:2<113::AID-MRD1>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Keohane AM, O’Neill LP, Belyaev ND, Lavender JS, Turner BM. X-Inactivation and histone H4 acetylation in embryonic stem cells. Dev. Biol. 1996;180:618–630. doi: 10.1006/dbio.1996.0333. [DOI] [PubMed] [Google Scholar]
- Kimura H, Cook PR. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 2001;153:1341–1353. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Lumelsky N, Studer L, Auerbach JM, McKay RD. Efficient generation of midbrain and hindbrain neurons from mouse embryonic stem cells. Nat. Biotechnol. 2000;18:675–679. doi: 10.1038/76536. [DOI] [PubMed] [Google Scholar]
- Lee JH, Hart SR, Skalnik DG. Histone deacetylase activity is required for embryonic stem cell differentiation. Genesis. 2004;38:32–38. doi: 10.1002/gene.10250. [DOI] [PubMed] [Google Scholar]
- Lever MA, Th’ng JP, Sun X, Hendzel MJ. Rapid exhange of histone H1.1 on chromatin in living human cells. Nature. 2000;408:873–876. doi: 10.1038/35048603. [DOI] [PubMed] [Google Scholar]
- Loebel DA, Watson CM, De Young R, Tam PP. Lineage choice and differentiation in mouse embryos and embryonic stem cells. Dev. Biol. 2003;264:1–14. doi: 10.1016/s0012-1606(03)00390-7. [DOI] [PubMed] [Google Scholar]
- Loring JF, Porter JG, Seilhammer J, Kaser MR, Wesselschmidt R. A gene expression profile of embryonic stem cells and embryonic stem cell-derived neurons. Restor. Neurol. Neurosci. 2001;18:81–88. [PubMed] [Google Scholar]
- McBurney MW. P19 embryonal carcinoma cells. Int. J. Dev. Biol. 1993;37:135–140. [PubMed] [Google Scholar]
- Misteli T, Gunjan A, Hock R, Bustin M, Brown DT. Dynamic binding of histone H1 to chromatin in living cells. Nature. 2000;408:877–881. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- Muller C, Leutz A. Chromatin remodeling in development and differentiation. Curr. Opin. Genet. Dev. 2001;11:167–174. doi: 10.1016/s0959-437x(00)00175-1. [DOI] [PubMed] [Google Scholar]
- O’Shea KS. Self-renewal vs. differentiation of mouse embryonic stem cells. Biol. Reprod. 2004;71:1755–1765. doi: 10.1095/biolreprod.104.028100. [DOI] [PubMed] [Google Scholar]
- Patterton D, Wolffe AP. Developmental roles for chromatin and chromosomal structure. Dev. Biol. 1996;173:2–13. doi: 10.1006/dbio.1996.0002. [DOI] [PubMed] [Google Scholar]
- Phair RD, Scaffidi P, Elbi C, Vecerova J, Dey A, Ozato K, Brown DT, Hager G, Bustin M, Misteli T. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol. 2004;24:6393–6402. doi: 10.1128/MCB.24.14.6393-6402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinney DF, Emerson CP., Jr. 10T1/2 cells: an in vitro model for molecular genetic analysis of mesodermal determination and differentiation. Environ. Health Perspect. 1989;80:221–227. doi: 10.1289/ehp.8980221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen TP. Embryonic stem cell differentiation: a chromatin perspective. Reprod. Biol. Endocrinol. 2003;1:100. doi: 10.1186/1477-7827-1-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray-Gallet D, Quivy JP, Scamps C, Martini EM, Lipinski M, Almouzni G. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell. 2002;9:1091–1100. doi: 10.1016/s1097-2765(02)00526-9. [DOI] [PubMed] [Google Scholar]
- Roberts C, Sutherland HF, Farmer H, Kimber W, Halford S, Carey A, Brickman JM, Wynshaw-Boris A, Scambler PJ. Targeted mutagenesis of the Hira gene results in gastrulation defects and patterning abnormalities of mesoendodermal derivatives prior to early embryonic lethality. Mol. Cell. Biol. 2002;22:2318–2328. doi: 10.1128/MCB.22.7.2318-2328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma K, Reinberg D. Histone variants meet their match. Nat. Rev. Mol. Cell Biol. 2005;6:139–149. doi: 10.1038/nrm1567. [DOI] [PubMed] [Google Scholar]
- Schmiedeberg L, Weisshart K, Diekmann S, Meyer Zu Hoerste G, Hemmerich P. Highand low-mobility populations of HP1 in heterochromatin of mammalian cells. Mol. Biol. Cell. 2004;15:2819–2833. doi: 10.1091/mbc.E03-11-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperger JM, Chen X, Draper JS, Antosiewicz JE, Chon CH, Jones SB, Brooks JD, Andrews PW, Brown PO, Thomson JA. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc. Natl. Acad. Sci. USA. 2003;100:13350–13355. doi: 10.1073/pnas.2235735100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani MA. Reprogramming of genome function through epigenetic inheritance. Nature. 2001;414:122–128. doi: 10.1038/35102186. [DOI] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- Weiss MJ, Orkin SH. In vitro differentiation of murine embryonic stem cells. New approaches to old problems. J. Clin. Invest. 1996;97:591–595. doi: 10.1172/JCI118454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiblin AE, Cui W, Clark AJ, Bickmore WA. Distinctive nuclear organisation of centromeres and regions involved in pluripotency in human embryonic stem cells. J. Cell Sci. 2005;118:3861–3868. doi: 10.1242/jcs.02500. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FRAP of H3.3-YFP in wt ES cell. Shown are a total of 12 frames taken at 5 sec intervals. Bleaching was after frame 3. Note immediate recovery at frame 4.
FRAP of H3.3-YFP in HirA-/- ES cell. Shown are a total of 12 frames taken at 5 sec intervals. Bleaching was after frame 3.