Abstract
Although serum total IgE is generally elevated in atopic conditions, it is an unreliable trait for dissecting the genetic and environmental components contributing to atopic immune responses, as it can be significantly confounded by demographic factors (age, gender, race) and clinical status (atopic vs. non-atopic). Allergen-specific IgE is a discontinuous trait found only in those with sensitivity to allergens. However, all people will produce allergen-specific IgG1, which is elevated among those atopically sensitized to specific allergens. We screened 91 Caucasian nuclear families (total N = 367) with medical histories of atopic diseases, and used variance components analysis to compare heritability estimates for total IgE and IgG1 produced against the common major allergen from house dust mite, Der p 1. An estimate of total IgE heritability was ≈ 48%, although this was significantly confounded by age, gender and clinical atopic status. In contrast, Der p 1-IgG1 showed a significant inherited component of ≈ 62% that was not influenced by age, gender or clinical status. For genetic studies of atopic humoral responses, allergen-specific IgG1 may be a more reliable quantitative trait than serum IgE. Moreover, atopy is an inherited deregulation of immune responses to non-infectious antigens involving antibody isotypes other than IgE.
Keywords: Allergen, Atopy, IgE, Allergen-IgG1, Variance Components
INTRODUCTION
Atopic disorders, such as allergic rhinitis or atopic bronchial asthma, are common, complex human diseases that result from adverse immune system responses to non-infectious environmental antigens (allergens). While it has been shown that there is an inherited component for these conditions, they are also influenced by numerous environmental factors. As a result, quantitative traits associated with these diseases show a great amount of phenotypic variability.
Genetic and environmental components can be dissected by comparisons of trait values among kinships by regression statistics or comparable analyses. Employing these methods assumes that the traits are continuously distributed, that there are detectable, measurable trait levels in all people screened and the sources of variation are known or can be reasonably assumed, Among the most frequently used quantitative traits for genetic studies of atopy is serum total IgE, as this is often elevated with atopic disease.
However, the use of total IgE as a quantitative trait for genetic studies of atopy has produced widely conflicting results, with evidence for co-dominant genetic effects [1], recessive effects [2–4], pleiotropic effects [5] and other modes of inheritance [6–10]. Further, it has not been clearly established that enhanced total IgE production is a predisposing cause for atopy, as it may only be a consequence that accompanies clinical conditions, so the physiological ‘sources’ of total IgE are unclear.
We have previously argued that serum total IgE consists of at least two physiologically distinct components [11]. One of these is a basal (‘non-cognate’) component apparently unrelated to atopic disease, while a second arises from atopic sensitization to allergens resulting in allergen-specific (‘cognate’) IgE. Yet, this cognate IgE fraction is not a reliable marker for genetic studies as it is a discontinuous trait that is only produced by those with atopic sensitivity to allergens. It is influenced by the number of allergens to which these individuals are sensitized [11], and the number and specific kinds of allergens to which people are sensitized are random variables [12]. Thus, specific IgE is not a useful marker for dissecting the genetic and environmental components related to atopic immune responses using regression based methods.
Aside from clinical associations, production of allergen-specific IgE is the result of an adaptive, memory-forming humoral immune response to non-infectious environmental antigens (allergens). Save for specific IgE production, it has been shown that it is equally likely for anyone to mount a T cell-dependent humoral response to allergens involving varying amounts of the IgA and IgG isotype subclasses [13–15]. In particular, both specific IgA and IgG1 are elevated among those with atopic sensitization to a particular allergen.
IgA production is chiefly confined to the mucosa, making it difficult to evaluate as a quantitative trait for genetic studies. In contrast, IgG1 derives from isotype class switch recombination mechanisms in lymph node germinal centers with the subsequent generation of long-lived memory B and plasma cells that are available to produce IgG1 that is released into the peripheral circulation [16, 17]. Specific IgE production presumably arises and is produced in a comparable fashion. And, it is possible that elevated specific IgG1 among those with atopic sensitivity to allergens parallels the production of specific IgE, as both are influenced by similar metabolic controls like the cytokine IL-4 [18] or neuro-endocrine modulators such as β2-adrenergic agonists [19].
To evaluate allergen-specific IgG1 as a quantitative marker for genetic studies of atopy-associated immune responses, we screened small nuclear families with medical history of atopic diseases. We employed variance components statistical analyses to evaluate heritability for serum total IgE and dust must-specific IgG1. We chose dust mite as a model, as dust mites are a source of a common indoor, non-seasonal allergen that is ubiquitously distributed and to which most (all) people are continually exposed. We also included an antigen-specific antibody to tetanus toxoid (TT), TT-IgG1, to demonstrate that inheritance of specific antibody responses is not confined to allergens alone.
The results show that specific IgG1 provides more parsimonious estimates of heritability for genetic studies on immune responses resulting from allergen sensitization than total IgE, as it is not significantly confounded by demographic co-factors or clinical atopic status unrelated to dust mite sensitivity. Also, the results suggest that the humoral response associated with atopy is a more generalized immune system deregulation involving antibody isotype production other than IgE alone.
METHODS
Study Population
Caucasian families residing in the Minneapolis/St. Paul, Minnesota metropolitan area that included two children with a physician's diagnosis of asthma were recruited through media advertisements as part of studies for the genetics of asthma. Greater than 2/3 of this population had evidence of allergen sensitivity by standard allergen-specific skin prick test (SPT). Each person, or legal guardian, provided written, informed consent according to the guidelines of the Institutional Review Board of the University of Minnesota.
We evaluated 367 people from 91 nuclear families that included both parents, the asthmatic children and their siblings. A total of 169 parents (mean age = 43.7 years; range = 22–75 years) and 198 children (mean age = 14.9 years; range = 6–42 years) were screened. There were 176 females and 191 males. All participants had been previously inoculated against tetanus toxoid antigen.
Asthma and Pulmonary Function Screens
Each subject was given a nurse-administered, standardized questionnaire, prepared according to American Thoracic Society guidelines, regarding history of asthma and other respiratory diseases as previously described [20]. Baseline spirometry measures for FEV1 (Forced Expiratory Volume at 1 second) were made according to American Thoracic Society guidelines, followed by provocative methacholine challenges [20]. Bronchial hyperreactivity (BHR) was recorded if there was a decrease of 20% or more in FEV1 after methacholine challenge (≤ 25 mg/ml maximum dosage), or reversibility of 15% or greater after administration of albuterol.
IgE Screens
(1.) Serum Total IgE Tests
Serum total IgE tests were done using the ACCESS Immunoassay System® (Beckman Coulter, Chaska, MN), according to manufacturer's specifications. All IgE results are reported as IU/ml (1 IU = 2.4 ng).
(2.) Skin Prick Tests (SPT)
Skin prick testing was done as previously described [20] using 14 standardized allergen extracts for dust mites (including Der p allergen extract) (2), ragweed, molds (3), trees (4), grasses (2), animal danders (2). A positive SPT result was recorded for a wheal area ≥ 9 mm2 above negative saline control.
Der p 1 Allergen
The major allergen Der p 1 from house dust mite was purified by methods previously described [21]. We chose Der p 1 allergen as a model, as we have previously shown that the atopic response to any particular allergen is not obligately inherited. Rather, it is a random variable independent of any family history of atopic allergen sensitivity [12]. Der p 1 is a common non-seasonal indoor allergen that, for this study population, induced the highest incidence of atopic sensitization among all of the common indoor allergens that were tested in the SPT screens.
ELISA’s for Serum Der p 1-IgG1 and Tetanus Toxoid (TT)-IgG1
Measures of Der p 1-specific IgG1 were done using modifications of ‘reverse sandwich’ ELISA assays previously described [15, 22]. Briefly, a “capture antibody” was an isotype-specific monoclonal antibody (mouse anti-human and anti-IgG1; Sigma, St. Louis, MO) that eliminates possible cross-reactivity with other isotypes. This was bound to wells of microtest plates overnight at 20 °C. Non-specific binding was blocked using 2% BSA in PBS. Serum or serially diluted standards were added for 2 hours at 37 °C. For serum test samples, “captured” IgG1 was evaluated for the presence of Der p 1- specific IgG1 by addition of biotinylated Der p 1 standard (Indoor Biotechnologies, Charlottesville, NC). Calibration of the assay was done using human serum immunoglobulin calibrators (The Binding Site, Birmingham, UK). “Detection antibody” for the calibrators was mouse, anti-human IgG1 (Sigma). Controls for assay validation were from a serum pool with known concentrations of Der p 1-IgG1 maintained by us. A biotinylated “detection antibody” was added for 1 hour at 37 °C. Optimized concentrations of streptavidin-conjugated alkaline phosphatase (Sigma) were added for 1 hour at room temperature. A chemiluminescent substrate (Lumi-Phos 550®, Beckman Coulter, Chaska, MN) was added followed by reading the test signals with a commercial luminometer (Tropix, Inc., Bedford, MA). This assay gave a linear response on a log-log scale over the range of 0.05 to 100 ng/ml.
For TT-IgG1 assays, we modified the assay described by Sedgwick et al [23] using a “direct binding” ELISA. Prior to use, tetanus toxoid (Sigma) was dialyzed (10,000 MW cut-off) against 50 mM borate buffer, pH 8.5 overnight. This preparation was diluted 1:1000 in 50 mM carbonate buffer, pH 9.6, and coated to microtest plates wells overnight. Non-specific binding sites were blocked with 2%BSA. Serum (1:100 dilution) was added for 1 hour @ 37 °C. “Detection antibody” was alkaline phosphatase-conjugated mouse, anti-human IgG1 mAb (Zymed, S. San Francisco, CA. Calculations were done as described by Sedgwick et al [23], which provided a linear response in arbitrary chemiluminescent units.
Statistics
Comparisons between groups categorized by demographics or clinical status were made by two-sided t-tests (significance level α = 0.05) or analysis of variance (ANOVA) using commercial software (Microsoft Excel® and SPSS®). Age effects for serum antibody concentrations were assessed by simple linear regression (SLR) and evaluated by ANOVA.
Variance components analysis was used to partition total phenotypic variance (VP) into additive genetic (VA) and non-genetic, environmental components (VE): VP = VA + VE. The environmental component was further partitioned into two components for common family environment, VCE, and common sibling effects, VCS. The narrow-sense heritability (h2N) was defined as the ratio of the variance due to additive genetic effects to all sources of phenotype variance: h2N = VA/( VA + VCE + VCS).
As previously described by Palmer et al [24], the variance components were determined from calculations of the covariances (COV) about common mean values for quantitative traits for three kinship pairings (spouse-spouse, parent-child and sib-sib). This results in three algebraic equations that can be simultaneously solved for three unknown values, namely:
COV (Spouse-Spouse) = VCE
COV (Parent-Child) = ½ VA + VCE
COV (Sibling-Sibling) = ½ VA + VCE + VCS
Implementation of the variance components analyses, and estimates of the narrow-sense heritability, including estimates for the effects of the co-factors of age and gender, were obtained using SOLAR, version 2.1.4, as first described by Almasy and Blangero [25] (Southwest Foundation for Biomedical Research, San Antonio, TX: http://www.sfbr.org/solar).
RESULTS
Serum Antibody Concentrations: Age and Gender Effects
A total of 367 individuals from 91 Caucasian nuclear families with history of atopic asthma were screened. On logarithmic scales, concentrations of serum total IgE, Der p 1-IgG1 and TT-IgG1 were approximately normally distributed in this study population.
By simple linear regression, there was a significant age effect for Log [Total IgE] values: Log [Total IgE] = 2.24 – 0.013 x Age; F (1,365) = 29.2; p ≪ 0.001.
In contrast, there were no significant age effects for either Log [Der p 1-IgG1] or Log [TT-IgG1] values.
Log [Der p 1-IgG1] = −0.75 + 0.001 x Age; F (1, 365) = 0.009; p = 0.93.
Log [TT-IgG1] = −0.17 – 0.002 x Age; F (1, 365) = 1.92; p = 0.17.
To account for gender effects for Log [Total IgE] values, subjects were first classified as being either skin prick test positive (SPT [+]) or SPT [−] because, as expected, there were significant differences in Log [Total IgE] values based upon these classifications. For those with any SPT [+] result, the mean value for Log [Total IgE] = 2.13 (S.D. = 0.64; N = 262). For those who were SPT [−], mean Log [Total IgE] = 1.28 (S.D. = 0.60; N = 105). By two-sided t-test, these mean values were significantly different: t(205) = 12.1; p ≪ 0.001.
For females who were SPT [−], mean Log [Total IgE] = 1.18 (S.D. = 0.62; N = 57). For males who were SPT [−], mean Log [Total IgE] = 1.41 (S.D. = 0.55; N = 48). These mean values were significantly different: t(103) = 2.01; p = 0.024.
For females who had any SPT [+] result, mean Log [Total IgE] = 2.05 (S.D. = 0.68; N = 119). For SPT [+] males, mean Log [Total IgE] = 2.20 (S.D. = 0.60; N = 143). These mean values were also significantly different: t(239) = 1.82; p = 0.035.
As shown in the next section of Results, mean Log [Der p1-IgG1] and Log [TT-IgG1] values were not affected by SPT results, save for Der p SPT results on Log [Der p 1-IgG1]. However, there were no gender effects for these antibody measures.
Log [Der p 1-IgG1]: Mean for females = −0.75 (S.D. = 0.50; N = 176). Mean for males = −0.75 (S.D. = 0.46; N = 191). t(365) = 0.04; p = 0.49.
Log [TT-IgG1]: Mean for females = −0.24 (S.D. = 0.36; N = 176). Mean for males = −0.20 (S.D. = 0.39; N = 191). t(365) = 0.91; p = 0.18.
To be used in genetic studies employing variance components analysis, serum total IgE values must be adjusted for age and sex, as well as evidence or not for sensitivity to allergens. In contrast, Der p 1-IgG1 and TT-IgG1 values are not appreciably confounded by age or sex.
Serum Antibody Concentrations: Effects of Clinical Status
In addition to age and gender effects, serum antibody concentrations can be influenced by clinical atopic status. For this study, each person was screened for asthma symptoms and bronchial hyperreactivity (BHR) by standardized spirometry. They were also screened for atopic sensitivity by percutaneous skin prick test (SPT) using a battery of 14 allergens, including house dust mite (Der p) extract.
We first subdivided the study population into four categories based upon results for asthma symptoms and BHR, or any SPT [+] result. Table 1 shows the results for the serum antibody concentrations for these categories.
Table 1.
Clinical Status And Serum Antibody Concentrations
| Asthma/BHR1 | SPT [+]2 | N | Total IgE3 | Der p 1-IgG13 | TT-IgG13 |
|---|---|---|---|---|---|
| Neg | Neg | 54 | 1.15 (0.52)4 | −0.72 (0.48) | −0.25 (0.31) |
| Neg | Pos | 71 | 1.84 (0.64) | −0.68 (0.49) | −0.23 (0.38) |
| Pos | Neg | 51 | 1.42 (0.64) | −0.78 (0.46) | −0.19 (0.43) |
| Pos | Pos | 191 | 2.24 (0.61) | −0.77 (0.47) | −0.22 (0.38) |
| F (3, 363)5 | 58.0 | 0.77 | 0.25 | ||
| p-value | ≪ 0.001 | 0.51 | 0.86 |
Asthma symptoms and bronchial hyperreactivity (BHR).
Any skin prick test (SPT) positive result.
Values are Base 10 Logarithms.
Mean (Std. Dev.)
F-ratio from ANOVA by Column.
By analysis of variance (ANOVA), the mean values for Log [Der p 1-IgG1] and Log [TT-IgG1] were not significantly different based upon these clinical categorizations. However, Log [Total IgE] values differed significantly.
As shown in Table 1, for those with any SPT [+] result, Log [Total IgE] values were different for those with or without asthma symptoms plus BHR [+] results. By two-sided t-test, these mean values were significantly different: t (119) = 4.50; p ≪ 0.001. A similar trend was observed for those who were SPT [−]: t (97) = 2.32; p = 0.011. Serum total IgE values are influenced by clinical respiratory status, while Der p 1-IgG1 and TT-IgG1 levels are not.
We next categorized subjects according to SPT results, and dust mite (Der p) sensitivity in particular. This resulted in three categories as shown in Table 2. By ANOVA, serum Log [Total IgE] values were significantly different for these categories, but Log [TT-IgG1] values were not.
Table 2.
Atopic Status And Serum Antibody Concentrations
| SPT [+]1 | Der p [+]2 | N | Total IgE3 | Der p 1-IgG13 | TT-IgG13 |
|---|---|---|---|---|---|
| Yes | Yes | 122 | 2.29 (0.59)4 | −0.64 (0.51) | −0.26 (0.35) |
| Yes | No | 140 | 2.00 (0.65) | −0.84 (0.43) | −0.19 (0.40) |
| No | No | 105 | 1.28 (0.60) | −0.75 (0.47) | −0.22 (0.37) |
| F (3, 363)5 | 78.4 | 6.16 | 0.99 | ||
| p-value | ≪ 0.001 | 0.002 | 0.37 |
Any skin prick test (SPT) positive result
SPT [+] for Der p allergen.
Values are Base 10 Logarithms.
Mean (Std. Dev.)
F-ratio from ANOVA by Column.
Log [Der p 1-IgG1] values were significantly different. However, unlike total IgE, this was not due to generalized sensitivity to allergens (ant SPT [+] result). For those who had any SPT [+] result, mean Log [Der p 1-IgG1] = −0.74 (std. dev. = 0.48; N = 262). For those who were SPT [−], mean Log [Der p 1-IgG1] = −0.75 (std. dev. = 0.47; N = 105). By two-sided t-test, the mean values were not significantly different: t (365) = 0.08; p = 0.93.
In contrast, for those who had a positive SPT result for the Der p dust mite extract, mean Log [Der p 1-IgG1] = −0.64 (std. dev. = 0.51; N = 122). For those who were SPT [−] for the Der p extract, mean Log [Der p 1-IgG1] = −0.80 (std. dev. = 0.45; N = 245). By two-sided t-test, these mean values were significantly different: t (365) = 3.17; p = 0.0008.
Figure 1 shows the distributions of the Log [Der p 1-IgG1] values, with the % of cases plotted against Log [Der p 1-IgG1] values. There is significant overlap of the values for those who were Der p [+] (solid circles, solid line) compared to those who were Der p [−] (open circles, dashed line). This situation is analogous to what we have observed previously for serum total IgE values for those who are SPT [+] compared to those who are SPT [−] (Figure 1 in ref. 11); there is considerable overlap in the expressions of the trait levels although they arise from separate mechanisms.
Figure 1.
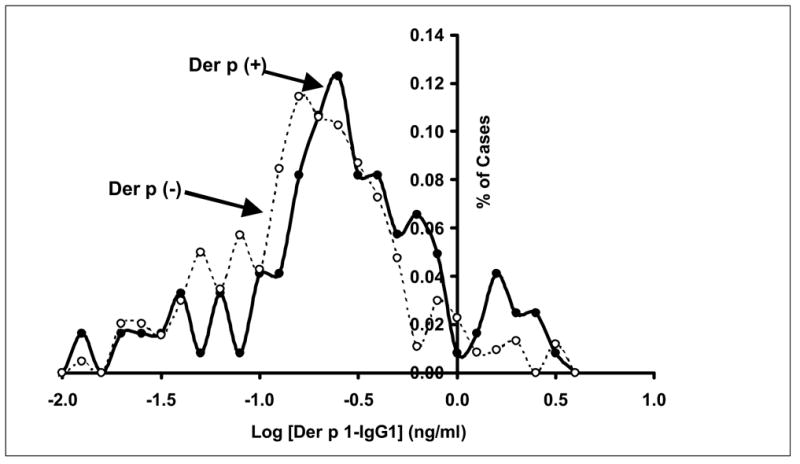
Serum Der p 1-IgG1 Distributions: Der p [+] vs. Der p [−]. The % of cases are plotted against serum Log10 [Der p 1-IgG1] for those who were SPT [+] to the Der p allergen extract (Der p (+); solid circles, solid line) or those who were SPT [−] to this extract (Der p (−); open circles, dashed line). For Der p (+): mean = −0.65 (std. dev. = 0.51; N = 122). For Der p (−): mean = −0.80 (std. dev. = 0.45; N = 245). Abbreviations: Der p = Dermatophagoides pteronyssinus (dust mite allergen extract); SPT = (percutaneous) skin prick test.
Serum total IgE levels are influenced by clinical respiratory status and atopic sensitivity to allergens, while Der p 1-IgG1 levels are only influenced by specific sensitivity to the dust mite allergen.
Heritability Estimates For Serum Antibody Concentrations
We used pedigree-based variance component analysis as implemented in SOLAR version 2.1.4 [25] to estimate the heritability of immune responses reflected by logarithmic values of serum total IgE, Der p 1-IgG1 and TT-IgG1. Heritability was calculated by comparing the covariance of each of these traits among related individuals with the overall variance of the trait. Each of the traits was adjusted for age and sex.
Table 3 gives the results of these analyses, with the estimated heritablity, its standard error of estimation and the probabilities that these estimates were influenced by age and gender. Statistically significant estimates of heritability were found for the three serum antibody concentrations, reflected by the p-values of these estimates. Both age and sex did not contribute significantly on Derp1-IgG1 and TT-IgG1 (Table 3) and hence were removed from the final analysis.
Table 3.
Heritability Estimates For Serum Antibody Concentrations
| Total IgE | Der p 1-IgG1 | TT-IgG1 | |
|---|---|---|---|
| A. Heritability | |||
| Estimated Value | 0.478 | 0.615 | 0.687 |
| S.E. of Estimate | 0.103 | 0.079 | 0.080 |
| p-Value of Estimate | 8.00x10−7 | 2.21x10−15 | 1.22x10−14 |
| B. Co-Factors | |||
| p-Value for Age Effect | 4.01x10−10 | 0.66 | 0.49 |
| p-Value for Gender Effect | 0.004 | 0.58 | 0.71 |
The estimated heritability of total IgE was lower than those for Der p 1-IgG1 and TT-IgG1. However, the source of production for IgE cannot be determined with any certainty. As shown above, serum total IgE values were significantly influenced in this study population by sensitivity or not to allergens and clinical respiratory status. Der p 1-IgG1 and TT-IgG1 values were independent of respiratory status, and Der p 1-IgG1 values were only elevated among those with evidence of sensitivity to the Der p allergen. Whereas the IgG1 values are reflective of humoral response capabilities to defined antigens, total IgE could have arisen from one or more sources (e.g. basal IgE, allergen-specific specific IgE or IgE associated with respiratory status).
We included heritability assessments for the tetanus toxoid antibody, TT-IgG1, to demonstrate that there can be a significant inherited component involving antigen-specific antibodies other than those produced against allergens. Although the heritability estimates for Der p 1- and TT-IgG1 were similar, these arose from independent physiological mechanisms as there was no association between these serum antibody concentrations as assessed by simple correlation; r = 0.050; t (366) = 0.93; p = 0.35. These responses do not arise from inter-human infectious transmission, and only the Der p 1 allergen is known to be associated with atopic immune responses.
Finally, the heritability for Der p 1-IgG1 responses were equally likely to arise from either parent, as parent-child correlations were similar for mothers and fathers. Mother-Child: r = 0.52; t (96) = 5.98; p ≪ 0.001. Father-Child: r = 0.53; t (97) = 6.19; p ≪ 0.001.
DISCUSSION
Although the results of this study may not directly relate to genetic factors contributing to atopic predisposition, two observations merit discussion. First, aside from genetic atopic predisposition, allergen-specific IgG1 provides parsimonious estimates for the genetic factors related to atopy-associated humoral immune responses to non-infectious environmental allergens, as it is not significantly effected by demographic co-factors. Its source is clearly defined, arising from a specific immune response to a defined antigen, whereas the physiological ‘sources’ of total IgE are unclear. Second, the genetic components of atopic sensitization to allergens contribute to a more generalized immune system deregulation influencing antibody isotype production other than IgE.
These findings are in contrast to those of other studies in which only IgE measures were used, particularly those in which it was implicitly assumed that atopic sensitization to allergens is an IgE-related phenomenon only. Using total IgE as a quantitative trait for genetic studies of atopy has produced widely conflicting results, particularly with regard to the proportions of the responses putatively attributed to inheritance [1–10]. Although controlled by tight metabolic regulation, the production of IgE involves multiple pathways [26–28], but the details of these interrelationships are not completely understood.
Epidemiological studies have shown that serum total IgE levels are significantly influenced by both demographic factors, like age, gender and race, as well as atopic clinical status [29–32], which undoubtedly confound the studies of inheritance. This was also observed in our study population, as shown in the results of Tables 1 and 2. Whereas the IgG1 values reflect immune response capabilities to defined antigens, it cannot be determined with any certainty what the sources are for IgE production.
The measured serum IgE values could conceivably arise from three, possibly interdependent, sources as either basal IgE production unrelated to atopic disease (“non-cognate fraction”), specific IgE directed against one or more allergens (“cognate fraction”) [11] or elevated IgE production due to atopic respiratory conditions (“ectopic fraction”). Attempting to account for one or more of these factors suggest that IgE production is influenced by multiple genes, some of which may not be related to immune system regulatory elements [24, 33, 34].
A comprehensive survey of genetic studies on atopy-related phenotypes showed that no less than 64 genes or chromosomal regions have been implicated in these disorders, of which roughly 50–60% were immune regulatory genes, including inflammatory and antibody production promoters, while the remainder had no apparent association with immune system regulation [34]. These complex scenarios argue against any obligatory inherited pathway(s) leading to an atopic outcome.
As we have previously argued [12], the atopic predisposition more likely arises from a “stochastic bias” model. In this scenario, it is equally likely for anyone to mount an immune response to allergens, but the specific outcome is a random event. The specific outcome is dependent upon the interplay of numerous genetic and environmental elements that, in certain circumstances, may “skew” a response towards atopy, while others experiencing similar conditions remain clinically unaffected. A first step toward unraveling this complexity is to understand the commonality of the atopic and non-atopic humoral responses, as opposed to attempting to clearly delineate the presumed differences.
We have previously shown among randomly selected individuals, that those with and without atopic sensitization to a particular allergen can mount an equally vigorous humoral immune response against that allergen [15]. The atopic response was characterized by the production of specific IgE with very high equilibrium binding affinity for the allergen and an attenuated, lower affinity specific IgG1 response. In contrast, the non-atopic response was characterized by specific IgG1 with an equilibrium binding affinity for the allergen comparable to that of the atopic IgE response.
We have also found that in the earliest years of life, when most atopic humoral responses develop, that there is a non-linear increase in the age-dependent ‘developmental trajectories’ of humoral responses involving specific IgE among atopic children and specific IgG1 among non-atopic children are remarkably similar [35]. Interestingly, this study also showed that the atopy-associated specific IgG1 response was ‘deregulated’ compared to the IgG1 response among non-atopic children, as there was no comparable non-linear increase with age. Rather, the vigor of the atopic IgG1 response remained relatively unchanged, suggesting again that stochastic features among immune regulators favored a vigorous IgE response at the expense of a vigorous IgG1 response among those children with an atopic predisposition.
From these results we have surmised that atopy may only be an ‘error’ in isotype class switching during an otherwise normal process of antigen recognition and humoral response development. If this is so, then as shown in Table 3, about 60% of the factors that contribute to this ‘error’ are inherited. This does not imply, however, that these factors will be common to all people with the atopic predisposition, or that families with similar atopic histories will also share the same genetic complement predisposing to atopy. Again, from the literature survey by Hoffjan et al [34], there are numerous genes that may contribute to an atopic immune response. By probability alone, it is extremely unlikely that all people with atopy or all families with atopic histories could possibly share the same combinations of these genes.
Recent evidence suggests that single nucleotide polymorphisms (SNP’s) in the promoter regions of immune regulatory genes can profoundly impact upon the specific details of an immune response to allergens [36]. Three specific examples include SNP’s in the CD14 endotoxin receptor gene that influences both allergen-specific IgE andIgG1 production among those with atopy [37], SNP’s in the promoter region for ICOS (inducible co-factor of stimulation) that mediates T cell-B cell interaction during humoral response development [38] and CTLA-4 gene promoter SNP’s that also impacts upon T cell-B cell interaction [39, 40].
Given that each of these gene promoter elements can have at least one of three genotypes (e.g. C/C, C/T or T/T for the CD14 promoter), then there are at least 3 x 3 x 3 = 27 possible combinations taking all three together. If only one of these combinations is the “correct” one for atopy, then in the total population there is about a 1/27 ≈ 4% chance of an atopic response, which clearly does not conform to the current atopy incidence rates in industrialized nations of about 25–40%. Of course, any number of the 27 possible combinations could be proposed to be “correct” to account for current atopy incidence, but assuming that any of these combinations will be exactly represented in all atopic families is, again, extremely unlikely.
What is more likely, however, is that IgE and IgG1 production may have more immune regulators in common with one another than are disparate, and a more thorough search among these regulators might provide clues to the probability of isotype switching ‘errors.’ For example, considerable attention has been paid to the genes in the “cytokine cluster” region on human chromosome 5q31 [41] that includes genes for IL-4, CD14 and the beta-2 adrenergic receptor (β2-AR) among others.
By focusing upon this region it was hoped that gene alleles would become evident that promote enhanced IgE production, and provide clues to the atopic predisposition. However, the cytokine IL-4 is a critical antibody isotype ‘switch factor’ for both IgG1 and IgE [18], CD14 modulates both allergen-specific IgG1 and IgE responses [37] and β2-AR agonists have been shown to enhance the amount of IgG1 and IgE produced per B cell in vitro [19]. Any one alone or in combination could account for enhanced production of both specific IgG1 and IgE.
However, the dynamics of humoral response development are not totally accounted for by isotype switching and the amount of the particular isotype that is produced. Somatic hypermutation of the genes encoding for the variable heavy and light chains of antibodies are also involved, which leads to enhanced antibody binding to antigen during ‘affinity maturation’ in germinal center reactions [42].
The fully developed humoral response is a function of both the concentration of the particular antibody isotype, [IgX], and the binding affinity, K, of this antibody for the antigen that induced its production. The mathematical production of these two variables, that we have called the total antibody binding capacity (CAP = [IgX] x K), is the variable that clearly distinguishes the atopic from the non-atopic humoral response [15]. It is the age-dependent, nonlinear increase in total IgG1 and IgE capacities that are comparable in non-atopic and atopic children [35].
Both the atopic and the non-atopic humoral responses are the result of the interactions among numerous genes, that control isotype class switching, the amount of the particular isotype that is produced and factors involved in affinity maturation. It has been argued that the mechanisms of isotype class switching and affinity maturation within lymph node germinal centers are independently regulated [43]. In effect, the particular antibody repertoire that develops in any given circumstance is a matter of most probable outcomes, akin to Darwinian competitive selection on a microscopic scale [15]. From the results of the present study, allergen-specific IgG1 is a robust, and convenient, quantitative marker to be used to unravel the genetic complexities of the atopy-associated humoral response to allergens.
Acknowledgments
This work was supported by N.I.H. Grant 2 RO1-HL049609-11, and the Asthma and Allergy Research Fund, Department of Medicine, University of Minnesota (Malcolm N. Blumenthal, M.D., Director). This is manuscript # MSP-003-2006 of the Asthma & Allergy Center, University of Minnesota. We gratefully acknowledge Andreas Rosenberg, Emeritus Professor, University of Minnesota, for a critical reading of the manuscript.
ABBREVIATIONS
- BHR
Bronchial Hyperreactivity
- Der p 1
Major house dust mite allergen (Dermatophagoides pteronyssinus)
- IgG1
Immunoglobulin G1
- IgE
Immunoglobulin E
- SPT
(percutaneous) Skin Prick Test
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Martinez FD, Holberg CJ, Halonen M, Morgan WJ, Wright AL, Taussig LM. Evidence for Mendelian inheritance of serum IgE levels in Hispanic and non-Hispanic white families. Am J Hum Genet. 1994;55:555. [PMC free article] [PubMed] [Google Scholar]
- 2.Meyers DA, Beaty TH, Colyer CR, Marsh DG. Genetics of total serum IgE: regressive model approach to segregation analysis. Genet Epidemiol. 1991;8:351. doi: 10.1002/gepi.1370080507. [DOI] [PubMed] [Google Scholar]
- 3.Dizier MH, Hill M, James A, Faux J, Ryan G, le Souef P, Lathrop M, Musk AW, Demenais F, Cookson W. Detection of a recessive major gene for high IgE levels acting independently of specific response to allergens. Genet Epidemiol. 1995;12:93. doi: 10.1002/gepi.1370120109. [DOI] [PubMed] [Google Scholar]
- 4.Palmer LJ, Cookson WOCM, James AL, Musk AW, Burton PR. Gibbs sampling-based segregation analysis of asthma-associated quantitative traits in a population-based sample of nuclear families. Genet Epidemiol. 2001;20:356. doi: 10.1002/gepi.6. [DOI] [PubMed] [Google Scholar]
- 5.Borecki IB, Rao DC, Lalouel JM, McGue M, Gerrard JW. Demonstration of a common major gene with pleiotropic effects in immunoglobulin E levels and allergy. Genet Epidemiol. 1985;2:327. doi: 10.1002/gepi.1370020402. [DOI] [PubMed] [Google Scholar]
- 6.Postma DS, Bleecker ER, Amelung PJ, Holroyd KJ, Xu J, Panhuysen CIM, Meyers DA, Levitt RC. Genetic susceptibility to asthma-bronchial hyperresponsiveness coinherited with a major gene for atopy. New Engl J Med. 1995;333:894. doi: 10.1056/NEJM199510053331402. [DOI] [PubMed] [Google Scholar]
- 7.Xu J, Levitt RC, Panhuysen CIM, Postma DS, Taylor EW, Amelung PJ, Holroyd KJ, Bleecker ER, Meyers DA. Evidence for two unlinked loci regulating total serum IgE levels. Am J Hum Genet. 1995;57:425. [PMC free article] [PubMed] [Google Scholar]
- 8.Blumenthal MN, Namboodiri K, Mendell N, Gleich G, Elston RC, Yunis E. Genetic transmission of serum IgE levels. Am J Med Genet. 1981;10:219. doi: 10.1002/ajmg.1320100304. [DOI] [PubMed] [Google Scholar]
- 9.Hanson B, McGue M, Roitman-Johnson B, Segal NL, Bouchard TJ, Jr, Blumenthal MN. Atopic disease and immunoglobulin E in twins reared apart and together. Am J Hum Genet. 1991;48:873. [PMC free article] [PubMed] [Google Scholar]
- 10.Gerrard JW, Rao DC, Morton NE. A genetic study of immunoglobulin. E Am J Hum Genet. 1978;30:46. [PMC free article] [PubMed] [Google Scholar]
- 11.Jackola DR, Blumenthal MN, Rosenberg A. Evidence for two independent distributions of serum immunoglobulin E in atopic families: Cognate and Non-Cognate IgE. Human Immunol. 2004;65:20. doi: 10.1016/j.humimm.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 12.Jackola DR, Liebeler CL, Blumenthal MN, Rosenberg A. Random outcomes of allergen-specific responses in atopic families. Clin Exp Allergy. 2004;34:540. doi: 10.1111/j.1365-2222.2004.1920.x. [DOI] [PubMed] [Google Scholar]
- 13.Batard T, Basuyaux B, Lambin P, Bremard-Oury C, Hamilton RG, David B, Peltre G. Isotypic analysis of grass pollen-specific immunoglobulins in human plasma. 1 Specialization of certain classes and subclasses in the immune response. Int Arch Allergy Immunol. 1993a;100:68. doi: 10.1159/000236390. [DOI] [PubMed] [Google Scholar]
- 14.Batard T, Basuyaux B, Laroze A, Lambin P, Bremard-Oury C, Aucouturier P, Hamilton RG, David B, Peltre G. isotypic analysis of grass-pollen-specific immunoglobulins in human plasma. 2 Quantification of the IgE, IgM, IgA class and the IgG subclass antibodies. Int Arch Allergy Immunol. 1993b;102:279. doi: 10.1159/000236537. [DOI] [PubMed] [Google Scholar]
- 15.Jackola DR, Pierson-Mullany LK, Liebeler CL, Blumenthal MN, Rosenberg A. Variable binding affinities for allergen suggests a ‘selective competition’ among immunoglobulins in atopic and non-atopic humans. Molec Immunol. 2002;39:367. doi: 10.1016/s0161-5890(02)00108-6. [DOI] [PubMed] [Google Scholar]
- 16.Slifka MK, Ahmed R. Long-lived plasma cells: a mechanism for maintaining persistent antibody production. Curr Opin Immunol. 1998;10:252. doi: 10.1016/s0952-7915(98)80162-3. [DOI] [PubMed] [Google Scholar]
- 17.McHeyzer-Williams MG, Ahmed R. B cell memory and the long-lived plasma cell. Curr Opin Immunol. 1999;11:172. doi: 10.1016/s0952-7915(99)80029-6. [DOI] [PubMed] [Google Scholar]
- 18.Stavnezer J. Molecular processes that regulate class switching. Curr Topics Micro Immunol. 2000;245:127. doi: 10.1007/978-3-642-59641-4_6. [DOI] [PubMed] [Google Scholar]
- 19.Kasprowicz DJ, Kohm AP, Berton MT, Chruscinski AJ, Sharpe A, Sanders VM. Stimulation of the B cell receptor, CD86 (B7-2), and the β2-adrenergioc receptor intrinsically modulates the level of IgG1 and IgE produced per B cell. J Immunol. 2000;165:680. doi: 10.4049/jimmunol.165.2.680. [DOI] [PubMed] [Google Scholar]
- 20.CSGA (Collaborative Study on the Genetics of Asthma) A genome-wide search for asthma susceptibility loci in ethnically diverse populations. Nature Genetics. 1997;15:389. doi: 10.1038/ng0497-389. [DOI] [PubMed] [Google Scholar]
- 21.Pierson LK, Allauzen S, Blumenthal MN, Rosenberg A. An automated method for determination of antibody affinity distribution functions with nanogram quantities. J Immunol Methods. 1998;211:97. doi: 10.1016/s0022-1759(97)00204-4. [DOI] [PubMed] [Google Scholar]
- 22.Pierson-Mullany LK, Jackola DR, Blumenthal MN, Rosenberg A. Evidence of an affinity threshold for IgE-allergen binding in the percutaneous skin test reaction. Clin Exp Allergy. 2002;32:107. doi: 10.1046/j.0022-0477.2001.01244.x. [DOI] [PubMed] [Google Scholar]
- 23.Sedgwick AK, Ballow M, Sparks K, Tilton RC. Rapid quantitative microenzyme-linked immunosorbent assay for tetanus antibodies. J Clin Microbiol. 1983;18:104. doi: 10.1128/jcm.18.1.104-109.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palmer LJ, Burton PR, Faux JA, James AL, Musk AW, Cookson WOCM. Independent inheritance of serum immunoglobulin E concentrations and airway responsiveness. Am J Respir Crit Care Med. 2000;161:1836. doi: 10.1164/ajrccm.161.6.9805104. [DOI] [PubMed] [Google Scholar]
- 25.Almasy L, Blangero J. Multipoint quantitative trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sutton BJ, Gould KJ. The human IgE network. Nature. 1993;366:421. doi: 10.1038/366421a0. [DOI] [PubMed] [Google Scholar]
- 27.Oettgen HC. Regulation of the IgE isotype switch; new insights on cytokine signals and the functions of ε germline transcripts. Curr Opin Immunol. 2000;12:619. doi: 10.1016/s0952-7915(00)00153-9. [DOI] [PubMed] [Google Scholar]
- 28.Geha RS, Jabara HH, Brodeur SR. The regulation of immunoglobulin E class-switch recombination. Nature Rev Immunol. 2003;3:721. doi: 10.1038/nri1181. [DOI] [PubMed] [Google Scholar]
- 29.Barbee RA, Halonen M, Lebowitz M, Burrows B. Distribution of IgE in a community population sample: correlations with age, sex, and allergen skin test reactivity. J Allergy Clin Immunol. 1981;68:106. doi: 10.1016/0091-6749(81)90167-6. [DOI] [PubMed] [Google Scholar]
- 30.Barbee RA, Halonen M, Kaltenbron W, Lebowitz M, Burrows B. A longitudinal study of serum IgE in a community cohort: correlations with age, sex, smoking, and atopic status. J Allergy Clin Immunol. 1987;79:919. doi: 10.1016/0091-6749(87)90241-7. [DOI] [PubMed] [Google Scholar]
- 31.Freidhoff LR, Meyers DA, Bias WB, Chase GA, Hussain R, Marsh DG. A genetic-epidemiologic study of human immune responsiveness to allergens in an industrial population: I Epidemiology of reported allergy and skin-test positivity. Am J Med Genet. 1981;9:323. doi: 10.1002/ajmg.1320090409. [DOI] [PubMed] [Google Scholar]
- 32.Burrows B, Martinez FD, Halonen M, Barbee RA, Cline MG. Association of asthma with serum IgE levels and skin-test reactivity to allergens. New Engl J Med. 1989;320:271. doi: 10.1056/NEJM198902023200502. [DOI] [PubMed] [Google Scholar]
- 33.Xu J, Postma DS, Howard TD, Koppelman GH, Zheng SL, Stine OC, Bleecker ER, Meyers DA. Major genes regulating total serum immunoglobulin E levels in families with asthma. Am J Hum Genet. 2000;67:1163. doi: 10.1086/321190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoffjan S, Nicolae D, Ober C. Association studies for asthma and atopic disease: a comprehensive review of the literature. Resp Res. 2003;4:14. doi: 10.1186/1465-9921-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jackola DR, Liebeler CL, Lin C-Y, Chiu Y-K, Blumenthal MN, Rosenberg A. Evidence that negative feedback between antibody concentration and affinity regulates humoral response consolidation to a non-infectious antigen in infants. Molec Immunol. 2005;42:19. doi: 10.1016/j.molimm.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 36.Palmer LJ, Cookson OCM. Using single nucleotide polymorphisms as a means to understanding the pathophysiology of asthma. Resp Res. 2001;2:102. doi: 10.1186/rr45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jackola DR, Basu S, Liebeler CL, Willaert R, Luah SS, Oetting W, King R, Blumenthal MN. CD14 polymorphisms in atopic families: implications for modulated allergen-specific IgE and IgG1 responses. Intl Archives Allergy Immunol. 2006;139:217. doi: 10.1159/000091167. [DOI] [PubMed] [Google Scholar]
- 38.Shilling RA, Pinto JM, Decker DC, Schneider DH, Bandukwala HS, Schneider JR, Camoretti-Mercado B, Ober C, Sperling AI. Cutting edge: polymorphisms in the ICOS promoter region are associated with allergic sensitization and Th2 cytokine production. J Immunol. 2005;175:2061. doi: 10.4049/jimmunol.175.4.2061. [DOI] [PubMed] [Google Scholar]
- 39.Hizawa N, Yamaguchi E, Jinushi E, Konno S, Kawakami Y, Nishimura M. Increased total serum IgE levels in patients with asthma and promoter polymorphisms at CTLA4 and FCER1B. J Allergy Clin Immunol. 2001;108:74. doi: 10.1067/mai.2001.116119. [DOI] [PubMed] [Google Scholar]
- 40.Howard TD, Postma DS, Hawkins GA, Koppelman GH, Zheng SL, Wysong AKS, Xu J, Meyers DA, Bleecker ER. Fine mapping of an IgE-controlling gene on chromosome 2q: analysis of CTLA4 and CD28. J Allergy Clin Immunol. 2002;110:743. doi: 10.1067/mai.2002.128723. [DOI] [PubMed] [Google Scholar]
- 41.Marsh DG, Neely JD, Breazeale DR, Ghosh B, Freidhoff LR, Ehrlich-Kautzky E, Schou C, Krishnaswamy G, Beaty TH. Linkage analysis of IL4 and other chromosome 5q311 markers and total serum immunoglobulin E concentrations. Science. 1994;264:1152. doi: 10.1126/science.8178175. [DOI] [PubMed] [Google Scholar]
- 42.Berek C, Ziegner M. The maturation of the immune response. Immunol Today. 1993;14:400. doi: 10.1016/0167-5699(93)90143-9. [DOI] [PubMed] [Google Scholar]
- 43.Tarlinton DM, Smith KGC. Dissecting affinity maturation: a model explaining selection of antibody-forming cells and memory B cells in the germinal center. Immunol Today. 2000;21:436. doi: 10.1016/s0167-5699(00)01687-x. [DOI] [PubMed] [Google Scholar]


