Abstract
Proteolytic processing of procaspase-9 is required for its activation, but processing in itself appears to be insufficient for its activity. We studied caspase activation in a cell-free system and found that incubation of cytosol from rat kidney proximal tubule cells with Cytochrome c (Cyt c) and dATP results in rapid autocatalytic processing of procaspase-9 from ~50 kD to ~38 kD size fragment. Moreover, Cyt c concentration influences the production of alternatively processed forms of caspase-9. At lower Cyt c concentration (0.01–0.05 mg/ml), two fragments of caspase-9 of the size 38 and 40 kD are produced. In contrast, at higher concentrations of Cyt c (>0.1 mg/ml) only 38 kD fragment will prevail. However, our failure to capture processed caspase-9 by affinity labeling or co-elution with Apaf-1 suggested that caspase-9 undergoes a conformational change during its enzymatic action on effector caspases, resulting in its release from the apoptosome complex and inactivation. In support of this hypothesis, catalytic inhibitors of caspase-9 prevented its release from the apoptosome complex without affecting its auto-processing and allowed successful capture of active caspase-9 (38 kD) and its complex by affinity labeling. These observations suggest that complex allosteric interactions with the apoptosome complex influence caspase-9 activity and function by controlling not only the induction of its enzymatic activity, but also its rapid termination.
Keywords: Apaf-1, Apoptosis, Apoptosome, Caspase-9, Caspase-3, Cytochrome C, Inhibitor, Research Article
2. INTRODUCTION
Based on the length of their pro-domains and their order of activation, caspases, the cysteine proteases that cleave their cellular targets after an aspartic acid residue, are classified as either initiator caspases or effector caspases (1–3). Activation of effector caspases is initiated by intra-chain cleavage by an initiator caspase by separating the zymogen into large and small subunits (4). In contrast, activation of the initiator caspase-9 requires docking and proximity dependent autocatalytic intra-chain processing on the apoptosome, a multimeric complex of Apaf-1, Cyt c and dATP (5–7). However, intra-chain cleavage alone is insufficient since fully processed caspase-9 is inactive or only marginally active unless it is associated with the apoptosome (8). Once activated, caspase-9 cleaves and activates downstream effector caspases like caspase-3 and caspase-7, which target many cellular proteins for degradation leading to cell death. However, the exact mechanisms for the activation or inactivation of the initiator caspase-9 still remained elusive.
A recent hypothesis of proximity-induced dimerization, based on the earlier proximity induced model of activation, suggests that dimerization of the initiator caspases drives their activation. According to this model, initiator caspases autoprocess themselves when brought into close proximity of each other (9, 10), and the formation of an oligomeric complex of apoptosome merely facilitates dimerization via their intrinsic dimerization interface. This model was supported by studies that used heterologous dimerization domains to induce caspase activation as well as by the work that identified in addition to monomers, a minor fraction of homodimers of caspase-9 during gel filtration analysis of activated enzyme (11–13). However, this hypothesis was questioned by studies that failed to detect dimers by analytical ultracentrifugation (14). Alternative models, based on interactions between Apaf-1 and caspase-9, suggest that caspase-9 is activated upon its binding to the apoptosome as a monomer or as a homooligomer by stabilizing the required conformation of the active site (15, 16). In support of the induced conformation model for initiator caspase activation, studies by Chao et al have shown that caspase-9 activity on the apoptosome complex is several folds higher compared to its activity as a dimer (17). We have examined the mechanisms that govern the initiation and decay of the initiator caspase-9 in a cell-free system using cytosol derived from kidney proximal tubule cells. Our data show that association and dissociation of the apoptosome and caspase-9 have far reaching effects on initiation and termination of the activity of this initiator caspase possibly through conformational changes.
3. MATERIALS AND METHODS
We obtained synthetic fluorogenic substrates (Peptides International, Louisville, KY), caspase inhibitors (ICN Pharmaceuticals, Aurora, OH), monoclonal anti-caspase-9 (LabVision Corporation, Fremont, CA), monoclonal anticaspase-2 (Alexis Biochemicals, San Diego, CA), monoclonal anti-caspase-3 and polyclonal anticaspase-9 (Cell Signaling Technology, Beverly, MA), monoclonal anti-XIAP (BD Biosciences, San Diego, CA), monoclonal anti-caspase-7 (Novus, Littleton, CO), peroxidase-conjugated goat anti-rat and anti-rabbit secondary antibodies (Jackson Immunoresearch Laboratories, Westgrove, PA) and goat anti-mouse antibody (Sigma, St.Louis, MO). Polyclonal anti-Apaf-1 and caspase-3 antibodies were kindly provided by Dr. X. Wang (University of Texas at Dallas) and Dr. Y. Lazebnik (Coldspring Harbor Laboratory, NY) respectively.
3.1. Preparation and activation of cytosolic extracts
RPTC and HK-2 cells were cultured as described before (29,30) Cytosol was collected by permeabilizing cells with 0.02% digitonin in buffer A (10 mM HEPES pH 7.4, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 5 mM dithiothreitol (DTT), 250 mM sucrose), containing protease inhibitors at room temperature for 2 min and concentrated using Ultrafree (10-kDa cut-off) concentrators (Millipore Corporation, Bedford) to obtain a final protein concentration of 10–15 mg/ml. Unless indicated otherwise, cytosolic extracts (10 mg/ml) were activated in vitro by incubation at 30 °C with 400 microM dATP, 0.2 mg/ml horse heart Cyt c and 2 mM Mg2+ with or without caspase inhibitors (10 microM). All the inhibitors used are fmk-derivatives even though aldehyde-derivatives gave identical inhibition patterns. Caspase activities are measured by spectroflurometric assays as described before (31) and proteins were analyzed by SDS-PAGE and western blotting (32).
3.2. Gel Filtration
Control and activated cytosolic extracts (~1 mg of protein) were analyzed by gel filtration chromatography in AKTA purifier system using Superose 6 (10/30) high-resolution column (Amersham Biosciences, Piscataway, NJ) equilibrated in column buffer (20 mM HEPES pH 7.0, 50 mM NaCl, 0.1% CHAPS, 5 mM DTT, 5% sucrose). Column was eluted at a flow rate of 0.4 ml/min and 1 ml fractions were collected and assayed for caspase activity. Aliquots of each fraction (200 microL) were precipitated with trichloroacetic acid and analyzed by Western blotting. The column was calibrated with Amersham Pharmacia Biotech gel filtration protein standards.
3.3. Affinity capture of activated caspases
Activated cytosolic extracts were incubated with 10 microM bio-VAD in binding buffer (50 mM HEPES pH 7.0, 50 mM NaCl, 0.1% CHAPS, 2mM EDTA, 10% sucrose) at 25°C for 30 min to label active sites. Biotinylated proteins were captured by incubating with strep-MP (CPG Inc., Lincoln Park, NJ). After several washes with binding buffer, bounded proteins were removed from the beads by boiling in SDS sample buffer and analyzed by Western blotting. For gel filtration analysis of activated extracts without bio-VAD, to an aliquot (250 microL) of each fraction, bio-VAD was added to a final concentration of 1 microM, incubated for 30 min at 25°C and then captured by strep-MP as above. When extracts were activated in the presence of biotinylated inhibitor and fractionated, fractions were directly incubated with strep-MP without adding bio-VAD.
4. RESULTS
4.1. Rapid processing of rat kidney procaspase-9 is not blocked by caspase inhibitors
Cell-free activation of cytosolic extracts from RPTC showed that both Cyt c and dATP are required to trigger DEVDase (caspase-3) activity (Figure 1A). In order to confirm the requirement of caspase-9 to process and activate effector caspases, reconstitution of normal and caspase-9 depleted cytosol with dATP and Cyt c was carried out. Incubation of cytosol with Cyt c and dATP resulted in proteolytic processing of caspases -2, 3 and -7 (Figure 1B, lane 2 in all panels). In contrast, caspase-9 depleted cytosol (Figure 1B, compare lanes 1 and 2 with lanes 3 and 4 in top left panel) did not proteolytically process caspases -2, -3 and -7 after reconstitution without or with dATP and Cyt c (Figure 1B, lanes 3 and 4 in right and bottom left panels). Kinetic analysis of procaspase-9 processing by immunoblotting revealed maximal cleavage (~80%) from ~50 kD to ~ 38 kD size within 2 min of activation (Figure 1C, top left panel). In contrast, procaspase-3 was processed at a slower rate (Figure 1C, top right panel). Interestingly, presence of a general caspase-inhibitor VAD or a sequence based inhibitor LEHD did not prevent but slowed caspase-9 processing during in vitro activation (Figure 1C left middle and bottom panels). In contrast, caspase-3 processing was completely blocked by both VAD (Figure 1C, lower right panel) and LEHD (not shown). In essence, the presence of inhibitors during reconstitution modestly delayed but did not block autoproteolytic processing of procaspase-9.
Figure 1.
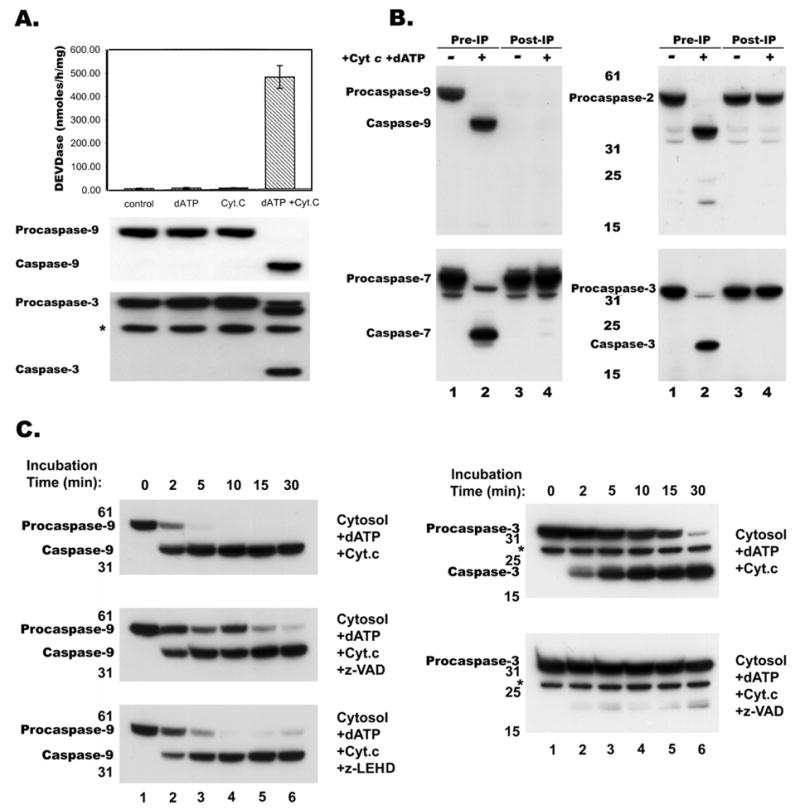
Proteolytic processing of procaspase-9. A, Cytosolic extract (10mg/ml) from RPTC was incubated with dATP (0.4 mM) or Cyt c (0.2 mg/ml) or both for 30 min at 30°C and were assayed for caspase processing activity by measuring DEVDase activity and immunoblotting as described in Materials and Methods. B, Control (lines 1 and 2) and caspase-9 immunodepleted (lines 3 and 4) cytosolic extracts from RPTC were concentrated and activated in presence of dATP and Cyt c for 30 min at 30°C, then proteins were separated by SDS-PAGE and analyzed by immunoblotting with anti-caspase-9, -2, -7 and -3 antibodies. C, Cytosolic extract was incubated at 30°C in presence of dATP and Cyt c for 2, 5, 10, 15, 30 min, the proteins were separated by SDS-PAGE and analyzed by immunoblotting with a monoclonal anti-caspase-9 (left panels) and a polyclonal anti-caspase-3 (right panels) antibody; * indicates a non-specific band detected by polyclonal anti-caspase-3 antibody. Presence of a caspase inhibitors z-VAD or z-LEHD during activation did not prevent but only slowed caspase-9 processing (middle and bottom left panels). In contrast, z-VAD completely blocked caspase-3 processing (bottom right panel).
4.2. Limited Cyt c concentration and alternative cleavage of caspase-9
Incubation of human and rat kidney tubular cell extracts with varying amounts of Cyt c and 0.4 mM dATP resulted in the increase of DEVDase activity with increased Cyt c concentrations (Figure 2A). Caspase activation in rat kidney cell extracts showed an allosteric response to Cyt c concentration, whilst caspase activation in human extracts had an almost linear response to Cyt c concentration (Figure 2A). These differences in Cyt c response could be attributed to the variations in the primary structures of human and rodent caspase-9 molecules (human caspase-9 has thirty eight fewer amino acids in the N-terminus than rodent caspase-9). Previous studies have noted that human procaspase-9 can be cleaved at two different sites generating 37 or 35 kD size large subunits (6, 7, and 18). It has been shown that autocatalytic processing produced a 35 kD subunit while a 37 kD subunit of caspase-9 is produced by caspase-3 mediated cleavage (6, 7, and 18). If so, at limited concentrations of Cyt c, only fewer procaspase-9 molecules will be recruited to the apoptosome complex. In turn, some of the free procaspase-9 molecules may be secondarily cleaved by effector caspases that were already activated by caspase-9 in the apoptosome complex.
Figure 2.
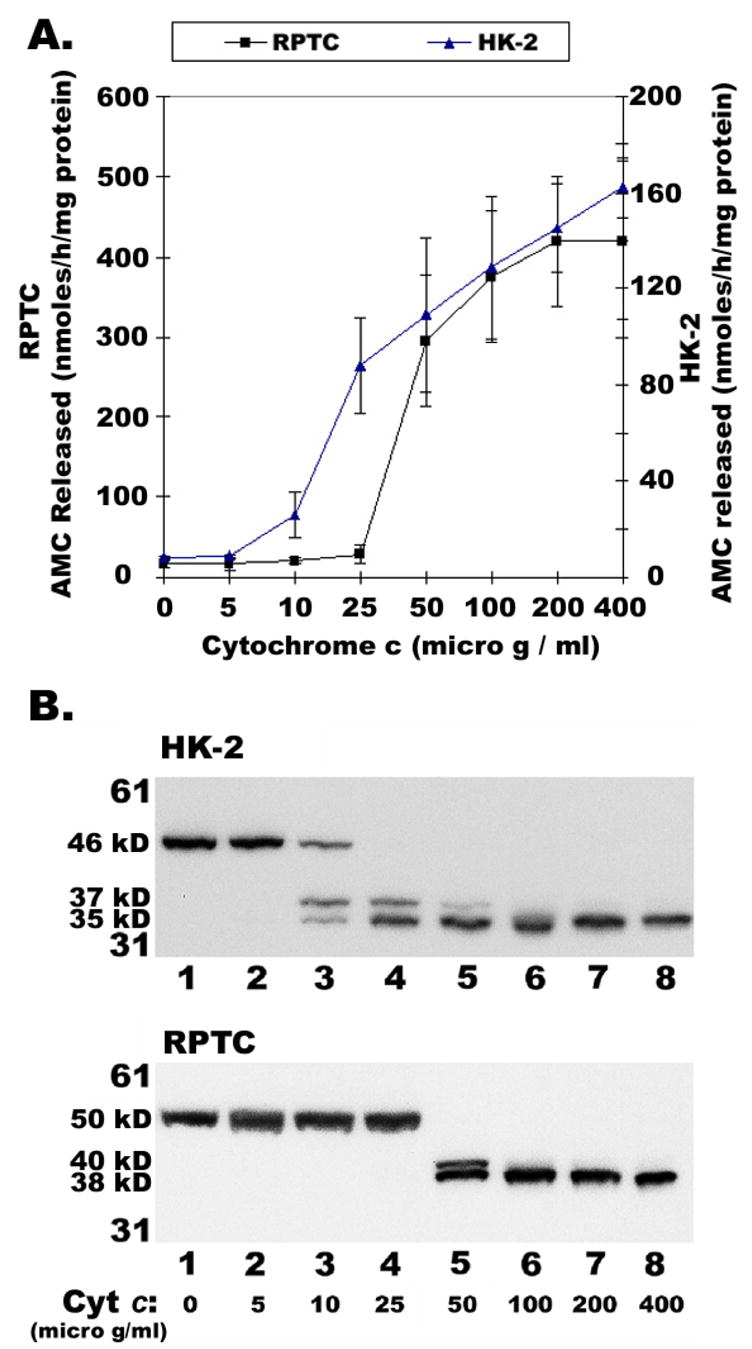
Effect of Cytochrome c on DEVDase activity and Caspase-9 processing. Cytosolic extracts (10mg/ml) from RPTC and HK-2 were incubated with dATP (0.4 mM) and varying concentrations of Cyt c at 30°C for 30 min and were assayed for caspase processing activity by measuring DEVDase activity (A), an average of 4 different experiments and immunoblotting for Caspase-9 (B), as described in Materials and Methods.
As expected, at lower Cyt c concentration (0.01 mg/ml), procaspase-9 was cleaved into both 37 and 35 kD fragments in HK-2 cell extracts, but with progressive increase in Cyt c concentration, the 37 kD fragment gradually decreased and eventually disappeared with concomitant increase in the 35 kD fragment (compare lanes 1–8 in Figure 2B, top panel). On the other hand, RPTC extracts required relatively higher amounts of Cyt c (0.05 mg/ml) to induce procaspase-9 cleavage into 40 and 38 kD fragments. With further increase in Cyt c concentration, the 40 kD fragment disappeared (compare lanes 1–8 in Figure 2B, bottom panel). Overall, the allosteric or linear response of Cyt c in caspase activation (Figure 2A) corresponds to caspase-9 processing in extracts from RPTC or HK-2, respectively (Figure 2B). Moreover, these results support the hypothesis that at lower, non-saturating, concentration of Cyt c, effector caspase mediated cleavage (37 kD in human and 40 kD in rodent) of procaspase-9 becomes predominant along with autocatalytic processing; while at higher saturating concentration of Cyt c, only autocatalytic processing of caspasae-9 would prevail.
4.3. Processed caspases-9 is the active enzyme but is rapidly inactivated
To evaluate whether processed caspase-9 is the active enzyme, we employed affinity labeling strategy to capture active caspases by treatment with biotin-VAD-fluromethylketone since enzymatic analysis by fluorescent peptide substrate cleavage revealed the lack of absolute specificity. This reagent binds to the active site of caspases and becomes covalently attached to the enzymes (19). Examination of cytosolic extract activated by incubation at 30°C with Cyt c and dATP for 30 min followed by incubation with bio-VAD and streptavidin-magnetic particles revealed that little or no processed caspase 9 had reacted with the affinity reagent (Figure 3, see lane 8 in top panels). In contrast, processed caspase-3 was fully captured by bio-VAD from this extract (Figure 3, see lane 8 in bottom panels). Failure to capture processed caspase-9 prompted us to hypothesize that inactivation of caspase-9 might have occurred by dissociation from the apoptosome complex while enzymatically acting on effector caspases. Since inhibitors did not block caspase-9 processing (Figure 1C), cytosolic extracts were incubated at 30°C simultaneously with Cyt c, dATP and bio-VAD for 30 min and then captured by streptavidin-magnetic particles. In support of our hypothesis, processed caspase-9 was captured when its activity was blocked by bio-VAD (Figure 3, see lane 9 in top panels). As expected, inclusion of bio-VAD during activation blocked the processing of caspase-3 (Figure 3, see lanes 3, 6 and 9 in bottom panels). Requirement of the affinity reagent to be present during the early activation steps for effective capture suggests that caspase-9 may be rapidly activated at the apoptosome complex but becomes inactivated soon thereafter. These results also suggested the possibility that inactivated enzyme is released from the apoptosome.
Figure 3.
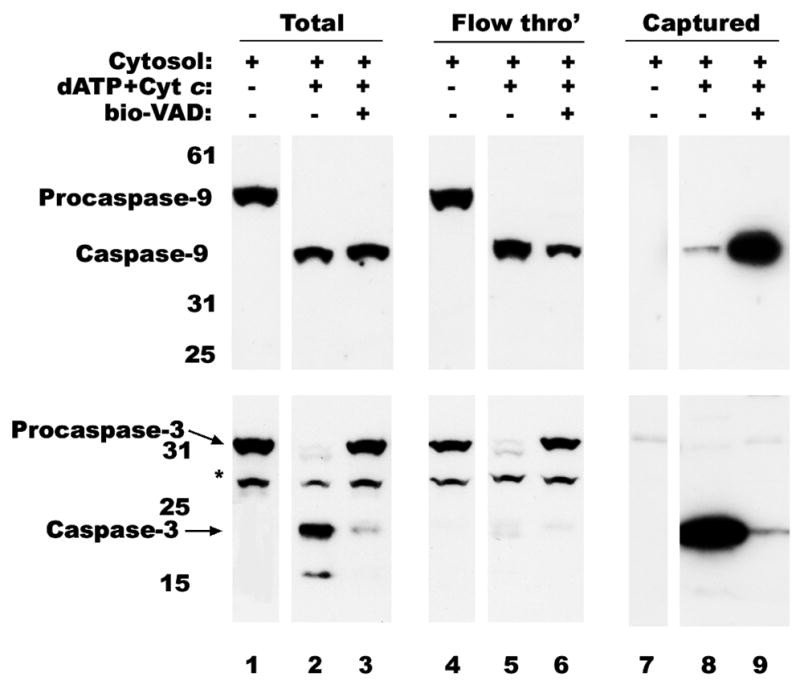
Capture of active caspase-9. Cytosolic extract from RPTC was activated, as described in Figure 1, without (lines 2, 5 and 8) or with bio-VAD (lines 3, 6 and 9). If no bio-VAD was added during activation it was added later and incubated for 30 min at RT. Biotinylated proteins were captured on strep-MP, eluted, separated by SDS-PAGE and immunoblotted with monoclonal anti-caspase-9 (top panel) or polyclonal anti-caspase-3 (bottom panel) antibodies as described in materials and methods. Lines 1, 4 and 7 correspond to untreated extract. Nonspecific band detected by anti-caspase-3 antibody is indicated with an asterisk (*).
4.4. Caspase inhibitors block dissociation of processed caspases-9 from the apoptosome complex
To further confirm our hypothesis that active processed caspase-9 on the apoptosome complex may undergo conformational change during its enzymatic action, resulting in its release from the apoptosome complex, in vitro reconstituted samples were subjected to size sieving gel filtration chromatography on a superose 6 column. The results presented in Figure 4A show that cytosolic Apaf-1, a 130 kD adapter protein involved in caspase activation, eluted with a peak elution volume corresponding to 158 kD (Figure 4, panel I, top). Incubation of cytosol with dATP alone did not change elution pattern of Apaf-1 (Figure 4, panel II, top). Incubation of cytosol with Cyt c alone significantly shifted Apaf-1 elution volumes to a higher molecular weight range (Figure 4, panel III, top). However, in the presence of both Cyt c and dATP, the peak elution volume of Apaf-1 was completely shifted to a size corresponding to ~1.34×106 daltons (Figure 4, panel IV, top). These results confirm the formation of an apoptosome complex reported by several laboratories (7; 16–18). Procaspase-9, a 50 kD protein, eluted at a peak volume between 158 kD and 67 kD in the untreated cytosol suggesting the possible existence of procaspase-9 as a dimer in the cytosol (Figure 4, panel I, bottom). Addition of dATP or Cyt c alone did not change the size or elution profile of procaspase-9 (Figure 4, panels II and III, bottom). However, incubation with both Cyt c and dATP for 2 min resulted in reduction of apparent molecular weight by SDS-PAGE (to ~38 kD) as well as modest shift in elution profile towards fractions containing larger proteins (Figure 4, panel IV, bottom). Although the major peak of elution remained between 67 and 158 kD for both processed and unprocessed enzymes (Figure 4, panel IV, bottom, lane 15), fractions with apparently larger sized enzyme were found, including a minor peak corresponding to 1.34×106 daltons coeluting with Apaf-1 and containing processed caspase-9 (Figure 4, panel IV, bottom, lane 10). However, gel filtration analysis of cytosol incubated with Cyt c and dATP for 30 min showed complete processing of procaspase-9 with a single elution peak between sizes 67 and 158 kD (Figure 4, panel V, bottom). In contrast, Apaf-1 still remained as part of a high molecular weight complex with some proteolysis (Figure 4, panel V, top). These observations suggested that caspase-9 associated with Apaf-1 in a large apoptosome complex early during activation but is rapidly released. To examine whether inhibition of caspase-9 activity might influence the physical state of the enzyme, LEHD, VAD or DQMD were included during activation. As illustrated in Figure 4, inclusion of LEHD or VAD completely shifted the elution pattern of processed caspase-9 to that seen for Apaf-1 with a peak elution at 1.34×106 daltons (panels VI and VII). A caspase-3 inhibitor (DQMD) only partially blocked the shift in caspase-9 elution profile (Figure 4A, panel VIII). It could be argued based on earlier reports (20, 21) that proteolytic destruction of Apaf-1 by caspase-3 could result in the release of bound caspase-9. However, inhibitors that are mainly targeted towards the effector caspase-3 such as DQMD (Figure 4A, panel VIII) or DEVD (not shown) inhibited the shift in caspase-9 elution partially, although they blocked Apaf-1 proteolysis. This prompted us to evaluate the effect of various caspase inhibitors on the processing of caspase-9 and caspase-3. Different inhibitors were added at 10 microM concentrations to the activation mixture of RPTC cytosol, dATP and Cyt c. As show in Figure 5A, all the inhibitors failed to prevent autocatalytic processing of Procaspase-9.
Figure 4.
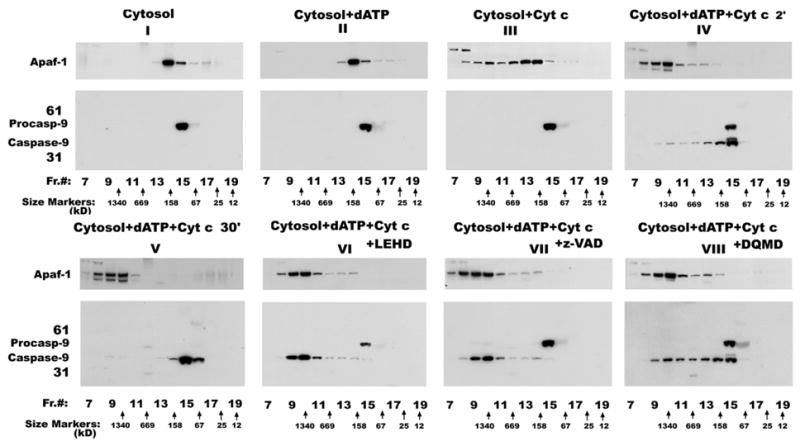
Inhibition of caspase-9 activity blocks its dissociation from the apoptosome complex. RPTC cytosolic extract was incubated at 30°C with dATP (panel II) or Cyt c (panel III) or both for 2 min (panels IV, VI and VII) or 30 min (panel V) and then loaded on to Superose-6 FPLC column. Panel I corresponds to untreated extract. To demonstrate that inhibition of caspase-9 activity prevents its dissociation from the high molecular weight complex, z-LEHD (panel VI), z-VAD (panel VII) or z-DQMD (panel VIII) were included during activation. Equal volumes of the column fractions after gel filtration chromatography were separated by SDS-PAGE and immunoblotted with anti-Apaf-1 and anti-caspase-9 antibodies. Nonspecific bands are indicated with an asterisk (*). The peak eluted fractions of protein standards on Superose-6 FPLC column are indicated at the bottom.
Figure 5.
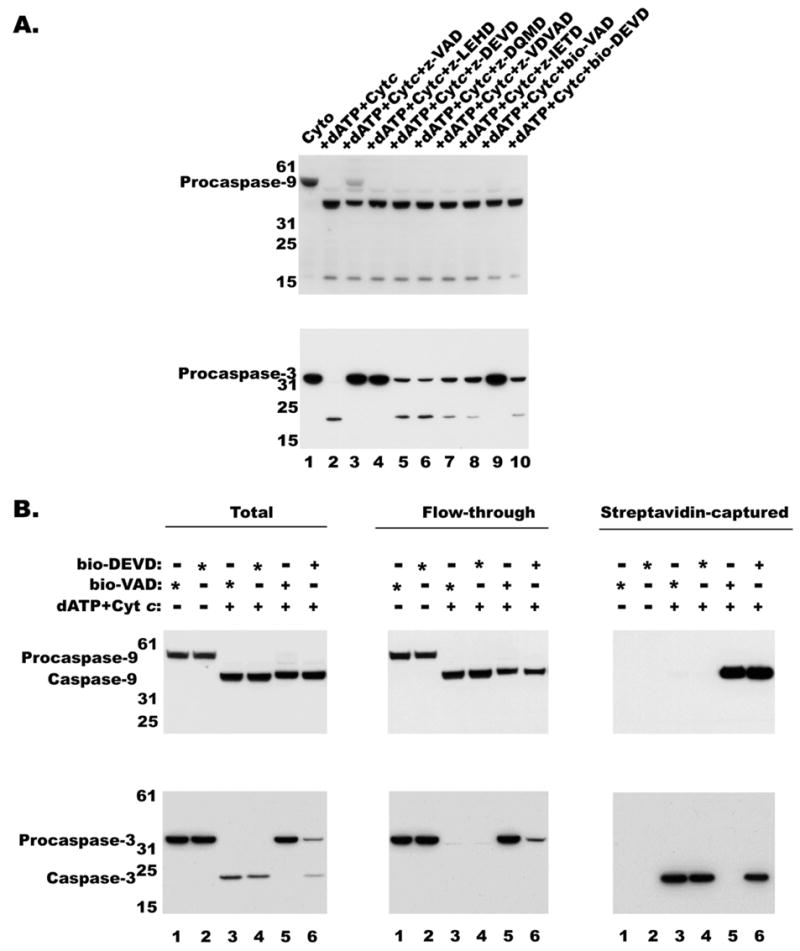
Effect of various caspase Inhibitors on caspase processing A, Cytosolic extracts (10mg/ml) from RPTC were incubated with dATP (0.4 mM) and Cyt c in the presence of various caspase inhibitors at 10 microM concentration at 30°C for 30 min and were assayed for caspase-9 and -3 processing by immunoblotting as described in Materials and Methods. B, Cytosolic extract from RPTC was activated, as described in Figure 1, activated without (lanes 3 and 4), with bio-VAD (lane 5) or with bio-DEVD (lane 6). If no bio-VAD or bio-DEVD was added during activation either one was added later as indicated with an asterisk and incubated for 30 min at RT. Biotinylated proteins were captured on strep-MP, eluted, separated by SDS-PAGE and immunoblotted with monoclonal anticaspase-9 (top panel) or monoclonal anti-caspase-3 (bottom panel) antibodies as described in materials and methods. Lanes 1 and 2 correspond to untreated extract.
However, from the processing of caspase-3 it appears that some of the inhibitors fully and others partially inhibited activated caspase-9. As shown in Figure 5A (bottom panel), the VAD derivatives and LEHD have completely inhibited caspase-3 processing, while other caspase inhibitors have partially blocked caspase-3 processing. Nevertheless, LEHDase and DEVDase activities could not be detected in the presence of any of the caspase inhibitors (not shown). It appears that DQMD or DEVD may have partially inhibited caspase-3 processing by binding to activated caspase-9 albeit with lower affinity. To test whether DEVD can bind caspase-9, we used biotinylated DEVD in the reconstitution mix along with dATP and Cyt c and used streptavidin-MP to separate biotin labeled proteins and analyzed for caspase-9. As shown in Fig 5B, both caspase-9 and caspase-3 were pulled down by streptavidin-MP from bio-DEVD incubated samples while only caspase-9 was pulled down by bio-VAD incubated samples. Thus suggesting in the presence of VAD no activation of caspase-3 would occur, while in the presence of DEVD, activation of both caspase-9 and caspase-3 could take place. Binding of bio-DEVD to caspase-9 and caspase-3 suggests lack of selective specificity for these so-called specific inhibitors. In fact, these results are in agreement with an earlier report that DEVD could also bind and inhibit caspase-9 (22) and explain why caspase-9 is partially retained on the apoptosome complex when incubated with DQMD and DEVD (Figure 4).
4.5. Capture of Apoptosome Complex by Biotinylated Caspase Inhibitors
Inclusion of bio-VAD during incubation with dATP and Cyt c resulted in a similar shift of processed caspase-9 elution that matched the elution of Apaf-1 (Figure 6, top and middle panel II) following size-sieving chromatography. Since co-elution of Apaf-1 and caspase-9 alone may not necessarily indicate complex formation between these two proteins, we attempted to capture these complexes with bio-VAD/strep-MP from all the eluted fractions. If no bio-VAD was included during initial incubation, bio-VAD was added to gel filtration fractions before capturing with strep-MP as described in the Materials and Methods section. Untreated RPTC cytosol did not yield any signal for the proteins tested after fractionation and affinity capture (not shown). Terminal addition of bio-VAD to the gel filtration fractions of cytosol activated with dATP and Cyt c did not yield any signal for caspase-9 or Apaf-1 (Figure 6, top and middle panel IV) but captured both caspase-7 (not shown) and caspase-3 (Figure 6, bottom panel IV). On the other hand, analysis of gel filtration fractions of cytosol activated in the presence of bio-VAD, dATP and Cyt c showed capture of both caspase-9 and Apaf-1 (see Figure 6, panel V), indicating the close association of these two proteins in the 1.34×106 dalton complex. Co-precipitation of both caspase-9 and Apaf-1 even after high dilution during gel filtration suggests a strong association between these two molecules, not only confirming previously published data (5,23–25) that oligomerized Apaf-1 is a functional complex that recruits procaspase-9, but also that caspase-9 associated with the apoptosome exists in the active form. Taken together, the failure of terminally added bio-VAD to capture processed caspase-9 in the low molecular weight fractions (Figure 6, panel IV), and the time course of shifts in elution profiles (Figure 4, panels IV and V), these results suggest that the sojourn of activated caspase-9 on the apoptosome is brief, and that inactivation and release follow rapidly while activating effector caspases.
Figure 6.
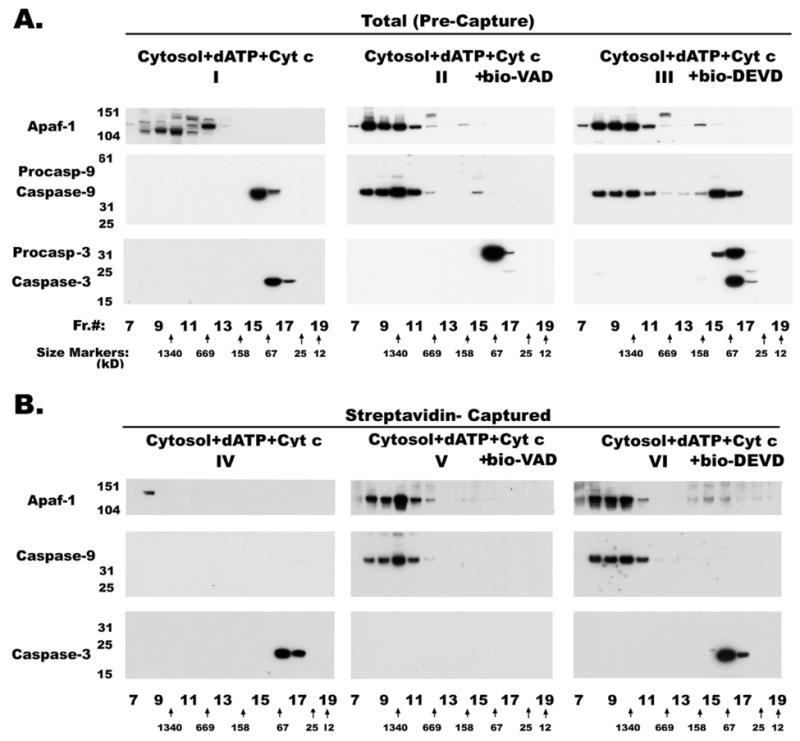
Affinity capture of caspase-9 and Apaf-1 complexes Elution profiles of cytosolic extracts from control (not shown) and activated without (panels I and IV), with bio-VAD (panels II and V) or with bio-DEVD (panels III and VI) subjected to gel filtration chromatography. Equal volumes of the column fractions were separated by SDS-PAGE and immunoblotted with anti-Apaf-1, anti-caspase-9 and anti-caspase-3 antibodies. Proteins from the same column fractions were captured on strep-MP (panels IV and VI), eluted, separated by SDS-PAGE and immunoblotted with anti -Apaf-1 (polyclonal), -caspase-9 (monoclonal) and -caspase-3 (monoclonal) antibodies. If no bio-VAD was included during initial incubation, 0.25mM bio-VAD was added to gel filtration fractions before capturing. The peak eluted fractions of protein standards on Superose-6 FPLC column are indicated at the bottom.
On the other hand, inclusion of bio-DEVD during incubation with dATP and Cyt c resulted in two pools of caspase-9 from the reconstituted mixture with a bound pool, coeluting with Apaf-1 at a high molecular weight, and a free pool co-eluting with proteins of lower molecular weight (Figure 6, panel III). It appears that the free pool of caspase-9 is present only when caspase-3 is being cleaved (Figure 6, panels I and III). In the absence of caspase-3 processing, almost all of the caspase-9 remained bound to the apoptosome complex (Figure 6, panel II). These results clearly support our hypothesis that caspase-9 undergoes a conformational change while processing caspase-3 resulting in its release from the apoptosome complex. Here, the question of proteolytic destruction of Apaf-1, resulting in the release of caspase-9, by caspase-3 does not arise because of the presence of bio-DEVD throughout incubation. Moreover, released caspase-9 is catalytically inactive and fails to bind the biotinylated inhibitor (Figure 6, panels IV and VI). If DEVD manages to bind active caspase-9, it prevented the release of caspase-9 from the apoptosome complex (Figure 6, panel VI).
5. DISCUSSION
The z-VAD non-inhibitable processing of caspase-9 clearly indicates that proximity-dependent interactions between procaspase-9 molecules brought close together in the “apoptosome” complex result in their processing (9). Only processing that may occur by direct action of active caspases was prevented by the inhibitors. Mutagenesis studies have indicated that although cysteine residue in the active site is required for both autoproteolytic processing and cleavage of effector caspases but aspartic acid residue at which processing occurs is not critical for its activity (6, 26). However, Srinivasula et al (7) had observed that proteolytic processing in advance is required for XIAP binding to inhibit caspase-9 activity. Consistent with this observation, the inhibitor studies presented here indicate that even z-VAD binding does not take place until the processing of caspase-9 occurs at the apoptosome complex. Bratton et al suggested that non-cleavable caspase-9 was as efficient as wild type caspase-9 in recruiting and activating caspase-3 based on the reasonable intensity of a band recognized by the caspase-3 antibody in the high molecular weight range corresponding to cleaved caspase-3 (26). However, the results presented by Bratton et al (26) showed that a significant amount of uncleaved and unbound endogenous caspase-3 being present in the reconstituted lysate with non-cleavable caspase-9. In contrast, with endogenous caspase-9 almost all of endogenous caspase-3 was completely cleaved. In reality, these reconstitution experiments are suggesting that unprocessed caspase-9 is not as efficient as processed caspase-9 in cleaving effector caspases. Moreover, unlike previous studies by Bratton et al., (26, 27), we could not detect caspase-3 in the apoptosome complex suggesting that its interaction with caspase-9 is only transient and can not form a stable complex. Even under conditions that stabilized Caspase-9 and Apaf-1 complex, we could not detect caspase-3 in the apoptosome complex (Figure 6). The co-migration of caspase-3 with high molecular weight Apaf-1 observed by Bratton et al (26) does not necessarily warrant its association with apoptosome complex unless proven by co-precipitation.
Based on the results presented, we suggest that caspase-9 may undergo at least three different conformational transitions on the apoptosome complex. In the initial conformational state VAD and LEHD may not effectively bind to caspase-9. Following binding to the apoptosome complex, procaspase-9 undergoes a rapid conformational change along with auto-proteolysis and attains a new conformation that allows binding of its substrates and inhibitors more readily. In this conformation caspase-9 appears to have a higher affinity for the apoptosome complex as well. If frozen in this state with VAD, even extreme dilutions that occur during gel filtration could not disrupt these interactions between caspase-9 and Apaf-1 (Figure 4). Finally, while processing effector caspase substrates, caspase-9 may attain a third inactive conformation in which it looses its ability to interact with the apoptosome complex and becomes free. This hypothesis is strongly supported by our results where inhibition of caspase-9 catalytic activity prevented caspase-9 release from the apoptosome complex without inhibiting its initial processing.
In contrast to our results with cytosol from rat kidney proximal tubule cells, several studies have shown association of caspase-9 with the apoptosome complex by gel filtration of activated cytosol from cells of human origin (8). Based on our findings, we hypothesized that the presence of natural caspase inhibitors in human cells may have a role in blocking caspase-9 release from the apoptosome complex. In fact, we found large quantities of XIAP being present in HEK cells than in RPTC or HK-2 cells (Mikhailova et al, unpublished results). However, presence of XIAP does not block caspase-9 processing, which is consistent with the observation that XIAP does not inhibit activation of procaspase-9 but inhibits the activity of the processed caspase-9 (28).
On the whole, our results are consistent with a model (Figure 7) of caspase activation in which procaspase-9 is activated through recruitment into apoptosome complex and processed. Although our results suggest that caspase-3 may not form a stable complex with apoptosome for its activation but they are still consistent with the idea that caspase-9 on the apoptosome complex may have enhanced affinity for effector caspases (procaspase-3 and -7) for their activation (32). Our results clearly indicate a necessary and important role for apoptosome complex in regulating caspase-9 activity. Therefore, the suggestion that role of apoptosome is only to present a platform for dimerization (10) of caspase-9 is an overstatement. Overall, the results presented in this study suggest that allosteric mechanisms that occur during caspase activation are important in regulating caspase-9 activity.
Figure 7.
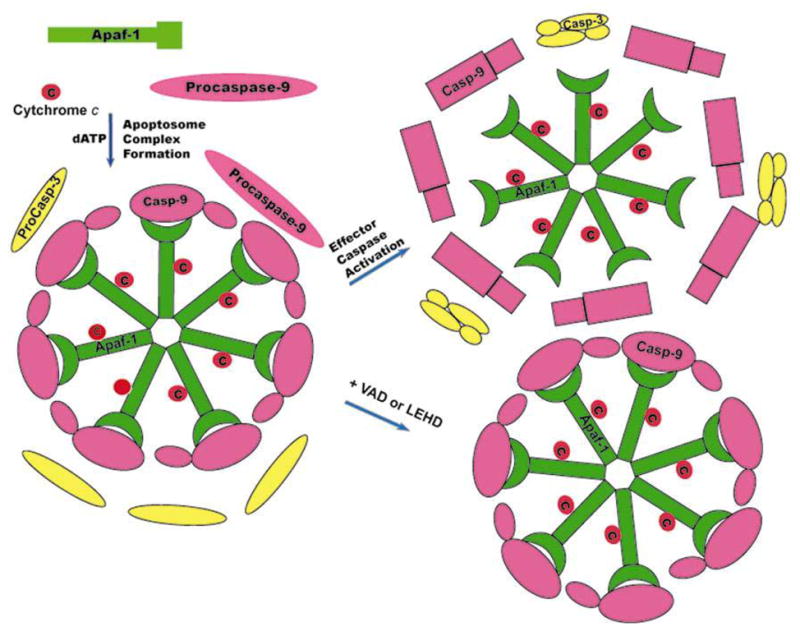
A hypothetical model for caspase-9 activation and inactivation Apaf-1 undergoes conformational change by binding to Cytochrome c into the cytosol and forms heptameric apoptosome complex and recruits procaspase-9 to this complex in the presence of dATP. Upon recruitment to this complex procaspase-9 undergoes conformational change and auto-catalytic processing. In this conformation processed caspase-9 (Casp-9) has stronger affinity to Apaf-1 and is enzymatically active to process effector caspases like procaspase-3. During catalytic action, Caspase-9 undergoes further conformational change and dissociates from the apoptosome complex resulting in its inactivation. Inhibition of catalytic activity prevents the release of caspase-9 from apoptosome complex.
Acknowledgments
This work is supported by National Institutes of Health Grant DK54472 and San Antonio Area Foundation grant (to P.S.). We gratefully acknowledge the generous gifts of Apaf-1 and caspase-3 antibodies from Dr. X. Wang and Dr. Y. Lazebnik respectively and caspase-9(−/−) and caspase-3(−/−) MEFs from Dr. Richard Flavell to test antibody specificities. We thank Anupama Surapaneni for technical assistance.
Abbreviations
- RPTC
Rat kidney proximal tubule cells
- Cyt c
Cytochrome c
- VAD
Benzyloxycarbonyl-Val-Ala-Asp (OMe) fluoromethylketone
- bio-VAD
Biotinyl-Val-Ala-Asp (OMe) fluoromethylketone
- LEHD
Benzyloxycarbonyl-Leu-Glu-His-Asp (OMe) fluoromethylketone
- DEVD
Benzyloxycarbonyl-Asp-Glu-Val-Asp (OMe) fluoromethylketone
- DQMD
Benzyloxycarbonyl-Asp-Gln-Met-Asp (OMe) fluoromethylketone
- Ac-LEHD-AMC
Acetyl-Leu-Glu-His-Asp-7-amino-4-methylcoumarin
- Ac-DEVD-AMC
Acetyl-Asp-Glu-Val-Asp-7-amino-4-methylcoumarin
- AMC
7 amino-4-methylcoumarin
- Strep-M
Strepatavidin-conjugated Magenetic Particles
References
- 1.Alnemri ES, Livingston DJ, Nicholson DW, Salvesen G, Thornberry NA, Wong WW, Yuan J. Human ICE/CED-3 protease nomenclature. Cell. 1996;87:171. doi: 10.1016/s0092-8674(00)81334-3. [DOI] [PubMed] [Google Scholar]
- 2.Thornberry NA. Caspases: key mediators of apoptosis. Chem Biol. 1998;5:R97–103. doi: 10.1016/s1074-5521(98)90615-9. [DOI] [PubMed] [Google Scholar]
- 3.Cohen GM. Caspases: the executioners of apoptosis. Biochem J. 1997;326 ( Pt 1):1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thornberry NA, Lazebnik Y. Caspases: enemies within. Science. 1998;281:1312–6. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 5.Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, Wang X. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–89. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 6.Stennicke HR, Deveraux QL, Humke EW, Reed JC, Dixit VM, Salvesen GS. Caspase-9 can be activated without proteolytic processing. J Biol Chem. 1999;274:8359–62. doi: 10.1074/jbc.274.13.8359. [DOI] [PubMed] [Google Scholar]
- 7.Srinivasula SM, Hegde R, Saleh A, Datta P, Shiozaki E, Chai J, Lee RA, Robbins PD, Fernandes-Alnemri T, Shi Y, Alnemri ES. A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO regulates caspase activity and apoptosis. Nature. 2001;410:112–6. doi: 10.1038/35065125. [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez J, Lazebnik Y. Caspase-9 and APAF-1 form an active holoenzyme. Genes Dev. 1999;13:3179–84. doi: 10.1101/gad.13.24.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salvesen GS, Dixit VM. Caspase activation: the induced-proximity model. Proc Natl Acad Sci U S A. 1999;96:10964–7. doi: 10.1073/pnas.96.20.10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boatright KM, Renatus M, Scott FL, Sperandio S, Shin H, Pedersen IM, Ricci JE, Edris WA, Sutherlin DP, Green DR, Salvesen GS. A unified model for apical caspase activation. Mol Cell. 2003;11:529–41. doi: 10.1016/s1097-2765(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 11.Chen M, Wang J. Initiator caspases in apoptosis signaling pathways. Apoptosis. 2002;7:313–9. doi: 10.1023/a:1016167228059. [DOI] [PubMed] [Google Scholar]
- 12.Renatus M, Stennicke HR, Scott FL, Liddington RC, Salvesen GS. Dimer formation drives the activation of the cell death protease caspase 9. Proc Natl Acad Sci U S A. 2001;98:14250–5. doi: 10.1073/pnas.231465798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang DW, Ditsworth D, Liu H, Srinivasula SM, Alnemri ES, Yang X. Oligomerization is a general mechanism for the activation of apoptosis initiator and inflammatory procaspases. J Biol Chem. 2003;278:16466–9. doi: 10.1074/jbc.C300089200. [DOI] [PubMed] [Google Scholar]
- 14.Shiozaki EN, Chai J, Rigotti DJ, Riedl SJ, Li P, Srinivasula SM, Alnemri ES, Fairman R, Shi Y. Mechanism of XIAP-mediated inhibition of caspase-9. Mol Cell. 2003;11:519–27. doi: 10.1016/s1097-2765(03)00054-6. [DOI] [PubMed] [Google Scholar]
- 15.Acehan D, Jiang X, Morgan DG, Heuser JE, Wang X, Akey CW. Three-dimensional structure of the apoptosome: implications for assembly, procaspase-9 binding, and activation. Mol Cell. 2002;9:423–32. doi: 10.1016/s1097-2765(02)00442-2. [DOI] [PubMed] [Google Scholar]
- 16.Shiozaki EN, Chai J, Shi Y. Oligomerization and activation of caspase-9, induced by Apaf-1 CARD. Proc Natl Acad Sci U S A. 2002;99:4197–202. doi: 10.1073/pnas.072544399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chao Y, Shiozaki EN, Srinivasula SM, Rigotti DJ, Fairman R, Shi Y. Engineering a dimeric caspase-9: a re-evaluation of the induced proximity model for caspase activation. PLoS Biol. 2005;3:e183. doi: 10.1371/journal.pbio.0030183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saikumar P, Dong Z, Patel Y, Hall K, Hopfer U, Weinberg JM, Venkatachalam MA. Role of hypoxia-induced Bax translocation and cytochrome c release in reoxygenation injury. Oncogene. 1998;17:3401–15. doi: 10.1038/sj.onc.1202590. [DOI] [PubMed] [Google Scholar]
- 19.Ryan MJ, Johnson G, Kirk J, Fuerstenberg SM, Zager RA, Torok-Storb B. HK-2: an immortalized proximal tubule epithelial cell line from normal adult human kidney. Kidney Int. 1994;45:48–57. doi: 10.1038/ki.1994.6. [DOI] [PubMed] [Google Scholar]
- 20.Nicholson DW, Ali A, Thornberry NA, Vaillancourt JP, Ding CK, Gallant M, Gareau Y, Griffin PR, Labelle M, Lazebnik YA, Munday NA, Raju SM, Smulson ME, Yamin T, Yu VL, Miller DK. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature. 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 21.Mikhailov V, Mikhailova M, Degenhardt K, Venkatachalam MA, White E, Saikumar P. Association of Bax and Bak homo-oligomers in mitochondria. Bax requirement for Bak reorganization and cytochrome c release. J Biol Chem. 2003;278:5367–76. doi: 10.1074/jbc.M203392200. [DOI] [PubMed] [Google Scholar]
- 22.Zou H, Yang R, Hao J, Wang J, Sun C, Fesik SW, Wu JC, Tomaselli KJ, Armstrong RC. Regulation of the Apaf-1/caspase-9 apoptosome by caspase-3 and XIAP. J Biol Chem. 2003;278:8091–8. doi: 10.1074/jbc.M204783200. [DOI] [PubMed] [Google Scholar]
- 23.Faleiro L, Kobayashi R, Fearnhead H, Lazebnik Y. Multiple species of CPP32 and Mch2 are the major active caspases present in apoptotic cells. Embo J. 1997;16:2271–81. doi: 10.1093/emboj/16.9.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bratton SB, Walker G, Roberts DL, Cain K, Cohen GM. Caspase-3 cleaves Apaf-1 into an approximately 30 kDa fragment that associates with an inappropriately oligomerized and biologically inactive approximately 1.4 MDa apoptosome complex. Cell Death Differ. 2001;8:425–33. doi: 10.1038/sj.cdd.4400834. [DOI] [PubMed] [Google Scholar]
- 25.Lauber K, Appel HA, Schlosser SF, Gregor M, Schulze-Osthoff K, Wesselborg S. The adapter protein apoptotic protease-activating factor-1 (Apaf-1) is proteolytically processed during apoptosis. J Biol Chem. 2001;276:29772–81. doi: 10.1074/jbc.M101524200. [DOI] [PubMed] [Google Scholar]
- 26.Del Bello B, Valentini MA, Mangiavacchi P, Comporti M, Maellaro E. Role of caspases-3 and -7 in Apaf-1 proteolytic cleavage and degradation events during cisplatin-induced apoptosis in melanoma cells. Exp Cell Res. 2004;293:302–10. doi: 10.1016/j.yexcr.2003.10.024. [DOI] [PubMed] [Google Scholar]
- 27.Srinivasula SM, Ahmad M, Fernandes-Alnemri T, Alnemri ES. Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Mol Cell. 1998;1:949–57. doi: 10.1016/s1097-2765(00)80095-7. [DOI] [PubMed] [Google Scholar]
- 28.Zou H, Li Y, Liu X, Wang X. An APAF-1. cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J Biol Chem. 1999;274:11549–56. doi: 10.1074/jbc.274.17.11549. [DOI] [PubMed] [Google Scholar]
- 29.Adams JM, Cory S. Apoptosomes: engines for caspase activation. Curr Opin Cell Biol. 2002;14:715–20. doi: 10.1016/s0955-0674(02)00381-2. [DOI] [PubMed] [Google Scholar]
- 30.Bratton SB, Walker G, Srinivasula SM, Sun XM, Butterworth M, Alnemri ES, Cohen GM. Recruitment, activation and retention of caspases-9 and -3 by Apaf-1 apoptosome and associated XIAP complexes. Embo J. 2001;20:998–1009. doi: 10.1093/emboj/20.5.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bratton SB, Lewis J, Butterworth M, Duckett CS, Cohen GM. XIAP inhibition of caspase-3 preserves its association with the Apaf-1 apoptosome and prevents CD95- and Bax-induced apoptosis. Cell Death Differ. 2002;9:881–92. doi: 10.1038/sj.cdd.4401069. [DOI] [PubMed] [Google Scholar]
- 32.Yin Q, Park H, Chung J, Lin S, Lo Y, da Graca L, Jiang X, Wu H. Caspase-9 holoenzyme is a Specific and optimal procaspase-3 processing machine. Mol Cell. 2006;22:259–268. doi: 10.1016/j.molcel.2006.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]


