Abstract
Molecular beacons are FRET-based target-activatable probes. They offer control of fluorescence emission in response to specific cancer targets, thus are useful tools for in vivo cancer imaging. Photodynamic therapy (PDT) is a cell-killing process by light activation of a photosensitizer (PS) in the presence of oxygen. The key cytotoxic agent is singlet oxygen (1O2). By combining these two principles (FRET and PDT), we have introduced a concept of photodynamic molecular beacons (PMB) for controlling the PS's ability to generate 1O2 and, ultimately, for controlling its PDT activity. The PMB comprises a disease-specific linker, a PS, and a 1O2 quencher, so that the PS's photoactivity is silenced until the linker interacts with a target molecule, such as a tumor-associated protease. Here, we report the full implementation of this concept by synthesizing a matrix metalloproteinase-7 (MMP7)-triggered PMB and achieving not only MMP7-triggered production of 1O2 in solution but also MMP7-mediated photodynamic cytotoxicity in cancer cells. Preliminary in vivo studies also reveal the MMP7-activated PDT efficacy of this PMB. This study validates the core principle of the PMB concept that selective PDT-induced cell death can be achieved by exerting precise control of the PS's ability to produce 1O2 by responding to specific cancer-associated biomarkers. Thus, PDT selectivity will no longer depend solely on how selectively the PS can be delivered to cancer cells. Rather, it will depend on how selective a biomarker is to cancer cells, and how selective the interaction of PMB is to this biomarker.
Keywords: activation, image-guided therapy, photodynamic therapy, matrix metalloproteinases, quencher
Photodynamic therapy (PDT) is an emerging cancer treatment modality involving the combination of light, a photosensitizer (PS), and molecular oxygen (1). It offers unique control in the PS's action, because the key cytotoxic agent, singlet oxygen (1O2), is produced only in situ upon irradiation. Therefore, PDT selectivity can be controlled at three different levels. The first level is to control how light is delivered to the disease tissue. This approach is the easiest to implement, because light can be readily manipulated and positioned, particularly through the judicious use of advanced fiber optics (e.g., prostate interstitial fibers) (2). However, it cannot achieve a high level of selectivity because of the limited tumor localization of existing PDT agents, which in turn cause treatment-related toxicity to surrounding normal tissues as well as sunlight-induced skin toxicity. The second level is to control how the PS is delivered to the tumor tissue. This approach has been actively pursued by many research groups, including ours, and has resulted in improvement of PDT selectivity (3–7). However, it is still vulnerable to collateral damage to surrounding normal tissues. A new direction is to exert control of the PS's ability to produce 1O2 (9–14). This third level control has the potential to achieve ultimate PDT selectivity to cancer cells while leaving normal cells unharmed.
We introduced the concept of photodynamic molecular beacons (PMB) that are capable of controlling 1O2 production that responds specifically to certain cancer biomarkers (9). This approach is an extension of the well known molecular beacons (MB) (15) that use the FRET principle for controlling fluorescence emission in response to target activation. By combining MB with PDT, we seek to enable cancer biomarker-controlled 1O2 production to achieve unprecedented PDT selectivity. The PMB comprises a disease-specific linker, a PS, and a 1O2 quencher, so that the PS's photoactivity is silenced until the linker interacts with a target molecule, such as a tumor-associated protease. In that initial study, a pyropheophorbide [(Pyro) as PS] and a carotenoid (as 1O2 quencher) were kept in close proximity by the self-folding of a caspase-3 specific peptide sequence, resulting in effective 1O2 quenching. Upon caspase-3-induced cleavage, the 1O2 production increased markedly in solution, as evidenced by direct 1O2 near-infrared (NIR) luminescence intensity and lifetime measurements. However, because caspase-3 is a cell apoptosis marker (16), it is difficult to further validate the concept in cancer cells by using this particular PMB, because of the difficulty of distinguishing PDT-induced toxicity in apoptotic cells. Hence, finding a specific cleavable peptide linker to target tumor-associated proteases is necessary.
Matrix metalloproteinases (MMPs) are known to be important in normal tissue remodeling but also play critical roles in many diseases (17), such as atherosclerosis, lung pulmonary fibrosis, and cancer. The presence of extracellular and membrane-bound MMPs in tumors not only aids the degradation of extracellular matrix by the neoplastic cells but also facilitates their motility and directs cell invasion (18). Thus, MMPs have long been of interest as pharmaceutical targets (17). Among these, matrix metalloproteinase-7 (MMP7) has been a particularly important target, because of its epithelial origin and its high expression in pancreatic, colon, breast, and nonsmall-cell lung cancer (19, 20).
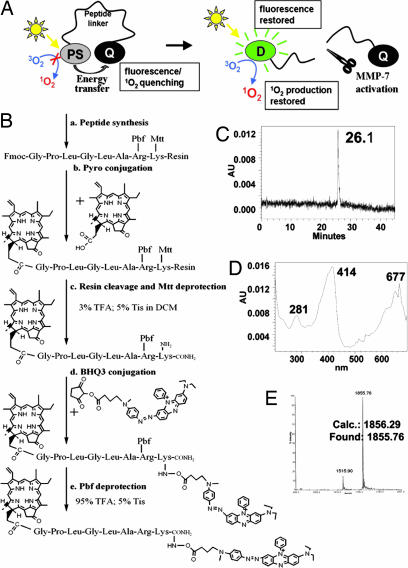
In this report, we describe the synthesis and characterization of an MMP7-triggered PMB, PPMMP7B, using: (i) Pyro as the PS, because of its excellent 1O2 quantum yield, NIR fluorescence emission, and high tumor affinity (21); (ii) black hole quencher 3 (BHQ3) as a dual fluorescence (22) and 1O2 quencher (10); and (iii) a short peptide sequence, GPLGLARK, as the MMP7-cleavable linker, with the cleavage site between G and L and recognition site, as indicated by italics (23). As depicted in Fig. 1A, Pyro and BHQ3 are conjugated to the opposite ends of the MMP7-specific cleavable peptide linker to keep them in close proximity, enabling FRET and 1O2 quenching to make this construct not only optically silent but also photodynamically inactive. We postulate that, when the beacon enters MMP7-expressing cells, MMP7 should induce specific cleavage of the peptide linker and remove the Pyro from the vicinity of BHQ3, restoring its fluorescence and photoreactivity. Subsequently, upon light irradiation, the targeted cells should be fluorescent and produce cytotoxic 1O2, while leaving normal cells undetectable and unharmed.
Fig. 1.
The PMB concept and the synthesis of PPMMP7B. (A) The concept of PMB. The synthesis and characteristics of PPMMP7B. (B) Synthesis protocol. (C) HPLC chromatography. (D) UV-vis spectrum. (E) MALDI-TOF mass spectrum of PPMMP7B.
To validate this approach, we first confirmed MMP7-specific activation of PPMMP7B in solution. Then, confocal microscopy and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays confirmed, respectively, the MMP7-specific fluorescence activation and photodynamic cytotoxicity in human nasopharyngeal epidermoid carcinoma (KB) cells (high MMP7 expression, MMP7+) vs. human breast carcinoma (BT20) cells (lack of MMP7 expression, MMP7−). In addition, a preliminary in vivo PDT study provides initial evidence of the MMP7-triggered PDT efficacy in KB tumor-bearing mice.
Results
PPMMP7B Synthesis.
Pyro-GPLGLARK(BHQ3) (4), PPMMP7B, was synthesized by the protocol shown in Fig. 1B. The final product was purified by HPLC (Fig. 1C), and its molecular structure was confirmed by UV-visible (UV-vis) spectroscopy (Pyro-specific absorbance was found at 414 nm and BHQ3-specific absorbance at 676 nm; Fig. 1D) and MALDI-TOF (calculated, 1,856.29; found, 1,855.76; Fig. 1E).
Validation of MMP7-Triggered PPMMP7B Activation and Its Corresponding 1O2 Activation in Solution.
Once the PPMMP7B was synthesized and its structure characterized, the next steps were to confirm that (i) MMP7 is able to specifically cleave the peptide linker GPLGLARK of PPMMP7B, (ii) both fluorescence and 1O2 production of Pyro are quenched in the native PPMMP7B construct, and (iii) the MMP7-induced peptide cleavage results in significant restoration of fluorescence and 1O2 production.
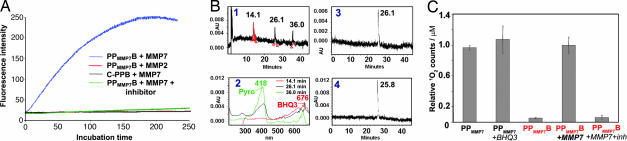
First, the fluorescence of PPMMP7B was compared with that of PPMMP7 (positive control) at the same concentration. PPMMP7B emitted 15-fold less fluorescence than PPMMP7, demonstrating that BHQ3 effectively quenches Pyro fluorescence in the intact PPMMP7B molecule. To evaluate the specificity of fluorescence activation by MMP7-induced peptide cleavage, fluorescence spectra were collected at 37°C on solutions of PPMMP7B+MMP7 (50:1 molar ratio), PPMMP7B+MMP7+MMP7 inhibitor (50:1:1,500 molar ratio), PPMMP7B+MMP2 (50:1 molar ratio), and Pyro-GDEVDGSGK-BHQ3 as a negative control for PPMMP7B (C-PPB)+MMP7 (50:1 molar ratio). As shown in Fig. 2A, we observed an immediate fluorescence increase in the case of PPMMP7B+MMP7, reaching a plateau at 3 h with a 12-fold fluorescence increase. However, this increase was prevented in the presence of the MMP7 inhibitor. No noticeable fluorescence increase was observed in the solution of PPMMP7B+MMP2 or C-PPB+MMP7. All these samples were then analyzed by HPLC. Fig. 2B1 shows that MMP7 cleaved PPMMP7B (at 26 min) into two fragments associated, respectively, with a Pyro moiety (at 36 min) and a BHQ3 moiety (at 14 min), based on their absorption characteristics (Fig. 2B2). This was further confirmed by MALDI-TOF, because the two fragments were identified as Pyro-GPLG-COO− (calculated, 857.44; found, 857.26) and NH2-LARK(BHQ3) (calculated, 1,014.62; found, 1,013.13). In addition, no cleavage was observed in the solution of PPMMP7B+MMP2 (Fig. 2B3) or C-PPB+MMP7 (Fig. 2B4). Taken together, these data demonstrate that the MMP7 specifically induced PPMMP7B cleavage at the known MMP7 cleavage site between G and L (24, 25).
Fig. 2.
The validation of MMP7-specific activation of PPMMP7B in solution. (A) Fluorescence kinetics of 0.4 μM PPMMP7B+MMP7 (50:1 molar ratio, blue line), 0.4 μM PPMMP7B+MMP7+inhibitor (50:1:1,500 molar ratio, green line), 0.4 μM PPMMP7B+MMP2 (50:1 molar ratio, red line), and 0.4 μM C-PPB+MMP7 (50:1 molar ratio, black line). (B) (1) HPLC spectrum of PPMMP7B+MMP7 and (2) corresponding UV-vis spectra. HPLC spectra of (3) PPMMP7B+MMP2 and (4) C-PPB+MMP7. All solutions were incubated at 37° for 2 h. (C) The relative 1O2 counts of PPMMP7, PPMMP7+BHQ3, PPMMP7B, PPMMP7B+MMP7, and PPMMP7B +MMP7+inhibitor.
To evaluate the efficiency of 1O2 quenching in intact PPMMP7B and the specificity of its 1O2 activation by MMP7, 1O2 NIR (1,270 nm) luminescence was measured directly in solutions of PPMMP7+BHQ3 (1:1 molar ratio), PPMMP7B, PPMMP7B+MMP7, and PPMMP7B+MMP7+MMP7 inhibitor. PPMMP7 alone was used as a positive control. As shown in Fig. 2C, compared with PPMMP7, 18-fold lower 1O2 production was observed in PPMMP7B, whereas a similar 1O2 production was found in the same concentration of PPMMP7+BHQ3, confirming that the close proximity of Pyro and BHQ3 by the self folding of the MMP7-cleavable peptide effectively inhibits the 1O2 production of Pyro. Moreover, adding MMP7 to the PPMMP7B (molar ratio: 1: 60; 3 h incubation) restored the quenched 1O2 production by 19-fold, an effect completely blocked by coincubation with the MMP7 inhibitor (molar ratio, 1:60:1,200 for MMP7; PPMMP7B, inhibitor), confirming that MMP7-induced separation of Pyro and BHQ3 allows the photoactivation of Pyro. All of the above experiments were repeated in triplicate and the 1O2 production of PPMMP7, PPMMP7+BHQ3, and PPMMP7B+MMP7 are all statistically different from that of PPMMP7B (P < 0.05), whereas none of those three is statistically different from the other.
Validation of MMP7-Triggered PPMMP7B Activation and Its Corresponding PDT Activation in Cancer Cells.
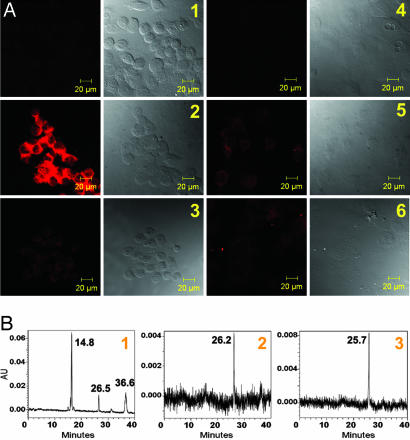
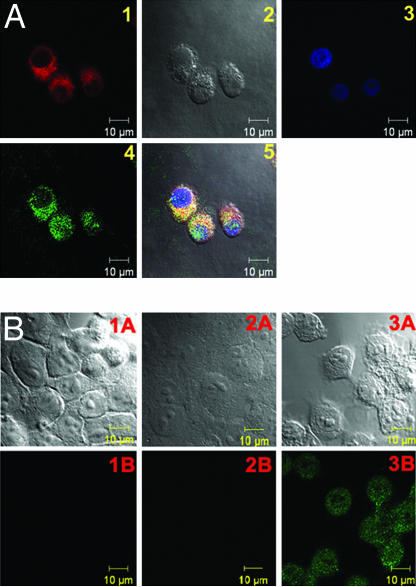
After we validated the MMP7-triggered PPMMP7B activation in solution, we evaluated this in cancer cells. To prove that PPMMP7B can be specifically activated by MMP7 in vitro, confocal fluorescence microscopy studies were performed on KB (MMP7+) and BT20 (MMP7−) (26) cells incubated with either PPMMP7B or the control C-PPB, with 633 nm excitation and >650 nm detection. As expected, confocal images showed a strong fluorescence signal in KB cells (MMP7+) incubated with PPMMP7B (Fig. 3A2), whereas control experiments yielded the following results: (i) BT20 cells (MMP7−) incubated with PPMMP7B showed minimal fluorescence (Fig. 3A5), and (ii) both KB and BT20 cells showed minimal fluorescence when incubated with C-PPB (Figs. 3A3 and 6). Further evidence was obtained by HPLC analysis of cell media collected at the end of drug incubation. Fig. 3B1 shows that PPMMP7B was cleaved by MMP7 in KB medium, generating two fragments consistent with the solution studies, whereas no cleavages were found in BT20 medium of PPMMP7B (Fig. 3B2) and KB medium of C-PPB (Fig. 3B3). These data clearly demonstrated that PPMMP7B can enter cells directly, and its cleavage was specifically mediated by MMP7 in cancer cells. Pyro serves in this case as a multifunctional module: (i) as a fluorescent dye, (ii) as a PS, and (iii) as a delivery vehicle, as shown (22).
Fig. 3.
In vitro validation of MMP7-specific activation. (A) Confocal images of PPMMP7B and control C-PPB in KB (MMP7+) and BT20 (MMP7−) cells showing fluorescence (Left) and bright field (Right) in each case. (1) KB cells alone, (2) KB cells plus 60 μM PPMMP7B, (3) KB cells plus 60 μM C-PPB, (4) BT20 cells alone, (5) BT20 cells plus 60 μM PPMMP7B, and (6) BT20 cells plus 60 μM C-PPB. (B) HPLC spectra of cell incubation media after 5-h incubation: (1) KB cells+PPMMP7B, (2) BT20 cells+PPMMP7B, and (3) KB cells+C-PPB.
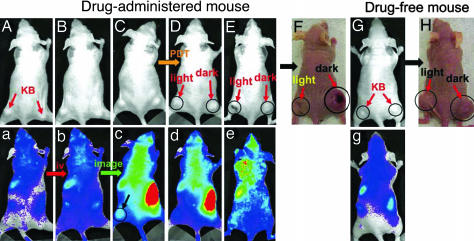
Fig. 6.
In vivo images of mice showing bright field (A–H) and fluorescence (a–e and g). (A–E) PPMMP7B-administered mouse (A, prescan; B, 10 min after i.v. injection; C, 3 h after i.v. injection; D, 5 h after i.v. injection and 1 h after PDT; E, 3 d after PDT). (F) Photograph of PPMMP7B-administered mouse (30 d after PDT). Light-treated tumors are marked as “light” and nonlight-treated tumors as “dark.” (G) Drug-free mouse (prescan). (H) Photograph of drug-free mouse (30 d after PDT) with light-treated tumor marked as “light” and nonlight-treated tumor as “dark.”
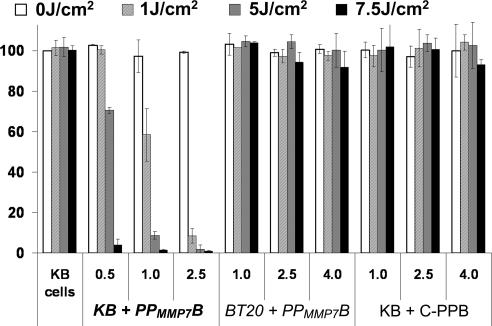
To verify that 1O2 production of PPMMP7B can be specifically activated by MMP7 in vitro, cell viability (MTT assay) was measured in KB (MMP7+) and BT20 (MMP7−) cells before and after PDT treatment, normalized to the viability of cells not treated with drug or light. Fig. 4 shows the following: (i) Neither PPMMP7B nor C-PPB (negative control) had noticeable dark toxicity in the range of concentrations used, the viability of cells incubated with even the highest PPMMP7B and C-PPB concentrations (4 μM) without light being the same as that of control cells without drug and light. (ii) Upon PDT treatment, PPMMP7B reduced the viability of only the KB cells (MMP7+), and its efficacy was drug- and light-dose-dependent. At a high light dose of 7.5 J/cm2, PPMMP7B was effective against KB cells at 0.5 μM, whereas it did not induce any photodynamic cytotoxicity in BT20 cells (MMP7−) even at 4 μM. (iii) At the same dose (4 μM, 7.5 J/cm2), C-PPB also did not reduce the cell viability of KB cells (MMP7+). Taken together, PPMMP7B is specifically photoactivated by MMP7 and its photodynamic cytotoxicity is MMP7 sequence-specific.
Fig. 4.
Photodynamic cytotoxicity determined by MTT assay as a function of PS and light doses, compared with untreated cells; means ± standard errors for triplicate experiments.
To determine the subcellular localization of the PPMMP7B, MitoTracker Green FM (Invitrogen, Carlsbad, CA) was used to stain the mitochondria of the KB cells incubated with PPMMP7B. Confocal microscopy showed a high degree of colocalization for Pyro and MitoTracker fluorescence (Fig. 5A), demonstrating that PPMMP7B was cleaved by MMP7 and that the Pyro fragment was internalized in the cells within or nearby mitochondria but was absent from the nucleus. This indicates that PPMMP7B could be a potent PDT agent, because mitochondria are known as effective targets for PDT damage (27).
Fig. 5.
Confocal images showing PPMMP7B localization and PPMMP7B-induced apo pto sis. (A) Confocal images of KB cells stained with 100 nM of MitoTracker Green FM for 30 min after 4-h incubation with 20 μM PPMMP7B: (1) Pyro image, (2) differential interference contrast image, (3) DAPI image, (4) MitoTracker image, and (5) overlaid image. (B) Confocal images showing fluorescence at 488 nm (Lower) and bright field (Upper). (1) KB cells stained with Apoptag; (2) KB cells incubated with 2 μM PPMMP7B for 3 h, kept in the dark for 3 h, and stained with Apoptag; (3) KB cells incubated with 2 μM PPMMP7B for 3 h, treated with PDT (5 J/cm2), and stained with Apoptag 3 h after PDT.
To determine whether the PPMMP7B-induced photodynamic cytotoxicity to KB cells is related to apoptosis, the Apoptag Plus in situ fluorescein detection kit S7111 (Chemicon, Temecula, CA) was used to stain KB cells in accordance with the manufacturer's protocol. As shown in Fig. 5B3, KB cells incubated with 2 μM PPMMP7B for 3 h, followed by PDT treatment (5 J/cm2), produced a strong apoptosis signal 3 h after PDT, as the cells lit up in the Apoptag fluorescein channel (excitation wavelength, 488 nm; emission wavelength, 497–580 nm), whereas KB cells incubated with the same concentration of PPMMP7B in the absence of light did not show any significant apoptosis (Fig. 5B2). This observation, together with the evidence of mitochondrial localization of the PMB, suggests that apoptosis is likely the primary mechanism of cell death.
Preliminary in Vivo PDT Response of PPMMP7B.
To evaluate the PDT efficacy of the PPMMP7B in vivo, the mouse bearing two KB tumors (one on each flank) was injected with 80 nmol PPMMP7B intravenously by the tail vein, whereas the second mouse was kept as a drug-free control. The activation of PPMMP7B in the first mouse was continuously monitored by in vivo fluorescence imaging. As shown in Fig. 6, immediately after injection (Fig. 6 B and b), no fluorescence increase was observed anywhere compared with the “prescan” images (Fig. 6 A and a) or the drug-free mouse (Fig. 6 G and g). This confirmed that PPMMP7B is optically silent in its native state because of fluorescence quenching by BHQ3. However, fluorescence signals started to increase in KB tumors 20 min after injection and reached the highest level at 3 h (Fig. 6 C and c), clearly indicating PPMMP7B activation presumably by MMP7. At this time point, the PDT treatment was given to the tumor on the left flank, whereas the tumor on the opposite side served as a dark control. The drug-free mouse was treated in the same way. One hour after PDT treatment (4 h after injection) (Fig. 6 D and d), the treated tumor in the drugged animal became edematous, whereas the untreated tumor in the drugged animal and both tumors in the drug-free mouse showed no changes in size or fluorescence signal. Three days after PDT, the treated tumor in the drugged mouse was reduced in size (Fig. 6 E and e), and 30 days after PDT, it completely regressed without any sign of regrowth (Fig. 6F), whereas the untreated tumor (Fig. 6F) and both tumors in the drug-free mouse (Fig. 6H) continued to grow. These data clearly demonstrate that PPMMP7B accumulates and can be photodynamically activated in MMP7+ tumors.
Discussion
The present report describes the full implementation of the PMB concept, including detailed synthesis of a MMP7-specific PMB (PPMMP7B), the systematic validation of its utility as an activatable PDT agent in vitro, and the initial demonstration of its feasibility in vivo. We have validated this PPMMP7B construct, because keeping Pyro and BHQ3 in close proximity using the self folding of an MMP7-specific peptide linker results in effective 1O2 quenching (94%), whereas the same concentration of PPMMP7 mixed with free BHQ3 (1:1 molar ratio) did not show any 1O2 quenching. Furthermore, when the peptide linker was specifically cleaved by MMP7, the quenched 1O2 was completely restored, and the 1O2 production was similar to that of mixture of PPMMP7 and BHQ3. Thus, this PMB provided a very high degree of control of the PS's ability to produce 1O2 in response specifically to activation by a cancer-associated biomarker (MMP7).
While this manuscript was under preparation, Choi et al. (28) reported that a chlorin e6-containing polymer sensitive to cathepsin B can be used for controlling 1O2 and PDT activity. However, this was not the first time that a protease-specific molecule has been used to control 1O2 (9), and the approach is distinctively different from ours in that it used PS self quenching to control 1O2 production, with both the 1O2 quenching and activation efficiencies inversely depending on the degree of PS substitution on the poly-l-lysine sequence. Thus, the higher degree of PS substitution leads to the higher 1O2 quenching efficiency. Meanwhile, the higher degree of PS substitution will also decrease the 1O2 activation efficiency, because it decreases the availability of the cathepsin B recognition sites.
NIR fluorescence imaging (NIRF) and PDT have become effective components in the armamentarium of cancer imaging and therapies, and their utility could be greatly increased if techniques can be devised to enhance the specificity of delivery of the dyes to the tumor tissue and to activate them, particularly in the NIR wavelength range, to treat deep-seated tumors. Our work is intended to achieve both goals by using a sequence-specific peptide as the triggering device for activating the NIRF and PDT only in targeted cells, thus significantly enhancing the tumor-to-tissue contrast of NIRF and potentially improving the therapeutic ratio of PDT. Thereby, the tumor selectivity of PDT treatment will no longer depend solely on how selectively the PS can be delivered to cancer cells. Rather, it will be based on how selective a specific biomarker is to cancer cells, and how selective the interaction of the PMB is to this biomarker.
The PMB described here is activated specifically by MMP7, a tumor-associated protease. However, the same principle could be applied to other activation schemes that could separate the PS from its quencher. For example, this concept could be applied to nucleic acid-based molecular beacons, in which a tumor-specific mRNA can hybridize with the complementary DNA sequence between the PS and a 1O2 quencher to trigger PDT-induced cell kill.‖ It also could be applied to DNA sequence-controlled on-and-off switchable 1O2 sensitizers (10), in which a PS and a 1O2 quencher are kept in close contact in the “off-state” by DNA-programmed assembly, and the PS will be activated after releasing from the quencher through a process of competitive DNA hybridization. Alternatively, it could be applied to a phospholipase-activated optical probe, in which a tumor-associated phospholipase is used as the triggering device.** Hence, this approach could have potential to generate a wide range of clinically useful strategies to enhance the specificity and efficacy of PDT to treat cancer and, by analogy, also other pathologies. In addition, our approach of achieving the high tumor selectivity of PDT treatment through controlling the PS's ability to produce 1O2 could significantly minimize PDT complications by protecting adjacent nontargeted tissues from photodamage, making PDT a safer and more selective clinical technique.
Materials and Methods
UV-vis and fluorescence spectra were recorded on a PerkinElmer (Boston, MA) Lambda 20 spectrophotometer and LS-50B spectrofluorometer, respectively. MALDI-TOF mass spectroscopy was performed on an Applied Biosystems (Foster City, CA) Voyager DE system. Reverse-phase analytical HPLC was performed on a Zorbax 300SB-C8 column using a Waters (Milford, MA) 600 Controller with a 2996 photodiode array detector (HPLC method; solvent A, 0.1% TFA and water; solvent B, acetonitrile; gradient, from 80% of A and 20% of B to 100% of B over 40 min; flow rate, 1.0 ml/min). Confocal microscopy images were acquired by using a Zeiss (Heidelberg, Germany) LSM510 META laser-scanning confocal microscope, and in vivo fluorescence images were acquired on a Xenogen IVIS imager (Hopkinton, MA).
The peptide synthesis reagents for activation, 1-hydroxybenzotriazole and O-(benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate, were purchased from ACROS (Morris Plains, NJ) and Sigma (St. Louis, MO), respectively. Sieber amide resins, and all of the N-α-Fmoc-protected amino acids were purchased from Novabiochem (San Diego, CA).
Synthesis of PPMMP7B (Fig. 1B).
Fmoc-GPLGLAR(Pbf)K(Mtt)-Sieber resin (1).
A peptide of Fmoc-GPLGLAR(Pbf)K(Mtt)-Sieber resin (1) was synthesized by manual Fmoc solid-phase peptide synthesis protocol by using commercially available N-α-Fmoc-protected amino acids, Sieber amide resin as a solid support, and O-(benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate/1-hydroxybenzotriazole as a carboxyl-group activating agent.
Pyro-GPLGLAR(Pbf)K(Mtt)-Sieber resin (2).
After the last Fmoc group was removed from the peptide-resin (1) with 20% piperidine in N,N-dimethylformamide, the resin was washed with 1-methyl-2-pyrrolidinone and pyropheophorbide α acid was coupled to the N-terminal glycine of the peptide-resin [Pyro acid/1-hydroxybenzotriazole/O-(benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate/peptide 3:3:3:1]. The reaction was shaken under argon overnight at room temperature, after which the green resin was washed and dried to give Pyro-GPLGLAR(Pbf)K(Mtt)-Sieber resin (2).
Pyro-GPLGLAR(Pbf)K(ε-NH2) (3).
This peptide resin (2) was treated with 3% TFA and 5% triisopropylsilane (Tis) in dichloromethane for 1 h at room temperature to cleave the peptide sequence from the Sieber resin and deprotect the Mtt group on the C-terminal of lysine. After removing the cleaved solid resin by filtration, the filtrate was concentrated and precipitated by adding anhydrous ether to give Pyro-GPLGLAR(Pbf)K(ε-NH2) (3). It was further washed by ether and dried under high vacuum. A part of Pyro-GPLGLAR(Pbf)K(ε-NH2) (3) was treated by 95% TFA and 5% Tis to remove the Pbf-protected group on arginine and was afterward purified by HPLC to obtain Pyro-GPLGLARK((ε-NH2) (PPMMP7), which was used as positive control (without quencher moiety).
Pyro-GPLGLARK(BHQ3) (4).
The Pyro peptide (3) (10 mg, 6 μmol) was dissolved in 200 μl of anhydrous DMSO with 1% of N,N-diisopropylethyl amine and reacted with BHQ3-NHS (4.7 mg, 6 μmol, Biosearch Technologies, Novato, CA) for 3 h under argon. The reaction was stopped by precipitation with 2 ml of ether to give Pyro-GPLGLAR(Pbf)K(BHQ3). This peptide was treated by 95% TFA and 5% triisopropylsilane to remove the Pbf-protected group on arginine and was afterward purified by HPLC to obtain Pyro-GPLGLARK(BHQ3) (PPMMP7B) (4).
PPMMP7B Activation by MMP7 in Buffer.
An MMP7 fluorescence assay kit, containing the active form of human MMP7 enzyme (Mr = 20,400), MMP7 inhibitor NNGH (8) (Mr = 316.4), and MMP7 fluorogenic substrate Ac-PLG-[2-mercapto-4-methyl-pentanoyl]-LG-OC2H5 (23), was purchased from Biomol (Plymouth Meeting, PA). The buffer used for cleavage contains 50 mM Hepes, 10 mM CaCl2, 0.05% Brij 35, pH 7.5. Before each cleavage study, the activity of MMP7 was confirmed by its ability to cleave the fluorogenic substrate Ac-PLG-[2-mercapto-4-methyl-pentanoyl]-LG-OC2H5. The PPMMP7B sample was prepared as 5 ml of 0.4 μM stock solution in the buffer. The composition of each sample was as follows: (i) PPMMP7B+MMP7: 1 ml of PPMMP7B stock solution with 1 μl of MMP7 (0.45 g/liter); (ii) PPMMP7B+MMP7+inhibitor: 1 ml of PPMMP7B stock solution with 2 μl of inhibitor (2 g/liter) and 1 μl of MMP7 (0.45 g/liter) (the molar ratio of PPMMP7B: MMP7: inhibitor was 50:1:1,500); (iii) PPMMP7B+MMP2: 1 ml of PPMMP7B stock solution with 1 μl of MMP2 (0.50 g/liter); and (iv) C-PPB+MMP7: 1 ml of 0.4 μM control C-PPB with 1 μl of MMP7 (0.45 g/liter). The fluorescence of these four solutions was monitored in real time by fluorescence spectroscopy at 37°C with an excitation wavelength of 660 nm and emission at 674 nm. After 2 h, the cleavage solutions were analyzed by HPLC to collect the cleaved fragments for identification by using MALDI-TOF.
1O2 Measurement.
PDT-generated 1O2 was quantified in solutions of PPMMP7, PPMMP7+BHQ3, PPMMP7B, PPMMP7B+MMP7, and PPMMP7B+MMP7+MMP7 inhibitor, by directly measuring its NIR luminescence at 1,270 nm by using an instrument and technique that have been described (29). Briefly, a 10 ns pulsed 523-nm laser excited the solution, and the luminescence spectrum was sampled, after rejection of PS fluorescence, by using a set of interference filters and a high-sensitivity NIR photomultiplier tube operating in the time-resolved single photon-counting mode.
Cell Lines.
KB cells (human nasopharyngeal epidermoid carcinoma cells, high MMP7 expression, MMP7+) and BT20 cells (a human breast cancer cell line, lack of MMP7 expression, MMP7−) (26) were purchased from American Type Culture Collection (Manassas, VA). Both KB and BT-20 cells were cultured in MEM supplemented with 2 mM l-glutamine/17.9 mM sodium bicarbonate/0.1 mM nonessential amino acids/1.0 mM sodium pyruvate/10% FBS. All cells were grown at 37°C in a humidified atmosphere containing 5% CO2.
Confocal Microscopy.
KB and BT20 cells were grown for 2 d in four-well Lab-Tek (Naperville, IL) chamber slides. MEM containing 60 μM PPMMP7B or control C-PPB was added, and the cells were incubated for 5 h at 37°C. The incubation medium was removed and run on HPLC, while the cells were washed five times with ice-cold PBS and then fixed for 20 min with 1% formaldehyde in PBS at room temperature. The chamber slides were then imaged by confocal microscopy with 633 nm excitation and >650 nm detection.
MTT Assay.
KB and BT20 cells were grown for 2 d in 96-well plates. Experiments were started, after one quick wash with ice-cold PBS buffer, by addition of 100 μl of cell growth medium containing the indicated amounts of PPMMP7B and control C-PPB. After 16 h incubation at 37°C with 5% CO2, the cells were washed twice with ice-cold PBS buffer, and then 100 μl of cell growth medium was added. PDT treatment was performed by using a 670-nm continuous wavelength laser with one of three different light fluences (1, 5, or 7.5 J/cm2). The cells were allowed to continue growth for 24 h, at which time the MTT tracer, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (Invitrogen) was added to the medium at 0.5 mg/ml. Two hours later, the medium was removed and replaced with 150 μl of 1:1 DMSO/70% isopropanol in 0.1 M HCl. The absorbance at 570 nm was measured on a Bio-Tek ELx model 800 (MTX Lab System, Vienna, VA).
In Vivo Imaging and PDT Treatment.
Two nude mice were inoculated with 107 KB cells on both left and right flanks, and the tumors were grown for 5 d with the mice maintained on a low-fluorescence diet. “Prescan” images were obtained on the Xenogen (Hopkinton, MA) imager with a Cy 5.5 filter (λex = 615–665 nm, λem = 695–770 nm). One mouse was anesthetized afterward and injected intravenously with 80 nmol PPMMP7B by tail vein, whereas the other was kept as a drug-free control. The former was then continually scanned to monitor the increase of fluorescence in tumor over time. Very strong fluorescence was observed in the KB tumor after 3 h. At this time, the left tumor on both the drug-administered and drug-free mice was treated by using surface irradiation (1.2 cm2) from a 670 nm laser to a light dose of 135 J/cm2 over 30 min, whereas the right tumors were kept as dark controls. The mice were then monitored daily in the fluorescence imager over 30 d.
Acknowledgments
This study was supported by U.S. DOD Breast Cancer Research Program DAMD17-03-1-0373 and National Institutes of Health Grant R21-CA95330, the Oncologic Foundation of Buffalo, the Joey and Toby Tanenbaum/Brazilian Ball Chair in Prostate Cancer Research Princess Margaret Hospital (G.Z.), and the National Cancer Institute of Canada (B.C.W.).
Abbreviations
- PDT
photodynamic therapy
- PMB
photodynamic molecular beacons
- NIR
near-infrared
- PS
photosensitizer
- 1O2
singlet oxygen
- Pyro
pyropheophorbide a
- BHQ3
black hole quencher 3
- MMP7
matrix metalloproteinase-7
- PPMMP7B
MMP7-triggered PMB
- C-PPB
control Pyro-Peptide-BHQ3
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- UV-vis
UV-visible
- KB
human nasopharyngeal epidermoid carcinoma cells
- BT20
human breast carcinoma cells.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Chen, J., Stefflova, K., Niedra, M.J., Wilson, B.C., Zheng, G. (2004) Mol Imag 3:277 (abstr).
Mawn, T.M., Popov, A.V., Milkevitch, M., Kim, S., Zheng, G., Delikatny, E.J. (2006) Mol Imag 5:315 (abstr).
References
- 1.Dougherty TJ, Gomer CJ, Henderson BW, Jori G, Kessel D, Korbelik M, Moan J, Peng Q. J Natl Cancer Inst. 1998;90:889–905. doi: 10.1093/jnci/90.12.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pinthus JH, Bogaards A, Weersink R, Wilson BC, Trachtenberg J. J Urol. 2006;175:1201–1207. doi: 10.1016/S0022-5347(05)00701-9. [DOI] [PubMed] [Google Scholar]
- 3.Chen B, Pogue BW, Hasan T. Exp Opin Drug Del. 2005;2:477–487. doi: 10.1517/17425247.2.3.477. [DOI] [PubMed] [Google Scholar]
- 4.Sharman WM, van Lier JE, Allen CM. Adv Drug Del Rev. 2004;56:53–76. doi: 10.1016/j.addr.2003.08.015. [DOI] [PubMed] [Google Scholar]
- 5.Zhang M, Zhang Z, Blessington D, Li H, Busch TM, Madrak V, Miles J, Chance B, Glickson JD, Zheng G. Bioconjug Chem. 2003;14:709–714. doi: 10.1021/bc034038n. [DOI] [PubMed] [Google Scholar]
- 6.Zheng G, Li H, Zhang M, Lund-Katz S, Chance B, Glickson JD. Bioconjug Chem. 2002;13:392–396. doi: 10.1021/bc025516h. [DOI] [PubMed] [Google Scholar]
- 7.Stefflova K, Li H, Chen J, Zheng G. Bioconjug Chem. 2007;18:379–388. doi: 10.1021/bc0602578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacPherson LJ, Bayburt EK, Capparelli MP, Carroll BJ, Goldstein R, Justice MR, Zhu L, Hu S, Melton RA, Fryer L, et al. J Med Chem. 1997;40:2525–2532. doi: 10.1021/jm960871c. [DOI] [PubMed] [Google Scholar]
- 9.Chen J, Stefflova K, Niedre MJ, Wilson BC, Chance B, Glickson JD, Zheng G. J Am Chem Soc. 2004;126:11450–11451. doi: 10.1021/ja047392k. [DOI] [PubMed] [Google Scholar]
- 10.Clo E, Snyder JW, Voigt NV, Ogilby PR, Gothelf KV. J Am Chem Soc. 2006;128:4200–4201. doi: 10.1021/ja058713a. [DOI] [PubMed] [Google Scholar]
- 11.Hirakawa K, Kawanishi S, Hirano T. Chem Res Toxicol. 2005;18:1545–1552. doi: 10.1021/tx0501740. [DOI] [PubMed] [Google Scholar]
- 12.McCarthy JR, Perez JM, Bruckner C, Weissleder R. Nano Lett. 2005;5:2552–2556. doi: 10.1021/nl0519229. [DOI] [PubMed] [Google Scholar]
- 13.McDonnell SO, Hall MJ, Allen LT, Byrne A, Gallagher WM, O'Shea DF. J Am Chem Soc. 2005;127:16360–16361. doi: 10.1021/ja0553497. [DOI] [PubMed] [Google Scholar]
- 14.Snyder JW, Lambert JD, Ogilby PR. Photochem Photobiol. 2006;82:177–184. doi: 10.1562/2005-05-30-RA-553. [DOI] [PubMed] [Google Scholar]
- 15.Matayoshi ED, Wang GT, Krafft GA, Erickson J. Science. 1990;247:954–958. doi: 10.1126/science.2106161. [DOI] [PubMed] [Google Scholar]
- 16.Thornberry NA, Lazebnik Y. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 17.Overall CM, Kleifeld O. Nat Rev. 2006;6:227–239. doi: 10.1038/nrc1821. [DOI] [PubMed] [Google Scholar]
- 18.Brooks PC, Stromblad S, Sanders LC, von Schalscha TL, Aimes RT, Stetler-Stevenson WG, Quigley JP, Cheresh DA. Cell. 1996;85:683–693. doi: 10.1016/s0092-8674(00)81235-0. [DOI] [PubMed] [Google Scholar]
- 19.Shiomi T, Okada Y. Cancer Metast Rev. 2003;22:145–152. doi: 10.1023/a:1023039230052. [DOI] [PubMed] [Google Scholar]
- 20.Leinonen T, Pirinen R, Bohm J, Johansson R, Ropponen K, Kosma VM. Lung Cancer. 2006;51:313–321. doi: 10.1016/j.lungcan.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 21.MacDonald IJ, Morgan J, Bellnier DA, Paszkiewicz GM, Whitaker JE, Litchfield DJ, Dougherty TJ. Photochem Photobiol. 1999;70:789–797. [PubMed] [Google Scholar]
- 22.Stefflova K, Chen J, Marotta D, Li H, Zheng G. J Med Chem. 2006;49:3850–3856. doi: 10.1021/jm060146u. [DOI] [PubMed] [Google Scholar]
- 23.Knight CG, Willenbrock F, Murphy G. FEBS Lett. 1992;296:263–266. doi: 10.1016/0014-5793(92)80300-6. [DOI] [PubMed] [Google Scholar]
- 24.Weingarten H, Feder J. Anal Biochem. 1985;147:437–440. doi: 10.1016/0003-2697(85)90294-5. [DOI] [PubMed] [Google Scholar]
- 25.Weingarten H, Martin R, Feder J. Biochemistry. 1985;24:6730–6734. doi: 10.1021/bi00344a064. [DOI] [PubMed] [Google Scholar]
- 26.Giambernardi TA, Grant GM, Taylor GP, Hay RJ, Maher VM, McCormick JJ, Klebe RJ. Matrix Biol. 1998;16:483–496. doi: 10.1016/s0945-053x(98)90019-1. [DOI] [PubMed] [Google Scholar]
- 27.Kessel D, Luguya R, Vicente MG. Photochem Photobiol. 2003;78:431–435. doi: 10.1562/0031-8655(2003)078<0431:lapeot>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 28.Choi Y, Weissleder R, Tung CH. Cancer Res. 2006;66:7225–7229. doi: 10.1158/0008-5472.CAN-06-0448. [DOI] [PubMed] [Google Scholar]
- 29.Niedre MJ, Secord AJ, Patterson MS, Wilson BC. Cancer Res. 2003;63:7986–7994. [PubMed] [Google Scholar]