Abstract
Background
Human cytomegalovirus (hCMV) is involved in the pathogenesis of atherosclerosis. We have previously shown in patients with atherosclerosis that antibodies directed against the hCMV-derived proteins US28 and UL122 are able to induce endothelial cell damage and apoptosis of non-stressed endothelial cells through cross-rection with normally expressed surface molecules. Our aim was to dissect the molecular basis of such interaction and to investigate mechanisms linking innate immunity to atherosclerosis.
Methodology/Principal Findings
We analysed the gene expression profiles in endothelial cells stimulated with antibodies affinity-purified against either the UL122 or the US28 peptides using the microarray technology. Microarray results were validated by quantitative PCR and by detection of proteins in the medium. Supernatant of endothelial cells incubated with antibodies was analysed also for the presence of Heat Shock Protein (HSP)60 and was used to assess stimulation of Toll-Like Receptor-4 (TLR4). Antibodies against UL122 and US28 induced the expression of genes encoding for adhesion molecules, chemokines, growth factors and molecules involved in the apoptotis process together with other genes known to be involved in the initiation and progression of the atherosclerotic process. HSP60 was released in the medium of cells incubated with anti-US28 antibodies and was able to engage TLR4.
Conclusions/Significance
Antibodies directed against hCMV modulate the expression of genes coding for molecules involved in activation and apoptosis of endothelial cells, processes known to play a pivotal role in the pathogenesis of atherosclerosis. Moreover, endothelial cells exposed to such antibodies express HSP60 on the cell surface and release HSP60 in the medium able to activate TLR4. These data confirm that antibodies directed against hCMV-derived proteins US28 and UL122 purified from patients with coronary artery disease induce endothelial cell damage and support the hypothesis that hCMV infection may play a crucial role in mediating the atherosclerotic process.
Introduction
Atherosclerosis is the primary cause of morbidity and mortality worldwide. It is a multifactorial disorder influenced by genetic [1] and environmental factors [2]. Indeed the genetic background is greatly influenced by classical environmental risk factors, such as cholesterol, diabetes, hypertension, obesity and smoking. In addition inflammation, infection and autoimmunity have been recently suggested to play a role in the pathogenesis of the disease [3]. Infectious agents seem to be involved in endothelium damage by inducing an autoimmune response to Heat Shock Proteins (HSPs) [4]. Antibodies against HSP60 are present in the majority of patients with coronary artery disease and their titre correlates with disease severity [5].
We have previously shown that autoantibodies against HSP60 are present in most atherosclerotic patients and that these autoantibodies are directed to an aminoacid sequence at position 153-163 of HSP60 that shows homology with two human cytomegalovirus (hCMV)-derived proteins, UL122 and US28 [ref. 6]. Anti-HCMV antibodies were present in a slightly higher percentage in CAD patients than in controls, however only the sera of patients with CAD showed antibodies against the identified viral peptides. Both viral peptides, UL122 and US28, show sequence similarity with molecules normally expressed on endothelial cell surface: US28 shows homology with integrin alpha 6 (CD49f) and UL122 with connexin 45 and CD151 [ref. 6]. IgG antibodies affinity purified against the HSP60 peptide from the sera of ten patients with coronary artery disease (CAD) bound endothelial cell derived HSP60, recombinant human HSP60 and recognized the hCMV-derived proteins UL122 and US28 [6]. Antibodies against the HSP60 peptide affinity purified from the serum samples of controls did not cross-react with the viral proteins. The IgG antibody component affinity purified against the viral peptides from the patients with CAD, recognized both the viral proteins and HSP60. The sera of patients with no evidence of CAD did not show antibodies against the viral peptides [6]. Antibodies directed against HSP60 and viral peptides bound endothelial cells upon interaction with the cell surface receptors sharing sequence homology with the viral peptides, through a mechanism of molecular mimicry, and induced apoptosis of non-stressed endothelial cells [6], considered a primary event in the pathogenesis of atherosclerosis [7], [8]. Moreover, HSP60 exposed on the surface of stressed endothelial cells could amplify the antibody aggression through cross-recognition of the anti-viral antibodies [6].
Taking advantage of the results previously obtained, we aimed at analyzing the molecular effects of the identified anti-hCMV antibodies on endothelial cells, using the DNA microarray approach, in order to ascertain whether the sets of genes modulated by anti-hCMV-derived proteins UL122 and US28 antibodies, are associated with the pathogenesis of atherosclerosis. We found indeed that most of the genes modulated are known to be associated with the atherosclerotic process. Moreover such antibodies induced release of HSP60 able to activate Toll-Like Receptor 4 (TLR4).
Material and Methods
Patients
The clinical characteristics of the ten patients with angiographic evidence of coronary artery disease, whose sera were used to obtain affinity purified antibodies against the peptides, have already been described elsewhere [6].
Purification of anti-US28 and anti-UL122 IgG antibodies
The UL122-derived peptide (GPKKKSKRIS), the US28-derived peptide (TDVLNQSKPVT) and the irrelevant control peptide (VTLPKDSDVELP) chosen on the basis of its absence of relevant homology with the viral peptides and with the endothelial molecules connexin 45, CD49f and CD151, were synthesized by solid phase synthesis with the 9-fluorenylmethoxycarbonyl strategy [6].
To affinity purify IgG antibodies against either the UL122 or the US28 peptide or the control peptide, each peptide (5 mg/g of dried sepharose powder) was coupled to sepharose 4B (Pharmacia Uppsala, Sweden), according to the manufacturer's instructions. Pooled sera from the 10 different patients analysed were diluted in PBS and bound immunoglobulins were eluted with 0,1 M glycine pH 2,5 and dialysed against PBS [6].
Cell culture and apoptosis
Human endothelial cells (HUVECs) and their growth media were purchased from Promocell Bioscience Alive (Heidelberg, Germany). HUVECs were used between passages 2 and 5. Cell monolayers were stimulated with antibodies purified against the UL122 or US28 peptide or the irrelevant control peptide (20 microgram/ml) and apoptosis evaluated at various intervals of time. Tumor necrosis factor alpha (TNFalpha 50 µg/L) was used as positive control to induce endothelial cell apoptosis. The extent of DNA fragmentation was quantified using a commercially available kit (Roche Biochemical, Basel, Switzerland). The enrichment of mono-and oligo-nucleosomes released into the cytoplasm is calculated as absorbance of sample cells (cells treated with antibodies)/absorbance of control cells (cells not exposed to antibodies). Enrichment factor is used as an index of apoptosis. An increase in the enrichment factor of 1.0 corresponded ap–proximately to 8–12% of apoptotic cells as determined in parallel by Fluorescence-activated cell sorting (FACS) analysis after staining with FITC-labeled annexin V (Alexis Biochemicals, San Diego, USA) (data not shown).
Detection of HSP60 and TLR4 activation
For FACS analysis, we incubated cells with specific or control antibodies for 30 minutes on ice. Antibody binding was revealed using fluorescein isothiocyanate (FITC)-conjugated secondary antibodies. Samples were run on a FACScan flow cytometer (Becton Dickinson, Mountain View, CA).
Soluble HSP60 released in the supernatant was evaluated using a commercially available kit (Stressgen, Victoria, BC, Canada). Surface expression of HSP60 was assessed by FACS analysis using a monoclonal antibody directed against HSP60 (clone KL-1, Stressgen). HUVEC cells stressed with 0,1% glutaraldehyde for 20 minutes on ice were used as positive control.
TLR signaling leads to translocation of NF-kB. To monitor the induction of TLR4 signaling in response to ligand stimulation, we have used the pNifty reporter plasmid (Invivogen, Ruschlikon, Switzerland), expressing the secreted embryonic alkaline phosphatase gene (SEAP) under the control of a NF-kB-inducible ELAM1 composite promoter. Engineered 293T cells stably transfected with TLR4 and the co-receptors MD2 and CD14 (Invivogen) were co-transfected with the pNifty plasmids. Cells were grown to 60–80% confluence in growth medium and then harvested and resuspended in HEK-Blue Detection medium (Invivogen). This medium is specifically designed for detection of NF-kB activation since it turns blue in the presence of phosphatase activity. 2,5×104 cells/well were plated in 96 well plates in the presence of the supernatant. Positive control for TLR4 activation was LPS (Invivogen). Supernatant was devoided of HSP60 using an anti-HSP60 monoclonal antibody (Stressgen) bound to a sepharose column. Cells were incubated for 24 hrs at 37°C in 5% CO2 and the blue color assessed by a spectrophotometer set at 620 nm. The supernatants used in this set of experiments were devoided of LPS contaminants using detoxy-gel endotoxin removing columns (Pierce Biotechnology, Rockford, IL, USA). Mock-transfected 293T cells were used as negative control.
Gene Array analysis
Cell pellets of endothelial cells cultured as previously described and obtained after 6 and 12 hours of incubation with affinity purified antibodies against US28, UL122 and an irrelevant peptide, were used for gene array experiments. Preparation of cRNA, hybridization, and scanning of probe arrays were performed according to the protocols of the manufacturer (Affymetrix Santa Clara, California, United States) by the Genopolis Consortium (University of Milano-Bicocca, Italy) using the Human Genome U133A GeneChip® (Affymetrix). The GeneChip® Human Genome U133A is a single array representing 14,500 well-characterized human genes and including more than 22,000 probe sets and 500,000 distinct oligonucleotide features.
The different gene expression patterns were analyzed using the Array Assist software version 2.0 (Stratagene, La Jolla, California, United States) that calculated a robust multi-array average of background-adjusted, normalized, and log-transformed intensity values applying the Robust Multi-Array Average algorithm (RMA). With this software the mean optical background level for each array was subtracted from the signal intensity for each probe. Finally, the normalized, background-corrected data were transformed to the log2 scale. A Signal Log2 Ratio of 1.0 indicates an increase of the transcript level by two fold change (2 F.C.) and−1.0 indicates a decrease by two fold (−2 F.C.). A Signal Log Ratio of zero would indicate no change [9].
In our study, we analyzed the gene expression profiles in endothelial cells stimulated with antibodies purified against either the UL122 or the US28 peptide (test samples) or with antibodies purified against an irrelevant peptide (control samples) for 6 and 12 hours. Genes were selected for final consideration when their expression (F.C.) was at least 1.5-fold different in the test sample versus control sample at least at one time point. Experiments were performed in duplicates. The microarray results have been reported according to the MIAME guidelines and deposited in the public repository Array-Express at http://www.ebi.ac.uk/arrayexpress.
To verify relationships from our gene list and the existing knowledge in the literature concerning genes and atherosclerosis, we used the automated literature mining tool, termed MedGene (hptt://hipseq.med.harvard.edu/MEDGENE/login.jsp), which summarizes and estimates the relative strength of all human gene disease relationships in Medline [10].
The software package Pathway Studio (Ariadne Genomics, Rockville USA) was used to identify functional interrelationships among the genes identified. These functional relationships were then graphically represented by the software as a network.
Validation of Gene Array results
Quantitative real-time polymerase chain reaction (Q-PCR) was carried out as described [9]. Briefly, total RNA has been extracted from HUVECs with TRIzol reagent (Invitrogen, Carlsbad, CA, USA), following manufacturer's instructions. One µg of total RNA from each sample was treated with amplification grade DNase I and then used as a template for the reverse transcription reaction, using random hexamers and SuperScript II Reverse Transcriptase (Invitrogen). All samples will be reverse transcribed under the same conditions and from the same reverse transcription master mix, in order to minimize differences in reverse transcription efficiency. Triplicate Q-PCR reactions for each sample have been performed. Control wells containing no template have been used to exclude the presence of contaminating template molecules and to identify potential primer-dimer products from the dissociation curve analysis. Amplification plots have been analyzed using Opticon Monitor software version 2.02 (MJ Research, Waltham, MA, USA) and data calculated with Q-Gene software (www.BioTechniques.com) and expressed as mean normalized expression (MNE). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) has been used as a normalizing gene according to its stable expression levels.
Detection of MCP-1 (Amersham Biosciences, Pisacataway, NJ, USA) and sE-Selectin (R&D Systems, Minneapolis, Minnesota, USA) in the supernatants were determined by ELISA.
Statistical analysis
Statistical testing was performed using StatsDirect (StatDirect, Cheshire, UK). Difference in the concentration of soluble molecules in the supernatants was performed using a parametric test. P-values less than 0.05 was considered significant.
Results
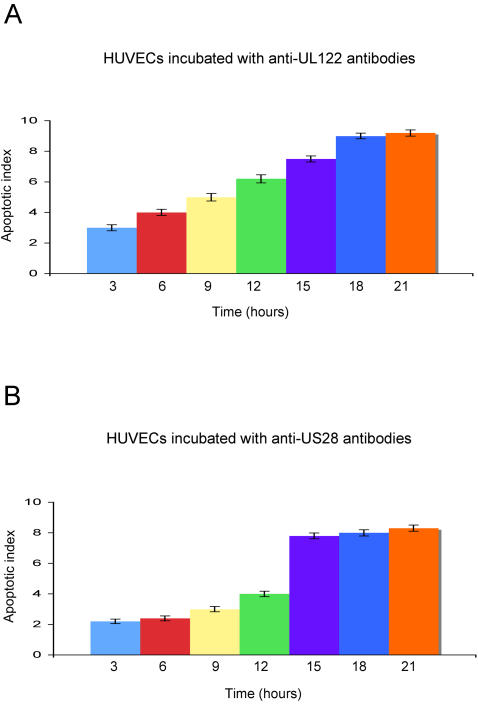
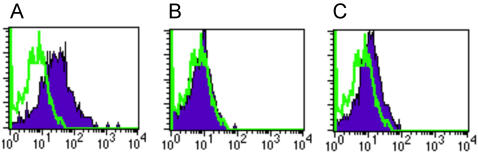
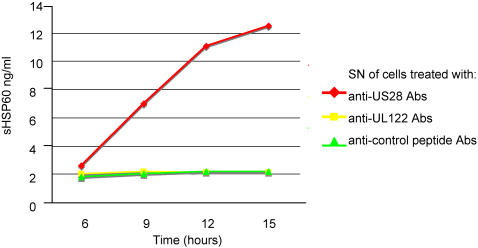
The IgG antibodies affinity purified against the HSP60 and the viral peptides obtained from patients with CAD had the same behaviour of the purified antibodies previously described both in the recognition of endothelial cell surface molecules and in the ability to induce endothelial cell apoptosis [6]. To more precisely analyze the mechanisms by which anti-hCMV-derived proteins UL122 and US28 antibodies induce apoptosis of HUVECs [6], a time course experiment was performed by incubating HUVECs with either anti-UL122 or anti-US28 or anti-irrelevant peptide antibodies obtained from the 10 patients with CAD. Figure 1, panels A and B, shows that apoptosis induced by anti-UL122 antibodies occurs at an earlier stage compared to the other anti-hCMV antibody subset. Moreover internucleosomal DNA fragmentation induced by anti-UL122 peptide antibodies reached 80% and that induced by anti-US28 reached 72% of the level of internucleosomal DNA fragmentation seen after exposure of endothelial cells to 50 µg/L of TNFalpha. This finding is in accordance with our previous report that the antibodies can trigger apoptosis in human endothelial cells upon interaction with different cell-membrane molecules through a mechanism of molecular mimicry [6]. Antibodies against HSP60 play a pivotal role in the cytotoxic damage of stressed endothelial cells; since antibodies against hCMV are able to crossreact with particular HSP60 epitopes thus amplifying the cell damage by binding surface HSP60, we evaluated the surface HSP60 expression in endothelial cells after exposure to anti-UL122 and US28 antibodies. Surprisingly only HUVECs incubated with anti-US28 antibodies translocated HSP60 on the cell surface (figure 2, panels A–C) and released the soluble molecule (sHSP60) in the medium (figure 3). These data suggest that the two antibody subsets induce apoptosis through different mechanisms: anti-UL122 antibodies induce apoptosis through the engagement of the surface molecules CD151 and connexin 45 (GJA7), whereas the apoptotic damage triggered by the engagement of CD49f (ITGA6) by anti-US28 can be further amplified by the antibody interaction with HSP60 expressed on the cell surface.
Figure 1. Anti-hCMV antibodies induce apoptosis of endothelial cells.
Apoptosis induced in HUVECs by antibodies directed against UL122 peptide (A) and against US28 peptide (B); X axis: apoptotic index; Y axis: time expressed in hours. Data are mean of three independent experiments. Error bars = SD. Apoptosis induced by antibodies directed against the irrelevant peptide gave an apoptotic index<1 at all time points, corresponding to less than 8–12% of apoptotic cells as determined in parallel by FACS analysis.
Figure 2. FACS analysis of the expression of HSP60 at the cell surface membrane.
(A) Positive control: HUVECs stressed with glutaraldehyde for 20 minutes; (B) HUVECs treated with anti-UL122 antibodies for 9 hours; (C) HUVECs treated with anti-US 28 antibodies for 9 hours. HUVEC cells were stained with a monoclonal antibody directed against HSP60. Percentage of positive cells: (A) 44,2%; (B) 1,3%; (C): 6,7%.
Figure 3. Soluble HSP60 in the supernatant of stimulated cells.
Soluble HSP60 in the supernatant (SN) of HUVECs after stimulation with anti-US28 and anti-UL122 antibodies (Abs) at different time points. X axis: time expressed in hours. Y axis: soluble HSP60 concentration expressed in ng/ml.
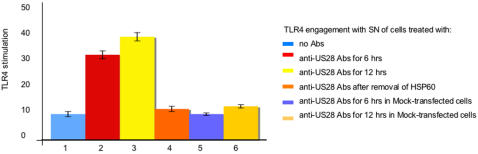
Since a link between innate immunity and atherosclerosis has been proposed and several data indicate that signaling through TLR4 promotes atherosclerosis [11], and since HSP60 has been proposed as an endogenous ligand for TLR4 [12], we next evaluated whether HSP60 present in the supernatant of endothelial cells stimulated with anti-US28 antibodies could engage TLR4. 293T cells transfected with TLR4 were activated by the supernatant that induced an activation of TLR4 similar to the one obtained with 40 ng of LPS (figure 4). Such activation was not present when the supernatant was deprived of HSP60 and when supernatant was used with mock-transfected 293T cells. These data suggest that HSP60 is indeed an endogenously generated ligand for TLR4.
Figure 4. TLR4 activation by the supernatant of treated cells.
TLR4 engagement by the SN of cells treated with the antibodies (Abs) described. Y axis: levels of TLR4 stimulation expressed as ng/mL of LPS. Data are mean of three independent experiments. Error bars = SD.
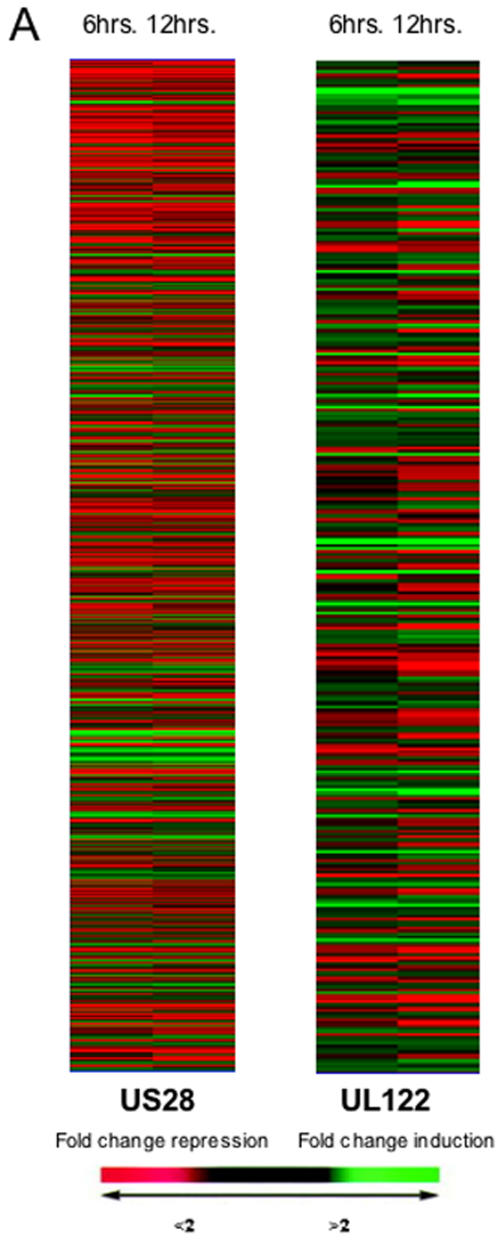
We next analyzed the gene expression profiles in endothelial cells treated with antibodies affinity purified against the hCMV-derived peptides in order to identify clusters of genes involved in the pathogenesis of vascular damage in atherosclerosis. Anti-US28 peptide antibodies up-regulated 907 transcripts (Table 1 and Table S1). Clustergrams of modulated genes are shown in figure 5. Anti-US28 antibodies up-regulated genes encoding for adhesion molecules, such as E selectin, VCAM-1 and ICAM-1 and genes encoding for chemokines including CCL2, CCL20, CXCL1, CXCL2, together with chemokine receptors such as CCR3, CXCR4.
Table 1. Gene expression in HUVEC after 6 and 12 huors of stimulation with anti-US28 IgG antibodies.
| Gene accession | Gene description | Gene Symbol | F.C. 6 h | F.C.12 h |
| Adhesion molecules | ||||
| NM_000450.1 | endothelial adhesion molecule 1 | SELE | 5,95 | 1,79 |
| NM_001078.2 | vascular cell adhesion molecule 1 | VCAM1 | 3,76 | 2,06 |
| NM_002203.2 | integrin, alpha 2 | ITGA2 | 2,61 | 2,69 |
| NM_000212.2 | integrin, beta 3 | ITGB3 | 1,72 | 1,72 |
| NM_000201.1 | intercellular adhesion molecule 1 (CD54) | ICAM1 | 1,60 | 1,33 |
| Inflammation | ||||
| S69738.1 | chemokine (C-C motif) ligand 2 | CCL2 | 2,46 | 1,98 |
| NM_004591.1 | chemokine (C-C motif) ligand 20 | CCL20 | 1,63 | 1,42 |
| NM_001511.1 | chemokine (C-X-C motif) ligand 1 | CXCL1 | 3,32 | 1,63 |
| M57731.1 | chemokine (C-X-C motif) ligand 2 | CXCL2 | 4,21 | 2,72 |
| NM_002090.2 | chemokine (C-X-C motif) ligand 3 | CXCL3 | 1,62 | 1,40 |
| NM_001837.2 | chemokine (C-C motif) receptor 3 | CCR3 | 1,49 | 1,77 |
| AJ224869 | chemokine (C-X-C motif) receptor 4 | CXCR4 | 2,24 | 1,99 |
| NM_017801.2 | chemokine-like factor super family 6 | CKLFSF6 | 2,07 | 1,69 |
| NM_003326.2 | tumor necrosis factor (ligand) superfamily, member 4 | TNFSF4 | 1,96 | 1,36 |
| NM_002184.2 | interleukin 6 signal transducer | IL6ST | 3,61 | 4,36 |
| NM_002185.2 | interleukin 7 receptor | IL7R | 2,04 | 2,01 |
| Immune responce | ||||
| NM_000593.3 | transporter 1, ATP-binding cassette, sub-family B | TAP1 | 1,98 | 1,85 |
| NM_005348.2 | heat shock 90kDa protein 1, alpha | HSPCA | 2,11 | 1,84 |
| NM_006260.2 | DnaJ (Hsp40) homolog, subfamily C, member 3 | DNAJC3 | 1,78 | 1,65 |
| Transcription | ||||
| NM_005239.4 | v-ets erythroblastosis virus E26 oncogene homolog 2 | ETS2 | 3,21 | 2,34 |
| NM_003199.1 | transcription factor 4 | TCF4 | 3,12 | 3,42 |
| NM_021964.1 | zinc finger protein 148 | ZNF148 | 3,53 | 2,92 |
| BC003566.1 | zinc finger protein 24 | ZNF24 | 3,44 | 2,76 |
| BC003070.2 | GATA binding protein 3 | GATA3 | 1,91 | 1,50 |
| NM_007348.2 | activating transcription factor 6 | ATF6 | 2,79 | 2,69 |
| U97103.1 | alpha thalassemia/mental retardation syndrome X-linked | ATRX | 4,68 | 4,41 |
| NM_013448.2 | bromodomain adjacent to zinc finger domain, 1A | BAZ1A | 3,98 | 4,62 |
| NM_013354.5 | CCR4-NOT transcription complex, subunit 7 | CNOT7 | 1,71 | 1,60 |
| NM_004459.5 | fetal Alzheimer antigen | FALZ | 2,97 | 3,00 |
| AF548661 | general transcription factor IIH, polypeptide 3, 34kDa | GTF2H3 | 3,33 | 3,68 |
| NM_003489.2 | nuclear receptor interacting protein 1 | NRIP1 | 11,86 | 8,12 |
| NM_004774.2 | PPAR binding protein | PPARBP | 2,88 | 2,67 |
| Inhibition of cell growth and apoptosis | ||||
| NM_001970.3 | eukaryotic translation initiation factor 5A | EIF5A | 22,93 | 17,78 |
| BC000076.2 | cyclin D1 (PRAD1: parathyroid adenomatosis 1) | CCND1 | 2,03 | 2,04 |
| NM_003810.2 | tumor necrosis factor (ligand) superfamily, member 10 | TNFSF10 | 2,03 | 2,53 |
| U20537.1 | caspase 6, apoptosis-related cysteine protease | CASP6 | 2,65 | 2,35 |
| NM_001227.3 | caspase 7, apoptosis-related cysteine protease | CASP7 | 1,61 | 1,31 |
| NM_005256.2 | growth arrest-specific 2 | GAS2 | 1,84 | 1,19 |
| BC001152.1 | growth arrest-specific 7 | GAS7 | 1,78 | 1,95 |
| NM_004401.2 | DNA fragmentation factor, 45kDa, alpha polypeptide | DFFA | 1,23 | 1,66 |
| NM_020529.1 | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | NFKBIA | 2,40 | 1,75 |
| AF091555.1 | C-terminal binding protein 1 | CTBP1 | 6,48 | 5,10 |
| NM_021111.1 | reversion-inducing-cysteine-rich protein with kazal motifs | RECK | 1,74 | 1,74 |
| NM_016272.3 | transducer of ERBB2, 2 | TOB2 | 1,92 | 1,99 |
| BC006325.2 | G-2 and S-phase expressed 1 | GTSE1 | 1,92 | 1,92 |
| Collagens and extracellular matrix | ||||
| NM_005713.1 | collagen, type IV, alpha 3 binding protein | COL4A3BP | 1,76 | 1,87 |
| NM_006665.2 | heparanase | HPSE | 1,74 | 1,33 |
| NM_021809.4 | TGFB-induced factor 2 (TALE family homeobox) | TGIF2 | 1,69 | 1,13 |
| NM_003243.2 | transforming growth factor, beta receptor III (betaglycan) | TGFBR3 | 1,55 | 1,61 |
| Metabolism | ||||
| NM_014508.2 | apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3C | APOBEC3C | 1,88 | 2,00 |
| BC015818.1 | lectin, galactoside-binding, soluble, 8 (galectin 8) | LGALS8 | 2,78 | 1,38 |
| NM_002858.2 | ATP-binding cassette, sub-family D (ALD), member 3 | ABCD3 | 1,86 | 1,50 |
| NM_017526.2 | leptin receptor | LEPR | 1,74 | 1,98 |
| AF285167.1 | ATP-binding cassette, sub-family A,1 | ABCA1 | 1,52 | 1,11 |
| NM_005506.2 | scavenger receptor class B, member 2 | SCARB2 | 1,42 | 1,88 |
| Growth factors | ||||
| NM_002632.4 | placental growth factor | PGF | 3,34 | 3,02 |
| Receptors | ||||
| NM_000875.2 | insulin-like growth factor 1 receptor | IGF1R | 4,41 | 3,99 |
| AW157070 | epidermal growth factor receptor precursor | EGFR | 3,11 | 2,57 |
| M34641 | fibroblast growth factor receptor 1 | FGFR1 | 2,47 | 2,61 |
| NM_000565.2 | interleukin 6 receptor | IL6R | 1,55 | 1,31 |
| NM_000808.2 | gamma-aminobutyric acid (GABA) A receptor, alpha 3 | GABRA3 | 1,43 | 1,58 |
| Enzymes | ||||
| NM_003358.1 | UDP-glucose ceramide glucosyltransferase | UGCG | 3,50 | 2,73 |
| AF065214.1 | phospholipase A2, group IVC (cytosolic, calcium-independent) | PLA2G4C | 3,36 | 3,64 |
| NM_001394.5 | dual specificity phosphatase 4 | DUSP4 | 3,06 | 2,69 |
| NM_024906.1 | stearoyl-CoA desaturase 4 | SCD4 | 2,45 | 1,57 |
| NM_006888.2 | calmodulin 1 | CALM1 | 5,48 | 5,56 |
| M61906.1 | phosphoinositide-3-kinase, regulatory subunit (p85 alpha) | PIK3R1 | 2,25 | 2,31 |
| L18964.1 | protein kinase C, iota | PRKCI | 2,21 | 2,60 |
| BC005365.1 | mitogen-activated protein kinase kinase 7 | MAP2K7 | 2 | 2,45 |
| Solute carriers | ||||
| NM_000112.2 | solute carrier family 26 (sulfate transporter), member 2 | SLC26A2 | 5,70 | 4,53 |
| NM_004731.3 | solute carrier family 16 (monocarboxylic acid transporters), member 7 | SLC16A7 | 2,92 | 2,11 |
| Small G proteins | ||||
| AI189609 | RAB2, member RAS oncogene family | RAB2 | 2,64 | 2,93 |
| NM_022157.2 | Ras-related GTP binding C | RRAGC | 2,19 | 1,25 |
| Miscellaneuos | ||||
| NM_001015.3 | ribosomal protein S11 | RPS11 | 119,98 | 40,70 |
| BC000603.2 | ribosomal protein L38 | RPL38 | 31,39 | 15,66 |
| BC000023.2 | ribosomal protein S19 | RPS19 | 3,08 | 3,08 |
| M25077 | Sjogren syndrome antigen A2 (60kDa, ribonucleoprotein autoantigen SS-A/Ro) | SSA2 | 2,38 | 2,12 |
| AF055585.1 | slit 2 | SLIT2 | 2,16 | 1,66 |
| NM_014399.3 | transmembrane 4 superfamily member 13 | TM4SF13 | 1,56 | 1,60 |
| AF065241.1 | thioredoxin (stress-induced) | TXN | 2,43 | 2,04 |
| NM_006287.3 | tissue factor inhibitor protein | TFPI | 6,64 | 7,77 |
| NM_033360.2 | v-Ki-ras2 Kirsten rat sarcoma 2 viral oncogene homolog | KRAS2 | 8,92 | 8,48 |
Figure 5. Gene array analysis of HUVECs stimulated with anti-hCMV antibodies.
Clustergrams of genes modulated by incubation of endothelial cells with antibodies against the US28 peptide or the UL122 peptide. Colours indicate the fold change.
Genes encoding for molecules involved in the apoptotic process were induced including eukaryotic translation initiation factor 5 (EIF5A) [13], which induces the expression of proteins essential for apoptosis of HUVECs, and other apoptosis-related genes such as caspase-6 (CASP6) and CASP7 and DNA fragmentation factor (DFFA). Up-regulated transcription factors included transcription factor 4 (TCF4) and v-ets erythroblastosis virus E26 oncogene homolog 2 (ETS2), involved in promoting apoptosis [14]. Moreover some ribosomal proteins were highly induced (up to 119,98 fold change in expression for RPS11) as already observed in HUVECs treated with asymmetrical dimethylarginine (ADMA), an endogenous inhibitor of nitric oxide synthase considered a risk factor for cardiovascular disease [15]. Interestingly ribosomal protein S19 is over-expressed during apoptotic cell death [16]. Genes involved in the mitogen-activated protein kinase (MAPK) signal pathway, which plays a pitoval role in transmitting transmembrane signals required for cell proliferation and apoptosis, were up-regulated by anti-US28 antibodies, including MAP2K7 (or MKK7), one of the major SAPK/JNK activating kinases [17], calmodulin 1 (CALM1) [18] and phosphatidylinositol 3-kinase regulatory 1 (PIK3R1). The activation of the MAPK pathway is responsible of the activation of other regulatory proteins, including the cytosolic enzyme phospholipase A2 (PLA2G4C), upregulated by anti-US28 antibodies, known to be induced in HUVECs in response to hypoxia, with consequent release of arachidonic acid [19].
Induction of growth factors is a common feature of endothelial cell activation; interestingly placental growth factor (PGF), known to promote atherosclerotic intimal thickening and macrophage accumulation [20], was induced by anti-US28 peptide antibodies. Moreover epidermal growth factor receptor (EGFR) and fibroblast growth factor receptor 1 (FGFR1), both induced by anti-US28 antibodies, are expressed in atherosclerotic plaques [21], [22]. Finally, several other genes were up-regulated including genes involved in lipid metabolism such as leptin receptor (LEPR), ATP-binding cassette (ABCA1) and peroxisome proliferator-activated receptor binding protein (PPARBP), and in extracellular matrix metabolism: collagen type IV alpha 3 binding protein (COL4A3BP), heparanase (HPSE), TGF beta-induced factor 2 (TGIF2).
Among the 1379 transcripts down-regulated by anti-US28 antibodies (Table S2) there were endothelial nitric oxide synthase (eNOS) and LDL receptor. The gene encoding for the anti-apoptotic molecule programmed cell death 4 protein (PDCD4) was repressed, accordingly to the observation that endothelial cells undergo apoptosis. Interestingly we could observe a down-regulation of HSP70, reported to exert a protective role against the development of atherosclerosis [23].
Anti-UL122 peptide antibodies up-regulated 186 transcripts including genes encoding for adhesion molecules, chemokines, transcription factors and molecules involved in apoptosis (Table 2 and Table S3). The highest increase in expression was observed among genes encoding for adhesion molecules: selectin E, VCAM-1 and ICAM-1. Another cluster of up-regulated genes was represented by genes encoding for inflammatory molecules including chemokines; CCL2, was highly increased, with a peak of 19-fold increase in expression at 6 hrs of stimulation. Others chemokine genes were highly induced, including CCL20, CXCL2, CXCL3 and CX3CL1. Genes encoding for extracellular matrix and matrix remodeling proteins [24] were also found up-regulated: matrix metalloproteinase-10 (MMP-10), laminin, nidogen, collagen type IV and cathepsin S. Several genes encoding for molecules involved in apoptosis such as CASP7, tumor necrosis factor alpha-induced protein 1 (TNFAIP1), and TNFAIP2 were upregulated. Similarly to what observed with anti-US28 peptide antibodies, anti-UL122 antibodies induced up-regulation of transcription factors, in particular genes involved in NF-kB activation pathway such as TANK and RELB, which have been implicated in atherosclerosis [25]. Moreover genes encoding for a variety of proteins that have been associated with atherosclerosis were up-regulated including phospholipase A2 (PLA2G4C), prostaglandin synthase 2 (PTGS2) also known as cyclooxygenase-2, thrombin receptor like-1 and the gene encoding for superoxide dismutase 2 (SOD2).
Table 2. Gene expression in HUVEC after 6 and 12 hours of stimulation with anti-UL122 IgG antibodies.
| Gene accession | Gene description | Gene Symbol | F.C. 6 h | F.C.12 h |
| Adhesion molecules | ||||
| NM_000450.1 | selectin E (endothelial adhesion molecule 1) | SELE | 31,15 | 15 |
| NM_001078.2 | vascular cell adhesion molecule 1 | VCAM1 | 18,8 | 6,1 |
| NM_000201.1 | intercellular adhesion molecule 1 (CD54) | ICAM-1 | 12,7 | 7,2 |
| NM_002210.2 | integrin, alpha V | ITGAV | 1,54 | 1,32 |
| Inflammation | ||||
| S69738.1 | chemokine (C-C motif) ligand 2 | CCL2 | 19 | 16 |
| NM_004591.1 | chemokine (C-C motif) ligand 20 | CCL20 | 7,6 | 4,7 |
| NM_001511.1 | chemokine (C-X-C motif) ligand 1 | CXCL1 | 6,2 | 4,5 |
| M57731.1 | chemokine (C-X-C motif) ligand 2 | CXCL2 | 9,5 | 6,8 |
| NM_002090.2 | chemokine (C-X-C motif) ligand 3 | CXCL3 | 10 | 6 |
| NM_002994.3 | chemokine (C-X-C motif) ligand 5 | CXCL5 | 2 | 2 |
| NM_002993.2 | chemokine (C-X-C motif) ligand 6 | CXCL6 | 2 | 1,8 |
| NM_000584.2 | interleukin 8 | IL8 | 2,8 | 2,3 |
| NM_005409.3 | chemokine (C-X-C motif) ligand 11 | CXCL11 | 5,5 | 6 |
| NM_002996.3 | chemokine (C-X3-C motif) ligand 1 | CX3CL1 | 3,11 | 2,9 |
| NM_002341.1 | lymphotoxin beta (TNF superfamily, member 3) | LTB | 1,67 | 1,6 |
| NM_005658.3 | TNF receptor-associated factor 1 (TRAF1) | TRAF1 | 1,6 | 1,37 |
| NM_003300.2 | TNF receptor-associated factor 3 (TRAF3) | TRAF 3 | 1,6 | 1,45 |
| NM_000395.1 | colony stimulating factor 2 receptor, beta | CSF2RB | 1,52 | 1,2 |
| AJ224869 | chemokine (C-X-C motif) receptor 4 | CXCR4 | 1,83 | 2,17 |
| NM_004221.4 | natural killer cell transcript 4 (interleukin 32) | NK4 | 1,6 | 4,3 |
| NM_003855.2 | interleukin 18 receptor 1 | IL18R | 2,02 | 1,3 |
| Immune response | ||||
| X56841.1 | major histocompatibility complex, class I, E | HLA-E | 1,2 | 1,5 |
| NM_000593.5 | transporter 1, ATP-binding cassette, sub-family B | TAP 1 | 1,8 | 2,14 |
| NM_006260.2 | DnaJ (Hsp40) homolog, subfamily C, member 3 | DNAJC3 | 1,19 | 1,63 |
| Transcription | ||||
| NM_003440.2 | zinc finger protein 140 | ZNF140 | 1,6 | 1,3 |
| NM_002448.2 | msh homeo box homolog 1 | MSX1 | 1,8 | 1,7 |
| NM_006509.2 | v-rel reticuloendotheliosis viral oncogene homolog B | RELB | 2 | 2 |
| NM_003199.1 | transcription factor 4 | TCF4 | 1,6 | 1,5 |
| NM_030751.2 | transcription factor 8 | TCF 8 | 1,4 | 1,8 |
| NM_004556.2 | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon | NFKBIE | 2 | 1,8 |
| NM_000222.1 | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene | KIT | 2,3 | 2,2 |
| U59863.1 | TRAF family member associated NFKB activator | TANK | 1,6 | 1,4 |
| NM_005239.4 | v-ets erythroblastosis virus E26 oncogene homolog 2 | ETS2 | 1,26 | 1,53 |
| NM_003489.2 | nuclear receptor interacting protein 1 | NRIP1 | 1,67 | 1,60 |
| Inhibition of cell growth and apoptosis | ||||
| NM_006291.2 | tumor necrosis factor, alpha-induced protein 2 | TNFAIP2 | 2,37 | 1,57 |
| NM_021137.3 | tumor necrosis factor, alpha-induced protein 1 (endothelial) | TNFAIP1 | 1,7 | 1,44 |
| NM_001227.3 | caspase 7, apoptosis-related cysteine protease | CAS P7 | 1,68 | 1,35 |
| NM_000546.2 | tumor protein p53 | TP53 | 1,2 | 1,7 |
| NM_006290.2 | tumor necrosis factor, alpha-induced protein 3 | TNFAIP3 | 2,7 | 2,44 |
| NM_003810.2 | tumor necrosis factor (ligand) superfamily, member 10 | TNFSF10 | 2,3 | 3,39 |
| NM_020529.1 | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor | NKFBIA | 2,7 | 2,3 |
| Collagens and extra-cellular matrix | ||||
| NM_007361.2 | nidogen 2 (osteonidogen) | NID2 | 1,8 | 2,9 |
| NM_001846.1 | collagen, type IV, alpha 2 | COL4A2 | 1,5 | 2,4 |
| NM_001845.3 | collagen, type IV, alpha 1 | COL4A1 | 1,75 | 1,85 |
| NM_002999.2 | syndecan 4 | SDC4 | 1,5 | 1,36 |
| NM_004079.3 | cathepsin S | CTSS | 1,2 | 1,58 |
| NM_002425.1 | matrix metalloproteinase 10 | MMP10 | 1,7 | 2,3 |
| NM_005562.1 | laminin,gamma 2 | LAMC2 | 1,5 | 2,3 |
| Metabolism | ||||
| NM_014349.2 | apolipoprotein L, 3 | APOL3 | 2,4 | 1,7 |
| AF065214.1 | phospholipase A2 | PLA2 G4C | 5,1 | 10,8 |
| NM_004915.3 | ATP-binding cassette, sub-family G (WHITE), member 1 | ABCG1 | 3 | 2,8 |
| Miscellaneous | ||||
| L07555.1 | CD69 antigen | CD 69 | 6 | 3,7 |
| NM_004428.2 | ephrin-A1 | EFNA1 | 2,1 | 1,8 |
| NM_000963.1 | prostaglandin-endoperoxide synthase 2 (COX-2) | PTGS2 | 1,9 | 1,3 |
| NM_005242.3 | Thrombin receptor like 1 | F2RL1 | 1,72 | 1,63 |
| NM_003277.2 | Claudin 5 | CLDN5 | 1,17 | 1,7 |
| AY267901.1 | superoxide dismutase 2, mitochondrial | SOD 2 | 5,5 | 4,6 |
| NM_021804.1 | angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 | ACE2 | 1,2 | 1,5 |
| NM_006398.2 | ubiquitin D | UBD | 2,7 | 3,8 |
| NM_002053.1 | guanylate binding protein 1, interferon-inducible, 67kDa | GBP1 | 1,8 | 1,7 |
A total number of 140 transcripts were down-regulated by anti-UL122 antibodies (Table S4). Some genes are involved in lipid metabolism: 24-dehydrocholesterol reductase (DHCR24), squalene epoxidase (SQLE), LDL receptor and low density lipoprotein receptor-related protein 8 (LRP8). The decreased expression of the gene encoding for eNOS has already been reported in endothelial cells isolated from patients with vascular damage indicating an intrinsic defect in the mechanism of nitric oxide production [26].
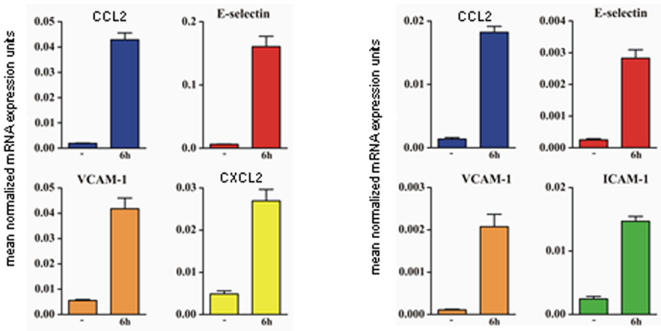
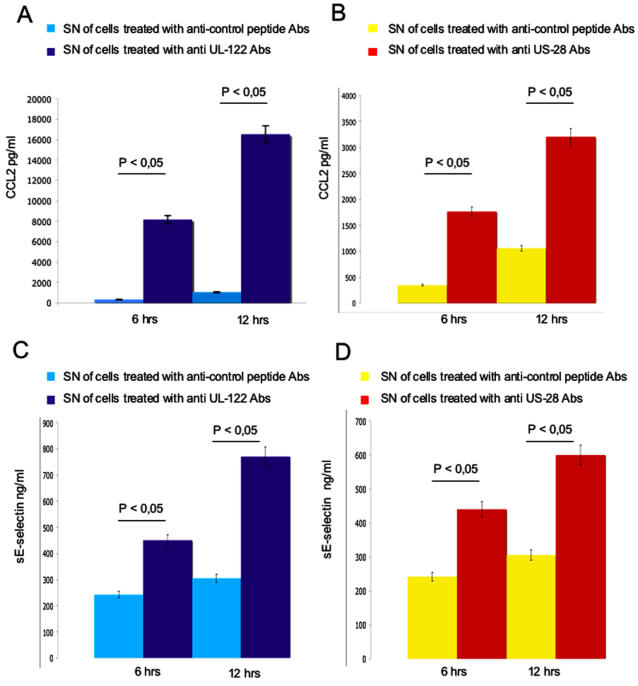
The gene array results were validated by real-time PCR of some of the genes up-regulated by both anti-US28 and UL122 antibodies (figure 6 A, B). These findings were paralleled by increased secretion of the corresponding molecules in the supernatant of stimulated cells (figure 7 A, B).
Figure 6. Validation of gene array by Q-PCR.
(A) Genes selected for validation by Q-PCR in endothelial cells treated with anti-US28 peptide antibodies. CCL2, E-selectin, VCAM-1 and CXCL2 transcripts were increased by 2,46-, 5,95-, 3,76-, and 4,21-fold, respectively, compared with control endothelial cells. (B) Genes selected for validation by Q-PCR in endothelial cells treated with anti-UL122 antibodies. CCL2, E-selectin, VCAM-1 and ICAM-1 transcripts were increased 19-, 31,15-, 18,8-, and 12,7-fold compared to control endothelial cells. The level of transcript expression is reported on the vertical axis. GAPDH was selected as endogenous gene.
Figure 7. Soluble mediators released in cell culture supernatants.
(A, B) Quantification of CCL2 released in the supernatant (SN) of HUVECs stimulated with antibodies (Abs) against the irrelevant peptide and with anti-UL122 and anti-US28 affinity purified antibodies. (C, D) Quantification of soluble E-selectin in the SN of HUVECs stimulated with abs against the irrelevant peptide and with anti–UL122 and anti-US28 affinity purified antibodies. Results are expressed in pg/mL and ng/mL, respectively. Results are expressed as mean of three independent experiments.
Using MedGene, the automated literature mining tool which summarizes and estimates the relative strength of all human gene disease relationships in Medline, the transcripts induced after stimulation with anti US28 antibodies were sorted into 3 classes, according to their relationship with the disease Atherosclerosis: 95 genes were directly associated to atherosclerosis, 348 were associated through others genes and 263 genes were new for this disease. Similarly, among the genes up-regulated by anti-UL122 antibodies, 40 genes resulted directly linked to atherosclerosis, 88 genes have been indirectly associated with the disease, and only 33 genes non associated to the atherosclerosis. Considering the down-regulated genes, 329 were not related to the disease after US28 antibody stimulation and 33 resulted not previously associated to the disease after UL122 antibody stimulation. A total of 25 genes were up-regulated by both anti-hCMV antibodies, while 26 genes were simultaneously down regulated by both anti-hCMV antibodies (Table S5 and Table S6). Interestingly all of these genes are classified as “directly related to atherosclerosis” when analyzed with MedGene.
We next wanted to identify the biological processes influenced by the engagement of CD49f and HSP60, connexin 45 and CD151 in HUVECs treated with either anti-US28 or anti-UL122 antibodies by using a pathway analysis software (Pathway Studio). To answer this question we generated connection pathways linking either CD151 and Connexin 45 or CD49f and HSP60 with the sets of modulated genes. The graphic outputs obtained showed that the different receptors use different pathways which ultimately lead to the same biological processes, including immune response, inflammation and apoptosis (Figure S1).
Discussion
We had previously reported that antibodies isolated from patients with atherosclerosis and directed against the hCMV-derived proteins UL122 and US28 are able to induce endothelial cell apoptosis following the engagement of cell surface molecules [6]. The apoptotic process was more evident when HUVEC cells were exposed to anti-UL122 antibodies and here we show that these antibodies induce apoptosis earlier than anti-US28 antibodies do. Anti-US28 antibodies induce membrane translocation and release in the supernatant of HSP60. Abnormal surface expression of HSP60 sensitizes endothelial cells to apoptosis and cross-recognition of HSP60 on stressed endothelial cells by such antibodies may amplify the process. HSP60 is released in the supernatant and is able to trigger signaling through TLR4, suggesting a possible a link between the atherosclerotic process and defence against both foreign pathogens and endogenously generated inflammatory ligands [11]. Indeed TLR4 may play a pivotal role in the early stages of atherosclerosis since it is upregulated on endothelial cell by low shear stress and oxidized lipoproteins. On the other hand TLR4 initiates an immune response after recognition of pathogen-associated molecular patterns inducing upregulation of proinflammatory cytokines. HSP60, released upon stress signals, acts as an endogenous ligand for TLR4, thus amplifying TLR4 activity and suggesting a HSP60-related autocrine process [27]. Moreover it has been reported that ligation of TLR strongly inhibits cholesterol efflux from macrophages [28]. Therefore TLR4 may bridge oxidized lipoproteins, infection, inflammation and low shear stress in the pathogenesis of the disease [29].
To investigate the gene modulation induced by anti-hCMV antibodies, we used a gene array approach, which allows the simultaneous detection of thousands of genes in a given sample. By this approach we found that the purified anti-hCMV antibodies modulate a vast array of genes encoding for molecules which play a pivotal role in endothelial cells activation and damage. Interestingly the genes associated to atherosclerosis VCAM1, SELE, ICAM1, CCL2, CCL20, CXCL2, TAP1, NFKBIA, TCF4 and ABCG1 are upregulated in cells treated with both UL122 and US28. The up-regulation of genes encoding for the adhesion molecules ICAM-1, VCAM-1, E-selectin is present also when endothelial cells are stimulated by proinflammatory cytokines such as tumor necrosis factor alpha [30]. These adhesion molecules are markers of endothelial dysfunction and damage and have been found elevated in the sera of patients affected by atherosclerosis [31]. Reflecting the inflammatory status of the atherosclerotic process [32], different chemokine-encoding genes have been reported to be activated in endothelial cells [33]. Also in our experimental model, some chemokine-endoding genes were up-regulated, in particular the gene encoding for CCL2 presented the highest fold change, mainly in the cells treated with anti-UL122 antibodies. This result was confirmed by the high increase in CCL2 transcripts by Q-PCR and by the detection of increased levels of the molecule in the cell culture supernatants.
Genes involved in the extracellular matrix metabolism were also up-regulated reflecting the extracellular matrix rearrangement within the vessel walls during the atherosclerotic process [24]. The gene expression profile in our cells resulted in the up-regulation of genes encoding for several transcription factors and proteins involved in the intracellular signaling. In particular in cells stimulated with anti-US28 antibodies the genes encoding for ETS2 and calmodulin 1 were highly induced. In this context it has to be mentioned that different stress factors, (i.e. mechanical force and thrombin) may initiate intracellular signaling relevant to vascular damage [34]. Moreover, TANK and RELB induced in anti-UL122 treated cells, as well as MAPK induced by anti-US28 antibodies, are involved in the NF-kB activation, known to play a pivotal role in the regulation of several genes related to atherogenesis [25]. Finally the diminished expression of endothelial eNOS is an early event in endothelial cell damage and a suppressed eNOS activity has been recently reported in hCMV infected endothelial cells [35].
It is noteworthy that all of the genes modulated by the two subsets of anti-hCMV antibodies encode molecules that have been already associated with atherosclerosis [36], supporting the possible role of hCMV in inducing endothelial cell damage that plays a pivotal role in the atherosclerotic process.
Taken together these data indicate that the anti-UL122 and -US28 antibodies induce endothelial cell damage through the activation of genes encoding for adhesion molecules, chemokines, for molecules involved in immune response, inflammation and extracellular matrix deposition. Moreover anti-US28 antibodies ampliphy their effect on endothelial cells through the traslocation of HSP60 on cell surface.
Since these subset of anti-hCMV antibodies have been detected in the majority of patients with atherosclerosis [6], our results give further support to the role played by hCMV in the pathogenesis of the disease.
Supporting Information
Functional pathway analysis of genes influenced by the engagement of CD151 and connexin 45 (A) and CD49f and HSP60 (B). The Pathway Studio software was used to identify connection pathways linking CD151 and connexin 45 or CD49f and HSP60 with the sets of modulates genes. Genes are represented as yellow ovals connected by arrows, major biologic processes related to these genes are represented as orange rectangles.
(0.29 MB TIF)
Genes up-regulated by anti-US28 antibodies.
(0.17 MB XLS)
Genes down-regulated by anti-US28 antibodies.
(0.26 MB XLS)
Genes up-regulated by anti-UL122 antibodies.
(0.04 MB XLS)
Genes down-regulated by anti-UL122 antibodies.
(0.03 MB XLS)
Genes up-regulated by both anti-UL28 and anti-US122 IgG antibodies.
(0.02 MB XLS)
Genes down-regulated by both anti-US28 and anti-UL122 antibodies.
(0.01 MB XLS)
Acknowledgments
We are indebted to Prof. Marco Cassatella for helpful suggestions in writing the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grants from the Italian Ministry for Scientific Research and Technology (AP and CL), and from the Italian Ministry of Health (RC). Funders had no role in the design and conduct of the study, in the collection, analysis, and interpretation of the data, and in the preparation, review, or approval of the manuscript.
References
- 1.Lusis AJ, Mar R, Pajukanta P. Genetics of atherosclerosis. Annu Rev Genomics Hum Genet. 2004;5:189–218. doi: 10.1146/annurev.genom.5.061903.175930. [DOI] [PubMed] [Google Scholar]
- 2.Wilson PW, D'Agostino RB, Levy D, Belanger AM, Silbershatz H, et al. Prediction of coronary heart disease using risk categories. Circulation. 1998;97:837–847. doi: 10.1161/01.cir.97.18.1837. [DOI] [PubMed] [Google Scholar]
- 3.Wich G, Knoflach M, Xu Q. Autoimmune and inflammatory mechanisms in atherosclerosis. Ann Rev Immunol. 2004;22:361–403. doi: 10.1146/annurev.immunol.22.012703.104644. [DOI] [PubMed] [Google Scholar]
- 4.Mayr M, Metzler B, Kiechl S, Willeit J, Schett Q, et al. Endothelial cytotoxicity mediated by serum antibodies to heat shock proteins of Escherichia coli and Chlamydia pneumoniae: immune reactions to heat shock proteins as a possible link between infection and atherosclerosis. Circulation. 1999;99:1560–1566. doi: 10.1161/01.cir.99.12.1560. [DOI] [PubMed] [Google Scholar]
- 5.Xiao Q, Mandal K, Schett G, Mayr M, Wick G, et al. Association of serum-soluble heat shock protein 60 with carotid atherosclerosis: clinical significance determined in a follow-up study. Stroke. 2005;12:2571–2576. doi: 10.1161/01.STR.0000189632.98944.ab. [DOI] [PubMed] [Google Scholar]
- 6.Bason C, Corrocher R, Lunardi C, Puccetti P, Olivieri O, et al. Interaction of antibodies against cytomegalovirus with heat shock protein 60 in pathogenesis of atherosclerosis. Lancet. 2003;362:1971–1977. doi: 10.1016/S0140-6736(03)15016-7. [DOI] [PubMed] [Google Scholar]
- 7.Mallat Z, Tedgui A. Apoptosis in the vasculature: mechanisms and functional importance. Br J Pharmacol. 2000;130:947–962. doi: 10.1038/sj.bjp.0703407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tedgui A, Mallat Z. Apoptosis as a determinant of atherothrombosis. Tromb Haemost. 2001;6:420–426. [PubMed] [Google Scholar]
- 9.Lunardi C, Dolcino M, Peterlana D, Bason C, Navone R, et al. Antibodies against human cytomegalovirus in the pathogenesis of systemic sclerosis: a gene array approach. PLoS Medicine. 2006;3:e2. doi: 10.1371/journal.pmed.0030002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu Y, Hines LM, Weng H, Zuo D, Rivera M, et al. Analysis of genomic and proteomic data using advanced literature mining. J Proteome Res. 2003;2:405–412. doi: 10.1021/pr0340227. [DOI] [PubMed] [Google Scholar]
- 11.Michelsen KS, Doherty TM, Shah PK, Arditi M. TLR signalling: an emerging bridge from innate immunity to atherogenesis. J Immunol. 2004;173:5901–5907. doi: 10.4049/jimmunol.173.10.5901. [DOI] [PubMed] [Google Scholar]
- 12.Ohashi K, Burkart V, Flohe S, Kolb H. Cutting edge: heat shock protein 60 is a putative endogenous ligand of the toll-like receptor-4 complex. J Immunol. 2000;164:558–561. doi: 10.4049/jimmunol.164.2.558. [DOI] [PubMed] [Google Scholar]
- 13.Lee Y, Kim HK, Park HE, Park MH, Joe YA. Effect of N1-guanyl-1,7-diaminoheptane, an inhibitor of deoxyhypusine synthase, on endothelial cell growth, differentiation and apoptosis. Mol Cell Biochem. 2002;237:69–76. doi: 10.1023/a:1016535217038. [DOI] [PubMed] [Google Scholar]
- 14.Wolvetang EJ, Wilson TJ, Sanij E, Busciglio J, Hatristavrou T, et al. ETS2 over expression in transgenic mouse models and in Down syndrome predisposes to apoptosis via the p53 pathway. Hum Mol Genet. 2003;12:247–255. doi: 10.1093/hmg/ddg015. [DOI] [PubMed] [Google Scholar]
- 15.Smith CL, Anthony S, Hubank M, Leiper JM, Vallance P. Effects of ADMA upon gene expression: an insight into the pathophysiological significance of raised plasma ADMA. PloS Med. 2005;2:e264. doi: 10.1371/journal.pmed.0020264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishiura H, Tanase S, Shibuya Y, Futa N, Sakamoto T, et al. S19 ribosomal protein dimer augments metal-induced apoptosis in a mouse fibroblastic cell line by ligation of the C5a receptor. J Cell Biochem. 2005;94:540–553. doi: 10.1002/jcb.20318. [DOI] [PubMed] [Google Scholar]
- 17.Foltz IN, Gerl RE, Wieler JS, Luckach M, Salmon RA, et al. Human mitogen-activated protein kinase kinase 7 (MKK7) is a highly conserved c-Jun N-terminal kinase/stress-activated protein kinase (JNK/SAPK) activated by environmental stresses and physiological stimuli. J Biol Chem. 1998;15:9344–9351. doi: 10.1074/jbc.273.15.9344. [DOI] [PubMed] [Google Scholar]
- 18.Spratt DE, Newman E, Mosher, Ghosh DK, Salerno JC, et al. Binding and activation of nitric oxide synthase isozymes by calmodulin EF hand pairs. FEBS J. 2006;273:1759–1771. doi: 10.1111/j.1742-4658.2006.05193.x. [DOI] [PubMed] [Google Scholar]
- 19.Lupo G, Nicotra A, Giurdanella G, Anfuso CD, Romeo L, et al. Activation of phospholipase A(2) and MAPK Kinases by oxidized low-density lipoproteins in immortalized GP8.39 endothelial cells. Biochim Biophys Acta. 2005;1735:135–150. doi: 10.1016/j.bbalip.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 20.Khurana R, Moons L, Shafi S, Luttun A, Collen D, et al. Placental growth factor promotes atherosclerotic intimal thickening and macrophage accumulation Circulation. 2005;111:2828–2836. doi: 10.1161/CIRCULATIONAHA.104.495887. [DOI] [PubMed] [Google Scholar]
- 21.Brogi E, Winkles JA, Underwood R, Clinton SK, Alberts GF, et al. Distinct patterns of expression of fibroblast growth factors and their receptors in human atheroma and nonatherosclerotic arteries: association of acidic FGF with plaque microvessels and macrophages. J Clin Invest. 1993;92:2408–2418. doi: 10.1172/JCI116847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daley SJ, Gotlieb AI. Fibroblast growth factor receptor-1 expression is associated with neointima formation in vitro. Am J Pathol. 1996;148:1193–1202. [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu J, Quyyumi AA, Wu H, Csako G, Rott D, et al. Increased serum levels of heat shock protein 70 are associated with low risk of coronary artery disease. Arterioscler Thromb Vasc Biol. 2003;23:1055–1059. doi: 10.1161/01.ATV.0000074899.60898.FD. [DOI] [PubMed] [Google Scholar]
- 24.Newby AC. Dual role of matrix metalloproteinases (matrixins) in intimal thickening and atherosclerotic plaque rupture. Physiol Rev. 2005;85:1–31. doi: 10.1152/physrev.00048.2003. [DOI] [PubMed] [Google Scholar]
- 25.Monaco C, Andreakos E, Kiriakidis S, Mauri C, Bicknell C, et al. Canonical pathway of nuclear factor kB activation selectively regulates proinflammatory and prothrombotic responses in human atherosclerosis. Proc Natl Acad Sci USA. 2004;101:5634–5639. doi: 10.1073/pnas.0401060101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Nigris F, Lerman LO, Ignarro SW, Sica G, Lerman A, et al. Beneficial effects of antioxidants and l-arginine on oxidation-sensitive gene expression and endothelial NO synthase activity at sites of disturbed shear stress. Proc Natl Acad Sci USA. 2003;100:1420–1425. doi: 10.1073/pnas.0237367100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Graaf R, Kloppenburg G, Kitslaar PJ, Bruggeman CA, Stassen F. Human heat shock protein 60 stimulates vascular smooth muscle cell proliferation through Toll-like receptors 2 and 4. Microbes Infect May 3 [Epub ahead of print]. 2006 doi: 10.1016/j.micinf.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 28.Vink A, de Kkeijn DPV, Pasterkamp G. Functional role of toll-like receptors in atherosclerosis and arterial remodelling. Curr Op Lipidol. 2004;15:515–521. doi: 10.1097/00041433-200410000-00004. [DOI] [PubMed] [Google Scholar]
- 29.Yang QW, Mou L, Lv FL, Wang JZ, Wang L, et al. Role of Toll-like receptor4/NF-kappaB pathway in monocyte-endothelial adhesion induced by low shear stress and ox-LDL. Biorheology. 2005;42:225–236. [PubMed] [Google Scholar]
- 30.Murakami T, Mataki C, Nagao C, Umetani M, Wada Y, et al. The gene expression profile of human umbilical vein endothelial cells stimulated by tumor necrosis factor alpha using DNA microarray analysis. J Atheroscler Thromb. 2000;7:39–44. doi: 10.5551/jat1994.7.39. [DOI] [PubMed] [Google Scholar]
- 31.Haught WH, Mansour M, Rothlein R, Kishimoto TK, Mainolfi EA, et al. Alterations in circulating intercellular adhesion molecule-1 and L-selectin: further evidence for chronic inflammation in ischemic heart disease. Am Heart J. 1996;132:1–8. doi: 10.1016/s0002-8703(96)90383-x. [DOI] [PubMed] [Google Scholar]
- 32.Ross R. Atherosclerosis: an inflammatory disease. N Engl J Med. 1999;340:115–126. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 33.Charo IF, Taubman MB. Chemokines in the pathogenesis of vascular disease. Circ Res. 2004;95:858–866. doi: 10.1161/01.RES.0000146672.10582.17. [DOI] [PubMed] [Google Scholar]
- 34.Shaw A, Xu Q. Biomechanical stress-induced signaling in smooth muscle cells: an up-date. Curr Vasc Pharmacol. 2003;1:41–58. doi: 10.2174/1570161033386745. [DOI] [PubMed] [Google Scholar]
- 35.Shen YH, Zhang I, Utama B, Wang J, Gan Y, et al. Human cytomegalovirus inhibits Akt-mediated eNOS activation through upregulating PTEN (phosphatase and tensin homolog deleted on chromosome 10). Cardiovasc Res. 2006;69:502–511. doi: 10.1016/j.cardiores.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 36.Tuomisto TT, Yla-Herttuala S. What have we learnt about microarray analysis of atherogenesis? Curr Opin Lipidol. 2005;16:201–205. doi: 10.1097/01.mol.0000162325.85290.e5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Functional pathway analysis of genes influenced by the engagement of CD151 and connexin 45 (A) and CD49f and HSP60 (B). The Pathway Studio software was used to identify connection pathways linking CD151 and connexin 45 or CD49f and HSP60 with the sets of modulates genes. Genes are represented as yellow ovals connected by arrows, major biologic processes related to these genes are represented as orange rectangles.
(0.29 MB TIF)
Genes up-regulated by anti-US28 antibodies.
(0.17 MB XLS)
Genes down-regulated by anti-US28 antibodies.
(0.26 MB XLS)
Genes up-regulated by anti-UL122 antibodies.
(0.04 MB XLS)
Genes down-regulated by anti-UL122 antibodies.
(0.03 MB XLS)
Genes up-regulated by both anti-UL28 and anti-US122 IgG antibodies.
(0.02 MB XLS)
Genes down-regulated by both anti-US28 and anti-UL122 antibodies.
(0.01 MB XLS)