Summary
Sphingolipids function as required membrane components of virtually all eukaryotic cells. Data indicate that members of the sphingolipid family of lipids, including sphingoid bases, sphingoid base phosphates, ceramides, and complex sphingolipids, serve vital functions in cell biology by both direct mechanisms (e.g., binding to G-protein coupled receptors to transduce an extracellular signal) and indirect mechanisms (e.g., facilitating correct intracellular protein transport). Because of the diverse roles these lipids play in cell biology, it is important to understand not only their biosynthetic pathways and regulation of sphingolipid synthesis, but also the mechanisms by which some sphingolipid species with specific functions are modified or converted to other sphingolipid species with alternate functions. Due to many factors including ease of culture and genetic modification, and conservation of major sphingolipid metabolic pathways, Saccharomyces cerevisiae has served as an ideal model system with which to identify enzymes of sphingolipid biosynthesis and to dissect sphingolipid function. Recent exciting developments in sphingolipid synthesis, transport, signaling, and overall biology continue to fuel vigorous investigation and inspire investigations in mammalian sphingolipid biology.
Keywords: sphingolipids, heat stress response, endocytosis, sphingoid bases, ceramide, sphingosine-1-phosphate
I. Introduction
Sphingolipids have emerged over the last several decades as a family of key signaling molecules including sphingosine, ceramide, and sphingosine-1-phosphate. Data indicate that these lipids regulate fundamental and diverse cell processes such as differentiation, migration, and apoptosis. Moreover, on the organismal level, sphingolipids play roles in higher order physiological processes including inflammation [1, 2] and vasculogenesis [3]. Most importantly, however, recent studies implicate sphingolipid involvement in many of the most common and currently relevant human diseases including diabetes [4–6], a range of cancers [7, 8], infection by microorganisms [9–12], Alzheimer’s disease [13, 14], diseases of the cardiovascular and respiratory systems including heart disease [15], an array of neurological syndromes [16, 17], and many others. Thus, it is increasingly important to understand the routes and regulation of sphingolipid metabolism, to elucidate the signaling targets for these lipids, to identify cell processes that they regulate, and to determine the sphingolipid-dependent component(s) of cellular, physiological, and disease processes.
Due to its relative simplicity as a model system, the yeast Saccharomyces cerevisiae has provided numerous opportunities for understanding fundamental cell processes in fields including cell cycle control [18], aging [19], and stress responses [20]. Moreover, the conservation of sphingolipid metabolic pathways between all eukaryotes has enabled utilization of yeast for studying the formation and function of these important lipid mediators. In fact, cloning of many of the mammalian genes of sphingolipid metabolism was possible based on their homology to yeast counterparts [21]. In general, yeast studies have provided key insights into sphingolipid biosynthesis, regulation, and function [22]. Though major developments in sphingolipid biology have already arisen from studies in yeast, recent and exciting data emphasize the need for continued vigorous investigation and hint at the depth and complexity of sphingolipid biology that awaits discovery.
II. Sphingolipid metabolism in yeast
a. De novo synthesis
De novo sphingolipid biosynthesis begins with the condensation of fatty acyl-coA (most commonly palmitoyl- and stearoyl coA, yielding C18 and C20 products, respectively) and serine via the serine palmitoyltransferase (SPT) complex (Scheme 1)[23]. This enzyme, which consists of two major subunits, encoded by LCB1 and LCB2 (long chain base), and a minor subunit, encoded by TSC3 (temperature-sensitive suppressor of csg2Δ, discussed below)[24], requires pyridoxal-5′-phosphate and bears sequence and functional homology to α-oxoamine synthases [25]. In vitro and in vivo data unequivocally indicate that both Lcb1p and Lcb2p are required for SPT activity [23], and deletion of either gene renders cells inviable unless they are supplied with exogenous sphingoid bases [26–28]. The precise nature of the interaction of these two subunits has yet to be determined; however, modeling studies utilizing alignments to a bacterial α-oxoamine synthase with known crystal structure indicated that the subunits form a heterodimer in a 1:1 stoichiometry [29]. An interesting observation is that Lcb2p requires interaction with Lcb1p for stability, though Lcb1p is relatively stable without Lcb2p [30].
Scheme 1.
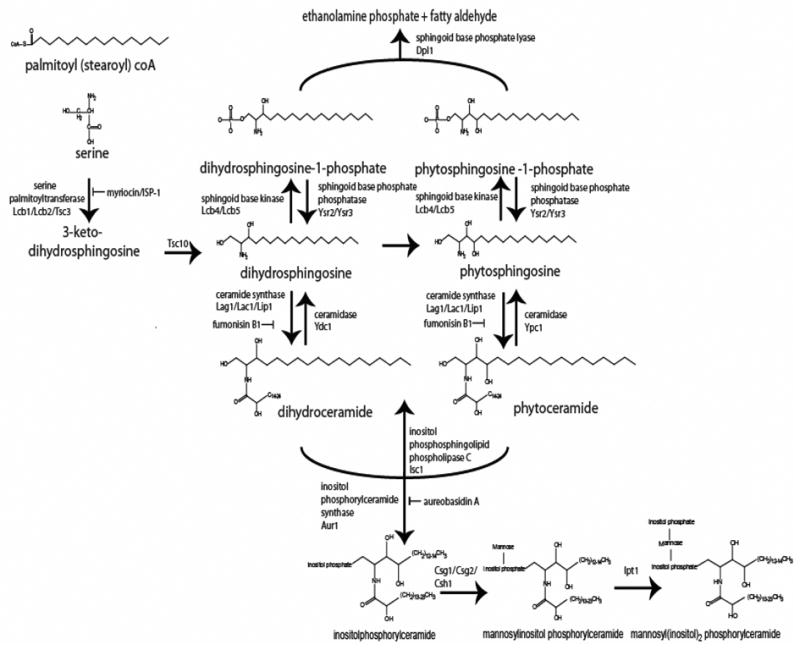
Major pathways of sphingolipid metapolism in Saccharomyces cerevisiae
Though Lcb1p and Lcb2p are sufficient for SPT activity, several reports indicate that a small peptide encoded by TSC3 interacts with the complex and stimulates activity [24]. Data suggest that this increased activity is required under certain stress conditions [24], however, the mechanism of interaction of Tsc3p with SPT as well as factors regulating the interaction remain to be determined. Interestingly, however, several point mutants in LCB2 increased SPT activity in the absence of TSC3, suggesting that Tsc3p mediates SPT activation possibly via interaction with the Lcb2p subunit [31].
The reaction of SPT produces 3-ketodihydrosphingosine, a short lived metabolite that is rapidly converted to dihydrosphingosine in an NADPH-dependent manner by the 3-ketodihydrospihgosine reductase encoded by TSC10 (temperature-sensitive suppressor of csg2Δ, discussed below) [32]. TLC methods routinely used to quantify sphingoid bases do not detect accumulation of 3-ketodihydrosphingosine, suggesting that it is not a biologically active species, but a transient intermediate in de novo sphingolipid synthesis [32]. Thus, the first biologically relevant sphingolipid produced in the de novo pathway is probably dihydrosphingosine. TSC10 was identified in a screen for suppressors of a mutation that caused the aberrant accumulation of a complex sphingolipid species, imparting a calcium-sensitive phenotype [32]. Thus, isolation of mutants that could override this defect revealed many genes that prevented accumulation of the complex lipid, and thus, genes likely to catalyze prior steps of sphingolipid synthesis, many of which are discussed below (and are similarly named ‘TSC’). This clever screen exemplified the usage of yeast genetics for discovery of biochemical pathways. Moreover, the screen identified several complementation groups that await characterization.
The product of Tsc10p, dihydrosphingosine, can be converted to phytosphingosine by the Sur2p/Syr2p hydroxylase (suppressor of rsv161/syringomycin resistance, discussed below)[33](Scheme 1). Deletion of this enzyme leads to an accumulation of dihydrosphingosine, but mutant cells grow and divide at normal rates and have otherwise normal complex sphingolipids, indicating that dihydrosphingosine probably serves as a suitable substrate for all subsequent sphingolipid synthesis necessary for growth under typical laboratory conditions [33]. Together, phyto- and dihydrosphingosine are termed ‘sphingoid bases’, and to date, no known functions unequivocally distinguish one from the other in yeast (though some activities of the bases, as discussed below, are mediated by lower concentrations of phytosphingosine). Furthermore, each sphingoid base can potentially undergo either phosphorylation at C1 or N-acylation, as described below.
The sphingoid bases can be phosphorylated to form phyto- or dihydrosphingosine phosphate by the sphingoid base kinases encoded by LCB4 and LCB5 [34]. Data indicate significant overlap in biochemical function of these enzymes, though by far the greatest source of sphingoid base kinase activity comes from Lcb4p, which contributes over 90% of total activity [34]. The role of Lcb5p remains unknown, but perhaps it mediates sphingoid base phosphorylation under specific conditions or in specific intracellular compartments. For most experimental purposes, depletion of sphingoid base kinase activity requires deletion of both isoforms. Interestingly, deletion of LCB4 together with LCB5 causes tremendous accumulation of sphingoid bases, indicating that the sphingoid base kinase pathway may function as a ‘bottleneck’ in the overall biochemical pathway[35]. The double deletion mutant has normal growth rates and few detectable phenotypes despite the aberrant sphingoid base content, indicating that, sphingoid bases do not inhibit yeast growth.
The sphingoid base phosphate lyase, Dpl1p, cleaves phyto- and dihydrosphingosine-1-phosphate, generating fatty aldehydes and ethanolamine phosphate [36]. Importantly, this pathway remains the only know biochemical route by which sphingolipids are converted to non-sphingolipid molecules, and thus, is the only route by which sphingolipids may ‘exit’ the pathway. Therefore, this degradation pathway holds a unique position in overall sphingolipid biology, as it may function as a key node to regulate overall sphingolipid levels. On the other hand, some data suggest that it may also serve to produce ethanolamine phosphate for phosphatidylethanolamine biosynthesis through the Kennedy pathway [37]. The relative importance of each of these possibilities for the lyase pathway remains to be elucidated, and a possibility exists that both functions, i.e., degradation of sphingolipids and production of ethanolamine phosphate, are important.
Interestingly, a deletion mutant lacking the DPL1 gene accumulated extremely high levels of sphingoid base phosphates, which were inhibitory for yeast growth [38]. This phenotype enabled a screen for suppressors of sphingoid base phosphate toxicity, which revealed the existence of an ATP-dependent sphingiod base transporter, Rsb1p [39]. Rsb1p may pump excess sphingoid bases outside the cell to preserve normal intracellular functions including correct membrane composition [39]. It is possible that this transporter is upregulated and/or posttranscriptionally modified to provide cells with a biologically viable solution to increased sphingoid base phosphates. In an exciting recent study, this pump was shown to be regulated by transcription factors known to mediate the retrograde and pleiotropic drug resistance responses, raising intriguing possibilities that sphingoid bases participate in these transcriptional programs [37].
Sphingoid base phosphates can be hydrolyzed back to sphingoid bases by the phosphatases encoded by YSR2 and YSR3 [40, 41]. Though enzymes encoded by both of these genes participate in similar reactions, Ysr3p, but not Ysr2p, was determined to facilitate utilization of exogenous sphingoid bases from media [40, 42]. This study and further studies with the Lcb4p sphingoid base kinase indicated that exogenous sphingoid bases taken up from media are likely phosphorylated (i.e., by Lcb4p) and then dephosphorylated by Ysr3p prior to their incorporation into cell sphingolipid pools [43].
In addition to conversion to phosphorylated derivatives, an alternative fate of sphingoid bases is their N-acylation to form phyto- or dihydroceramides. Like the SPT reaction, this reaction also requires acyl CoA as an acyl chain donor, but the preferred substrates are C24–C26 in length [44]. The lengthened acyl CoAs derive from the fatty acid elongases Elo2p and Elo3p [45], Tsc13p, which co-immunoprecipitated with Elo2p and Elo3p, and the protein encoded by the YBR159w gene [46, 47], which co-immunoprecipitated with Elo3p and Tsc13p, suggesting these proteins are subunits of a fatty acid elongation complex [47]. Deletion of these enzymes caused accumulation of short chain fatty acids as well as ceramides containing N-acyl groups much shorter than in parental strains [47]. The N-acylation of sphingoid bases to form ceramides requires the gene products encoded by LAG1 or LAC1 [48]. These genes are highly homologous and overlap in function, since both must be deleted to attenuate ceramide synthesis. Until very recently, whether these peptides actually catalyzed the ceramide synthase reaction had not been biochemically demonstrated; however, a recent study confirmed that the enzyme a mammalian homologue of LAG1 and LAC1, LASS5, is a bona fide ceramide synthase [49]. The LASS genes comprise a family of six members which catalyze ceramide synthesis in mammalian cells [50], and which are partially distinguished by fatty-acyl chain length specificity [51, 52]. In yeast, purification of Lac1p and Lag1p subunits, followed by electrophoretic resolution of immunoprecipitated complexes, revealed a novel component of the ceramide synthase complex, termed Lip1p [53]. In yeast and mammals, ceramide synthesis is inhibitable by the fungal toxin Fumonisin B1 [54, 55], making fumonisin quite a useful experimental tool for dissecting the biological relevance of various branches of sphingolipid synthesis.
The very long chain fatty acids incorporated into phytoceramide by Lag1p/Lac1p/Lip1p can undergo hydroxylation after their incorporation. This activity is encoded by SCS7/FAH1 [25, 56, 57]. The biological function of this hydroxylation remains unclear; however, similar to the syr2 deletion, a scs7 deletion strain is less sensitive to certain fungicides that function by causing pore formation in the plasma membrane [33, 57–59]. This suggests that both the hydroxylations mediated by Syr2p and Fah1p affect lipid packing in membranes. Potentially supporting this notion, the mammalian homologue of FAH1, FA2H, hydroxylates lipids that are highly abundant in the myelin sheath of the nervous system, perhaps enhancing structural stability of the membrane [60]. Interestingly, a deletion in scs7 suppressed the calcium sensitive phenotype of the csg2 mutant strain, indicating that part of the toxicity observed in this mutant may derive from accumulation of hydroxylated ceramides, thus suggesting specific functions for that subclass of ceramides [57].
After sphingoid bases convert to ceramides, they can be cleaved back into bases and free fatty acid by the ceramidases Ypc1p and Ydc1p [61, 62]. The roles for these enzymes in yeast remain unknown; however, their mammalian counterparts are key players in sphingolipid signaling, as they are required to generate sphingosine, the precursor for sphingosine-1-phosphate (sphingosine has also been reported to have biological activity in some situations). Interestingly, however, Ypc1p, which is specific for phytoceramide, catalyzed the reverse ceramidase reaction, or ceramide synthesis from free fatty acid (as opposed to fatty acyl-coA, which is required for Lag1p/Lac1p/Lip1p mediated ceramide synthesis) and phytosphingosine, and was not inhibitable by fumonisin B1 [62]. Thus, in experimental situations where ceramide synthase activity is detected in the presence of fumonisin and/or using free fatty acid as substrate, one should consider the likelihood of reverse ceramidase activity.
In yeast, ceramides serve as substrates for the synthesis of complex lipids that comprise up to 10% of total membrane lipids. The inositolphosphorylceramide (IPC) synthase Aur1p attaches a phosphoinositol headgroup to ceramide, forming IPC [63], which can then undergo mannosylation by the enzymes encoded by CSG1, CSG2, and CSH1 [64, 65]. Current evidence supports a model in which two enzyme complexes exist for inositol phosphosphingolipid mannosylation, Csg1p/Csg2p and Csg2p/Csh1p [65]. The relative importance of each complex, as well as the biological significance of having two complexes, remains obscure. Deletion of CSG1 and 2, however, imparted a calcium-sensitive phenotype to cells, suggesting that sphingolipids function to regulate calcium sensitivity, and thus, these genes also served as tools for identification of many genes involved in sphingolipid metabolism in a screen for suppressors of their defect, as described above [25] After mannosylation, the enzyme Ipt1p can then catalyze the addition of another inositol phosphate group, forming mannosyl(inositol)2phosphorylceramide [66]. Complex sphingolipids in yeast not only play important structural roles, but they also may function as a ‘sink’ for ceramide, since enzymes exist that catalyze the reverse of many of the reactions thus far described, producing sphingolipids through catabolic pathways.
b. Sphingolipid catabolism
The headgroups of complex sphingolipids are hydrolyzed by the enzyme encoded by the ISC1 gene to yield both phyto- and dihydroceramides [67]. Isc1p bears significant structural and functional homology to mammalian neutral sphingomyelinases, a fact that facilitated its cloning based on mammalian enzyme sequences [67]. Interestingly, this enzyme bears activity not only to yeast complex sphingolipids but also to the mammalian counterpart to yeast complex sphingolipids, sphingomyelin, though sphingomyelin is not present in yeast [67]. Whether this sphingomyelinases activity serves a biological role or is merely an evolutionary anomaly remains to be determined; however, recent studies in pathogenic strains of S. cerevisiae indicate that in order for yeast to survive in a host, it must adapt to low glucose concentrations in vivo [68], a role that Isc1 plays in yeast batch cultures, as discussed below. Thus, the possibility exists that Isc1 or its homologues in more virulent pathogenic yeast, such as Candida albicans, may act on host sphingomyelin during infection. In a similar vein, the IPC synthase enzyme encoded by AUR1 in Cryptococcus neoformans was recently demonstrated to function in pathogenesis of that virulent fungus [11].
Ceramides produced either de novo through Lag1p/Lac1p/Lip1p, or from complex sphingolipid hydrolysis through Isc1p, can undergo cleavage back to sphingoid bases by the ceramidases encoded by the YPC1 and YDC1 genes, as discussed above. The sphingoid bases produced by these enzymes can theoretically undergo all reactions that de novo produced lipids undergo, including phosphorylation and, interestingly, N-acylation back to ceramide. This process has been termed ‘recycling’, and seems to plays major roles in mammalian sphingolipid biology [69, 70]. Though re-acylation of a recently cleaved ceramide seems inefficient, evidence suggests that this process functions to alter N-acyl chain lengths of ceramides, and topology (i.e., compartmentalization of ceramide production) may also be a consideration in the recycling reaction.
II. Regulation of sphingolipid metabolism
For such an important biochemical pathway, puzzlingly little is understood about its regulation. In fact, in several situations where increased production of one or more of the products occurs, no evidence exists for changes in enzyme levels [71]. The advance of microarray technology has enabled the production of many yeast gene expression data sets under a myriad of conditions [20]; perplexingly, however, across a wide variety of conditions, little transcriptional regulation is observed for the key enzymes in the sphingolipid metabolic pathway (Cowart LA, unpublished observations). There are, however, a few situations where data suggest that regulation could occur by enzyme translocation, posttranslational modification, or transcriptional regulation.
The requirement of yeast for both Lcb1p and Lcb2p allowed isolation of suppressor strains that generated novel glycerophospholipids [72]. These lipids presumably serve some of the same functions as sphingolipids; however, not all functions were complemented by the novel lipids, as the suppressor strains displayed sensitivity to growth at low pH or high temperature [73]. Notably, however, the isolation of these suppressor strains inspired studies that demonstrated a flux through de novo sphingolipid synthesis after heat stress. This flux is characterized by an initial rise in sphingoid bases and their phosphorylated derivatives, followed by a decrease in these species as their downstream metabolites, ceramides, accumulate [35, 71, 74–77]. The driving force for this flux remains unknown; however, the generation of 3-ketodihydrosphingosine by serine palmitoyltransferase is the first and rate-limiting step of de novo sphingolipid synthesis and thus, studies into its mechanisms of regulation deserve special emphasis, as regulation at this initial step potentially regulates all anabolic pathways of sphingolipid biosynthesis.
Previous data indicate that sphingoid base phosphates are inhibitory for cell growth and thus, that regulation of their levels mediates cell cycling and/or viability [38]. Interestingly, a relatively recent study identified that the Lcb4p sphingoid base kinase is phosphorylated and utilized a kinase mutant yeast collection to identify the Pho85 cyclins Pcl1p and 2p as required for this modification [78]. Moreover, data in this study indicate that phosphorylation mediated the degradation of phosphorylated Lcb4p in the vacuole after its sorting through the mutli-vesicular body, thus indicating that those factors that regulate Pho85p, including nutrient deprivation, may also regulate sphingoid base kinase levels via degradation of the Lcb4p kinase. This is supported by the observation that Lcb4p levels significantly decreased during stationary phase, with a concomitant decrease in sphingoid base phosphate levels and a slowing of the cell cycle [78]. Moreover, in the dpl1Δ mutant, where sphingoid base phosphate levels are elevated, cell cycle arrest does not occur as cells approach stationary phase [79]. These findings support the notion that phosphorylation of Lcb4p regulates cell cycle arrest mediated by decreasing sphingoid base phosphates. Phosphorylation of Lcb4p is apparently related to its localization at the plasma membrane due to palmitoylation by Akr1p [80], as strain expressing Lcb4p mutated at its palmitoylation sites showed aberrant cytosolic localization of Lcb4p and also failed to downregulate Lcb4p during stationary phase. The mechanism by which phosphorylation initiates the degradation of Lcb4p remains unknown.
Though factors including heat stimulate ceramide synthesis in yeast and mammalian systems, mechanisms that potentially regulate the enzymes have been elusive. A very interesting study presented data that ceramide synthase is subject to regulation by casein kinase, as a deletion of a subunit of that complex encoded by CKA2 was demonstrated to have 70–75% reduction in ceramide synthase activity in isolated membranes, as well as an accumulation of ceramide precursors in intact cells [81]. The mechanism by which Cka2p regulates ceramide synthase could be direct, i.e., a casein kinase-mediated phosphorylation event is required to activate the enzyme, or indirect, e.g., regulation of transcription of a component of the ceramide synthase complex. In case of the former, that mechanism would represent a major development of the regulation of de novo synthesis. Importantly, these findings may relate to mammalian situations where factors including chemotherapy mediate the production of ceramide, which conducts many of their downstream actions including cell death (reviewed in [8, 82]).
In another set of exciting studies, it was found that transcriptional mediators of the pleiotropic drug response (PDR), Pdr1p and Pdr3p, bind to a functional PDR response element (PDRE) in the promoter of LAC1 and activate its transcription [83]. Interestingly, in this study, the promoter of the LAG1 gene did not contain a PDRE, perhaps providing some insight as to the apparent biochemical redundancy of these two enzymes. Perplexingly, hyperactivation of the PDR caused overall changes in sphingolipid levels that were not dependent on LAG1 or LAC1, and subsequently, the promoters of other sphingolipid metabolic enzymes were examined for PDREs. Indeed, active PDREs were found in the promoters of both the LCB2 and SUR2 genes, suggesting the aberrant sphingolipid profiles resulted from enzymes required for sphingoid base production and underscoring the importance of this regulatory node for the control of overall sphingolipid levels [83]. It should be taken into account, however, these specific transcriptional changes are not yet linked to major changes in sphingolipid profiles, perhaps reflecting the complex nature of sphingolipid biosynthesis. Despite this deficiency, however, involvement of the PDR in mediating multidrug resistance could potentially indicate a role for sphingolipids in this important pathway.
The above described regulatory routes for sphingolipid metabolism pertain primarily to de novo synthesis. However, several recent studies suggest regulation of the hydrolysis of complex lipids by the inositol phosphosphingolipid phospholipase C, Isc1p. Levels of this enzyme increased in mitochondria after the diauxic shift based on fractionation of yeast cells expressing a FLAG-tagged construct [84]. Moreover, Isc1p was demonstrated to require phosphatidylglycerol or cardiolipin as a lipid activator for optimum activity [85], and these lipids are synthesized in mitochondria by phosphatidylglycerol synthase Pgs1p. Supporting that Pgs1p activity is required to activate Isc1p, phenotypes of the pgs1Δ strain were highly overlapping those of the isc1Δ strain, and neither strain effectively utilizes non fermentable carbon sources, an activity requiring mitochondria [86].
These recent studies have shed some light on mechanisms of regulation of sphingolipid metabolism. All in all, however, many key issues remain unanswered, including the regulation of serine palmitoyltransferase in response to heat stress, which conditions signal alterations in sphingolipid metabolism, and the overall mechanisms by which the pathway shifts emphasis from one branch, for example, production of complex lipids to another, for example, hydrolysis of complex lipids to form ceramides.
III. Functions of yeast sphingolipids
a. Roles in Heat Stress
The finding that sphingolipid synthesis increases upon heat stress (above) laid ground for a flurry of studies of these lipids regarding their roles in stress responses. An observation that further fueled the field of yeast sphingolipids was the isolation of a temperature-sensitive mutant in endocytosis, end-8 [87], which was later mapped to the LCB1 locus [88]. The temperature sensitive allele of LCB1 (lcb1-100) conferred a heat sensitive phenotype, and thus, the lcb1-100 mutant strain has been used extensively to demonstrate sphingolipid-mediated regulation of nearly every major event of the yeast heat stress response (discussed below).
When yeast are cultured at elevated temperature (i.e., 37°C–42°C), they induce an array of cellular programs, the ultimate result of which is increased thermotolerance, or ability to continue to divide at elevated temperature. These programs include major changes in gene transcription, regulation of translation (especially of heat shock proteins), degradation of nutrient permeases and denatured proteins, synthesis of trehalose, and others [89]. Moreover, yeast undergo a transient cell cycle arrest in G0/G1 while these programs progress. After the original observation that heat increased sphingoid base production through SPT and the fortuitous finding that the increase could be selectively blocked with a heat-sensitive allele for LCB1, Jenkins et al. demonstrated a requirement for de novo sphingoid bases for the arrest. Whereas parental strains arrest by 1 hour of heat stress and then resume the cell cycle by 2 hours, lcb1-100 cells failed to arrest and subsequently died [90]. This finding emphasizes the importance of sphingolipids in stress responses. Interestingly, whereas the lcb4/5 double deletion strain, which accumulates high levels of sphingoid bases, showed a normal heat-induced arrest, it failed to re-enter the cell cycle, which prompted the speculation that degradation of bases via their conversion to phosphorylated derivatives was required to permit continuation of the cell cycle [90]. It is uncertain how these studies relate to those described above wherein buildup of sphingoid base phosphates inhibited cell cycle progression, thus, further study is required to dissect respective roles of sphingoid bases vs. their phosphates in the regulation of cell cycle progression. Moreover, another study demonstrated that ceramides increased at later time points in heat stress, after the initial rise in sphingoid bases and base phosphates [75]. Much of this ceramide increase was via de novo synthesis; however, a later study demonstrated an Isc1p-dependent component of ceramide production, and the Isc1p-dependent ceramides were predominantly long-chain (as would be expected from complex lipid hydrolysis) [91]. The relative importance and functions between de novo and catabolic ceramides remain to be fully elucidated.
In addition to their role in heat-induced cell cycle arrest, microarrays of the yeast genome were utilized to determine the degree of heat-mediated gene regulation that was dependent on sphingolipid metabolism. Interestingly, greater than 50% of all genes regulated during heat stress in a wild type strain were aberrantly regulated in the lcb1-100 mutant [92]. There is no doubt that many of these genes display altered regulation as an indirect consequence of other failed heat stress subprograms, however, some of these genes may be direct targets for sphingolipids and thus, could serve as important tools to determine mechanisms of sphingolipid-dependent gene regulation.
A perplexing finding emerging from that study was the failure of sphingoid bases to mediate transcriptional regulation of most heat shock proteins, as these mRNAs showed essentially equivalent induction during heat stress in the lcb1-100 mutant as in the parental strain. However, previous studies had demonstrated the lcb1-100 mutant failing to synthesize heat shock proteins [93]. Upon further investigation, however, a translational defect was demonstrated in that mutant. Specifically, both polysome analysis and 35S-methionine labeling showed a significantly lower rate of protein synthesis during heat stress in the lcb1-100 mutant [94]. Particularly noteworthy is that sphingolipid-dependent regulation of translation proceeded in part through the Pkh1p kinase, discussed below.
b. Sphingoid base-mediated signaling
One mechanism that seems to act in sphingolipid-mediated signal transduction is the activation of the Pkh1p/Pkh2p kinases. These highly redundant kinases bear significant homology to mammalian phosphoinositide-dependent kinase 1, PDK1, and thus, any insights into how yeast signal through Pkh kinases could possibly have direct implications to mammalian sphingolipid signaling. Sphingoid bases directly activate these kinases in vitro, though specific binding sites remain to be identified [95–97]; However, overexpression of Pkh1p ameliorated many of the heat-related phenotypes of the lcb1-100 strain, indicating a role for these kinases in mediating sphingoid base signaling during heat stress [97]. Functions identified for these kinases thus far include regulation of cell wall integrity signaling [98], translation initiation of heat shock proteins [94], and endocytic internalization [97]. Interestingly, sphingolipids have also been implicated in each of these processes.
Downstream signaling partners of the Pkh1/2 kinsases include members of the AGC kinase family (homologous to the SGK kinases in mammals) such as Pkc1p, Sch9p, and Ypk1p/Ypk2p. Of particular importance, Pkc1p as well may mediate many of the heat-induced programs that require sphingolipids [97, 99]. Furthermore, Sch9p, a kinase closely related to mammalian Akt/PKB, became phosphorylated in vitro in a sphingoid base and Pkh1p-dependent manner, though its roles in the heat stress response remain poorly defined [96]. Interestingly, a direct role for the Ypk1p/Ypk2p kinases during heat stress has not yet become apparent in any study to date, but it likely functions in regulating overall translation as it was shown to mediate levels of eIF4G [100], presenting the possibility that it does mediate some aspect of recovery from heat stress.
Perhaps one of the most interesting findings so far is the discovery that, while sphingoid bases positively regulate Pkh1p singaling through AGC kinases, two recently described proteins, Pil1p and Lsp1p, seem to allow negative regulation of Pkh1p-mediated signaling in a sphingolipid-dependent manner. Indeed, sphingoid bases reduced the phosphorylation of Pil1p and increased phosphorylation of Lsp1p in vitro by Pkh kinases [101]. Pil1p and Lsp1p then down-regulated Pkh activity including phosphorylation of Ypk1p [101]. This study provided new insights into signaling pathways regulated by sphingoid bases, and, moreover, gave scientists a glimpse of the complexity of sphingolipid-mediated signaling.
c. Endocytosis
In another set of studies, Pil1p and Lsp1p were identified as localizing to constitutive cellular points for initiation of endoycytosis, termed ‘eisosomes’, with Pil1p itself proposed to regulate formation of theses sites, and thus, to regulate spatial organization of endocytosis [102]. This finding returned focus to the original observation of a role for sphingoid bases in regulation of endocytosis due to mediation of cytoskeleton organization through actin dynamics. Interestingly, the endocytic defect observed in the lcb1-100 strain was overcome by overexpression of the yeast kinases Pkc1p or Yck2p, or deletion of a protein phosphatase 2A isoform [99]. These alterations also restored normal organization of the actin cytoskeleton in that study, indicating that sphingolipids facilitate, at least in part, the intimate relationships between cytoskeletal organization and efficient endocytosis. These studies demonstrated a sphingoid base requirement for endocytosis of the mating factor receptor and centered on cytoskeletal dynamics; however, subsequent studies clarified roles for sphingoid bases in endocytosis of nutrient permeases and discovered subsequent roles for sphingolipids in ubiquitin-mediated proteolysis [103, 104]. The role of permease degradation in the heat stress response remains to be determined; however, one possibility is that these proteins become damaged upon exposure to heat and must be cleared from the cell. A more interesting possibility is that degradation of these permeases sends an amino acid starvation signal to the cell. This hypothesis is particularly intriguing because many amino acid biosynthesis pathways were transcriptionally activated upon heat stress in a sphingolipid-dependent manner [92].
Taken together, these studies provide several insights into mechanisms by which sphingolipids mediate endocytosis including through modulation of signaling pathways, regulation of cytoskeletal dynamics, and regulation of the spatial organization of endocytosis via eisosome maintenance; however, many questions remain unanswered and thus, further investigation is needed to completely elucidate these roles.
d. Intracellular protein trafficking and assembly
Aside from these roles of sphingoid bases, many roles in protein trafficking, targeting to the plasma membrane, and assembly of protein complexes have previously been described and extensively reviewed. In brief, sphingolipids were identified as necessary for appropriate ER to golgi transport of GPI-anchored proteins [105, 106]. Sphingolipids have also been demonstrated as necessary for appropriate topology, cell surface delivery, stability, and/or plasma membrane association of major proteins including the plasma membrane ATPase, Pma1p [107, 108], the Vacuolar ATPase [109], and the uracil permease Fur4p[110]. It will be interesting to see how specific these roles are to these specific proteins, or if these findings represent a more general role for sphingolipids in mediating protein trafficking and assembly. In the case of Fur4p, however, sphingolipids may be a component of lipid microdomains necessary for correct association with the plasma membrane. These microdomains are probably composed of sterols and sphingolipids [111]. Thus, an important route of investigation should involve teasing apart the structural vs. signaling roles of yeast sphingolipids; however, the two roles may be related in the sense that microdomains could serve as signaling platforms, i.e., through the assembly of multi-protein signaling complexes.
IV. Conclusions
Though previous and current research continues to demonstrate the importance of sphingolipids for cell growth and responses to extracellular stressors, the mechanisms by which sphingolipid synthesis mediates programs of cell regulation remain largely undefined. The ability of these lipids to mediate both signaling through activation of protein kinases, protein trafficking and intracellular localization, and the structure of lipid microdomains and signaling platforms, suggest complex and integral roles for sphingolipids in cell regulation. Many of these sphingolipid functions may not necessarily proceed through previously established paradigms and thus, creativity may be required for progress in this area. Furthermore, several exciting new findings should inspire vigorous investigation into sphingolipid-mediated cell regulation, as many interesting roles for these key lipids likely await definition.
Table 1.
Major Functions of Yeast Sphingolipids
| Function | Implicated Lipid(s) | Implicated Gene(s) | Reference(s) |
|---|---|---|---|
| Endocytosis | Sphingoid bases | LCB1 | [88, 97, 99, 104] |
| Pkh1 activation | Sphingoid bases | LCB1/LCB2 | [96, 97, 101] |
| Heat Stress Response | Sphingoid bases, Sphingoid base phosphates, Phytoceramide | LCB1/2, LCB4/5, LAG1/LAC1, ISC1 | [35, 41, 74–77, 90–92, 94, 104] |
| Regulation of Translation | Sphingoid bases | LCB1/2 | [94] |
| Regulation of transcription | Unknown | LCB1 | [92] |
| GPI-anchored protein trafficking | Very long chain Ceramides, complex sphingolipids | LAG1/LAC1, ELO2, ELO3 | [44, 105, 106, 112, 113] |
| Calcium sensitivity | Complex sphingolipids | TSC3, TSC10, TSC13, CSG1, CSG2 | [29, 46, 56, 57, 64, 66, 114–116] |
| Cell cycle regulation | Sphingoid bases, Sphingoid base phosphates | LCB1/2, LCB4/5, DPL1, YSR2, YSR3 | [42, 79, 90] |
Acknowledgments
The authors would like to acknowledge Stefanka Spassieva for a thorough reading of the manuscript and helpful suggestions. This work was supported in part by a MREP award (to L.A.C.) and by a MERIT Award (to L.M.O.) by the Office of Research and Development, Department of Veterans Affairs, Ralph H. Johnson VA Medical Center, Charleston, South Carolina. L.M.O. would also like to acknowledge support from NIH Grants AG16583 and GM 62887.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.El Alwani M, Wu BX, Obeid LM, Hannun YA. Bioactive sphingolipids in the modulation of the inflammatory response. Pharmacol Ther. 2006 doi: 10.1016/j.pharmthera.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 2.Lamour NF, Chalfant CE. Ceramide-1-phosphate: the “missing” link in eicosanoid biosynthesis and inflammation. Mol Interv. 2005;5:358–367. doi: 10.1124/mi.5.6.8. [DOI] [PubMed] [Google Scholar]
- 3.Argraves KM, Wilkerson BA, Argraves WS, Fleming PA, Obeid LM, Drake CJ. Sphingosine-1-phosphate signaling promotes critical migratory events in vasculogenesis. J Biol Chem. 2004;279:50580–50590. doi: 10.1074/jbc.M404432200. [DOI] [PubMed] [Google Scholar]
- 4.Summers SA. Ceramides in insulin resistance and lipotoxicity. Prog Lipid Res. 2006;45:42–72. doi: 10.1016/j.plipres.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Summers SA, Nelson DH. A role for sphingolipids in producing the common features of type 2 diabetes, metabolic syndrome X, and Cushing’s syndrome. Diabetes. 2005;54:591–602. doi: 10.2337/diabetes.54.3.591. [DOI] [PubMed] [Google Scholar]
- 6.Unger RH, Orci L. Lipoapoptosis: its mechanism and its diseases. Biochim Biophys Acta. 2002;1585:202–212. doi: 10.1016/s1388-1981(02)00342-6. [DOI] [PubMed] [Google Scholar]
- 7.Modrak DE, Gold DV, Goldenberg DM. Sphingolipid targets in cancer therapy. Mol Cancer Ther. 2006;5:200–208. doi: 10.1158/1535-7163.MCT-05-0420. [DOI] [PubMed] [Google Scholar]
- 8.Ogretmen B, Hannun YA. Biologically active sphingolipids in cancer pathogenesis and treatment. Nat Rev Cancer. 2004;4:604–616. doi: 10.1038/nrc1411. [DOI] [PubMed] [Google Scholar]
- 9.Rawat SS, Johnson BT, Puri A. Sphingolipids: modulators of HIV-1 infection and pathogenesis. Biosci Rep. 2005;25:329–343. doi: 10.1007/s10540-005-2894-5. [DOI] [PubMed] [Google Scholar]
- 10.Rawat SS, Viard M, Gallo SA, Rein A, Blumenthal R, Puri A. Modulation of entry of enveloped viruses by cholesterol and sphingolipids (Review) Mol Membr Biol. 2003;20:243–254. doi: 10.1080/0968768031000104944. [DOI] [PubMed] [Google Scholar]
- 11.Luberto C, Toffaletti DL, Wills EA, Tucker SC, Casadevall A, Perfect JR, Hannun YA, Del Poeta M. Roles for inositol-phosphoryl ceramide synthase 1 (IPC1) in pathogenesis of C. neoformans. Genes Dev. 2001;15:201–212. doi: 10.1101/gad.856001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heung LJ, Kaiser AE, Luberto C, Del Poeta M. The role and mechanism of diacylglycerol-protein kinase C1 signaling in melanogenesis by Cryptococcus neoformans. J Biol Chem. 2005;280:28547–28555. doi: 10.1074/jbc.M503404200. [DOI] [PubMed] [Google Scholar]
- 13.Sawai H, Domae N, Okazaki T. Current status and perspectives in ceramide-targeting molecular medicine. Curr Pharm Des. 2005;11:2479–2487. doi: 10.2174/1381612054367463. [DOI] [PubMed] [Google Scholar]
- 14.Han X. Lipid alterations in the earliest clinically recognizable stage of Alzheimer’s disease: implication of the role of lipids in the pathogenesis of Alzheimer’s disease. Curr Alzheimer Res. 2005;2:65–77. doi: 10.2174/1567205052772786. [DOI] [PubMed] [Google Scholar]
- 15.Park TS, Panek RL, Rekhter MD, Mueller SB, Rosebury WS, Robertson A, Hanselman JC, Kindt E, Homan R, Karathanasis SK. Modulation of lipoprotein metabolism by inhibition of sphingomyelin synthesis in ApoE knockout mice. Atherosclerosis. 2006 doi: 10.1016/j.atherosclerosis.2005.12.029. [DOI] [PubMed] [Google Scholar]
- 16.Raas-Rothschild A, Pankova-Kholmyansky I, Kacher Y, Futerman AH. Glycosphingolipidoses: beyond the enzymatic defect. Glycoconj J. 2004;21:295–304. doi: 10.1023/B:GLYC.0000046272.38480.ef. [DOI] [PubMed] [Google Scholar]
- 17.Ginzburg L, Kacher Y, Futerman AH. The pathogenesis of glycosphingolipid storage disorders. Semin Cell Dev Biol. 2004;15:417–431. doi: 10.1016/j.semcdb.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 18.Humphrey T, Pearce A. Cell cycle molecules and mechanisms of the budding and fission yeasts. Methods Mol Biol. 2005;296:3–29. doi: 10.1385/1-59259-857-9:003. [DOI] [PubMed] [Google Scholar]
- 19.Sinclair DA, Lin SJ, Guarente L. Life-span extension in yeast. Science. 2006;312:195–197. doi: 10.1126/science.312.5771.195d. author reply 195–197. [DOI] [PubMed] [Google Scholar]
- 20.Gasch AP, Werner-Washburne M. The genomics of yeast responses to environmental stress and starvation. Funct Integr Genomics. 2002;2:181–192. doi: 10.1007/s10142-002-0058-2. [DOI] [PubMed] [Google Scholar]
- 21.Obeid LM, Okamoto Y, Mao C. Yeast sphingolipids: metabolism and biology. Biochim Biophys Acta. 2002;1585:163–171. doi: 10.1016/s1388-1981(02)00337-2. [DOI] [PubMed] [Google Scholar]
- 22.Dickson RC, Sumanasekera C, Lester RL. Functions and metabolism of sphingolipids in Saccharomyces cerevisiae. Prog Lipid Res. 2006 doi: 10.1016/j.plipres.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 23.Nagiec MM, Baltisberger JA, Wells GB, Lester RL, Dickson RC. The LCB2 gene of Saccharomyces and the related LCB1 gene encode subunits of serine palmitoyltransferase, the initial enzyme in sphingolipid synthesis. Proc Natl Acad Sci U S A. 1994;91:7899–7902. doi: 10.1073/pnas.91.17.7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gable K, Slife H, Bacikova D, Monaghan E, Dunn TM. Tsc3p is an 80-amino acid protein associated with serine palmitoyltransferase and required for optimal enzyme activity. J Biol Chem. 2000;275:7597–7603. doi: 10.1074/jbc.275.11.7597. [DOI] [PubMed] [Google Scholar]
- 25.Zhao C, Beeler T, Dunn T. Suppressors of the Ca(2+)-sensitive yeast mutant (csg2) identify genes involved in sphingolipid biosynthesis. Cloning and characterization of SCS1, a gene required for serine palmitoyltransferase activity. J Biol Chem. 1994;269:21480–21488. [PubMed] [Google Scholar]
- 26.Dickson RC, Wells GB, Schmidt A, Lester RL. Isolation of mutant Saccharomyces cerevisiae strains that survive without sphingolipids. Mol Cell Biol. 1990;10:2176–2181. doi: 10.1128/mcb.10.5.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinto WJ, Wells GW, Lester RL. Characterization of enzymatic synthesis of sphingolipid long-chain bases in Saccharomyces cerevisiae: mutant strains exhibiting long-chain-base auxotrophy are deficient in serine palmitoyltransferase activity. J Bacteriol. 1992;174:2575–2581. doi: 10.1128/jb.174.8.2575-2581.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wells GB, Lester RL. The isolation and characterization of a mutant strain of Saccharomyces cerevisiae that requires a long chain base for growth and for synthesis of phosphosphingolipids. J Biol Chem. 1983;258:10200–10203. [PubMed] [Google Scholar]
- 29.Gable K, Han G, Monaghan E, Bacikova D, Natarajan M, Williams R, Dunn TM. Mutations in the yeast LCB1 and LCB2 genes, including those corresponding to the hereditary sensory neuropathy type I mutations, dominantly inactivate serine palmitoyltransferase. J Biol Chem. 2002;277:10194–10200. doi: 10.1074/jbc.M107873200. [DOI] [PubMed] [Google Scholar]
- 30.Han G, Gable K, Yan L, Natarajan M, Krishnamurthy J, Gupta SD, Borovitskaya A, Harmon JM, Dunn TM. The topology of the Lcb1p subunit of yeast serine palmitoyltransferase. J Biol Chem. 2004;279:53707–53716. doi: 10.1074/jbc.M410014200. [DOI] [PubMed] [Google Scholar]
- 31.Monaghan E, Gable K, Dunn T. Mutations in the Lcb2p subunit of serine palmitoyltransferase eliminate the requirement for the TSC3 gene in Saccharomyces cerevisiae. Yeast. 2002;19:659–670. doi: 10.1002/yea.864. [DOI] [PubMed] [Google Scholar]
- 32.Beeler T, Bacikova D, Gable K, Hopkins L, Johnson C, Slife H, Dunn T. The Saccharomyces cerevisiae TSC10/YBR265w gene encoding 3-ketosphinganine reductase is identified in a screen for temperature-sensitive suppressors of the Ca2+-sensitive csg2Delta mutant. J Biol Chem. 1998;273:30688–30694. doi: 10.1074/jbc.273.46.30688. [DOI] [PubMed] [Google Scholar]
- 33.Grilley MM, Stock SD, Dickson RC, Lester RL, Takemoto JY. Syringomycin action gene SYR2 is essential for sphingolipid 4-hydroxylation in Saccharomyces cerevisiae. J Biol Chem. 1998;273:11062–11068. doi: 10.1074/jbc.273.18.11062. [DOI] [PubMed] [Google Scholar]
- 34.Nagiec MM, Skrzypek M, Nagiec EE, Lester RL, Dickson RC. The LCB4 (YOR171c) and LCB5 (YLR260w) genes of Saccharomyces encode sphingoid long chain base kinases. J Biol Chem. 1998;273:19437–19442. doi: 10.1074/jbc.273.31.19437. [DOI] [PubMed] [Google Scholar]
- 35.Ferguson-Yankey SR, Skrzypek MS, Lester RL, Dickson RC. Mutant analysis reveals complex regulation of sphingolipid long chain base phosphates and long chain bases during heat stress in yeast. Yeast. 2002;19:573–586. doi: 10.1002/yea.861. [DOI] [PubMed] [Google Scholar]
- 36.Saba JD, Nara F, Bielawska A, Garrett S, Hannun YA. The BST1 gene of Saccharomyces cerevisiae is the sphingosine-1-phosphate lyase. J Biol Chem. 1997;272:26087–26090. doi: 10.1074/jbc.272.42.26087. [DOI] [PubMed] [Google Scholar]
- 37.Panwar SL, Moye-Rowley WS. Long chain base tolerance in Saccharomyces cerevisiae is induced by retrograde signals from the mitochondria. J Biol Chem. 2006;281:6376–6384. doi: 10.1074/jbc.M512115200. [DOI] [PubMed] [Google Scholar]
- 38.Kim S, Fyrst H, Saba J. Accumulation of phosphorylated sphingoid long chain bases results in cell growth inhibition in Saccharomyces cerevisiae. Genetics. 2000;156:1519–1529. doi: 10.1093/genetics/156.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kihara A, Igarashi Y. Identification and characterization of a Saccharomyces cerevisiae gene, RSB1, involved in sphingoid long-chain base release. J Biol Chem. 2002;277:30048–30054. doi: 10.1074/jbc.M203385200. [DOI] [PubMed] [Google Scholar]
- 40.Mao C, Wadleigh M, Jenkins GM, Hannun YA, Obeid LM. Identification and characterization of Saccharomyces cerevisiae dihydrosphingosine-1-phosphate phosphatase. J Biol Chem. 1997;272:28690–28694. doi: 10.1074/jbc.272.45.28690. [DOI] [PubMed] [Google Scholar]
- 41.Mandala SM, Thornton R, Tu Z, Kurtz MB, Nickels J, Broach J, Menzeleev R, Spiegel S. Sphingoid base 1-phosphate phosphatase: a key regulator of sphingolipid metabolism and stress response. Proc Natl Acad Sci U S A. 1998;95:150–155. doi: 10.1073/pnas.95.1.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mao C, Saba JD, Obeid LM. The dihydrosphingosine-1-phosphate phosphatases of Saccharomyces cerevisiae are important regulators of cell proliferation and heat stress responses. Biochem J. 1999;342(Pt 3):667–675. [PMC free article] [PubMed] [Google Scholar]
- 43.Funato K, Lombardi R, Vallee B, Riezman H. Lcb4p is a key regulator of ceramide synthesis from exogenous long chain sphingoid base in Saccharomyces cerevisiae. J Biol Chem. 2003;278:7325–7334. doi: 10.1074/jbc.M209925200. [DOI] [PubMed] [Google Scholar]
- 44.Guillas I, Kirchman PA, Chuard R, Pfefferli M, Jiang JC, Jazwinski SM, Conzelmann A. C26-CoA-dependent ceramide synthesis of Saccharomyces cerevisiae is operated by Lag1p and Lac1p. Embo J. 2001;20:2655–2665. doi: 10.1093/emboj/20.11.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oh CS, Toke DA, Mandala S, Martin CE. ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J Biol Chem. 1997;272:17376–17384. doi: 10.1074/jbc.272.28.17376. [DOI] [PubMed] [Google Scholar]
- 46.Han G, Gable K, Kohlwein SD, Beaudoin F, Napier JA, Dunn TM. The Saccharomyces cerevisiae YBR159w gene encodes the 3-ketoreductase of the microsomal fatty acid elongase. J Biol Chem. 2002;277:35440–35449. doi: 10.1074/jbc.M205620200. [DOI] [PubMed] [Google Scholar]
- 47.Kohlwein SD, Eder S, Oh CS, Martin CE, Gable K, Bacikova D, Dunn T. Tsc13p is required for fatty acid elongation and localizes to a novel structure at the nuclear-vacuolar interface in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:109–125. doi: 10.1128/MCB.21.1.109-125.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schorling S, Vallee B, Barz WP, Riezman H, Oesterhelt D. Lag1p and Lac1p are essential for the Acyl-CoA-dependent ceramide synthase reaction in Saccharomyces cerevisae. Mol Biol Cell. 2001;12:3417–3427. doi: 10.1091/mbc.12.11.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lahiri S, Futerman AH. LASS5 is a bona fide dihydroceramide synthase that selectively utilizes palmitoyl-CoA as acyl donor. J Biol Chem. 2005;280:33735–33738. doi: 10.1074/jbc.M506485200. [DOI] [PubMed] [Google Scholar]
- 50.Venkataraman K, Riebeling C, Bodennec J, Riezman H, Allegood JC, Sullards MC, Merrill AH, Jr, Futerman AH. Upstream of growth and differentiation factor 1 (uog1), a mammalian homolog of the yeast longevity assurance gene 1 (LAG1), regulates N-stearoyl-sphinganine (C18-(dihydro)ceramide) synthesis in a fumonisin B1-independent manner in mammalian cells. J Biol Chem. 2002;277:35642–35649. doi: 10.1074/jbc.M205211200. [DOI] [PubMed] [Google Scholar]
- 51.Pewzner-Jung Y, Ben-Dor S, Futerman AH. When do Lasses (Longevity Assurance Genes) become CerS (Ceramide Synthases)? Insights into the regulation of ceramide synthesis. J Biol Chem. 2006 doi: 10.1074/jbc.R600010200. [DOI] [PubMed] [Google Scholar]
- 52.Mizutani Y, Kihara A, Igarashi Y. Mammalian Lass6 and its related family members regulate synthesis of specific ceramides. Biochem J. 2005;390:263–271. doi: 10.1042/BJ20050291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vallee B, Riezman H. Lip1p: a novel subunit of acyl-CoA ceramide synthase. Embo J. 2005;24:730–741. doi: 10.1038/sj.emboj.7600562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu WI, McDonough VM, Nickels JT, Jr, Ko J, Fischl AS, Vales TR, Merrill AH, Jr, Carman GM. Regulation of lipid biosynthesis in Saccharomyces cerevisiae by fumonisin B1. J Biol Chem. 1995;270:13171–13178. doi: 10.1074/jbc.270.22.13171. [DOI] [PubMed] [Google Scholar]
- 55.Wang E, Norred WP, Bacon CW, Riley RT, Merrill AH., Jr Inhibition of sphingolipid biosynthesis by fumonisins. Implications for diseases associated with Fusarium moniliforme. J Biol Chem. 1991;266:14486–14490. [PubMed] [Google Scholar]
- 56.Dunn TM, Haak D, Monaghan E, Beeler TJ. Synthesis of monohydroxylated inositolphosphorylceramide (IPC-C) in Saccharomyces cerevisiae requires Scs7p, a protein with both a cytochrome b5-like domain and a hydroxylase/desaturase domain. Yeast. 1998;14:311–321. doi: 10.1002/(SICI)1097-0061(19980315)14:4<311::AID-YEA220>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 57.Haak D, Gable K, Beeler T, Dunn T. Hydroxylation of Saccharomyces cerevisiae ceramides requires Sur2p and Scs7p. J Biol Chem. 1997;272:29704–29710. doi: 10.1074/jbc.272.47.29704. [DOI] [PubMed] [Google Scholar]
- 58.Hama H, Young DA, Radding JA, Ma D, Tang J, Stock SD, Takemoto JY. Requirement of sphingolipid alpha-hydroxylation for fungicidal action of syringomycin E. FEBS Lett. 2000;478:26–28. doi: 10.1016/s0014-5793(00)01821-4. [DOI] [PubMed] [Google Scholar]
- 59.Cliften P, Wang Y, Mochizuki D, Miyakawa T, Wangspa R, Hughes J, Takemoto JY. SYR2, a gene necessary for syringomycin growth inhibition of Saccharomyces cerevisiae. Microbiology. 1996;142(Pt 3):477–484. doi: 10.1099/13500872-142-3-477. [DOI] [PubMed] [Google Scholar]
- 60.Alderson NL, Rembiesa BM, Walla MD, Bielawska A, Bielawski J, Hama H. The human FA2H gene encodes a fatty acid 2-hydroxylase. J Biol Chem. 2004;279:48562–48568. doi: 10.1074/jbc.M406649200. [DOI] [PubMed] [Google Scholar]
- 61.Mao C, Xu R, Bielawska A, Szulc ZM, Obeid LM. Cloning and characterization of a Saccharomyces cerevisiae alkaline ceramidase with specificity for dihydroceramide. J Biol Chem. 2000;275:31369–31378. doi: 10.1074/jbc.M003683200. [DOI] [PubMed] [Google Scholar]
- 62.Mao C, Xu R, Bielawska A, Obeid LM. Cloning of an alkaline ceramidase from Saccharomyces cerevisiae. An enzyme with reverse (CoA-independent) ceramide synthase activity. J Biol Chem. 2000;275:6876–6884. doi: 10.1074/jbc.275.10.6876. [DOI] [PubMed] [Google Scholar]
- 63.Nagiec MM, Nagiec EE, Baltisberger JA, Wells GB, Lester RL, Dickson RC. Sphingolipid synthesis as a target for antifungal drugs. Complementation of the inositol phosphorylceramide synthase defect in a mutant strain of Saccharomyces cerevisiae by the AUR1 gene. J Biol Chem. 1997;272:9809–9817. doi: 10.1074/jbc.272.15.9809. [DOI] [PubMed] [Google Scholar]
- 64.Beeler TJ, Fu D, Rivera J, Monaghan E, Gable K, Dunn TM. SUR1 (CSG1/BCL21), a gene necessary for growth of Saccharomyces cerevisiae in the presence of high Ca2+ concentrations at 37 degrees C, is required for mannosylation of inositolphosphorylceramide. Mol Gen Genet. 1997;255:570–579. doi: 10.1007/s004380050530. [DOI] [PubMed] [Google Scholar]
- 65.Uemura S, Kihara A, Inokuchi J, Igarashi Y. Csg1p and newly identified Csh1p function in mannosylinositol phosphorylceramide synthesis by interacting with Csg2p. J Biol Chem. 2003;278:45049–45055. doi: 10.1074/jbc.M305498200. [DOI] [PubMed] [Google Scholar]
- 66.Dickson RC, Nagiec EE, Wells GB, Nagiec MM, Lester RL. Synthesis of mannose-(inositol-P)2-ceramide, the major sphingolipid in Saccharomyces cerevisiae, requires the IPT1 (YDR072c) gene. J Biol Chem. 1997;272:29620–29625. doi: 10.1074/jbc.272.47.29620. [DOI] [PubMed] [Google Scholar]
- 67.Sawai H, Okamoto Y, Luberto C, Mao C, Bielawska A, Domae N, Hannun YA. Identification of ISC1 (YER019w) as inositol phosphosphingolipid phospholipase C in Saccharomyces cerevisiae. J Biol Chem. 2000;275:39793–39798. doi: 10.1074/jbc.M007721200. [DOI] [PubMed] [Google Scholar]
- 68.Kingsbury JM, Goldstein AL, McCusker JH. Role of nitrogen and carbon transport, regulation, and metabolism genes for Saccharomyces cerevisiae survival in vivo. Eukaryot Cell. 2006;5:816–824. doi: 10.1128/EC.5.5.816-824.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sultan I, Senkal CE, Ponnusamy S, Bielawski J, Szulc Z, Bielawska A, Hannun YA, Ogretmen B. Regulation of the sphingosine-recycling pathway for ceramide generation by oxidative stress, and its role in controlling c-Myc/Max function. Biochem J. 2006;393:513–521. doi: 10.1042/BJ20051083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ogretmen B, Pettus BJ, Rossi MJ, Wood R, Usta J, Szulc Z, Bielawska A, Obeid LM, Hannun YA. Biochemical mechanisms of the generation of endogenous long chain ceramide in response to exogenous short chain ceramide in the A549 human lung adenocarcinoma cell line. Role for endogenous ceramide in mediating the action of exogenous ceramide. J Biol Chem. 2002;277:12960–12969. doi: 10.1074/jbc.M110699200. [DOI] [PubMed] [Google Scholar]
- 71.Jenkins GM, Cowart LA, Signorelli P, Pettus BJ, Chalfant CE, Hannun YA. Acute activation of de novo sphingolipid biosynthesis upon heat shock causes an accumulation of ceramide and subsequent dephosphorylation of SR proteins. J Biol Chem. 2002;277:42572–42578. doi: 10.1074/jbc.M207346200. [DOI] [PubMed] [Google Scholar]
- 72.Lester RL, Wells GB, Oxford G, Dickson RC. Mutant strains of Saccharomyces cerevisiae lacking sphingolipids synthesize novel inositol glycerophospholipids that mimic sphingolipid structures. J Biol Chem. 1993;268:845–856. [PubMed] [Google Scholar]
- 73.Patton JL, Srinivasan B, Dickson RC, Lester RL. Phenotypes of sphingolipid-dependent strains of Saccharomyces cerevisiae. J Bacteriol. 1992;174:7180–7184. doi: 10.1128/jb.174.22.7180-7184.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Skrzypek MS, Nagiec MM, Lester RL, Dickson RC. Analysis of phosphorylated sphingolipid long-chain bases reveals potential roles in heat stress and growth control in Saccharomyces. J Bacteriol. 1999;181:1134–1140. doi: 10.1128/jb.181.4.1134-1140.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wells GB, Dickson RC, Lester RL. Heat-induced elevation of ceramide in Saccharomyces cerevisiae via de novo synthesis. J Biol Chem. 1998;273:7235–7243. doi: 10.1074/jbc.273.13.7235. [DOI] [PubMed] [Google Scholar]
- 76.Dickson RC, Nagiec EE, Skrzypek M, Tillman P, Wells GB, Lester RL. Sphingolipids are potential heat stress signals in Saccharomyces. J Biol Chem. 1997;272:30196–30200. doi: 10.1074/jbc.272.48.30196. [DOI] [PubMed] [Google Scholar]
- 77.Jenkins GM, Richards A, Wahl T, Mao C, Obeid L, Hannun Y. Involvement of yeast sphingolipids in the heat stress response of Saccharomyces cerevisiae. J Biol Chem. 1997;272:32566–32572. doi: 10.1074/jbc.272.51.32566. [DOI] [PubMed] [Google Scholar]
- 78.Iwaki S, Kihara A, Sano T, Igarashi Y. Phosphorylation by Pho85 cyclin-dependent kinase acts as a signal for the down-regulation of the yeast sphingoid long-chain base kinase Lcb4 during the stationary phase. J Biol Chem. 2005;280:6520–6527. doi: 10.1074/jbc.M410908200. [DOI] [PubMed] [Google Scholar]
- 79.Gottlieb D, Heideman W, Saba JD. The DPL1 gene is involved in mediating the response to nutrient deprivation in Saccharomyces cerevisiae. Mol Cell Biol Res Commun. 1999;1:66–71. doi: 10.1006/mcbr.1999.0109. [DOI] [PubMed] [Google Scholar]
- 80.Kihara A, Kurotsu F, Sano T, Iwaki S, Igarashi Y. Long-chain base kinase Lcb4 Is anchored to the membrane through its palmitoylation by Akr1. Mol Cell Biol. 2005;25:9189–9197. doi: 10.1128/MCB.25.21.9189-9197.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kobayashi SD, Nagiec MM. Ceramide/long-chain base phosphate rheostat in Saccharomyces cerevisiae: regulation of ceramide synthesis by Elo3p and Cka2p. Eukaryot Cell. 2003;2:284–294. doi: 10.1128/EC.2.2.284-294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ogretmen B, Hannun YA. Updates on functions of ceramide in chemotherapy-induced cell death and in multidrug resistance. Drug Resist Updat. 2001;4:368–377. doi: 10.1054/drup.2001.0225. [DOI] [PubMed] [Google Scholar]
- 83.Kolaczkowski M, Kolaczkowska A, Gaigg B, Schneiter R, Moye-Rowley WS. Differential regulation of ceramide synthase components LAC1 and LAG1 in Saccharomyces cerevisiae. Eukaryot Cell. 2004;3:880–892. doi: 10.1128/EC.3.4.880-892.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vaena de Avalos S, Okamoto Y, Hannun YA. Activation and localization of inositol phosphosphingolipid phospholipase C, Isc1p, to the mitochondria during growth of Saccharomyces cerevisiae. J Biol Chem. 2004;279:11537–11545. doi: 10.1074/jbc.M309586200. [DOI] [PubMed] [Google Scholar]
- 85.Okamoto Y, Vaena de Avalos S, Hannun YA. Functional analysis of ISC1 by site-directed mutagenesis. Biochemistry. 2003;42:7855–7862. doi: 10.1021/bi0341354. [DOI] [PubMed] [Google Scholar]
- 86.Vaena de Avalos S, Su X, Zhang M, Okamoto Y, Dowhan W, Hannun YA. The phosphatidylglycerol/cardiolipin biosynthetic pathway is required for the activation of inositol phosphosphingolipid phospholipase C, Isc1p, during growth of Saccharomyces cerevisiae. J Biol Chem. 2005;280:7170–7177. doi: 10.1074/jbc.M411058200. [DOI] [PubMed] [Google Scholar]
- 87.Munn AL, Riezman H. Endocytosis is required for the growth of vacuolar H(+)-ATPase-defective yeast: identification of six new END genes. J Cell Biol. 1994;127:373–386. doi: 10.1083/jcb.127.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zanolari B, Friant S, Funato K, Sutterlin C, Stevenson BJ, Riezman H. Sphingoid base synthesis requirement for endocytosis in Saccharomyces cerevisiae. Embo J. 2000;19:2824–2833. doi: 10.1093/emboj/19.12.2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Piper PW. Molecular events associated with acquisition of heat tolerance by the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev. 1993;11:339–355. doi: 10.1111/j.1574-6976.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 90.Jenkins GM, Hannun YA. Role for de novo sphingoid base biosynthesis in the heat-induced transient cell cycle arrest of Saccharomyces cerevisiae. J Biol Chem. 2001;276:8574–8581. doi: 10.1074/jbc.M007425200. [DOI] [PubMed] [Google Scholar]
- 91.Cowart LA, Okamoto Y, Lu X, Hannun YA. Distinct roles for de novo versus hydrolytic pathways of sphingolipid biosynthesis in Saccharomyces cerevisiae. Biochem J. 2006;393:733–740. doi: 10.1042/BJ20050643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cowart LA, Okamoto Y, Pinto FR, Gandy JL, Almeida JS, Hannun YA. Roles for sphingolipid biosynthesis in mediation of specific programs of the heat stress response determined through gene expression profiling. J Biol Chem. 2003;278:30328–30338. doi: 10.1074/jbc.M300656200. [DOI] [PubMed] [Google Scholar]
- 93.Friant S, Meier KD, Riezman H. Increased ubiquitin-dependent degradation can replace the essential requirement for heat shock protein induction. Embo J. 2003;22:3783–3791. doi: 10.1093/emboj/cdg375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Meier KD, Deloche O, Kajiwara K, Funato K, Riezman H. Sphingoid base is required for translation initiation during heat stress in Saccharomyces cerevisiae. Mol Biol Cell. 2006;17:1164–1175. doi: 10.1091/mbc.E05-11-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu K, Zhang X, Sumanasekera C, Lester RL, Dickson RC. Signalling functions for sphingolipid long-chain bases in Saccharomyces cerevisiae. Biochem Soc Trans. 2005;33:1170–1173. doi: 10.1042/BST20051170. [DOI] [PubMed] [Google Scholar]
- 96.Liu K, Zhang X, Lester RL, Dickson RC. The sphingoid long chain base phytosphingosine activates AGC-type protein kinases in Saccharomyces cerevisiae including Ypk1, Ypk2, and Sch9. J Biol Chem. 2005;280:22679–22687. doi: 10.1074/jbc.M502972200. [DOI] [PubMed] [Google Scholar]
- 97.Friant S, Lombardi R, Schmelzle T, Hall MN, Riezman H. Sphingoid base signaling via Pkh kinases is required for endocytosis in yeast. Embo J. 2001;20:6783–6792. doi: 10.1093/emboj/20.23.6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schmelzle T, Helliwell SB, Hall MN. Yeast protein kinases and the RHO1 exchange factor TUS1 are novel components of the cell integrity pathway in yeast. Mol Cell Biol. 2002;22:1329–1339. doi: 10.1128/mcb.22.5.1329-1339.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Friant S, Zanolari B, Riezman H. Increased protein kinase or decreased PP2A activity bypasses sphingoid base requirement in endocytosis. Embo J. 2000;19:2834–2844. doi: 10.1093/emboj/19.12.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gelperin D, Horton L, DeChant A, Hensold J, Lemmon SK. Loss of ypk1 function causes rapamycin sensitivity, inhibition of translation initiation and synthetic lethality in 14-3-3-deficient yeast. Genetics. 2002;161:1453–1464. doi: 10.1093/genetics/161.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang X, Lester RL, Dickson RC. Pil1p and Lsp1p negatively regulate the 3-phosphoinositide-dependent protein kinase-like kinase Pkh1p and downstream signaling pathways Pkc1p and Ypk1p. J Biol Chem. 2004;279:22030–22038. doi: 10.1074/jbc.M400299200. [DOI] [PubMed] [Google Scholar]
- 102.Walther TC, Brickner JH, Aguilar PS, Bernales S, Pantoja C, Walter P. Eisosomes mark static sites of endocytosis. Nature. 2006;439:998–1003. doi: 10.1038/nature04472. [DOI] [PubMed] [Google Scholar]
- 103.Chung N, Mao C, Heitman J, Hannun YA, Obeid LM. Phytosphingosine as a specific inhibitor of growth and nutrient import in Saccharomyces cerevisiae. J Biol Chem. 2001;276:35614–35621. doi: 10.1074/jbc.M105653200. [DOI] [PubMed] [Google Scholar]
- 104.Chung N, Jenkins G, Hannun YA, Heitman J, Obeid LM. Sphingolipids signal heat stress-induced ubiquitin-dependent proteolysis. J Biol Chem. 2000;275:17229–17232. doi: 10.1074/jbc.C000229200. [DOI] [PubMed] [Google Scholar]
- 105.Skrzypek M, Lester RL, Dickson RC. Suppressor gene analysis reveals an essential role for sphingolipids in transport of glycosylphosphatidylinositol-anchored proteins in Saccharomyces cerevisiae. J Bacteriol. 1997;179:1513–1520. doi: 10.1128/jb.179.5.1513-1520.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Horvath A, Sutterlin C, Manning-Krieg U, Movva NR, Riezman H. Ceramide synthesis enhances transport of GPI-anchored proteins to the Golgi apparatus in yeast. Embo J. 1994;13:3687–3695. doi: 10.1002/j.1460-2075.1994.tb06678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gaigg B, Timischl B, Corbino L, Schneiter R. Synthesis of sphingolipids with very long chain fatty acids but not ergosterol is required for routing of newly synthesized plasma membrane ATPase to the cell surface of yeast. J Biol Chem. 2005;280:22515–22522. doi: 10.1074/jbc.M413472200. [DOI] [PubMed] [Google Scholar]
- 108.Lee MC, Hamamoto S, Schekman R. Ceramide biosynthesis is required for the formation of the oligomeric H+-ATPase Pma1p in the yeast endoplasmic reticulum. J Biol Chem. 2002;277:22395–22401. doi: 10.1074/jbc.M200450200. [DOI] [PubMed] [Google Scholar]
- 109.Chung JH, Lester RL, Dickson RC. Sphingolipid requirement for generation of a functional v1 component of the vacuolar ATPase. J Biol Chem. 2003;278:28872–28881. doi: 10.1074/jbc.M300943200. [DOI] [PubMed] [Google Scholar]
- 110.Hearn JD, Lester RL, Dickson RC. The uracil transporter Fur4p associates with lipid rafts. J Biol Chem. 2003;278:3679–3686. doi: 10.1074/jbc.M209170200. [DOI] [PubMed] [Google Scholar]
- 111.Bagnat M, Keranen S, Shevchenko A, Shevchenko A, Simons K. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc Natl Acad Sci U S A. 2000;97:3254–3259. doi: 10.1073/pnas.060034697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Watanabe R, Funato K, Venkataraman K, Futerman AH, Riezman H. Sphingolipids are required for the stable membrane association of glycosylphosphatidylinositol-anchored proteins in yeast. J Biol Chem. 2002;277:49538–49544. doi: 10.1074/jbc.M206209200. [DOI] [PubMed] [Google Scholar]
- 113.Sutterlin C, Doering TL, Schimmoller F, Schroder S, Riezman H. Specific requirements for the ER to Golgi transport of GPI-anchored proteins in yeast. J Cell Sci. 1997;110(Pt 21):2703–2714. doi: 10.1242/jcs.110.21.2703. [DOI] [PubMed] [Google Scholar]
- 114.Birchwood CJ, Saba JD, Dickson RC, Cunningham KW. Calcium influx and signaling in yeast stimulated by intracellular sphingosine 1-phosphate accumulation. J Biol Chem. 2001;276:11712–11718. doi: 10.1074/jbc.M010221200. [DOI] [PubMed] [Google Scholar]
- 115.Dunn TM, Gable K, Monaghan E, Bacikova D. Selection of yeast mutants in sphingolipid metabolism. Methods Enzymol. 2000;312:317–330. doi: 10.1016/s0076-6879(00)12918-0. [DOI] [PubMed] [Google Scholar]
- 116.Beeler T, Gable K, Zhao C, Dunn T. A novel protein, CSG2p, is required for Ca2+ regulation in Saccharomyces cerevisiae. J Biol Chem. 1994;269:7279–7284. [PubMed] [Google Scholar]


