Abstract
sonic hedgehog (shh) is expressed in anterior endoderm, where it is required to repress pancreas gene expression and to pattern the endoderm, but the pathway controlling endodermal shh expression is unclear. We find that expression of meis3, a TALE class homeodomain gene, coincides with shh expression in the endoderm of zebrafish embryos. Using a dominant negative construct or anti-sense morpholino oligos (MOs) to disrupt meis3 function, we observe ectopic insulin expression in anterior endoderm. This phenotype is also observed when meis3 MOs are targeted to the endoderm, suggesting that meis3 acts within the endoderm to restrict insulin expression. We also find that meis3 is required for endodermal shh expression, indicating that meis3 acts upstream of shh to restrict insulin expression. Loss of pbx4, a TALE gene encoding a Meis cofactor, produces the same phenotype as loss of meis3, consistent with Meis3 acting in a complex with Pbx4 as reported in other systems. Lastly, we observe a progressive anterior displacement of endoderm-derived organs upon disruption of meis3 or pbx4, apparently as a result of underdevelopment of the pharyngeal region. Our data indicate that meis3 and pbx4 regulate shh expression in anterior endoderm, thereby influencing patterning and growth of the foregut.
Keywords: homeodomain transcription factor, endoderm patterning, pancreas development: hedgehog signaling, foxa2, meis, pbx, lazarus, sox32
INTRODUCTION
The vertebrate pancreas is made up of islets of Langerhans that contain endocrine cells and control blood glucose levels by releasing hormones (e.g. insulin and glucagon) into the bloodstream, as well as of acinar tissue that contains exocrine cells and secretes digestive enzymes (e.g. Amylase and Carboxypeptidase A) into the duodenum and the digestive tract. The exocrine and endocrine cells of the vertebrate pancreas share common cellular progenitors that arise within the duodenal epithelium. Initial differentiation of these cells occurs in a highly stereotyped position within the duodenal epithelium, in close proximity to the notochord. Subsequent emigration of pancreatic cells, branching morphogenesis, fusion of dorsal and ventral lobes, and endocrine islet formation all occur within a restricted domain of the endoderm. Abnormalities in positioning and growth of the pancreas cause a number of human disease conditions. For instance, nesidioblastosis (hyperplastic and hypertrophic islets), annular pancreas and ectopic pancreas underlie fetal and adult cases of hyperinsulinemia and hypoglycemia (Hill and Lebenthal, 1993; Kaczirek and Niederle, 2004).
The genetic mechanisms that determine normal positioning of the pancreas and the other gut-associated organs along the anteroposterior axis of the embryonic endoderm include inductive and inhibitory signals from adjacent tissues. For instance, retinoic acid (likely produced by lateral mesoderm) is necessary and sufficient to induce pancreas-specific gene expression both in vivo and in explanted tissue (Kumar et al., 2003; Stafford and Prince, 2002). Similarly, bone morphogenetic proteins (particularly BMP4 and BMP7) are produced in the lateral mesoderm and have been implicated in the induction of pancreas-specific gene expression, at least in explanted endoderm (Kumar et al., 2003). These inductive signals induce expression of the orphan homeodomain transcription factor pdx1 (an early marker of the pancreas primordium) within the duodenal epithelium. Pancreas development is negatively regulated by signals that restrict pancreas-specific gene expression within the endoderm, thereby defining the precise position of the pancreas primordium. In particular, Hedgehog signaling is thought to prevent expression of pancreas-specific genes in the anterior endoderm (reviewed in (Hebrok, 2003)). Thus, mice homozygous mutant for the Indian hedgehog gene develop annular pancreas (Hebrok et al., 2000), while disruption of Hedgehog signaling by application of cyclopamine promotes ectopic expression of pancreatic genes (diIorio et al., 2002; Kim and Melton, 1998) and the development of hyperplastic islets within the gastric endoderm (Kim and Melton, 1998). Intriguingly, such nesidioblastotic islets have also been observed in human patients with Smith-Lemli-Opitz (SLO) syndrome (Lachman et al., 1991). Since SLO patients have defective cholesterol biosynthesis (as a result of defective 7-dehydrocholesterol reductase function; reviewed in (Wolf, 1999)) and hedgehog proteins are modified with cholesterol (Porter et al., 1996), this finding suggests a role for Hedgehog signaling in negatively regulating pancreas formation also in humans. In spite of the likely role for Hedgehog signaling in positioning the pancreas primordium and restricting ectopic pancreas formation, it is unclear what other genes act in this pathway to control Hedgehog activity in the anterior endoderm.
Members of the homeodomain super-family of DNA-binding proteins regulate transcription of many essential developmental genes during embryogenesis. The TALE (three amino acid loop extension) subclass of homeodomain proteins includes members of the meis/prep and pbx gene families. TALE class homeodomain proteins have been implicated in formation of the nervous system and the limbs, as well as various internal organs during vertebrate development (e.g. (Brendolan et al., 2005; Choe et al., 2002; Deflorian et al., 2004; Ferretti et al., 2006; Hisa et al., 2004; Maeda et al., 2002; Mercader et al., 2005; Pöpperl et al., 2000; Selleri et al., 2001; Waskiewicz et al., 2001; Waskiewicz et al., 2002)). We have now examined meis gene expression in the developing endoderm and we find that meis3 is expressed in the endoderm immediately anterior to the pancreas primordium of zebrafish embryos. This meis3 expression domain overlaps with the shh expression domain, suggesting that meis3 may act in the shh-dependent pathway to repress expression of pancreas-specific genes in the anterior endoderm. Using a dominant negative construct and anti-sense morpholino oligos (MOs) to disrupt meis3 function, we find ectopic expression of insulin and reduced expression of shh and foxa2 in the anterior endoderm, suggesting that meis3 acts upstream of shh to restrict expression of pancreas-specific genes in the foregut. We also find that the liver and pancreas become displaced anteriorly in older embryos with disrupted meis3 function. This effect takes place at the expense of the pharyngeal endoderm and likely results from underdevelopment of the pharyngeal region in the absence of shh. Lastly, interfering with the function of Pbx4, a known cofactor to Meis proteins, causes a similar phenotype, suggesting that Meis3 and Pbx4 act together to control shh expression in the anterior endoderm. Our data are consistent with the proposed role for shh in demarcating the boundaries of the pancreas primordium (reviewed in (Hebrok, 2003)), and begin delineating the pathway by which shh expression is regulated during organogenesis in the anterior endoderm.
MATERIALS AND METHODS
Fish Maintenance
Wild type, gutGFP (Field et al., 2003) and lazarus (lzrb557; (Pöpperl et al., 2000)) embryos were collected from natural matings and reared in 1/3 Ringer’s. Embryos were staged using morphological criteria up to 24 hours post fertilization (hpf) and by time of development at 28.5°C thereafter (Kimmel et al., 1995).
mRNA and morpholino injections
The dominant negative Meis construct (ΔCPbx4) has been reported previously (Choe et al., 2002) and ~1 nl of a 300μg/ml ΔCPbx4 mRNA stock was injected at the 1–2 cell stage. Two antisense morpholino oligos designed to block translation of the meis3 mRNA (tMO1 5′ATCCATGCGATACGGAAGCCGAGCT3′ complementary to position −19 to +6 and tMO2 5′CACACACTCACTGACGGAGGACAAC 3′ complementary to position −44 to −19, where +1 indicates the first nucleotide of the AUG codon) and one control morpholino (MOCO 5′ATCgATGCcATACcGAAcCCGAcCT3′ that has 5 mismatches relative to tMO1) were obtained from Gene Tools. ~1nl of MO at various concentrations (see text; note that 100uM MO corresponds to ~0.84 ng/nl) was injected at the 1–2 cell stage. For targeting of MOs to the endoderm, a stock containing 100 pg/nl sox32 mRNA, 0.0125 pg/nl tetramethylrhodamine dextran (10,000 MW) and various concentrations of MO (see text) was injected into one of the four central blastomeres on the 8-cell side of the 8 × 4-cell array at the 32-cell stage. sox32 encodes a transcription factor that is sufficient to drive cells to an endodermal fate (Fig. 2G–I; (Alexander and Stainier, 1999; Kikuchi et al., 2001)) and injecting sox32 mRNA into a single cell at the 32-cell stage does not perturb endoderm development (Supplemental Fig. 1). In a control experiment, 500uM fluorescein-tagged control MO was co-injected with sox32 mRNA and rhodamine dextran at the 32-cell stage. We find that the rhodamine and fluorescein signals co-localize during cleavage and blastula stages (Supplemental Fig. 2), as well as at 24 hpf (Fig. 2L–N), demonstrating that MOs are co-distributed with the rhodamine dextran lineage label in the endoderm. We also spiked the sox32 mRNA with 200 pg GFP mRNA and co-injected with rhodamine dextran. Again, we find that the fluorescein and rhodamine signals co-localize at cleavage stages (Supplemental Fig. 3) and at 24hpf (Fig. 2J, K), demonstrating that mRNAs also co-distribute with the lineage label. Taken together, these experiments demonstrate that mRNA, MOs and rhodamine dextran co-localize in the endoderm following co-injection at the 32-cell stage. In a typical experiment (injecting sox32 mRNA, MO and rhodamine dextran) ~50–60% of surviving embryos displayed rhodamine in 2 or 4 cells at the 64-cell stage and went on to express rhodamine primarily in the endoderm at 24 hpf (70–99% of embryos depending on the experiment; see Fig. 2G–N) and were morphologically normal. The remaining ~40–50% of embryos displayed broad rhodamine expression at the 64-cell stage and went on to develop with morphological abnormalities of the trunk and/or tail. These embryos were excluded from further analysis. mRNA and MO injected embryos were fixed in 4% paraformaldehyde at various developmental stages for in situ hybridization.
Figure 2.

meis3 and pbx4 are required to prevent insulin expression in the anterior endoderm. (A–F) Ectopic insulin-expressing cells arise in the anterior endoderm of meis3 and pbx4-deficient embryos. Wild type embryos (A, D), lzr mutant embryos (B, E) and embryos injected with the dominant negative Meis construct (ΔCPbx4; C, F) were assayed for insulin expression at 24hpf (A–C) and 48hpf (D–F). Ectopic insulin expression is indicated by arrows. Note that all insulin positive cells become shifted anteriorly by 48hpf (see text for details). (G–I) Cells injected with sox32 mRNA at the 32-cell stage primarily populate the endoderm. Wild type (G) or GutGFP transgenic (H, I; (Field et al., 2003)) embryos were injected with sox32 mRNA and rhodamine dextran at the 32 cell stage. At 24hpf, the rhodamine signal is observed primarily in a deep layer of the embryo adjacent to the yolk (panel G shows a confocal stack of ~125um that extends about halfway through the embryo). This region corresponds to the endoderm, since the rhodamine signal coincides with the GFP-expressing endoderm (white arrows in H, I), but not the GFP-expressing hatching gland (white asterisks in H, I) of GutGFP embryos. White arrows point to endoderm. (J, K) mRNA and rhodamine dextran co-localize in the endoderm following co-injection at the 32-cell stage. Wild type embryos were injected with sox32 mRNA, GFP mRNA and rhodamine dextran at the 32-cell stage and monitored for GFP (J) and rhodamine (K) expression at 24hpf. White arrows point to the endoderm. (L–N) MOs co-localize with the rhodamine lineage label in endoderm following co-injection at the 32-cell stage. Wild type embryos were injected with sox32 mRNA, rhodamine dextran and fluorescein-tagged MO at the 32-cell stage. Embryos were raised to 24hpf and monitored for fluorescein (L) and rhodamine (M) expression. Panel N is an overlay of panels L and M. White arrows point to endoderm. (O, P) Targeting of a meis3 MO (tMO1) to the endoderm produces ectopic anterior insulin expression (arrow in P). All panels are lateral views with anterior to the left. s1–s4 denotes the position of somites 1–4 as determined using Nomarski optics.
In situ hybridization
Antisense digoxigenin- and fluorescein-labeled probes were produced by standard methods. The meis3, insulin, sid4, carboxypeptidase A, pdx1 and shh probes used were described previously (diIorio et al., 2005; Sagerström et al., 2001). In addition, probes to hoxb6a and hoxb6b (Davidson et al., 2003), foxa2 (Strähle et al., 1993) and dlx2 (Akimenko et al., 1994) were used. One- and two-color in situ hybridization was carried out as described previously (Sagerström et al., 1996; Sagerström et al., 2001).
RESULTS
Restricted expression of meis3 in the zebrafish endoderm
We examined expression of meis family genes in the developing endoderm. Of the four meis genes identified in the zebrafish (Sagerström et al., 2001; Waskiewicz et al., 2001; Zerucha and Prince, 2001), we find that only meis3 shows a restricted expression pattern in the endoderm at 24hpf (Fig. 1D, E). Specifically, meis3 (purple stain; black arrow in Fig. 1D, E) is expressed in the endoderm anterior to the forming pancreas (identified by insulin expression; red stain; red arrow in Fig. 1D, E) and there is little overlap between the meis3 and insulin expression domains. At 48hpf, meis3 expression can be seen in the endoderm immediately adjacent to the pancreas, in a region that also expresses pdx1 and shh (compare Fig. 1I to 1J, K). In contrast, meis1, meis2 and meis4 are not expressed at high levels in the endoderm at 24 hpf (Fig. 1A–C) or 48 hpf (Fig. 1F–H), although at least two of these genes (meis1 and meis2) are expressed in the neural tube (white asterisks in Fig. 1A, B). Previous work has identified two divergent meis family genes (prep1and prep2; (Choe et al., 2002; Deflorian et al., 2004; Waskiewicz et al., 2001)), but these are reportedly expressed in broad diffuse patterns throughout the embryo and we have therefore not examined them further.
Figure 1.

meis3 is expressed in the endoderm anterior to the developing islet. (A–C) 24hpf zebrafish embryos labeled by double in situ hybridization (both probes detected in purple) for insulin (black arrows) together with meis1 (A), meis2 (B) or meis4 (C) reveal that these meis genes are not expressed in the anterior endoderm. Both meis1 and meis2 are expressed in the neural tube (white asterisks). (D, E) At 24hpf, meis3 (purple stain; black arrow) is expressed in endoderm just anterior to the insulin-expressing islet (red stain; red arrow). (F–H) Expression of meis1, meis2 and meis4 cannot be detected in the gut at 48hpf (embryos co-labeled for insulin expression [black arrow]; both probes detected in purple). (I) meis3 (purple stain; black arrow) is expressed adjacent to the insulin-expressing islet (red stain, red arrow) at 48hpf. (J) At 48 hpf, pdx1 expression is seen in duodenal cells and putative duct progenitors as well as the islet (yellow outline). (K) shh (purple stain; black arrow) is expressed adjacent to the insulin-expressing islet (red stain; red arrow) at 48hpf. Developmental stage is indicated at lower left, probes are indicated at lower right. Lateral views in AD; dorsal views in E, H–K; ventrolateral views in F, G. Anterior is to the left in all panels.
meis3 is required for expression of shh and repression of insulin in the foregut
We have previously reported a construct (ΔCPbx4) that sequesters Meis proteins in the cytoplasm of zebrafish embryos (Choe et al., 2002). This prevents Meis proteins from regulating transcription in the nucleus and produces a dominant negative phenotype (Choe et al., 2002; Choe and Sagerstrom, 2004; Choe and Sagerstrom, 2005).
We find that ΔCPbx4-mediated disruption of meis function leads to ectopic expression of insulin in the anterior endoderm. Specifically, 70% (51/77) of 24hpf and 89% (64/72) of 48hpf ΔCPbx4-injected embryos display ectopic anterior clusters of insulin-expressing cells (Fig. 2C, F). Since the ΔCPbx4 construct is likely to interfere with all meis family members (Choe et al., 2002; Choe and Sagerstrom, 2004), we next employed antisense morpholino oligos (MOs) to selectively target meis3. Two separate MOs produce ectopic insulin expression in the anterior endoderm in a dose-dependent manner (Table 1), suggesting that meis3 is required to repress insulin expression in the anterior endoderm. The greatest effect was observed with 200uM tMO1 (74% of embryos affected) and 150uM tMO2 (48% affected), whereas a 5 bp mis-matched control MO (MOCO) had minimal effect. To specifically address whether meis3 is required in the endoderm, we next co-injected meis3 tMO1 and sox32 mRNA (which encodes a transcription factor that is sufficient to drive cells to an endodermal fate; (Alexander and Stainier, 1999; Kikuchi et al., 2001)) together with rhodamine-dextran into a single cell at the 32-cell stage (see Materials and Methods). The injected material co-segregates during embryonic development and descendants of the injected cell populate primarily the endoderm (Fig. 2G–N; see Materials and Methods). We find that 67% (14/21) of 24hpf and 52% (16/31) of 48hpf embryos with tMO1 targeted to the endoderm display ectopic anterior insulin-positive cell clusters (Table 2; Fig. 2P), while embryos injected with a 5bp mismatch control MO are normal (59/59; Fig. 2O). We note that the number of ectopic insulin-expressing clusters in MO- and ΔCPbx4-injected embryos varies somewhat, such that while we usually observe one ectopic cluster (e.g. Fig. 2C, F, P), we occasionally observe two or more ectopic clusters (e.g. Fig. 5C). The position of the ectopic clusters is also somewhat variable, but we never observe insulin-expressing cells caudal to somite 4 or rostral to the level of the pericardium. We also note that in 24hpf MO- or ΔCPbx4- injected embryos, the caudal-most cluster of insulin-expressing cells is correctly positioned adjacent to somite 4 and the ectopic patches of expression are found further anteriorly, but at later stages all insulin-positive clusters shift anteriorly relative to the somites (e.g. 48hpf embryo in Fig. 2F; see also Figs. 5, 6).
Table 1.
Dose-response data for meis3 MOs
| Morpholino1 | Embryos with ectopic insulin clusters2 | Deformed Embryos3 |
|---|---|---|
| tMO1 | ||
| 100uM | 22/38 (58%) | 0/38 (0%) |
| 200uM | 86/117 (74%) | 1/117 (1%) |
| 300uM | 22/31 (71%) | 3/28 (11%) |
| 400uM | 29/52 (56%) | 5/56 (9%) |
| tMO2 | ||
| 100uM | 22/63 (35%) | 0/63 (0%) |
| 150uM | 46/96 (48%) | 0/96 (0%) |
| MOCO | ||
| 100uM | 6/48 (12%) | 0/48 (0%) |
| 200uM | 18/240 (7%) | 0/240 (0%) |
| 300uM | 3/25 (12%) | 2/25 (8%) |
| 400uM | 2/29 (7%) | 3/29 (10%) |
Embryos were injected with the indicated MO concentration at the 1-cell stage, raised to 72hpf and scored for insulin expression by in situ hybridization.
Embryos with more than one patch of insulin expression were scored as having ectopic insulin clusters.
Deformed embryos showed convergence/gastrulation defects and were not scored for insulin expression.
Table 2.
Dose response data for targeting of MOs to the endoderm
| Morpholino1 | Embryos with ectopic insulin clusters2 | Embryos with anterior shift of insulin clusters3 |
|---|---|---|
| 500uM tMO1 | ||
| 24hpf | 14/21 (67%) | 0/21 (0%) |
| 48hpf | 16/31 (52%) | 31/31 (100%) |
| 72hpf | 47/89 (53%) | 89/89 (100%) |
| 1mM tMO1 | ||
| 24hpf | 9/27 (33%) | 0/27 (0%)4 |
| 48hpf | 4/36 (11%) | 0/36 (0%)4 |
| 72hpf | 27/59 (46%) | 59/59 (100%) |
| 500uM MOCO | ||
| 24hpf | 0/5 (0%) | 0/5 (0%) |
| 48hpf | 0/54 (0%) | 0/54 (0%) |
| 72hpf | 0/30 (0%) | 0/30 (0%) |
| 1mM MOCO | ||
| 48hpf | 1/15 (6%) | 0/15 (0%) |
| 72hpf | 4/33 (12%) | 0/33 (0%) |
MOs were injected at the indicated concentration and targeted to the endoderm as outlined in Materials and Methods. insulin expression was detected by in situ hybridization at the stage indicated.
Embryos with more than one patch of insulin expression were scored as having ectopic insulin clusters.
Embryos where the predominant cluster of insulin positive cells was found anterior to the level of somite 4 were scored as having an anterior shift.
The predominant cluster of insulin positive cells showed an anterior ‘smear’ in many of these embryos (18/27 at 24hpf; 32/36 at 48hpf), possibly indicating a mild anterior shift.
Figure 5.

Disruption of meis3 and pbx4 function causes an anterior shift of insulin-positive cells. Control (A), lzr mutant (B), ΔCPbx4-injected (C), MOCO-injected (D), tMO1-injected (E) and tMO2-injected (F) embryos, as well as embryos where MOCO (G) or tMO1 (H) was targeted to the endoderm, were assayed for insulin expression by in situ hybridization at 72hpf. Note that all insulin positive cells are found anterior to somite 4 in experimental embryos (B, C, E, F, H), but not in control embryos (A, D, G). s1–s4 indicate the position of somites 1–4. All embryos are in lateral view with anterior to the left.
Figure 6.

Multiple organs are displaced anteriorly in meis- and pbx4-deficient embryos. 72hpf lzr mutant and meis-deficient embryos exhibit anterior displacement of exocrine pancreas (carboxypeptidase A, A–C) and liver (sid4, D–F) gene expression. (G–I) Anterior shifts in meis-deficient embryos can result in loss of liver-, but not islet-specific gene expression. (J, K) Posterior, dlx2-expressing arches are absent from meis-deficient embryos. A–I are in lateral view; J, K are in dorsal view. Anterior is to the left in all panels.
hedgehog expressed in the anterior endoderm is thought to prevent expression of pancreas genes in this region of zebrafish (diIorio et al., 2002; Sun and Hopkins, 2001), chick (Kim and Melton, 1998) and mouse (Hebrok et al., 2000) embryos. We therefore examined shh expression in 48hpf ΔCPbx4-injected embryos. We find that the shh-expression domain in the anterior endoderm is reduced (Fig. 3C–E), as is shh expression in the fin buds, while CNS expression appears unaffected (Fig. 3H). We next examined the relationship between insulin and shh expression in ΔCPbx4-injected embryos. Using double in situ hybridization to simultaneously detect insulin and shh expression, we find insulin positive cells (red arrowhead in Fig. 4A) caudal to the posterior margin of the shh expression domain (black arrowhead in Fig. 4A) of wild type embryos. In ΔCPbx4-injected embryos, the shh expression domain is reduced and ectopic clusters of insulin positive cells are found in regions of low shh expression (red arrow in Fig. 4C, D). Lastly, we examined foxa2, which is expressed in the anterior endoderm and gut in a pattern similar to that of shh ((Strähle et al., 1993), Fig. 3I), and find that the foxa2 expression domain in the anterior endoderm of ΔCPbx4-injected embryos is reduced in a similar fashion to shh expression (Fig. 3K).
Figure 3.

shh and foxa2 expression is disrupted in the anterior endoderm of pbx4- and meis-deficient embryos. (A) shh is expressed in the anterior endoderm (ae) and fin buds (fb) of 48hpf wild type embryos. (B) lzr mutants (as identified by the absence of pectoral fin buds) exhibit reduced shh expression in the anterior endoderm. (C–E) shh expression is reduced in the anterior endoderm and fin buds of embryos injected with the dominant negative Meis construct (ΔCPbx4). Variations in the severity of the ΔCPbx4-mediated phenotype are likely due variable distribution of the injected mRNA, as routinely seen with this type of injections. (F–H) Prominent sites of neurectodermal shh expression (black arrows) are unaltered in lzr mutant and meis-deficient embryos. (I) foxa2 is expressed in the anterior endoderm of 48hpf embryos. (J, K) Disruption of foxa2 expression in lzr mutant and meis-deficient embryos is similar to that seen for shh. Dashed black lines outline the normal expression domains of shh (A–E) and foxa2 (I–K) in the endoderm. Dorsal views in A–E, I–K; lateral views in F–H. Anterior is to the left in all panels.
Figure 4.
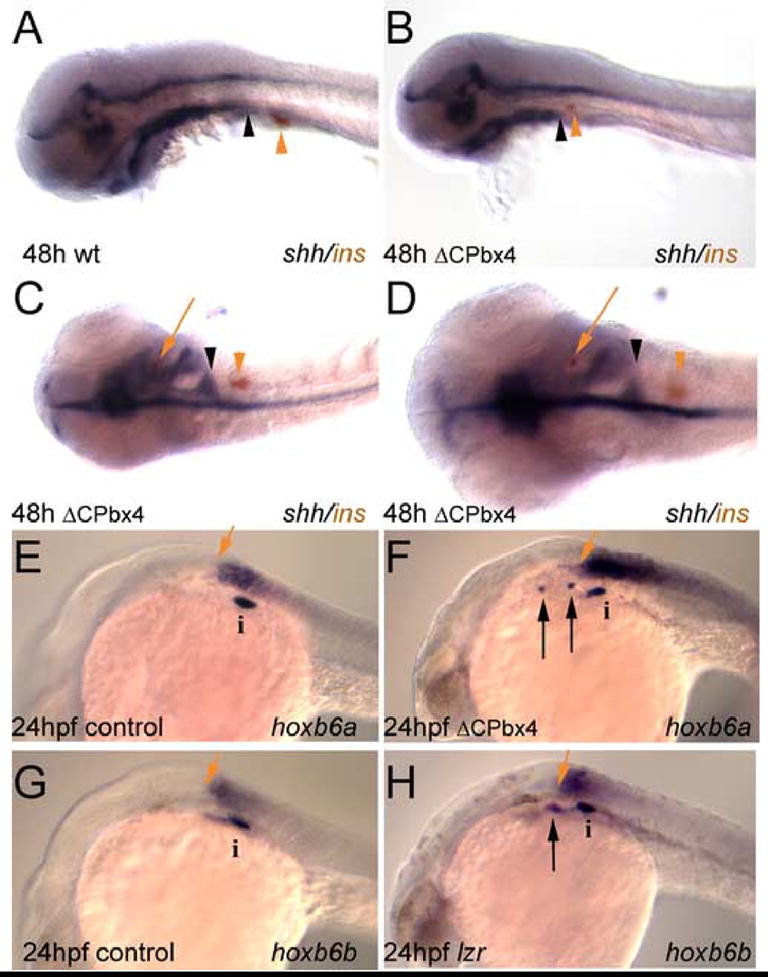
Ectopic insulin expression in meis-deficient embryos coincides with loss of shh expression. (A) insulin expressing cells (orange arrowhead) develop just caudal to the posterior margin of pharyngeal shh expression (black arrowhead). (B, C) This spatial relationship is conserved in meis-deficient embryos exhibiting caudally reduced shh expression (compare the position of arrowheads in panel A with the position in panel B, C). (D) Higher magnification of meis-deficient embryo in panel C demonstrating the appearance of insulin expressing cells (red arrow) in a shh negative region of the anterior endoderm. (E–H) hox gene expression in mesoderm is unaffected in meis-deficient embryos and lzr mutants. Mesodermal expression of hoxb6a (E) and hoxb6b (G) normally terminates just anterior to the principal islet (i). Ectopic insulin expressing cells develop anterior to the rostral limits of hox expression in meis-deficient (F) and lzr (H) embryos. In both classes of embryos the anterior limit of hox expression is unchanged relative to the principal islet. A, B, E–H are in lateral view; C and D are in dorsal view. Anterior is to the left in all panels. i, islet; black arrow indicates ectopic insulin-expressing cells; red arrows in E–H indicate anterior limit of hox gene expression.
Our findings demonstrate that meis3 regulates gene expression in the anterior endoderm. First, meis3 is required for repression of insulin expression in the anterior endoderm and targeted MO-injections suggest that this requirement is within the endoderm. Accordingly, we also do not observe defects in the CNS (shh expression in Fig. 3H) or mesoderm (hoxb6a expression in Fig 4F) of ΔCPbx4-injected embryos. Second, meis3 is required for shh and foxa2 expression in the anterior endoderm. Since shh is known to repress pancreas gene expression (diIorio et al., 2002; Hebrok et al., 2000; Kim and Melton, 1998), we conclude that meis3 act upstream of shh in an endodermal pathway that represses pancreas gene expression in the anterior endoderm. Although foxa2 appears to directly regulate shh expression in the floor plate of the CNS (reviewed in (Strahle et al., 2004)), it is not yet clear if it occupies the same position in the endodermal pathway.
Reduced shh expression and ectopic insulin expression in embryos with disrupted pbx4 function
In many systems, Meis proteins do not control transcription by themselves, but function in regulatory complexes that contain additional transcription factors. The predominant class of Meis-interaction partners are the Pbx proteins (reviewed in (Mann and Affolter, 1998)). Like Meis proteins, Pbx proteins belong to the TALE subfamily of homeodomain proteins. At least four pbx proteins have been identified in zebrafish (Pöpperl et al., 2000; Vlachakis et al., 2000; Waskiewicz et al., 2002), but pbx2 and pbx4 are the predominant pbx family members during early zebrafish development (Waskiewicz et al., 2002) and both are expressed ubiquitously (Pöpperl et al., 2000; Vlachakis et al., 2000; Waskiewicz et al., 2002). To examine whether pbx function may also be required for development of the anterior endoderm, we next examined zebrafish lazarus (lzrb557) embryos that lack zygotic pbx4 function as a result of a point mutation in the pbx4 gene (Pöpperl et al., 2000). Similar to the phenotype of ΔCPbx4- or meis3 MO-injected embryos, we find ectopic insulin expression in ~25% of embryos derived from in-crosses of lzr heterozygous parents (an embryo from a representative experiment where 21% [12/56] of embryos displayed ectopic insulin expression is shown in Fig. 2B). Furthermore, similar to ΔCPbx4-injected embryos, lzr embryos exhibit reduced shh-expression in the anterior endoderm, absence of shh expression in the fin buds, normal shh expression in the CNS and reduced endodermal foxa2 expression (Fig. 3B, G, J). The similar phenotypes of pbx4-deficient and meis3-deficient embryos are consistent with Meis3 and Pbx4 proteins cooperating to control gene expression in the anterior endoderm.
Progressive displacement of gut-associated organs in embryos with disrupted pbx4 or meis3 function
We note that disruption of meis3 or pbx4 function leads to an anterior displacement of insulin positive cells at later stages of development. In particular, 24hpf ΔCPbx4-injected, tMO1-injected and lzr mutant embryos display a prominent cluster of insulin positive cells adjacent to somite 4 (where the pancreas is normally located), as well as ectopic clusters further anteriorly (Fig. 2B, C, P). However, by 48hpf the posterior-most cluster is found further anteriorly (e.g. at the level of somite 1 in Fig. 2E and at somite 2 in Fig. 2F) and the more anterior clusters are located beneath the hindbrain. The same effect is observed in embryos where tMO1 was targeted to the endoderm (Table 2). These embryos do not display the anterior shift by 24hpf, but by 72hpf insulin expression is displaced anteriorly in 100% of embryos. This effect also correlates with the size of the shh expression domain such that in 48hpf ΔCPbx4-injected embryos, where the shh expression domain is truncated posteriorly, the predominant cluster of insulin expressing cells has been shifted anteriorly to a corresponding degree (black and red arrowheads in Fig. 4B–D).
We find that this anterior displacement is very noticeable by 72hpf. In particular, the anterior-most insulin-expressing cells are frequently seen in multiple clusters just caudal to the pericardium in lzr mutant, ΔCPbx4-injected, tMO1-injected and tMO2-injected embryos, as well as in embryos where tMO1 was targeted to the endoderm (Fig. 5A–H). To determine if the anterior displacement is specific to insulin-expressing β cells, we next examined expression of carboxypeptidase A (a marker of exocrine pancreas) and sid4 (a marker of the developing liver). At 72hpf, carbA expression is concentrated around the islet at the level of somite 4 and tapers caudally along the gut tube of wild type embryos (Fig. 6A). This caudal expression is lost in lzr and ΔCPbx4-injected embryos and remaining expression is seen anterior to somite 1 (Fig. 6B, C). Further, in 72hpf wild type embryos, sid4 expression in the developing liver extends from somite 1 caudally to somite 4 (Fig. 6D), but this expression is displaced anteriorly in lzr mutant and ΔCPbx4-injected embryos (Fig. 6E, F). Notably, in these embryos the anterior edge of the liver is in close proximity to the caudal pericardium and the posterior boundary of sid4 expression is at the level of somite 1. Lastly, double in situ analysis of insulin and sid4 gene expression in ΔCPbx4-injected embryos suggests that the anterior shift in organ development occasionally results in the loss of anterior foregut gene expression (sid4 expression in the liver), while posterior foregut gene expression (insulin expression in the pancreas) is never lost (Fig. 6G–I). Thus, disruption of pbx4 or meis3 function leads to a progressive anterior-ward shift in the position of endodermal gene expression domains and occasionally results in the loss of gene expression marking the anterior gut.
As a result of this anterior-ward shift, the endodermal organs, as well as the somitic mesoderm, appear to move closer to the eye, indicating that tissues in the intervening pharyngeal region may not develop properly. In particular, endoderm in the pharyngeal region forms pouches that interact with neural crest-derived cells to generate the branchial arches. We therefore reasoned that the anterior displacement of endodermal organs upon disruption of meis3 and pbx4 function might be due to underdevelopment of the branchial arches. Indeed, we find that the posterior-most expression of dlx2 (arrow in Fig. 6J), a marker of the pharyngeal arches, is lost in ΔCPbx4-injected embryos (Fig 6K). This reduction in branchial arches might stem in part from neural crest defects, since meis3 and pbx4 also function in the hindbrain (Choe et al., 2002; Pöpperl et al., 2000; Waskiewicz et al., 2001) where neural crest cells originate. However, the pharyngeal arches are underdeveloped also in smoothened (smu) mutants, indicating that Hh signaling is directly required for their formation (Chen et al., 2001). Thus, loss of shh expression in the pharyngeal endoderm of pbx4 and meis3 deficient embryos likely underlies both the ectopic insulin expression (loss of shh mediated repression) and the subsequent anterior displacement of the pancreas and liver (failure of pharyngeal growth).
DISCUSSION
Previous work indicates that shh acts to exclude pancreas-specific gene expression from the anterior endoderm (reviewed in (Hebrok, 2003)). Here we demonstrate that TALE homeodomain proteins (meis3 and pbx4) are required for shh expression in the endoderm anterior to the pancreas. In particular, reduced meis3 and pbx4 function leads to loss of shh expression and enables ectopic insulin expression in the anterior endoderm of zebrafish embryos. We also a observe a progressive anterior-ward displacement of endodermal organs in embryos with reduced meis3 and pbx4 function and we conclude that this effect results from underdevelopment of the pharyngeal region in the absence of shh. While targeted deletion of meis genes has not been reported to affect pancreas development in the mouse (Azcoitia et al., 2005; Ferretti et al., 2006; Hisa et al., 2004), pbx function has been implicated in pancreas development. In particular, targeted deletion of pbx1 in the mouse causes pancreas hypoplasia with significantly reduced numbers of endocrine and exocrine cells, but does not appear to cause ectopic expression of pancreas-specific genes (Kim et al., 2002). This discrepancy may be explained by other pbx genes (e.g. the broadly expressed pbx2 gene), being required to maintain shh expression in the anterior endoderm of the mouse.
Tissue specific regulation of shh expression
Our results indicate that meis3 and pbx4 are required for shh expression in the anterior endoderm and the fin buds, but not in the CNS, suggesting that the pathway regulating shh expression differs between tissues. This is of particular interest since shh produced in different tissues and at different stages of development, may have different effects on pancreas development. On the one hand, Hedgehog signaling appears to restrict pancreas formation within the endoderm (reviewed in (Hebrok, 2003)). Accordingly, exposing chick embryos to cyclopamine (a drug that blocks Hedgehog signaling) leads to ectopic differentiation of endocrine cells in the anterior endoderm (Kim and Melton, 1998) and treating endoderm explants with anti-Shh antibodies increases pancreas-specific gene expression (Hebrok et al., 1998). On the other hand, zebrafish embryos with mutations in components of the Hedgehog signaling cascade lack a pancreas and over-expression of shh expands the pancreas (diIorio et al., 2002; Roy et al., 2001), suggesting that Hedgehog signaling is required for pancreas formation. To reconcile these results, it has been proposed that Hedgehog protein is produced in two distinct locations and is required at two stages of pancreas development. According to this model, hedgehog genes expressed in dorsal mesoderm initially provide an inductive signal required for pancreas formation and subsequently shh expressed within the anterior endoderm restricts the size of the pancreas primordium (diIorio et al., 2002). We interpret our results to mean that meis3 and pbx4 are required for the second, but not the first, shh-dependent step. Notably, shh is likely to play additional roles in pancreas development at even later stages (reviewed in (Hebrok, 2003)).
Although the shh regulatory pathways appear to differ between the endoderm and the CNS, they may share some features. For instance, foxa2 is a known transcriptional regulator of shh expression in the floor plate and foxa2 is expressed in the anterior endoderm in a domain similar to shh (Fig. 2; reviewed in (Strahle et al., 2004)). Furthermore, we find that foxa2 expression is disrupted in the absence of meis and pbx function. These findings are consistent with foxa2 acting downstream of pbx4 and meis3 to regulate shh expression in the endoderm. However, initial analyses of foxa2 (monorail) mutant zebrafish embryos did not report any endoderm defects (Norton et al., 2005), although these defects may be subtle, especially since other foxa genes may act redundantly in the endoderm.
Meis3 and Pbx4 may act in multimeric complexes to regulate endodermal shh transcription
Meis and Pbx proteins belong to the homeodomain super family of DNA-binding proteins and have been shown to form heterodimers both in solution and on DNA templates (reviewed in (Mann and Affolter, 1998)). Since we observe a similar phenotype in the endoderm upon disruption of meis3 or pbx4 function, it is likely that Meis3 and Pbx4 act in a complex to regulate shh expression in the endoderm. Meis proteins reportedly contain a transcription activation domain that recruits the CBP coactivator (Huang et al., 2005) and Pbx proteins have been shown to interact with NCoR, SMRT and HDAC co-repressors (Asahara et al., 1999; Saleh et al., 2000), suggesting that a complex consisting of Meis3 and Pbx4 might be sufficient to regulate gene expression in the endoderm. However, in many situations Meis and Pbx interact with additional transcription factors (e.g. MyoD, Engrailed, Hox proteins (Chan et al., 1994; Knoepfler et al., 1999; Peltenburg and Murre, 1996; van Dijk and Murre, 1994)), raising the possibility that additional proteins may cooperate with Meis3 and Pbx4 to regulate endodermal gene expression. One candidate for such a factor is the homeodomain transcription factor pdx1, which is expressed early in the duodenum/pancreas primordium. Targeted deletion of pdx1 in the mouse and antisense-mediated knockdown of pdx1 in zebrafish causes pancreatic agenesis without completely blocking formation of pancreatic precursors (Jonsson et al., 1994; Offield et al., 1996; Yee et al., 2001), illustrating the importance of pdx1 in early pancreas development. Pbx and Meis proteins form complexes with Pdx1 in vitro (Peers et al., 1995; Swift et al., 1998) and a form of Pdx1 that cannot bind Pbx does not restore normal pancreas development in pdx1 mutant mouse embryos (Dutta et al., 2001). Furthermore, pbx1+/−/pdx1+/− trans-heterozygous mice are predisposed to develop diabetes (Kim et al., 2002), revealing a genetic interaction between these genes. However, the phenotype we observe upon disruption of meis and pbx function in zebrafish does not resemble the phenotype resulting from loss of pdx1 function, suggesting that Meis3 and Pbx4 do not cooperate with Pdx1 to control shh expression. Importantly, this does not exclude the possibility that Meis/Pbx/Pdx1 complexes function at later stages of pancreas development.
Supplementary Material
sox32 mRNA does not perturb normal endoderm development. A single cell was injected with a mixture of sox32 mRNA and rhodamine dextran at the 32-cell stage as outlined in Materials and Methods. Embryos were fixed and assayed for expression of insulin at 24 hpf (A, B), p48 at 30 hpf (C, D), sid4 at 48 hpf (E, F), pdx1 at 48 hpf (G, H), insulin at 72 hpf (I, J) and carbA at 72 hpf (K, L) by whole mount in situ hybridization. All embryos are in lateral view with anterior to the left.
Morpholinos and rhodamine dextran co-localize following co-injection. A single cell was injected with a mixture of sox32 mRNA, rhodamine dextran and fluorescein-tagged control MO at the 32-cell stage as outlined in Materials and Methods. Embryos were monitored by bright field (A–D) and for rhodamine (E–H) and fluorescein (I–L) signals at the 64-cell stage (~2 hpf), the 128-cell stage (~2.5 hpf), the 512-cell stage (~3 hpf) and the oblong stage (~3.5 hpf).
GFP mRNA and rhodamine dextran co-localize following co-injection. A single cell was injected with a mixture of sox32 mRNA, GFP mRNA and rhodamine dextran at the 32-cell stage as outlined in Materials and Methods. Embryos were monitored by bright field (A, B) and for rhodamine (C, D) and fluorescein (E, F) signals at the 256-cell stage (~2 hpf; A, C, E) and the oblong stage (~3.5 hpf; B, D, F).
Acknowledgments
CGS and PJD wish to thank Dr Aldo Rossini for his enthusiastic support during this project. The authors are grateful to Dr Cecilia Moens for the gift of meis1, 2, 4 probes and the lazarus line and to Drs Alan Davidson and Len Zon for the hoxb6a and hoxb6b probes. This work was supported by JDRF grant 1-2004-600 and NIH grants DK068237 and 5P30 DK32520. PJD wishes to acknowledge support from the Iacocca Foundation and the Worcester foundation for Biomedical Research. The contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akimenko MA, Ekker M, Wegner J, Lin W, Westerfield M. Combinatorial expression of three zebrafish genes related to distal-less: part of a homeobox gene code for the head. J Neurosci. 1994;14:3475–86. doi: 10.1523/JNEUROSCI.14-06-03475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander J, Stainier DY. A molecular pathway leading to endoderm formation in zebrafish. Curr Biol. 1999;9:1147–57. doi: 10.1016/S0960-9822(00)80016-0. [DOI] [PubMed] [Google Scholar]
- Asahara H, Dutta S, Kao HY, Evans RM, Montminy M. Pbx-Hox heterodimers recruit coactivator-corepressor complexes in an isoform-specific manner. Mol Cell Biol. 1999;19:8219–25. doi: 10.1128/mcb.19.12.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azcoitia V, Aracil M, Martinez AC, Torres M. The homeodomain protein Meis1 is essential for definitive hematopoiesis and vascular patterning in the mouse embryo. Dev Biol. 2005;280:307–20. doi: 10.1016/j.ydbio.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Brendolan A, Ferretti E, Salsi V, Moses K, Quaggin S, Blasi F, Cleary ML, Selleri L. A Pbx1-dependent genetic and transcriptional network regulates spleen ontogeny. Development. 2005;132:3113–26. doi: 10.1242/dev.01884. [DOI] [PubMed] [Google Scholar]
- Chan SK, Jaffe L, Capovilla M, Botas J, Mann RS. The DNA binding specificity of Ultrabithorax is modulated by cooperative interactions with extradenticle, another homeoprotein. Cell. 1994;78:603–15. doi: 10.1016/0092-8674(94)90525-8. [DOI] [PubMed] [Google Scholar]
- Chen W, Burgess S, Hopkins N. Analysis of the zebrafish smoothened mutant reveals conserved and divergent functions of hedgehog activity. Development. 2001;128:2385–96. doi: 10.1242/dev.128.12.2385. [DOI] [PubMed] [Google Scholar]
- Choe S-K, Vlachakis N, Sagerström CG. Meis family proteins are required for hindbrain development in the zebrafish. Development. 2002;129:585–595. doi: 10.1242/dev.129.3.585. [DOI] [PubMed] [Google Scholar]
- Choe S-K, Sagerstrom CG. Paralog group 1 hox genes regulate rhombomere 5/6 expression of vnhf1, a repressor of rostral hindbrain fates, in a meis-dependent manner. Developmental Biology. 2004;271:350–361. doi: 10.1016/j.ydbio.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Choe S-K, Sagerstrom CG. Variable meis-dependence among paralog group-1 hox proteins. Biochemical and Biophysical Research Communications. 2005;331:1384–1391. doi: 10.1016/j.bbrc.2005.04.063. [DOI] [PubMed] [Google Scholar]
- Davidson AJ, Ernst P, Wang Y, Dekens MP, Kingsley PD, Palis J, Korsmeyer SJ, Daley GQ, Zon LI. cdx4 mutants fail to specify blood progenitors and can be rescued by multiple hox genes. Nature. 2003;425:300–6. doi: 10.1038/nature01973. [DOI] [PubMed] [Google Scholar]
- Deflorian G, Tiso N, Ferretti E, Meyer D, Blasi F, Bortolussi M, Argenton F. Prep1.1 has essential genetic functions in hindbrain development and cranial neural crest cell differentiation. Development. 2004;131:613–27. doi: 10.1242/dev.00948. [DOI] [PubMed] [Google Scholar]
- diIorio PJ, Moss JB, Sbrogna JL, Karlstrom RO, Moss LG. Sonic hedgehog is required early in pancreatic islet development. Dev Biol. 2002;244:75–84. doi: 10.1006/dbio.2002.0573. [DOI] [PubMed] [Google Scholar]
- diIorio PJ, Runko A, Farrell CA, Roy N. Sid4: A secreted vertebrate immunoglobulin protein with roles in zebrafish embryogenesis. Dev Biol. 2005;282:55–69. doi: 10.1016/j.ydbio.2005.02.036. [DOI] [PubMed] [Google Scholar]
- Dutta S, Gannon M, Peers B, Wright C, Bonner-Weir S, Montminy M. PDX:PBX complexes are required for normal proliferation of pancreatic cells during development. Proc Natl Acad Sci U S A. 2001;98:1065–70. doi: 10.1073/pnas.031561298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti E, Villaescusa JC, Di Rosa P, Fernandez-Diaz LC, Longobardi E, Mazzieri R, Miccio A, Micali N, Selleri L, Ferrari G, Blasi F. Hypomorphic mutation of the TALE gene Prep1 (pKnox1) causes a major reduction of Pbx and Meis proteins and a pleiotropic embryonic phenotype. Mol Cell Biol. 2006;26:5650–62. doi: 10.1128/MCB.00313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field HA, Ober EA, Roeser T, Stainier DY. Formation of the digestive system in zebrafish. I. Liver morphogenesis. Dev Biol. 2003;253:279–90. doi: 10.1016/s0012-1606(02)00017-9. [DOI] [PubMed] [Google Scholar]
- Hebrok M. Hedgehog signaling in pancreas development. Mech Dev. 2003;120:45–57. doi: 10.1016/s0925-4773(02)00331-3. [DOI] [PubMed] [Google Scholar]
- Hebrok M, Kim SK, Melton DA. Notochord repression of endodermal Sonic hedgehog permits pancreas development. Genes Dev. 1998;12:1705–13. doi: 10.1101/gad.12.11.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebrok M, Kim SK, St Jacques B, McMahon AP, Melton DA. Regulation of pancreas development by hedgehog signaling. Development. 2000;127:4905–13. doi: 10.1242/dev.127.22.4905. [DOI] [PubMed] [Google Scholar]
- Hill ID, Lebenthal E. Congenital abnormalities of the exocrine pancreas. In: Go V, Dimango E, Gardner J, Lebenthal E, Reber H, Scheele G, editors. The Pancreas: Biology, Pathobiology and Disease. Raven; New York: 1993. pp. 1029–1040. [Google Scholar]
- Hisa T, Spence SE, Rachel RA, Fujita M, Nakamura T, Ward JM, Devor-Henneman DE, Saiki Y, Kutsuna H, Tessarollo L, Jenkins NA, Copeland NG. Hematopoietic, angiogenic and eye defects in Meis1 mutant animals. Embo J. 2004;23:450–9. doi: 10.1038/sj.emboj.7600038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Rastegar M, Bodner C, Goh SL, Rambaldi I, Featherstone M. MEIS C-termini harbor transcriptional activation domains that respond to cell signaling. J Biol Chem. 2005 doi: 10.1074/jbc.M413963200. [DOI] [PubMed] [Google Scholar]
- Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–9. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- Kaczirek K, Niederle B. Nesidioblastosis: an old term and a new understanding. World J Surg. 2004;28:1227–30. doi: 10.1007/s00268-004-7598-7. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y, Agathon A, Alexander J, Thisse C, Waldron S, Yelon D, Thisse B, Stainier DY. casanova encodes a novel Sox-related protein necessary and sufficient for early endoderm formation in zebrafish. Genes Dev. 2001;15:1493–505. doi: 10.1101/gad.892301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Melton DA. Pancreas development is promoted by cyclopamine, a hedgehog signaling inhibitor. Proc Natl Acad Sci U S A. 1998;95:13036–41. doi: 10.1073/pnas.95.22.13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Selleri L, Lee JS, Zhang AY, Gu X, Jacobs Y, Cleary ML. Pbx1 inactivation disrupts pancreas development and in Ipf1-deficient mice promotes diabetes mellitus. Nat Genet. 2002;30:430–5. doi: 10.1038/ng860. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullman B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dynamics. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Knoepfler PS, Bergstrom DA, Uetsuki T, Dac-Korytko I, Sun YH, Wright WE, Tapscott SJ, Kamps MP. A conserved motif N-terminal to the DNA-binding domains of myogenic bHLH transcription factors mediates cooperative DNA binding with pbx-Meis1/Prep1. Nucleic Acids Res. 1999;27:3752–61. doi: 10.1093/nar/27.18.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Jordan N, Melton D, Grapin-Botton A. Signals from lateral plate mesoderm instruct endoderm toward a pancreatic fate. Dev Biol. 2003;259:109–22. doi: 10.1016/s0012-1606(03)00183-0. [DOI] [PubMed] [Google Scholar]
- Lachman MF, Wright Y, Whiteman DA, Herson V, Greenstein RM. Brief clinical report: a 46,XY phenotypic female with Smith-Lemli-Opitz syndrome. Clin Genet. 1991;39:136–41. doi: 10.1111/j.1399-0004.1991.tb03000.x. [DOI] [PubMed] [Google Scholar]
- Maeda R, Ishimura A, Mood K, Park EK, Buchberg AM, Daar IO. Xpbx1b and Xmeis1b play a collaborative role in hindbrain and neural crest gene expression in Xenopus embryos. Proc Natl Acad Sci U S A. 2002;99:5448–53. doi: 10.1073/pnas.082654899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann RS, Affolter M. Hox proteins meet more partners. Curr Opin Genet Dev. 1998;8:423–9. doi: 10.1016/s0959-437x(98)80113-5. [DOI] [PubMed] [Google Scholar]
- Mercader N, Tanaka EM, Torres M. Proximodistal identity during vertebrate limb regeneration is regulated by Meis homeodomain proteins. Development. 2005;132:4131–42. doi: 10.1242/dev.01976. [DOI] [PubMed] [Google Scholar]
- Norton WH, Mangoli M, Lele Z, Pogoda HM, Diamond B, Mercurio S, Russell C, Teraoka H, Stickney HL, Rauch GJ, Heisenberg CP, Houart C, Schilling TF, Frohnhoefer HG, Rastegar S, Neumann CJ, Gardiner RM, Strahle U, Geisler R, Rees M, Talbot WS, Wilson SW. Monorail/Foxa2 regulates floorplate differentiation and specification of oligodendrocytes, serotonergic raphe neurones and cranial motoneurones. Development. 2005;132:645–58. doi: 10.1242/dev.01611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CV. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–95. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- Peers B, Sharma S, Johnson T, Kamps M, Montminy M. The pancreatic islet factor STF-1 binds cooperatively with Pbx to a regulatory element in the somatostatin promoter: importance of the FPWMK motif and of the homeodomain. Mol Cell Biol. 1995;15:7091–7. doi: 10.1128/mcb.15.12.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltenburg LT, Murre C. Engrailed and Hox homeodomain proteins contain a related Pbx interaction motif that recognizes a common structure present in Pbx. Embo J. 1996;15:3385–93. [PMC free article] [PubMed] [Google Scholar]
- Pöpperl H, Rikhof H, Chang H, Haffter P, Kimmel CB, Moens CB. Iazarus is a novel pbx gene that globally mediates hox gene function in zebrafish. Mol Cell. 2000;6:255–67. doi: 10.1016/s1097-2765(00)00027-7. [DOI] [PubMed] [Google Scholar]
- Porter JA, Young KE, Beachy PA. Cholesterol modification of hedgehog signaling proteins in animal development. Science. 1996;274:255–9. doi: 10.1126/science.274.5285.255. [DOI] [PubMed] [Google Scholar]
- Roy S, Qiao T, Wolff C, Ingham PW. Hedgehog signaling pathway is essential for pancreas specification in the zebrafish embryo. Curr Biol. 2001;11:1358–63. doi: 10.1016/s0960-9822(01)00402-x. [DOI] [PubMed] [Google Scholar]
- Sagerström CG, Grinblat Y, Sive H. Anteroposterior patterning in the zebrafish, Danio rerio: an explant assay reveals inductive and suppressive cell interactions. Development. 1996;122:1873–1883. doi: 10.1242/dev.122.6.1873. [DOI] [PubMed] [Google Scholar]
- Sagerström CG, Kao B, Lane ME, Sive H. Isolation and characterization of posteriorly expressed genes in the zebrafish gastrula. Developmental Dynamics. 2001;220:402–408. doi: 10.1002/dvdy.1119. [DOI] [PubMed] [Google Scholar]
- Saleh M, Rambaldi I, Yang XJ, Featherstone MS. Cell signaling switches HOX-PBX complexes from repressors to activators of transcription mediated by histone deacetylases and histone acetyltransferases. Mol Cell Biol. 2000;20:8623–33. doi: 10.1128/mcb.20.22.8623-8633.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selleri L, Depew MJ, Jacobs Y, Chanda SK, Tsang KY, Cheah KS, Rubenstein JL, O’Gorman S, Cleary ML. Requirement for Pbx1 in skeletal patterning and programming chondrocyte proliferation and differentiation. Development. 2001;128:3543–57. doi: 10.1242/dev.128.18.3543. [DOI] [PubMed] [Google Scholar]
- Stafford D, Prince VE. Retinoic acid signaling is required for a critical early step in zebrafish pancreatic development. Curr Biol. 2002;12:1215–20. doi: 10.1016/s0960-9822(02)00929-6. [DOI] [PubMed] [Google Scholar]
- Strähle U, Blader P, Henrique D, Ingham PW. Axial, a zebrafish gene expressed along the developing body axis, shows altered expression in cyclops mutant embryos. Genes Dev. 1993;7:1436–1446. doi: 10.1101/gad.7.7b.1436. [DOI] [PubMed] [Google Scholar]
- Strahle U, Lam CS, Ertzer R, Rastegar S. Vertebrate floor-plate specification: variations on common themes. Trends Genet. 2004;20:155–62. doi: 10.1016/j.tig.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Sun Z, Hopkins N. vhnf1, the MODY5 and familial GCKD-associated gene, regulates regional specification of the zebrafish gut, pronephros, and hindbrain. Genes Dev. 2001;15:3217–29. doi: 10.1101/gad946701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift GH, Liu Y, Rose SD, Bischof LJ, Steelman S, Buchberg AM, Wright CV, MacDonald RJ. An endocrine-exocrine switch in the activity of the pancreatic homeodomain protein PDX1 through formation of a trimeric complex with PBX1b and MRG1 (MEIS2) Mol Cell Biol. 1998;18:5109–20. doi: 10.1128/mcb.18.9.5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk MA, Murre C. extradenticle raises the DNA binding specificity of homeotic selector gene products. Cell. 1994;78:617–24. doi: 10.1016/0092-8674(94)90526-6. [DOI] [PubMed] [Google Scholar]
- Vlachakis N, Ellstrom DR, Sagerström CG. A novel pbx family member expressed during early zebrafish embryogenesis forms trimeric complexes with Meis3 and Hoxb1b. Dev Dyn. 2000;217:109–19. doi: 10.1002/(SICI)1097-0177(200001)217:1<109::AID-DVDY10>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Waskiewicz AJ, Rikhof HA, Hernandez RE, Moens CB. Zebrafish Meis functions to stabilize Pbx proteins and regulate hindbrain patterning. Development. 2001;128:4139–51. doi: 10.1242/dev.128.21.4139. [DOI] [PubMed] [Google Scholar]
- Waskiewicz AJ, Rikhof HA, Moens CB. Eliminating zebrafish pbx proteins reveals a hindbrain ground state. Dev Cell. 2002;3:723–33. doi: 10.1016/s1534-5807(02)00319-2. [DOI] [PubMed] [Google Scholar]
- Wolf G. The function of cholesterol in embryogenesis. J Nutr Biochem. 1999;10:188–192. doi: 10.1016/s0955-2863(98)00102-8. [DOI] [PubMed] [Google Scholar]
- Yee NS, Yusuff S, Pack M. Zebrafish pdx1 morphant displays defects in pancreas development and digestive organ chirality, and potentially identifies a multipotent pancreas progenitor cell. Genesis. 2001;30:137–40. doi: 10.1002/gene.1049. [DOI] [PubMed] [Google Scholar]
- Zerucha T, Prince VE. Cloning and developmental expression of a zebrafish meis2 homeobox gene. Mech Dev. 2001;102:247–50. doi: 10.1016/s0925-4773(01)00299-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
sox32 mRNA does not perturb normal endoderm development. A single cell was injected with a mixture of sox32 mRNA and rhodamine dextran at the 32-cell stage as outlined in Materials and Methods. Embryos were fixed and assayed for expression of insulin at 24 hpf (A, B), p48 at 30 hpf (C, D), sid4 at 48 hpf (E, F), pdx1 at 48 hpf (G, H), insulin at 72 hpf (I, J) and carbA at 72 hpf (K, L) by whole mount in situ hybridization. All embryos are in lateral view with anterior to the left.
Morpholinos and rhodamine dextran co-localize following co-injection. A single cell was injected with a mixture of sox32 mRNA, rhodamine dextran and fluorescein-tagged control MO at the 32-cell stage as outlined in Materials and Methods. Embryos were monitored by bright field (A–D) and for rhodamine (E–H) and fluorescein (I–L) signals at the 64-cell stage (~2 hpf), the 128-cell stage (~2.5 hpf), the 512-cell stage (~3 hpf) and the oblong stage (~3.5 hpf).
GFP mRNA and rhodamine dextran co-localize following co-injection. A single cell was injected with a mixture of sox32 mRNA, GFP mRNA and rhodamine dextran at the 32-cell stage as outlined in Materials and Methods. Embryos were monitored by bright field (A, B) and for rhodamine (C, D) and fluorescein (E, F) signals at the 256-cell stage (~2 hpf; A, C, E) and the oblong stage (~3.5 hpf; B, D, F).


