Abstract
Type I interferon (IFN) contributes significantly to innate immune responses to pathogen infections in macrophages. Our previous studies demonstrate that Ubp43, an ISG15-specific isopeptidase, is highly expressed in macrophages and noncatalytically inhibits Type I IFN signaling. To understand the effect of Type I IFN and Ubp43 in macrophage activation, we analyzed the expression of IFN-β stimulated genes in wild-type and Ubp43−/− bone marrow derived macrophages (BMMs). Here, we show that Ubp43 regulates IFN-β stimulated genes at genome level. IFN hypersensitivity of Ubp43−/− BMMs resulted in the identification of 749 unique genes that are upregulated by IFN-β, including a large group of previously unidentified IFN-stimulated genes. Functional analyses of these genes showed that Type I IFN strongly induced the expression of a group of immune response related genes, including genes for antigen presentation, antiviral responses, and chemokine and cytokine production. These results provide excellent biochemical support for the high resistance of viral and bacterial infection of Ubp43 knockout mice, suggesting that Ubp43 is a potential therapeutic target for the enhancement of immune responses against infections.
INTRODUCTION
Ubp43 (Usp18) is a member of the ubiquitin specific protease (USP) family [1]. A substrate specificity study indicated that Ubp43 preferentially removes ISG15 from its conjugates, compared to ubiquitin, SUMO, and Nedd8 conjugates [2]. The generation and analysis of Ubp43−/− mice showed a severe phenotype including premature death [3], hypersensitivity to double stranded RNA Poly(I-C) treatment [4], and resistance to certain viral and bacterial infections [5; 6]. We reported previously that Ubp43 deficient cells are hypersensitive to Type I IFN stimulation [4] and Ubp43 inhibits IFN signaling through the interaction with IFN receptor and independently of its ISG15 isopeptidase activity [7].
Among various hematopoietic cell lines tested, monocyte/macrophage lines exhibited the highest level of Ubp43 expression [1]. In normal adult mice, the highest level of murine Ubp43 expression was detected in thymus and macrophages, indicating that Ubp43 plays an important role in macrophages [1]. Recent study showed that IFN-β contributes significantly to LPS mediated signaling [8] and it is believed that Type I IFN also contributes to LPS induced macrophage activation. To understand the effects of Ubp43 on IFN signaling at genome level and to investigate the role of Type I IFN in macrophage immune responses, we treated wild-type and Ubp43−/− BMMs with IFN-β and compared IFN induced transcriptional profiling. We not only observed the IFN hypersensitivity in Ubp43−/− BMMs, which has broad spectrum effects on most, if not all, IFN stimulated genes, but also revealed significant numbers of previously unrecognized IFN-β-stimulated genes. Most importantly, functional analyses of these genes showed that a group of immune response related genes, including genes involved in antigen presentation, antiviral responses, and chemokine and cytokine production, were strongly induced by Type I IFN in Ubp43−/− BMMs. These results provide strong biochemical support of the high resistance to viral and bacterial infection of Ubp43 knockout mice, suggesting that Ubp43 is a potential therapeutic target for the regulation of immune responses.
MATERIALS AND METHODS
Mice and Cell Culture
The generation of Ubp43−/− mice and the culture of primary BMMs has been described previously [3; 5].
Microarray Analysis
BMMs were treated with mouse IFN-β (ICN Pharmaceuticals, Cleveland, OH) (100 U/ml) for different times as indicated in the figure legends. RNA was extracted using RNA Bee reagent according to the manufacturer’s instructions (Tel-Test), purified using Qiagen RNeasy Mini Kit (Qiagen, Valencia, CA). Purified RNAs were converted to double-stranded cDNA with a SuperScript kit (GibcoBRL) and an oligo-dT primer containing a T7 RNA polymerase promoter (Genset). Biotin-labeled cRNAs were generated from the cDNA samples by in vitro transcription with T7 RNA polymerase (Enzo kit, Enzo Diagnostics). The labeled cRNAs were fragmented to an average size of 35 to 200 bases by mild alkaline treatment at 94 °C for 35 min. Labeled cRNA were hybridized to Affymetrix Mouse Genome 430A 2.0 arrays (Affymetrix, Santa Clara, CA) in accordance with the procedures established by Affymetrix (Affymetrix Gene ChipR Expression Analysis Technical Manual). The experiment was performed in duplicate to minimize artifact. Robust Multichip Average (RMA) was used to convert the intensity values to expression values [9; 10]. RMA consists of a three-step approach that uses a background correction on the PM data (Perfect Match), a quantitative normalization and a summarization of the probe set information by using Tukey's median polish algorithm. “Present” and “Absent” calls were calculated in the R software package as implemented in the Affymetrix Microarray Suite version 5. This algorithm performs a Wilcoxon signed rank-based gene expression to make “Present” and “Absent” calls. Genes were filtered out which have “Absent” calls for all samples. The expression values of the remaining probe sets were analyzed using dChip2006 software, which is freely available to academic users (http://biosun1.harvard.edu/~cli/dchip2006.exe). To identify IFN stimulated genes, the genes with a difference in expression values between IFN treated and untreated groups > 100, a fold change >2 and a P value <0.05 were considered as genes showing significant differential expression. The hierarchical clustering algorithm was performed among the standardized expression values of identified genes in wild-type and Ubp43−/− BMMs together. The identified genes were annotated to unique genes using ingenuity pathway analysis (Ingenuity Systems, Mountain View, CA) (www.analysis.ingenuity.com/). If a single gene is presented by multiple probes, only one probe was randomly picked and used for subsequent analysis. Grouping of genes into different biological functions was performed using the David database (http://david.abcc.ncifcrf.gov), Ingenuity Pathway Analysis and literature search.
Northern Blotting
Total RNA from BMMs was isolated using RNA Bee reagent according to the manufacturer’s instructions (Tel-Test). This set of RNA is independently prepared and different from the RNA used in Microarray analysis. Ten micrograms of total RNA from each time point was separated in an agarose/formaldehyde gel (0.22 M), blotted on Hybond N+ membrane (Amersham Biosciences), and probed with 32P-labeled cDNAs, which were labeled with Prime-It II Random Primer Labeling Kit (Stratagene) using partial cDNA fragments as a template. The partial cDNA fragments were obtained by PCR amplification using a cDNA library prepared from IFN treated mouse BMMs as a template. The primers used were IRF7: 5’atggctgaag tgaggggggt3’and 5’tcaaggccac tgacccaggt3’; OAS2: 5’atgggaaact ggctgactgg3’ and 5’ttgagatcca catagacaca3’ GBP1: 5’atggcctcag agatccacat3’ and 5’ccattgactg tgatgcctcc3’; IRF1: 5’atgccaatca ctcgaatgcg3’ and 5’ctatggtgca caaggaatgg3’; Mx1: 5’agctgaatga gggagaggag3’ and 5’gaagaactct gaaatgaggg3’; Ly86: 5’gggaacatct gtccctggagcta3’ and 5’ccattgacca gtgttccaagcag3’; HDAC1: 5’ccctcctcat ctgagtccgagaa3’ and 5’gtttcaactt gcccatgctgatg3’; Gadd45γ: 5’atgactctgg aagaagtccg3’ and 5’tcactcggga agggtgatgc3’; Lipg: 5’ccagtcaacc accacgacgttag3’ and 5’aaagcccaaa ccaaaaacctgct3’; P8: 5’gttcttttgg gggctgtcttcct3’ and 5’cccttcccag caacctctaaacc3’; 5-Mar: 5’aacagactcg tcaggatgagagg3’ and 5’atgccggacc aagcccttcaaca3’; Gnb4: 5’atgagcgagc tggagcagct3’ and 5’tcaattccag attctaagaa3’; Serpina3γ:5’ggatgagaag aggtctgtga3’ and 5’ataaagaggg caatgtgagc3’. As a loading control, 28 S rRNA of the same set of experiments was shown.
RESULTS
Ubp43−/− BMM showed higher and extended expression of IFN stimulated genes
Macrophages play important roles in immune response and are the major defense against different pathogens [11]. Our previous study showed that Ubp43 was highly expressed in macrophages, suggesting that Ubp43 plays an important role in macrophage function [1]. To understand the effect of Type I IFN on macrophage activation and to analyze the expression of IFN stimulated genes altered in the absence of Ubp43, we performed GeneChip analysis to study IFN-β induced transcriptional profiling in BMMs from wild-type and Ubp43−/− mice. We treated macrophages from wild-type and Ubp43−/− mice with IFN-β for different times and performed Affymetrix Mouse Genome 430A 2.0 array, which can simultaneously examine the expression of 45,000 different probe sets. First, we compared the data from the IFN-β untreated and treated BMMs from wild-type mice. According to the criteria outlined in Materials and Methods, dChip2006 Compare Sample procedure allowed the identification of 110 significant probe sets upregulated in wild-type BMMs treated with 100 U/ml IFN-β for 6 hours. These 110 probe sets represent 77 unique known genes. As shown in figure 1, most of the 77 unique genes showed the highest expression at 6 hours in the IFN-β treated wild-type cells, followed by a decrease in expression by 24 hours and a significant decease at 48 hours. In contrast, Ubp43−/− BMM showed significantly higher upregulation of all of these identified IFN stimulated genes at 6 hours as compared to the wild-type cells. Induction of these genes was also prolonged, reaching its highest expression level at 24 hours or even 48 hours (for some genes). In summary, Ubp43 deficiency in BMMs leads to a significant enhancement of overall expression of ISGs when compared to the wild-type BMMs.
Figure 1. Hierarchical clustering analyses.
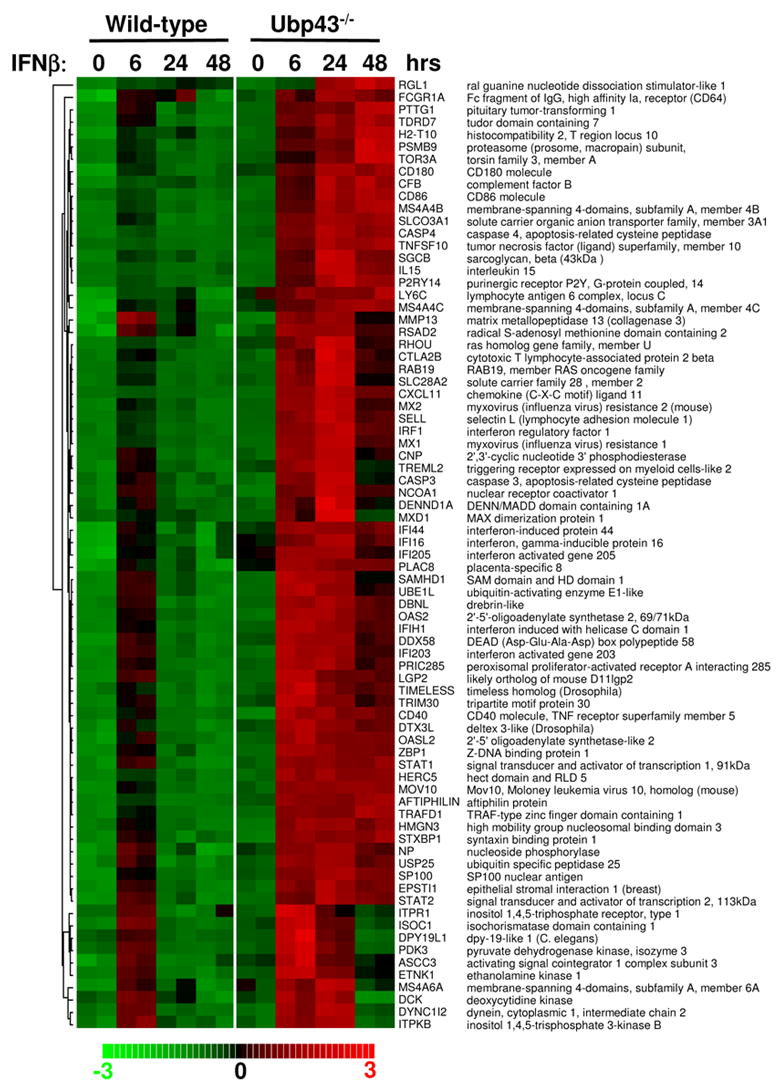
Seventy-seven unique known genes were found to be up-regulated by IFN-β in wild-type BMMs at 6 hours after 100 U/ml IFN treatment. The genes are ordered by clustering tightness. Each row represents a single Affymetrix probe set. Each column represents a sample treated with 100 U/ml IFN for indicated time.
Identification of IFN stimulated genes in Ubp43−/− BMMs
The gene expression profiles in figure 1 demonstrated that Ubp43 deficiency in BMMs generally enhanced the expression of genes that are induced by IFN in wild-type cells. It was hypothesized that analyzing the transcriptional profile of IFN treated Ubp43−/− BMMs would show clearer effects of IFN on BMMs. We compared the data from IFN-β treated Ubp43−/− BMMs with the one from untreated Ubp43−/− BMMs. dChip2006 Compare Sample procedure identified 1,270 different probe sets, which were upregulated by IFN-β treatment in Ubp43−/− BMMs at least at one time point (Fig. 2A). This total of 1,270 probes out of 45,000 probes, or roughly 3% of total probes, was a higher percentage than the ratio estimated in other cell lines [12]. On the basis of the annotation information from Ingenuity system, these 1,270 probes were mapped to 749 known unique genes (See supplemental data table Part 1). The expression of all 749 genes was induced by IFN treatment in Ubp43−/− BMMs. Some genes also showed IFN induction in wild-type cells, however, to a less degree than in Ubp43−/− cells (Fig. 2B). These 749 genes include many previously reported IFN stimulated genes, such as OAS2, IFIT1, IFIT3, ISG15, STAT1, ISG20, and UBE1L. We searched the IFN Stimulated Gene Database (http://www.lerner.ccf.org/labs/williams/oligo.cgi) [12; 13] and found that at least 100 different genes overlapped between our list and this database. We ranked all 749 genes according to the clustering tightness of hierarchical clustering analysis (Fig. 2B), which ordered the same pattern of expression together, and marked the overlapped genes in the list (Fig. 2B and supplemental data table Part 1). The overlapped genes, which can be used as "guideposts", are sporadically distributed in the whole table, validating the IFN induction of other genes in the list.
Figure 2. IFN inducible gene expression in Ubp43−/− macrophages.
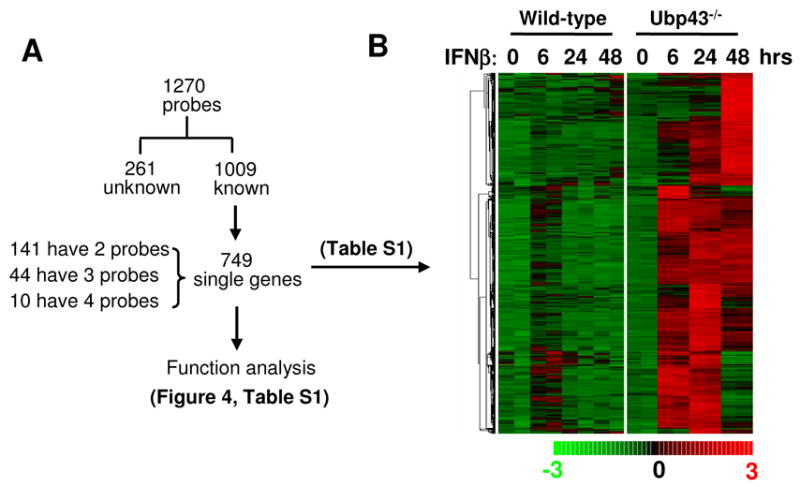
(A) A summary of the subsequent analysis for the identified 1,270 probe sets, which were found to be up-regulated in Ubp43−/− BMMs after 100 U/ml IFN-β treatment for 6, 24, or 48 hours. (B) Hierarchical clustering of 749 unique genes from 1,270 probes, which are found to be up-regulated by IFN-β in Ubp43−/− BMMs at 6, 24, or 48 hours after 100 U/ml IFN treatment. The genes are ordered by clustering tightness. Each row represents a single Affymetrix probe set. Each column represents a sample treated with 100 U/ml IFN-β for indicated time.
Confirmation of gene induction
Comparing IFN treated wild-type murine embryonic fibroblasts (MEFs) with Ubp43−/− cells , Knobeloch et al. detected prolonged expression of two ISGs, IRF7 and 2′,5′ oligoadenylate synthetase [14]. However, the expression of three other IFN-induced genes, GBP1, IRF1, and Mx1, remained unaffected [14]. All five genes were in our list as ISGs in Ubp43−/− BMM. To confirm the Microarray data, we did Northern blot analysis to examine the expression of 13 different genes. Firstly, we examined the expression of these five genes by Nothern blot analysis. As shown in figure 3A, all five genes were induced by IFN in wild-type cells and the expression of all five genes was significantly increased and prolonged in Ubp43−/− BMMs. The difference between the data obtained in this study and that previously reported by Knobeloch et al. could possibly be from the difference in cell lineages used for the analysis. Both studies used 100 U/ml IFN-β and it is believed that the IFN sensitivity of macrophages is higher than fibroblasts. At the same time, other unknown reasons could possibly explain the observed discrepancies, such as cell culture conditions.
Figure 3. Verification of the IFN hypersensitivity of gene expression in Ubp43−/− BMMs by Northern blotting analysis.
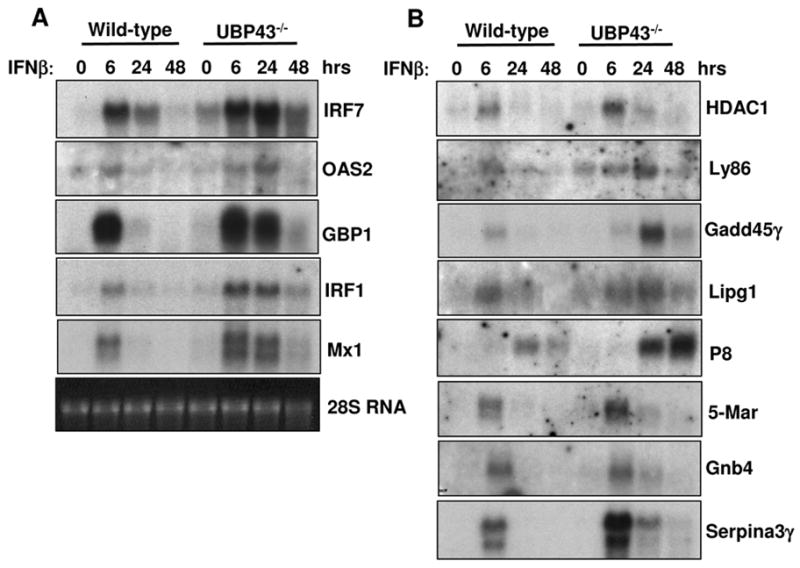
Wild-type and Ubp43−/− BMMs were either untreated or treated with 100 U/ml IFN-β for 6, 24 and 48 hours. Total RNA was isolated and subjected to Northern blot analysis using cDNA probes for IRF7, OAS2, GBP1, IRF1, Mx1, HDAC1, Ly86, Gadd45γ, Lipg, P8, 5-Mar, Gnb4 and Serpina3γ. Relative RNA loading was evaluated by the amount of 28S rRNA.
We also checked the expression of eight additional genes, which were randomly picked from our ISG list. Among them, only HDAC1 is present in the IFN Stimulated Gene Database [12; 13]. The other 7 genes, Serpina3γ, Gadd45γ, Gnb4, Ly86, Lipg, P8, and 5-Mar were identified as IFN-stimulated genes in Ubp43−/− cells in this study and were absent from the IFN Stimulated Gene Database. As shown in figure 3B, the expression of all tested genes was highly induced and prolonged in Ubp43−/− cells. Interestingly, according to our standard of Microarray analysis, the IFN induction of some genes is only identified in Ubp43−/− BMM but not in wild-type cells. However, the Northern blot analysis revealed that the expression of these genes also appears to be IFN-inducible in wild-type cells. As shown in figure 2B, most of identified genes were also slightly induced by IFN-β in wild type cells and the fold changes of expression values might be less than 2. However, the IFN hypersensitivity of Ubp43−/− cells makes it possible to detect these genes by using our reasonably strict standard.
Type I IFN strongly increases the expression of immune-response related genes in Ubp43 deficient macrophages
Ubp43 deficiency significantly increased gene expression in response to IFN stimulation and allowed the identification of a large group of previously unrecognized genes involved in the IFN response. To understand the effect of Type I IFN on Ubp43−/− BMMs, we used these genes (see supplemental table Part 1) as a starting point and performed global function analysis using gene ontology built from experimental evidence compiled in the David 2006 database [15]. The most significant functional group of genes was found to lie within immune or host defense response (82 genes, p value = 2.17E-15), in accordance with the known primary functions of macrophages, that play an important role in immune responses and in inflammatory diseases [16]. We further classified 82 immune response-related genes into different groups based on the gene ontology of the David 2006 database (see supplemental data table Part 2), which include 14 genes involved in the antigen presentation category, 14 genes directly related to the response to viral infection, and a variety of genes related to chemokine and cytokine activity (Fig. 4).
Figure 4. Functional classification and analyses of immune responses related genes induced by IFN-β in Ubp43−/− macrophages.
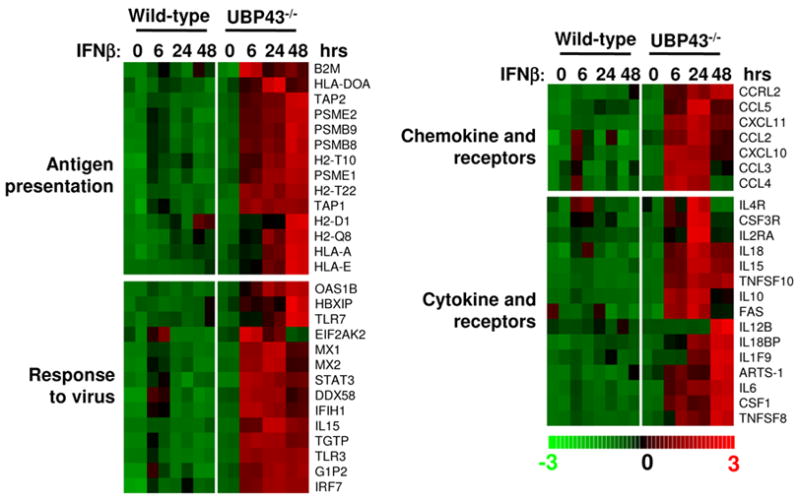
Each row represents a single Affymetrix probe set. Each column represents a sample treated with 100 U/ml IFN-β for indicated time.
DISCUSSION
In this study, large-scale analyses of ISGs expression demonstrated that Ubp43 deficiency in BMMs generally enhanced and prolonged the expression of genes that are induced by IFN in wild-type cells. The IFN hypersensitivity of Ubp43−/− BMMs made it possible to detect a spectrum of IFN inducible genes, including a large group of previously unidentified IFN stimulated genes. Functional analyses demonstrated that in Ubp43−/− BMMs Type I IFN can strongly induce the expression of immune response related genes including genes for antigen presentation, anti-viral function, and chemokine/cytokine activity. It is known that macrophages participate in the initiation of many antigen-specific immune responses via antigen presentation to T and B lymphocytes [11]. MHC proteins play important roles in macrophage antigen presentation and IFN-γ induce the expression of MHC proteins [17]. However, the role of Type I IFN in the expression of MHC proteins is less known. Our data showed that IFN-β also induce the expression of MHC proteins including MHC class two proteins H2-D1, H2-Q8, H2-T10, H2-T22, HLA-DOA and class I proteins, HLA-A and HLA-E, suggesting that IFN-β could also be involved in macrophage activation and supporting a role for IFN-β in virus and bacteria induced macrophage activation.
Ubp43−/− mice demonstrated greater resistance to the cytopathic effects caused by a number of viruses including lymphocytic choriomeningitis virus (LCMV), vesicular stomatitis virus (VSV), and Sindbis virus (SNV) [18]. Besides 14 antigen presentation related genes, there are 14 genes directly related to the response to viral infection (Fig. 4 and supplemental data table Part 2). Among these 14 genes, IL-15 is well known to be induced by IFN-α/β and required for the maintenance and/or accumulation of proliferating NK cells during murine cytomegalovirus infection [19]. Mx1 and Mx2 have been shown to inhibit viral transcription by interacting with RNA polymerase subunit PB2 [20]. Another protein is ISG15 (G1P2), which has been shown to be an antiviral protein against Sindbis virus and HIV-1 [21; 22], although the molecular mechanism remains unknown. TLR3 (Toll like receptor 3) participates in the innate immune response via recognition of double stranded RNA. It has been reported that Type I IFNs enhance TLR3 mediated anti-viral cytokine expression by up-regulating TLR3 expression [23]. The expression level and duration of all anti-viral genes mentioned above were dramatically augmented in Ubp43−/− BMMs (Fig. 4), explaining the greater degree of viral-resistance found in Ubp43−/− mice [18]. Importantly, some new antiviral genes have been identified as IFN-β stimulated genes in our study, such as TLR7 (Toll like receptor 7). TLR7 recognizes single-stranded RNA and is involved in innate antiviral responses by mediating influenza virus A induced IFN production [24]. In this study, we showed that IFN can induce TLR7 expression, which in its turn may cause positive feedback during single stranded RNA viral infection and expand the anti-viral functions of IFN.
Chemokines and cytokines play a critical role in the immunomodulatory function of macrophages. There are two types of chemokines; the CXC type in which a non-conservative amino acid is located between the first two cysteines and the CC type where these residues are juxtaposed [25]. CXC chemokines are chemotactic for neutrophils and CC chemokines recruit T cells and monocytes [25]. Our data showed that a variety of CXC (CXCL10, CXCL11) and CC chemokines (CCL2, 3, 4 and 5) as well as CC chemokine receptor (CCRL2) are induced by IFN-β in Ubp43−/− BMMs (Fig. 4 and supplemental data table Part 2). Besides chemokines, several members of the tumor necrosis factor (TNF) family (TNFSF8, TNFS10, FAS) and interleukin family members (IL6, ILF9, IL10, IL12B, IL15, IL18) are induced by IFN-β in Ubp43−/− macrophages. Their roles in IFN-β mediated macrophage function remains to be elucidated.
Although there are arguments about the effect of Ubp43 deficiency on the IFN signaling pathway and the expression of IFN stimulated genes [14], our present study using Microarray analysis reveals that Ubp43 deficiency in BMMs generally enhanced the expression of genes that are induced by IFN in wild-type cells. The IFN hypersensitivity of Ubp43−/− cells and subsequent higher induction of immune response related genes in Ubp43−/− cells (Fig. 4) provide a biochemical mechanism in support of the strong resistance to viral and bacterial infections observed in Ubp43−/− mice [5; 6]. A recent study showing that knock down of UBP43 by siRNA in human cells enhances the ability of IFN to inhibit HCV-RNA replication and infectious virus particle production also supports the enhanced anti-viral effects of Ubp43−/− mice [26]. In conclusion, Ubp43 is a potential therapeutic target for the enhancement of immune responses.
Supplementary Material
Acknowledgments
We thank L. Schaffer for microarray data analysis, O. Malakhova for valuable discussions and critical reading of the manuscript, J. Biggs for editing the manuscript. This work is supported by National Institutes of Health grant CA79849 to D.E.Z. W.G.Z is a postdoctoral fellow of Leukemia and Lymphoma Society. The Stein Endowment Fund has partially supported the Molecular and Experimental Medicine Departmental Molecular Biology Service Laboratory for DNA Sequencing and Oligonucleotide Synthesis. This is manuscript 18594-MEM from The Scripps Research Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Liu LQ, Ilaria R, Jr, Kingsley PD, Iwama A, van Etten RA, Palis J, Zhang DE. A novel ubiquitin-specific protease, UBP43, cloned from leukemia fusion protein AML1-ETO-expressing mice, functions in hematopoietic cell differentiation. Mol Cell Biol. 1999;19:3029–3038. doi: 10.1128/mcb.19.4.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malakhov MP, Malakhova OA, Kim KI, Ritchie KJ, Zhang DE. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J Biol Chem. 2002;277:9976–9981. doi: 10.1074/jbc.M109078200. [DOI] [PubMed] [Google Scholar]
- 3.Ritchie KJ, Malakhov MP, Hetherington CJ, Zhou L, Little MT, Malakhova OA, Sipe JC, Orkin SH, Zhang DE. Dysregulation of protein modification by ISG15 results in brain cell injury. Genes Dev. 2002;16:2207–2212. doi: 10.1101/gad.1010202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malakhova OA, Yan M, Malakhov MP, Yuan Y, Ritchie KJ, Kim KI, Peterson LF, Shuai K, Zhang DE. Protein ISGylation modulates the JAK-STAT signaling pathway. Genes Dev. 2003;17:455–460. doi: 10.1101/gad.1056303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim KI, Malakhova OA, Hoebe K, Yan M, Beutler MB, Zhang DE. Enhanced antibacterial potential in UBP43-deficient mice against Salmonella typhimurium infection by up-regulating type I IFN signaling. J Immunol. 2005;175:847–854. doi: 10.4049/jimmunol.175.2.847. [DOI] [PubMed] [Google Scholar]
- 6.Ritchie KJ, Hahn CS, Kim KI, Yan M, Rosario D, Li L, de la Torre JC, Zhang DE. Role of ISG15 protease UBP43 (USP18) in innate immunity to viral infection. Nat Med. 2004;10:1374–1378. doi: 10.1038/nm1133. [DOI] [PubMed] [Google Scholar]
- 7.Malakhova OA, Kim KI, Luo JK, Zou W, Kumar KG, Fuchs SY, Shuai K, Zhang DE. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 2006;25:2358–2367. doi: 10.1038/sj.emboj.7601149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas KE, Galligan CL, Newman RD, Fish EN, Vogel SN. Contribution of Interferon-beta to the Murine Macrophage Response to the Toll-like Receptor 4 Agonist, Lipopolysaccharide. J Biol Chem. 2006;281:31119–31130. doi: 10.1074/jbc.M604958200. [DOI] [PubMed] [Google Scholar]
- 9.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 10.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mosser DM. The many faces of macrophage activation. J Leukoc Biol. 2003;73:209–212. doi: 10.1189/jlb.0602325. [DOI] [PubMed] [Google Scholar]
- 12.Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM, Silverman RH, Williams BR. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol. 2001;69:912–920. [PubMed] [Google Scholar]
- 14.Knobeloch KP, Utermohlen O, Kisser A, Prinz M, Horak I. Reexamination of the role of ubiquitin-like modifier ISG15 in the phenotype of UBP43-deficient mice. Mol Cell Biol. 2005;25:11030–11034. doi: 10.1128/MCB.25.24.11030-11034.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 16.Li AC, Glass CK. The macrophage foam cell as a target for therapeutic intervention. Nat Med. 2002;8:1235–1242. doi: 10.1038/nm1102-1235. [DOI] [PubMed] [Google Scholar]
- 17.Kota RS, Rutledge JC, Gohil K, Kumar A, Enelow RI, Ramana CV. Regulation of gene expression in RAW 264.7 macrophage cell line by interferon-gamma. Biochem Biophys Res Commun. 2006;342:1137–1146. doi: 10.1016/j.bbrc.2006.02.087. [DOI] [PubMed] [Google Scholar]
- 18.Ritchie KJ, Zhang DE. ISG15: the immunological kin of ubiquitin Semin. Cell Dev Biol. 2004;15:237–246. doi: 10.1016/j.semcdb.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen KB, Salazar-Mather TP, Dalod MY, Van Deusen JB, Wei XQ, Liew FY, Caligiuri MA, Durbin JE, Biron CA. Coordinated and distinct roles for IFN-alpha beta, IL-12, and IL-15 regulation of NK cell responses to viral infection. J Immunol. 2002;169:4279–4287. doi: 10.4049/jimmunol.169.8.4279. [DOI] [PubMed] [Google Scholar]
- 20.Pavlovic J, Haller O, Staeheli P. Human and mouse Mx proteins inhibit different steps of the influenza virus multiplication cycle. J Virol. 1992;66:2564–2569. doi: 10.1128/jvi.66.4.2564-2569.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lenschow DJ, Giannakopoulos NV, Gunn LJ, Johnston C, O'Guin AK, Schmidt RE, Levine B, Virgin HW. Identification of interferon-stimulated gene 15 as an antiviral molecule during Sindbis virus infection in vivo. J Virol. 2005;79:13974–13983. doi: 10.1128/JVI.79.22.13974-13983.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okumura A, Lu G, Pitha-Rowe I, Pitha PM. Innate antiviral response targets HIV-1 release by the induction of ubiquitin-like protein ISG15. Proc Natl Acad Sci USA. 2006;103:1440–1445. doi: 10.1073/pnas.0510518103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tissari J, Siren J, Meri S, Julkunen I, Matikainen S. IFN-alpha enhances TLR3-mediated antiviral cytokine expression in human endothelial and epithelial cells by up-regulating TLR3 expression. J Immunol. 2005;174:4289–4294. doi: 10.4049/jimmunol.174.7.4289. [DOI] [PubMed] [Google Scholar]
- 24.Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 25.Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 26.Randall G, Chen L, Panis M, Fischer AK, Lindenbach BD, Sun J, Heathcote J, Rice CM, Edwards AM, Mcgilvray ID. Silencing of USP18 potentiates the antiviral activity of interferon against hepatitis C virus infection. Gastroenterology. 2006;131:1584–1591. doi: 10.1053/j.gastro.2006.08.043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


