Abstract
H1 linker histones stabilize the nucleosome, limit nucleosome mobility and facilitate the condensation of metazoan chromatin. Here, we have combined systematic mutagenesis, measurement of in vivo binding by photobleaching microscopy, and structural modeling to determine the binding geometry of the globular domain of the H1° linker histone variant within the nucleosome in unperturbed, native chromatin in vivo. We demonstrate the existence of two distinct DNA-binding sites within the globular domain that are formed by spatial clustering of multiple residues. The globular domain is positioned via interaction of one binding site with the major groove near the nucleosome dyad. The second site interacts with linker DNA adjacent to the nucleosome core. Multiple residues bind cooperatively to form a highly specific chromatosome structure that provides a mechanism by which individual domains of linker histones interact to facilitate chromatin condensation.
The basic subunit of eukaryotic chromatin is the nucleosome, which consists of the chromatosome and additional linker DNA1.The chromatosome is comprised of ∼168 bp of DNA, an octamer of core histones and one molecule of linker or H1 histone2,3. Linker histones act to stabilize the nucleosome and to facilitate folding of nucleosomal arrays into higher-order structures4-7. In higher organ-isms, linker histones have a conserved structure consisting of a central globular domain flanked by a long, lysine-rich C-terminal tail and a shorter, often basic N-terminal extension8. Specific subdomains of the C-terminal tail are crucial for H1-linker DNA binding and for stabilizing folded chromatin structures9-11. The central globular domain protects the chromatosome from nuclease digestion, and specific interactions of this domain within the chromatosome are likely to substantially influence the structural geometry of condensed chromatin2,12,13. Binding of the globular domain also has direct effects on gene expression at the nucleosomal level14-16. The central globular domain is believed to bind at or near the nucleosome dyad and to contact at least two strands of DNA17-20. Inspection of the crystal structure of the globular domain of the H5 linker histone variant (GH5) in the absence of DNA has suggested the existence of two well-separated DNA-binding sites21. Accordingly, simultaneous mutation of the residues in these sites abolishes the ability of GH5 to bind linker histone-depleted oligonucleosomes19 or reconstituted mononucleosomes in vitro20. However, the precise interaction surface and the position of H1 on the nucleosome in vivo remain unknown, which makes it difficult to determine the way in which H1 functionally influences chromatin structure and gene expression12,13.
Fluorescent recovery after photobleaching (FRAP) has recently been developed as a powerful tool to quantitatively measure the binding of proteins to unperturbed chromatin in living cells22-25. These analyses have shown that both mouse and human H1 isoforms bind dynamically to chromatin in vivo with a residence time on the order of minutes and that both the globular and C-terminal domains are required for high-affinity H1 binding22,23. Here, we have applied FRAP to a comprehensive collection of H1 mutants to identify specific residues within the globular domain of the mouse H1° linker histone variant that contribute to nucleosomal binding. Mapping of these residues onto the atomic structure of the globular domain defined the in vivo interaction surface of H1 with the nucleosome, and using molecular modeling, we determined the position of the linker histone within the chromatosome. The observed binding geometry suggests a mechanism wherein the globular and C-terminal domains of H1 interact to facilitate chromatin condensation.
RESULTS
Identification of H1 binding residues using FRAP
To identify the residues within the globular domain that are involved in nucleosomal contacts in vivo, we generated a comprehensive set of H1-green fluorescent protein (GFP) fusion constructs containing individual point mutations in the murine H1° variant. Within the globular domain, this subtype is 97% homologous to the well-characterized avian H5 variant26. First, simultaneous point mutations were introduced at each of the putative DNA-contact sites within the globular domain that have been suggested by in vitro analyses20(Fig. 1a). The basic residues in the primary site (Lys69, Lys73 andLys85), secondary site (Lys40, Arg42, Lys52 and Arg94) or both were simultaneously replaced with alanine, and the binding of the mutants was determined by FRAP in stable cell lines. All experiments were performed using an optimized FRAP protocol and a standardized bleach region in euchromatin (see Methods for details). These multisite mutants showed very rapid recovery after photobleaching (Fig. 1b), indicating severely compromised binding22. The half-time for recovery, t50, was used as a quantitative indicator of binding strength and was less than 1 s for all mutants but more than 50 s for the wild-type protein (Fig. 1b and Table 1). For comparison, t50 for nonbinding GFP alone is on the order of 380 ms under these conditions.
Figure 1.
FRAP analysis of mutant and wild-type (WT) H1-GFP. (a) Sequence of the globular domain of H1°. Residues are numbered from the initiator methionine to allow direct comparison to the H5 sequence. Asterisks indicate amino acids mutated in this study. Boxes and arrows below the sequence indicate α-helices and β-sheets, respectively, as determined from the crystal structure of H5 (ref. 21). (b) Quantitative FRAP analysis of stable transfectants expressing H1°-GFP constructs containing multiple mutations in putative binding sites. S1, site 1 mutations, K69A K73A K85A; S2, site 2 mutations, K40A R42A K52A R94A; S1S2, all seven of these mutations. (c,d) Quantitative FRAP analysis of stable transfectants expressing H1-GFP constructs containing single mutations in putative binding residues. (e) FRAP results upon mutation of each basic residue in the globular domain to a neutral residue. Transfectants expressing these H1°-GFP constructs were analyzed by FRAP to determine t50 and t80 (see Table 1). Values are averages from at least ten cells from three experiments. For clarity, error bars are omitted; typical standard deviations were below 10%.
Table 1.
Quantitative FRAP analysis of mutant H1 constructs
| Genotype | t50 (sec)a | t80 (sec)a | Genotype | t50 (sec) | t80 (sec) |
|---|---|---|---|---|---|
| Wild type | 52 ± 3.2 | 270 ± 7.3 | D65Kb | 8 ± 0.3 | 46 ± 2.4 |
| Site 1 | 1 ± 0.3 | 6 ± 1.1 | K55D D65Kb | 47 ± 3.6 | 189 ± 12.5 |
| Site 2 | 1 ± 0.4 | 8 ± 1.4 | H57A | 34 ± 2.8 | 169 ± 11.6 |
| S12 | <1 | 4.2 ± 0.8 | H57E | 28 ± 2.6 | 170 ± 10.8 |
| K20A | 37 ± 3.1 | 160 ± 5.2 | K59A | 37 ± 4.5 | 142 ± 8.2 |
| K21A | 38 ± 3.4 | 159 ± 6.5 | K59D | 37 ± 3.4 | 139 ± 9.6 |
| H25G | 12 ± 0.8 | 53 ± 2.3 | E62H | 76 ± 5.7 | 280 ± 10.2 |
| H25E | 10 ± 1.2 | 47 ± 4.1 | K69A | 4 ± 0.3 | 22 ± 1.6 |
| H25K | 32 ± 4.3 | 190 ± 8.6 | K69R | 23 ± 3.2 | 119 ± 7.5 |
| K27T | 53 ± 4.2 | 208 ± 12.1 | K73A | 8 ± 0.4 | 40 ± 2.8 |
| K40A | 39 ± 2.8 | 152 ± 9.5 | K73E | 4 ± 0.3 | 20 ± 3.1 |
| K40E | 34 ± 2.5 | 132 ± 9.1 | K73R | 49 ± 3.9 | 201 ± 11.9 |
| R42A | 16 ± 1.5 | 65 ± 3.2 | R74A | 14 ± 1.8 | 50 ± 3.1 |
| R42E | 10 ± 1.1 | 35 ± 2.2 | K82V | 63 ± 4.6 | 208 ± 12.7 |
| R42K | 45 ± 3.3 | 181 ± 9.2 | Q83D | 4 ± 0.3 | 17 ± 2.3 |
| R47A | 5 ± 0.2 | 28 ± 1.2 | K85A | 10 ± 1.1 | 72 ± 5.7 |
| R47E | 2 ± 0.3 | 20 ± 2.1 | K85E | 4 ± 0.3 | 18 ± 1.9 |
| R47K | 22 ± 2.2 | 121 ± 8.7 | K85R | 49 ± 4.2 | 197 ± 8.6 |
| R47L | 8 ± 0.5 | 41 ± 3.1 | A89D | 3 ± 0.2 | 19 ± 3.1 |
| K52A | 28 ± 1.6 | 120 ± 7.3 | S90D | 3 ± 0.2 | 21 ± 2.7 |
| K55Ab | 12 ± 1.2 | 52 ± 3.8 | R94A | 17 ± 1.4 | 56 ± 2.6 |
| K55Eb | 3 ± 0.2 | 20 ± 1.7 | R97A | 20 ± 1.3 | 84 ± 3.8 |
| K55Db | 5 ± 0.2 | 24 ± 1.3 |
Values for t50 and t80 were determined as previously described24.
See Supplementary Figure 1.
To determine the contributions of individual residues to binding, stable cell lines expressing constructs containing single point mutations of each residue were generated and analyzed by FRAP (Fig. 1c,d). Each mutant showed a recovery rate that was considerably faster than that of the wild-type protein (Fig. 1c,d). The strongest contributions to binding came from Lys69 and Lys73 (t50=4 s and 8 s, respectively), with intermediate contributions from Arg42 and Arg94 (t50=16 s and 17 s, respectively) and relatively weak contributions from Lys40 and Lys52 (t50=39s and 28 s, respectively) (Fig. 1c,d and Table 1). Thus, the FRAP approach confirms the involvement of several residues previously implicated in DNA binding on the basis of in vitro assays, and it extends the in vitro assays by determining the relative contributions of single residues to binding in vivo.
To generate a complete map of the interaction surface of the globular domain of H1 within the nucleosome and to identify new residues that contribute to binding, we systematically generated mutants in each of the basic residues, including histidines, within the entire globular domain (Fig. 1a). Residues conserved between H1° and other variants were substituted with alanine, whereas nonconserved residues were replaced with the residue most commonly found in the other variants (Table 1). Analysis of the crystal structure of the H5 globular domain indicated that, with the exception of Lys55 (see Supplementary Fig. 1 online), all of the mutated positively charged residues are oriented on or near the surface of the globular domain, where they could potentially interact with nucleosomal or linker DNA. For each mutant, stable cell lines were generated and their in vivo binding properties determined by FRAP (Fig. 1e and Table 1). Nine mutants had dramatically faster recovery rates with t50 20 s and t80 o 90 s (Fig. 1e). This group includes the previously identified lysines at positions 69, 73 and 85 in the putative primary interaction site and arginines 42 and 94 in the putative secondary site, as well as His25, which has been suggested to bind near the end of the chromatosome27. However, the data also identified three residues, Arg47, Arg74 and Lys97, that have not previously been implicated as contributing to nucleosomal binding. Identical results were obtained for multiple clonal cell lines of each mutant construct (data not shown).
To validate this mutagenesis approach, we created additional mutants for the residues that showed the greatest effects (His25, Arg42, Arg47, Lys73 and Lys85). These basic residues were replaced one by one, either with glutamic acid to further weaken their potential for electrostatic interaction or with another basic residue, to determine whether their contributions to binding were due to electrostatic interaction. In all cases, replacement with glutamic acid further reduced binding (Fig. 2). In contrast, replacement with another basic residue resulted in greater binding activity than that observed after the neutral mutation and restored partial (His25, Arg47) or, in several cases (Arg42, Lys73, Lys85), nearly wild-type binding (Table 1 and Fig. 2). The observed correlation between charge and binding behavior for all mutants is most consistent with these residues contributing electrostatic interactions with DNA to promote nucleosomal binding.
Figure 2.
Electrostatically conservative mutations have much smaller effects on binding affinity. (a,b) FRAP analysis of cell lines expressing the indicated construct. (c) Selected FRAP results (Table 1). Values are averages from at least ten cells from three experiments. Error bars show s.d.
Biochemical confirmation of positioning
We wished to exclude the possibility that the measured FRAP kinetics of H1 mutants were due to aberrant binding to nonspecific sites resulting from tagging, mutagenesis or structural aberration of the protein. The overall properties of GFP-tagged H1 are indistinguishable from endogenous H1 by salt extraction and icrococcal nuclease (MNase) digestion analyses from reconstituted and isolated chromatin22. We con-firmed that all H1 mutant proteins were structurally intact, in that all had circular dichroism spectra similar to that of the wild-type H1° globular domain (Supplementary Fig. 2 online). Finally, we recently showed H1-GFP to be fully functional in somatic-cell nuclear transfer28. Therefore, these mutations and fusion to GFP do not introduce major changes in the structure or function of the globular domain of H1.
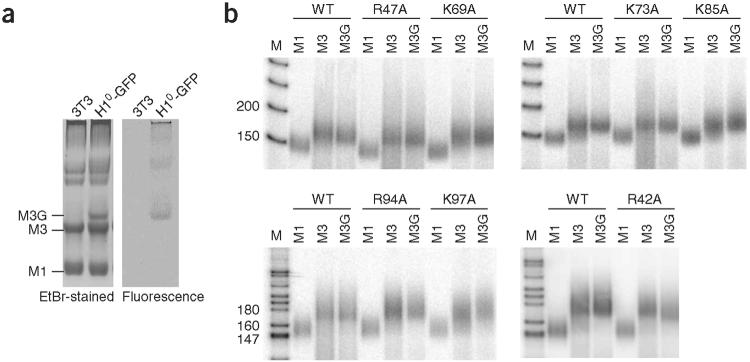
To conclusively demonstrate that mutant H1-GFP constructs are properly positioned on the nucleosome, we applied in vivo biochemical analysis. The most stringent assay for validating proper binding of H1 is protection of the chromatosome, a ∼168-bp kinetic intermediate generated during MNase digestion of native chromatin2,3. Proper in vivo positioning of H1-GFP can be assayed and directly compared to that of untagged, endogenous H1 by separating mononucleosomes containing H1-GFP from those containing endogenous H1 on nucleoprotein gels after MNase digestion (Fig. 3a)29. Mononucleosome particles denoted as M3G consist of mononucleosomes containing H1°-GFP, as demonstrated by the fluorescent signal, by the decreased migration expected to result from the increased mass of H1°-GFP relative to H1 and by the absence of any DNA-containing material in the corresponding M3G region of ethidium bromide-stained mononucleosome samples from control cells not expressing H1°-GFP (Fig. 3a). As expected in the case of proper in vivo positioning, the mononucleosomes containing H1-GFP contained ∼168 bp of DNA, as did mononucleosomes containing endogenous H1 (Fig. 3b, M3). Mononucleosomes lacking H1 (M1), used as a control, contained DNA of ∼148 bp. Notably, the mononucleosomes containing any of the H1-GFP mutants, with the exception of one (see below), contained DNA of ∼168 bp, indistinguishable in size from that isolated from mononucleosomes with endogenous H1 or wild-type H1-GFP (Fig. 3b). Therefore, H1-GFP mutants are properly positioned, albeit with lower affinity, at their specific sites in vivo. The DNA isolated from mononucleosomes containing mutant R42A was consistently slightly shorter, as confirmed by analysis of the DNA by free acrylamide electrophoresis on internally calibrated lab chips (Supplementary Fig. 3 online). Similar results have been obtained by in vitro studies of the binding of GH5 containing multiple mutations, including R42A, to oligonucleosomes20. Collectively, these results demonstrate that the mutant forms of H1-GFP confer protection of a discrete chromatosome-sized fragment of DNA. This would not be expected if these proteins were binding nonspecifically or atypically to linker DNA. We conclude that the mutant H1-GFP fusion proteins are specifically positioned on the nucleosome in the same manner as is the native protein and that their increased FRAP recovery rates reflect altered binding affinity.
Figure 3.
Mutant and wild-type H1-GFP proteins protect chromatosome-sized DNA. (a) Nuclei from parental 3T3 cells and transfectants expressing wild-type H1°-GFP were digested with MNase. Soluble chromatin was separated into nucleoprotein particles and stained with ethidium bromide (left). Particles containing H1-GFP were visualized by fluorescence excitation (right). M3G, mononucleosomes containing H1-GFP; M3, mononucleosomes containing H1; M1, mononucleosomes lacking H1. The more slowly migrating material represents di- and higher-order chromatosomes29. (b) Nucleoprotein particles from cell lines expressing mutant and wild-type H1-GFP were separated on nucleoprotein gels, then excised and electroeluted, and purified, labeled DNA was separated by PAGE. For all mutants, the pattern and mobility of mononucleosome particles as visualized by fluorescence and ethidium bromide staining was identical to that of material from cells expressing wild-type H1-GFP.
Definition of two distinct binding sites
To define the interaction surface of the H1° globular domain with the nucleosome, we mapped the positions of the crucial residues onto the atomic structure of the H5 globular domain21 (Fig. 4). The residues involved in binding are spatially clustered to form two distinct binding sites (Fig. 4). The larger, site 1, comprises His25, Arg47, Lys69, Lys73, Arg74 and Lys85; the smaller, site 2, comprises Arg42, Arg94 and Lys97. Notably, all of the binding residues in site 2 are located on flexible domains, possibly affording some latitude in binding geometry. Furthermore, all but one of the residues identified as nonbinding or as contributing little to the overall binding affinity were found to be located on a broad surface well separated from either binding site, on the opposite face of the domain (Fig. 4).
Figure 4.
Map of the interaction surface of H1° based on data from Table 1. Yellow, binding residues in site 1; green, site 2; blue, nonbinding residues.
Positioning of H1 onto the nucleosome
To determine the geometry of H1 binding to the nucleosome by molecular modeling, we took advantage of the availability of the 2.8-Å crystal structure of the core nucleosome30. The globular domain of H5 was modeled onto the 2.8-Å crystal structure of the 147 base pairs of DNA associated with the nucleosome core30 plus additional DNA to account for linker DNA associated with the chromatosome. To avoid potential bias regarding the nature of this additional DNA, modeling was performed on both straight and curved B-form DNA. The only assumptions in this approach are the experimentally verified properties that H1 binds at or near the nucleosome dyad12,13, contacts the DNA in at least two sites20 and protects 15-20 base pairs of chromatosomal DNA2,3.
Using the surface map of the globular domain, we were able to generate a model that fits all of our experimental FRAP data as well as the published information on positioning (Fig. 5). The resulting H1-nucleosome model was then stereochemically refined using 200 rounds of coordinate energy minimization. In the resulting consensus model, binding site 1 interacts with one side of the DNA approximately one helical turn away from the nucleosome dyad. Helix 3, containing binding residues Lys69, Lys73 and Arg74, fits into the major groove in a manner similar to the recognition helices of several DNA-binding proteins with which H1 shares structural similarity21,31. The additional binding residues in site 1 (His25, Lys47 and Lys85) are positioned to interact favorably with the DNA backbone. These interactions are facilitated by the curvature of DNA as it wraps around the nucleosome. By contrast, attempts to accommodate all binding residues of the primary site, site 1, in a favorable binding orientation with straight or even slightly bent DNA were unsuccessful. Site 2, consisting of Arg42, Arg94 and Arg97, interacts with one of the entering-and-exiting linker DNA strands. The site of interaction is about 15 base pairs from the end of the nucleosomal core DNA, consistent with protection of chromatosomal DNA. Finally, all residues determined to play little or no role in binding are oriented away from DNA.
Figure 5.
Molecular model of the location of H1 within the nucleosome. Blue, chromatosomal DNA; yellow, nucleosome dyad; red, the globular domain of H1.
To experimentally validate this model, we selectively mutated several residues in the β-turn structure located at the C-terminal end of the globular domain. This region is highly conserved in sequence and structure among H1 variants in many species. Our model predicts that mutation of residues within the β-turn would severely compromise binding. Indeed, mutation of Ala89 or Ser90 to aspartate resulted in a t50 of 3 s for both mutants, compared to more than 50 s for the wild-type protein, in FRAP assays (Table 1). This loss of binding is consistent with the location of these residues near site 1 at the dyad.
DISCUSSION
We have derived the interaction surface and binding geometry of the globular domain of linker histone H1 bound to chromatin, on the basis of experimental binding data from living cells combined with molecular modeling approaches. These studies have defined two distinct binding sites on the H1 globular domain, site 1 and site 2, and we propose that they interact with the major groove near the dyad axis and with the minor groove on the linker DNA, respectively, an orientation that was confirmed by targeted mutagenesis.
Our model is compatible with available information regarding the organization of nucleosomes into higher-order chromatin structures. Specifically, the binding residues in site 1 would be expected to be structurally well organized and therefore best suited for binding the conserved DNA structure found at the nucleosome dyad. In contrast, residues in site 2 are located in flexible domains that would allow some latitude in binding, to accommodate the more heterogeneous structures expected for linker DNA. This orientation of the globular domain of H1 would also place the N- and C-terminal tails of the molecule in favorable positions to interact with linker DNA distal to the chromatosome. An X-ray structure of a tetranucleosome, albeit reconstituted in the absence of linker histone, has recently been reported32. Remarkably, within the limits of resolution, our independently derived chromatosome structure is completely compatible with that of the tetranucleosome (Supplementary Fig. 4 online).
On the basis of in vitro binding experiments, models for the position of GH5 in the nucleosome have suggested that a primary binding site (Lys69, Lys73 and Lys85) interacts with the entering-and-exiting DNA and that a secondary site (Lys40, Arg42, Lys52 and Arg94) interacts with DNA near the dyad33,34. In such a scenario, residues Lys40 and Lys52 are important for binding and orienting GH5 near the dyad. However, the in vivo FRAP assays indicate that these residues do not make major contributions to binding. The much larger size and shape of site 1 identified herein is very difficult to reconcile with a model in which this site binds the entering-and-exiting DNA, requiring rather drastic distortions of the path of the DNA. Indeed, such distortions are not compatible with the architecture of linker DNA-H1 interactions as seen in EM images of chromatin6. Finally, the slightly smaller size of the chromatosomal DNA of the R42A mutant is most consistent with the location of this residue near the end of one of the entering-and-exiting strands.
Our observations also suggest the intriguing hypothesis that the globular domains of individual H1 variants might have distinct binding geometries within the nucleosome, thereby contributing to chromatin heterogeneity35,36. In particular, residues His25, Arg47 and Arg74 are strong contributors to site 1 binding in H1°, yet these residues are replaced by neutral and conserved residues in the other somatic H1 variants H1a through H1e37. Further, substitutions of the H1° binding residues with the conserved amino acids found in other H1 variants severely compromise H1° binding (Table 1). Finally, analysis of binding residues in variant H1c has revealed that several sites not conserved between H1c and H1° are indeed important for H1c binding (D.T.B., unpublished data).
The quantitative data obtained herein also suggest multiple cooperative interactions between numerous residues within the globular domain of H1 and nucleosomal DNA and/or proteins, including core histones. H1° mutants harboring multiple substitutions (Table 1), as well as those bearing deletions of entire binding domains22, have t50 values of approximately 1 s, a value approaching that of free GFP in the nucleus. Therefore, they may be considered ‘null’ or nonbinding mutants. Nonetheless, there are multiple key residues that are extremely important to binding, as mutation of, for example, Arg47, Lys69 or Lys73 alone also results in a t50 value of only ∼3 s, which approaches the null phenotype. This indicates that the contributions of these residues to binding in the wild-type protein are not additive, but rather they act in a synergistic fashion to facilitate binding.
Deletion of the entire C terminus or specific single-residue substitutions within this domain result in severely reduced residence times of human H1.1, indicating that the C terminus of H1 also contributes to binding10. These results are consistent with a biophysical analysis of a collection of deletion mutants of H1° suggesting that specific subdomains of the C terminus stabilize the structures of locally folded chromatin11. After the initial low-specificity electrostatic binding of the C terminus to linker DNA, these subdomains acquire specific structures that contribute to chromatin condensation. Notably, the subdomain most important for this process is located immediately downstream from the secondary binding site of the globular domain and, in our model, would be well positioned to induce formation of an apposed linker DNA stem6. We suggest that the specific binding geometry of the globular domain may guide or orient the C-terminal domain into a structure conducive to chromatin condensation. In this view, structural pliability resulting from the intrinsic disorder of the H1 C terminus allows for efficient but nonspecific initial capture of H1 through binding to linker DNA. This then places the globular domain near the nucleosome and, perhaps through one-dimensional scanning, promotes efficient and highly stable specific binding. Establishing stable binding of the globular domain would then promote the acquisition of structure within the C-terminal domain that facilitates chromatin condensation.
METHODS
Constructs and cell lines
Plasmid MTH1°GFPneo has been previously described29. In this plasmid, the coding sequence for enhanced GFP is fused to the C terminus of the coding region for H1°, and expression is under control of the mouse metallothionein promoter. Point mutations were introduced with the QuikChange mutagenesis kit (Stratagene) or by introduction of annealed oligonucleotides between restriction sites. Constructs were introduced into mouse BALB/c 3T3, and multiple stable transfectants were isolated and analyzed as described in ref. 22. For FRAP assays, cultures were grown in the absence of inducer. Under these conditions, H1-GFP comprises less than 5% of the total H1 population22.
FRAP assays
For FRAP, cells were plated and observed in LabTekII chambers (Nalgene). FRAP was performed on a Zeiss LSM 510 confocal microscope using the 488-nm line of an argon laser (nominal output 40 W, beam width at specimen 0.2 μm). All experiments were done at 37°C, and imaging was done with a ×100 objective, NA1.3. Scanning was bidirectional at the highest possible rate using a ×3 zoom with a pinhole of 1 Airy unit. Bleaching was achieved using two consecutive bleach scans of 49 ms duration at maximal laser power. The bleached region was 69 × 69 pixels and excluded heterogeneous nuclear features such as nucleoli and heterochromatin foci. During the recovery period, 100 images (512 × 512 pixels) were recorded until the recovery signal reached a plateau. To avoid erroneous measurements due to nonspecific bleaching during normal imaging, intervals between images during the post-bleach phase were spaced so as to maintain a constant number of images. Identical imaging settings (50% laser intensity, 0.1% attenuation) were used for all pre- and postbleach images. Normalized recovery rates were determined as described in ref. 38. All datasets consisted of at least ten cells per experiment and experiments were performed in triplicate.
Nucleoprotein analysis
Parental 3T3 cells and transfectants expressing H1°-GFP constructs were treated with ZnCl2 for 24-48 h before nuclei were isolated as previously described29. Nuclei were digested with 4 U ml-1 MNase (Sigma) for 15 min at 25 1C. The reaction was stopped and nuclei were lysed by incubation in 1 mM EDTA and 2 mM EGTA on ice for 15 min. Nuclear debris was pelleted by centrifugation at 13,000g for 15 min. Soluble chromatin was separated into nucleoprotein particles on 3.5% acrylamide/0.5% agarose gels containing 30% (v/v) glycerol exactly as described in ref. 39. Individual particles were excised from the nucleoprotein gels and electroeluted into dialysis bags. DNA was purified by phenol-chloroform extraction, labeled with [32P]ATP (ICN) and polynucleotide kinase (NEB) and separated on 6% neutral polyacrylamide gels.
Molecular modeling
The crystal structures of the nucleosome30 and the globular domain of the H5 linker histone variant (GH5)21, together with our in vivo photobleaching microscopy and systematic mutagenesis data, were used as the starting point to generate a model of the H1-nucleosome complex. A homology model of the globular domain of H1° was built from the GH5 crystal structure by homology modeling using LOOK 3.0 (Molecular Applications Group). The DNA of the nucleosome was extended using O40 to account for entering-and-exiting DNA associated with the chromatosome. The resulting H1-nucleosome model was then stereochemically refined by 200 rounds of coordinate energy minimization using CNS41.
Supplementary Material
ACKNOWLEDGMENTS
We thank D. Yellajoshyula and E. George for experimental assistance and M. Bustin and D. Sittman for discussions and critical reading of the manuscript. This work was supported by grants from the US National Science Foundation to D.T.B. (MCB0235800) and from the NIH to T.I. T.M. is a Fellow of the Keith Porter Endowment for Cell Biology. This research was in part supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research and by NIH grant RR016476 from the MFGN INBRE Program of the National Center for Research Resources.
Footnotes
COMPETING INTERESTS STATEMENT The authors declare that they have no competing financial interests.
References
- 1.Wolffe AP. Chromatin: Structure and Function. 3rd edn Academic Press; San Diego, USA: 1995. [Google Scholar]
- 2.Noll M, Kornberg RD. Action of microccoccal nuclease on chromatin and the location of histone H1. J. Mol. Biol. 1977;109:393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- 3.Simpson RT. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978;17:5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- 4.Thoma F, Koller T, Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt dependent superstructures of chromatin. J. Cell Biol. 1979;83:403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramakrishnan V. Histone H1 and chromatin higher order structure. Crit. Rev. Eukaryot. Gene Expr. 1997;7:215–230. doi: 10.1615/critreveukargeneexpr.v7.i3.20. [DOI] [PubMed] [Google Scholar]
- 6.Bednar J, et al. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc. Natl. Acad. Sci. USA. 1998;95:14173–14178. doi: 10.1073/pnas.95.24.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carruthers LM, Hansen JC. The core histone N termini function independently of linker histones during chromatin condensation. J. Biol. Chem. 2000;275:37285–37290. doi: 10.1074/jbc.M006801200. [DOI] [PubMed] [Google Scholar]
- 8.Allan J, Hartman PG, Crane-Robinson C, Aviles FX. The structure of histone H1 and its location in chromatin. Nature. 1980;288:675–679. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- 9.Allan J, Mitchell T, Harborne N, Bohm L, Crane-Robinson C. Roles of H1 domains in determining higher order chromatin structure and H1 location. J. Mol. Biol. 1986;187:591–601. doi: 10.1016/0022-2836(86)90337-2. [DOI] [PubMed] [Google Scholar]
- 10.Hendzel MJ, Lever MA, Crawford E, Th’ng JP. The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J. Biol. Chem. 2004;279:20028–20034. doi: 10.1074/jbc.M400070200. [DOI] [PubMed] [Google Scholar]
- 11.Lu X, Hansen JC. Identification of specific functional subdomains within the linker histone H1° C-terminal domain. J. Biol. Chem. 2004;279:8701–8707. doi: 10.1074/jbc.M311348200. [DOI] [PubMed] [Google Scholar]
- 12.Thomas JO. Histone H1: location and role. Curr. Opin. Cell Biol. 1999;11:312–317. doi: 10.1016/S0955-0674(99)80042-8. [DOI] [PubMed] [Google Scholar]
- 13.Travers AA. The location of linker histone in the nucleosome. Trends Biochem. Sci. 1999;24:4–7. doi: 10.1016/s0968-0004(98)01339-5. [DOI] [PubMed] [Google Scholar]
- 14.Brown DT, Gunjan A, Alexander BT, Sittman DB. Differential effect of H1 variant overproduction on gene expression is due to differences in the central globular domain. Nucleic Acids Res. 1997;25:5003–5009. doi: 10.1093/nar/25.24.5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vermaak D, Steinback OC, Dimitrov S, Rupp RA, Wolffe AP. The globular domain of histone H1 is sufficient to direct specific gene repression in early Xenopus embryos. Curr. Biol. 1998;8:533–536. doi: 10.1016/s0960-9822(98)70206-4. [DOI] [PubMed] [Google Scholar]
- 16.Cirillo LA, et al. Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. EMBO J. 1998;17:244–254. doi: 10.1093/emboj/17.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Staynov DZ, Crane-Robinson C. Footprinting of linker histones H5 and H1 on the nucleosome. EMBO J. 1988;7:3685–3691. doi: 10.1002/j.1460-2075.1988.tb03250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lambert S, et al. Neutron scattering studies of chromatosomes. Biochem. Biophys. Res. Commun. 1991;179:810–816. doi: 10.1016/0006-291x(91)91889-k. [DOI] [PubMed] [Google Scholar]
- 19.Goytisolo FA, et al. Identification of two DNA-binding sites on the globular domain of histone H5. EMBO J. 1996;15:3421–3429. [PMC free article] [PubMed] [Google Scholar]
- 20.Duggan MM, Thomas JO. Two DNA binding sites on the globular domain of histone H5 are required for binding to both bulk and 5 S reconstituted nucleosomes. J. Mol. Biol. 2000;304:21–33. doi: 10.1006/jmbi.2000.4205. [DOI] [PubMed] [Google Scholar]
- 21.Ramakrishnan V, Finch JT, Graziano V, Lee PL, Sweet RM. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature. 1993;362:219–223. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- 22.Misteli T, Gunjan A, Hock R, Bustin M, Brown DT. Dynamic binding of histone H1 to chromatin in living cells. Nature. 2000;408:877–881. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- 23.Lever MA, Th’ng JP, Sun X, Hendzel MJ. Rapid exchange of histone H1.1 on chromatin in living cells. Nature. 2000;408:873–876. doi: 10.1038/35048603. [DOI] [PubMed] [Google Scholar]
- 24.Phair RD, et al. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol. 2004;24:6393–6402. doi: 10.1128/MCB.24.14.6393-6402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheutin T, et al. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science. 2003;299:721–725. doi: 10.1126/science.1078572. [DOI] [PubMed] [Google Scholar]
- 26.Pehrson JR, Cole RD. Bovine H1° histone subfractions contain an invariant sequence which matches histone H5 rather than H1. Biochemistry. 1981;20:2298–2301. doi: 10.1021/bi00511a035. [DOI] [PubMed] [Google Scholar]
- 27.Mirzabekov AD, Pruss DV, Ebralidse KK. Chromatin superstructure-dependent crosslinking with DNA of the histone H5 residues Thr1, His25 and His62. J. Mol. Biol. 1990;211:479–491. doi: 10.1016/0022-2836(90)90366-T. [DOI] [PubMed] [Google Scholar]
- 28.Becker M, et al. Differential in vivo binding dynamics of somatic and oocyte-specific linker histones in oocytes and during ES cell nuclear transfer. Mol. Biol. Cell. 2005;16:3887–3895. doi: 10.1091/mbc.E05-04-0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gunjan A, Alexander BT, Sittman DB, Brown DT. Effects of H1 histone variant overexpression on chromatin structure. J. Biol. Chem. 1999;274:37950–37956. doi: 10.1074/jbc.274.53.37950. [DOI] [PubMed] [Google Scholar]
- 30.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 31.Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 32.Schalch T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 33.Zhou Y-B, Gerchman SE, Ramakrishnan V, Travers A, Muyldermans S. Position and orientation of the globular domain of linker histone H5 on the nucleosome. Nature. 1998;395:402–405. doi: 10.1038/26521. [DOI] [PubMed] [Google Scholar]
- 34.Bharath MM, Chandra NR, Rao MR. Molecular modeling of the chromatosome particle. Nucleic Acids Res. 2003;31:4264–4274. doi: 10.1093/nar/gkg481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alami R, et al. Mammalian linker-histone subtypes differentially affect gene expression in vivo. Proc. Natl. Acad. Sci. USA. 2003;100:5920–5925. doi: 10.1073/pnas.0736105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee H, Habas R, Abate-Shen C. Msx1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science. 2004;304:1675–1678. doi: 10.1126/science.1098096. [DOI] [PubMed] [Google Scholar]
- 37.Parseghian MH, Hamkalo BA. A compendium of the histone H1 family of somatic subtypes: an elusive cast of characters and their characteristics. Biochem. Cell Biol. 2001;79:289–304. [PubMed] [Google Scholar]
- 38.Phair RD, Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- 39.Huang S-Y, Garrard WT. Electrophoretic analyses of nucleosomes and other protein-DNA complexes. Methods Enzymol. 1979;170:116–142. doi: 10.1016/0076-6879(89)70044-6. [DOI] [PubMed] [Google Scholar]
- 40.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 41.Brunger AT, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.