Abstract
Amplification of the cyclin-dependent kinase 4 (CDK4) gene, located at 12q13-q14, has been found as an alternative genetic alteration to CDKN2A inactivation in various human tumors including malignant gliomas and sarcomas. In the present study, we have evaluated the frequency of the CDK4 gene amplification in sporadic breast cancer by applying a nonradioactive quantitative differential polymerase chain reaction based on fluorescent DNA technology. Fluorescent-labeled polymerase chain reaction products were analyzed with an automated DNA sequencer. Amplification of CDK4 gene was detected in 15 (15.8%) of 95 breast cancers. All tumors with CDK4 gene amplification showed high CDK4 protein expression determined by immunohistochemistry. Furthermore, the mean Ki-67 labeling index in tumors with CDK4 gene amplification was significantly higher than in those without CDK4 gene amplification. No significant associations were observed between CDK4 gene amplification and any specific histopathological parameter. The findings of this study provide the first evidence of CDK4 gene amplification in breast cancer and suggest that CDK4 gene amplification appears to be of importance in the pathogenesis of a subset of sporadic breast cancer.
Critical transitions in the different phases of the cell cycle are regulated by sequential activation of cyclins and their catalytic subunits, the cyclin-dependent kinases (CDKs). Disruption of the cell cycle machinery might enhance genomic instability, contribute to uncontrolled cell growth, and lead to the development of cancer. 1-3 When activated by cyclin D1, CDK4 is able to phosphorylate the retinoblastoma gene product (pRB) and can promote cell cycle progression through G1-phase into S-phase. 3,4 The activation of CDK4, however, can be constrained by binding the p16 protein, a cyclin-dependent kinase inhibitor encoded by the CDKN2A gene. 3,5 Alterations of these individual components have been implicated in the pathogenesis of many tumor types. The involvement of CDK4 gene in tumorigenesis has been suggested by the findings that suppression of CDK4 can lead to terminal differentiation of erythroleukemia cells, whereas overexpression of CDK4 can induce uncontrolled cell growth and eventual malignant transformation. 5 Furthermore, amplification and consequent overexpression of the CDK4 gene, located in the 12q13-q14 region, have been found in various cancers including different types of sarcomas and glioblastomas. 6-8 A somatic point mutation (R24C) of CDK4 gene was identified in human melanomas, causing a tumor-specific antigen and disrupting the interaction between CDK4 and its inhibitor p16. 9
Alterations of cell cycle control genes, such as amplification of cyclin D1 10,11 and inactivation of RB and CDKN2A, 12-14 have been well documented in human breast cancer. However, there has been no report on amplification and overexpression of CDK4 in breast cancer so far, although amplification of 12q13 has been detected by cytogenetic analyses in breast cancer. 15,16 Moreover, increased expression of CDK4 was found to be common in carcinogen-induced rat mammary tumors. 17
To delineate the role of the CDK4 gene in the genesis of breast cancer, we investigated CDK4 gene amplification in breast cancer by fluorescent differential polymerase chain reaction (PCR) followed by fragment analysis on an automated DNA sequencer. This approach enables rapid, nonradioactive quantitative analysis of gene amplification on small tumor samples. 18 The biological relevance of CDK4 gene amplification was examined by immunohistochemical staining for expression of CDK4 protein in breast cancer. CDK4 gene amplification and expression were correlated to relevant clinical and tumor characteristics.
Materials and Methods
Tumor Samples and DNA Extraction
Snap-frozen samples were obtained from 95 patients treated by surgery for primary breast cancer (80 ductal and 15 lobular breast carcinomas) at the Department of Obstetrics and Gynecology, Heinrich-Heine-University Düsseldorf, Germany. None of the breast carcinoma patients had a positive family history (at least two cases of breast or ovarian cancer, one case below the age of 60 years). None of the patients had distant metastases at the time of primary surgery. In case of axillary dissection (at least 10 lymph nodes), the number of lymph node metastases was determined. Tumors were classified according to the TNM classification (Union Internationale Contre le Cancer). The histological grade was determined according to the criteria of Elston and Ellis. 19 Histological evaluation of frozen tumor sections assured that all specimens studied contained at least 60% tumor cells. High molecular weight DNA was prepared from tumor tissues and blood lymphocytes as described previously. 20 DNA from 20 malignant gliomas with CDK4 gene copy number determined previously by Southern blot analysis 8 were used as controls to establish the differential PCR assay.
Fluorescent Differential PCR
The gene dosage of the CDK4 gene was analysed by differential PCR with fluorescein-labeled primers. 18 Two different reference loci were used: glyceraldehyde-3-phosphate dehydrogenase (GAPDH) on 12p13 and adenine phosphoribosyltransferase (APRT) on 16q23. Primer sequences for CDK4 and the control loci were as follows: 5′-CATGTAGACCAGGACCTAAGG (sense) and 5′-AACTGGCGCATCAGATCCTAG (antisense) for CDK4 resulting in a 206-bp PCR product, 5′-F-AACGTGTCAGTGGTGGACCTG (sense) and 5′-AGTGGGTGTCGCTGTTGAAGT (antisense) for GAPDH generating a 160-bp PCR product, and 5′-TGGGAAAGCTGTTTACTGCG (sense) and 5′-CAGGGAACACATTCCTTTGC (antisense) for APRT generating a 134-bp PCR product. One primer of each primer pair was labeled with fluorescein at the 5′-end. PCR amplification was carried out in a 50-μl volume containing 50 ng of genomic DNA, 1× PCR buffer (10 mmol/L Tris/HCl, pH 9.0, 50 mmol/L KCl, 1.5 mmol/L MgCl2, 0.001% gelatine), 150 μmol/L dNTPs, 0.3 μmol/L primers for CDK4 and GAPDH or APRT, and 2.0 units Taq DNA polymerase (Pharmacia, Freiburg, Germany). The reaction consisted for 25 cycles of 94°C for 1 minute, 60°C for 1 minute, and 72°C for 1 minute followed by a final extension for 8 minutes at 72°C. In each PCR experiment, DNA extracted from 1) placenta and peripheral blood leukocytes as reference tissue with normal CDK4 gene copy number and 2) two primary gliomas with known CDK4 gene amplification were included as controls. The fluorescein-labeled PCR products were separated with an automated fluorescent DNA sequencer (A. L. F.™, Pharmacia) on 6% denaturing polyacrylamide gels. Quantitative analysis of the peak areas obtained for CDK4 and GAPDH or APRT was performed with the Fragment Manager™ (FM1.1) software (Pharmacia), and CDK4 gene dose was calculated relative to control blood as described. 18 Only increases in the CDK4/GAPDH and CDK4/APRT quotients of more than three times relative to control blood and normal placenta were considered as evidence of CDK4 gene amplification.
Southern Blot Analysis
DNA from tumors and normal leukocytes (2.5 μg) were digested with the restriction enzyme TaqI, separated on 0.8% agarose gels, and blotted. The membranes were sequentially hybridized with probes for CDK4 and the control locus ERBB3, as described. 8
Immunohistochemistry
Immunohistochemical stainings for CDK4 protein, the proliferation-associated nuclear antigen Ki-67, ER, and PgR were performed on paraffin-embedded formalin-fixed sections using the indirect avidin-biotin peroxidase method as described. 21 To enhance immunoreactivity for Ki-67, sections were pretreated by microwave heating in 10 mmol/L citrate buffer (pH 6.0) for 3 × 10 minutes. Sections to be stained for CDK4 protein were treated in 0.1% trypsin for 5 minutes at 37°C. Histological sections were incubated for 30 minutes in methanol with 1% hydrogen peroxide. The sections were incubated with goat anti-CDK4 polyclonal antibody (C-22, Santa Cruz Biotechnology, Inc., Santa Cruz, CA), MIB-1 mouse anti-Ki-67 monoclonal antibody (Dianova, Hamburg, Germany), mouse anti-ER monoclonal antibody, or mouse anti-PgR monoclonal antibody (Sigma, Deisenhofen, Germany) for 16 hours at 4°C. This was subsequently followed by biotinylated secondary antibodies and by avidin-biotin-peroxidase complex (Vector Laboratories, Burlingame, CA) according to the manufacturer’s instructions. As negative controls, irrelevant rabbit/mouse monoclonal IgG were used instead of the primary antibodies. ER and PgR immunoreactivities were analysed and scored (according to recommendations of Remmele and Stegner) as described previously. 20 Immunoreactivity for CDK4 and Ki-67 was quantified by counting positive and negative tumor cells. A minimal number of 1000 cells was counted for each case. For CDK4 expression, a semiquantitative scoring was applied as described. 22 The assigned score first reflects the staining intensity A (0, negative; 1, weak; 2, moderate; 3, high) and second the percentage of positive cells B (0, no positive cells; 1, <25% positive cells; 2, 25 to 50% positive cells; 3, >50% positive cells). The sum of A + B attained a maximum score of 6. A score more than 3 was considered as positive.
Statistical Analysis
Associations of CDK4 gene amplification and other factors were calculated by Fisher’s exact test or Student’s unpaired t-test.
Results
Validation of Fluorescent Differential PCR
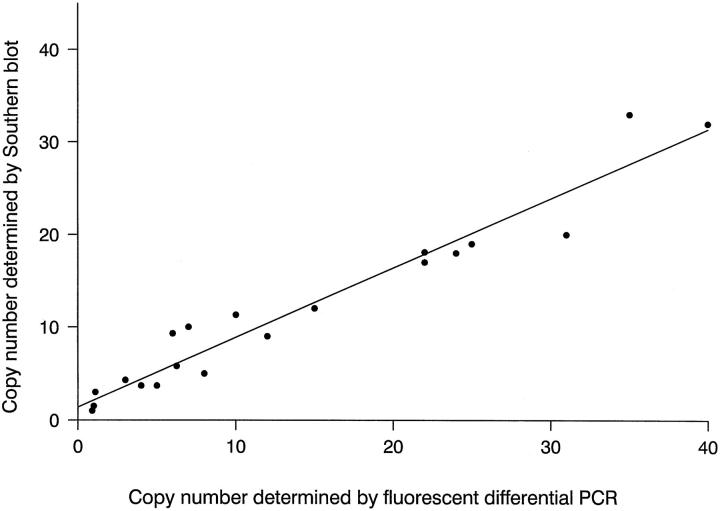
The fluorescent differential PCR method was established and optimized in terms of amplimers, enzyme concentration, magnesium concentration, and cycle number to establish conditions for exponential amplification of CDK4 and GAPDH or APRT specific sequences. To validate the sensitivity of differential PCR combined with detection of fluorescent-labeled PCR products by an automated DNA sequencer, CDK4 gene amplification of 20 malignant gliomas was determined by fluorescent differential PCR and compared with results from previous Southern blot analysis. 8 Results of gene copy numbers showed a linear relationship with a correlation coefficient of 0.86 between detection of CDK4 by fluorescent differential PCR and by Southern blot (Figure 1) ▶ .
Figure 1.
Correlation between Southern blot analysis and fluorescent differential PCR. Copy number of CDK4 in 20 human malignant gliomas was determined by fluorescent differential PCR (PCR conditions see Material and Methods) and compared with results from previous Southern blot analysis. 8 The correlation coefficient was 0.86, the correlation rate was highly significant (P < 0.01) according to Student’s t-test.
CDK4 Amplification in Breast Cancer
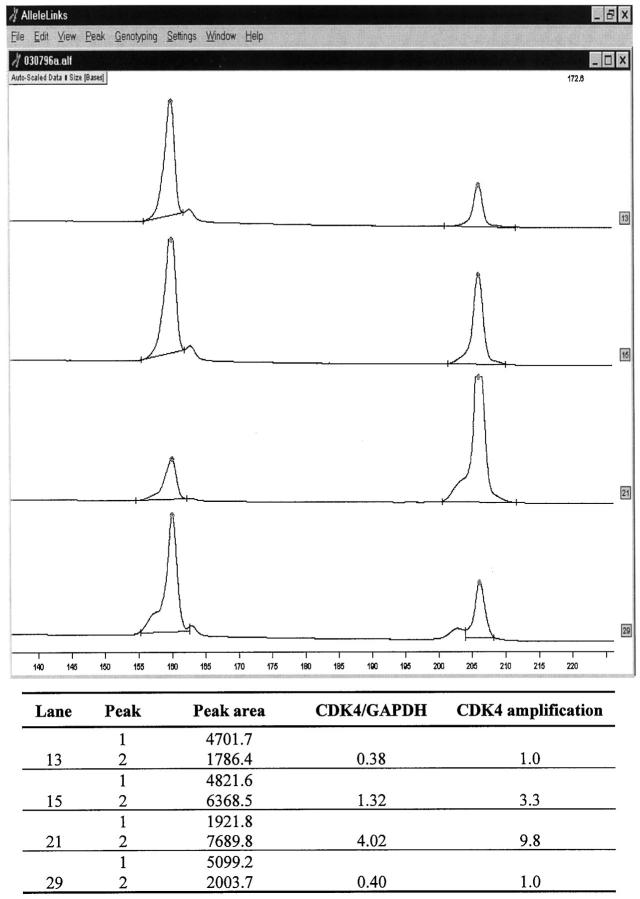
Ninety-five breast cancers were analyzed for CDK4 gene amplification by the fluorescent differential PCR. Among the 95 tumors analyzed, 15 (15.8%) were found to demonstrate CDK4 amplification ranging from 3- to 10-fold (12 tumors corresponded to a three to fivefold amplification, three tumors presented 5- to 10-fold amplification). Representative examples of CDK4 amplification are shown in Figure 2 ▶ .
Figure 2.
Analysis of CDK4 amplification by fluorescent differential PCR. Electrophoretogram of PCR-products of CDK4 (206 bp) and the control gene GAPDH (160 bp) in representative cases. The data were obtained with an automated fluorescent DNA sequencer (A. L. F., Pharmacia) and analyzed with the Fragment Manager™ software. The abscissa of the electrophoretogram shows the fragment size in bp. Lane 13, tumor DNA without CDK4 amplification; lane 15, tumor DNA with CDK4 amplification (threefold); lane 21, tumor DNA with CDK4 amplification (10-fold); lane 29, placenta DNA as normal control.
Expression of CDK4 Protein in Breast Cancer
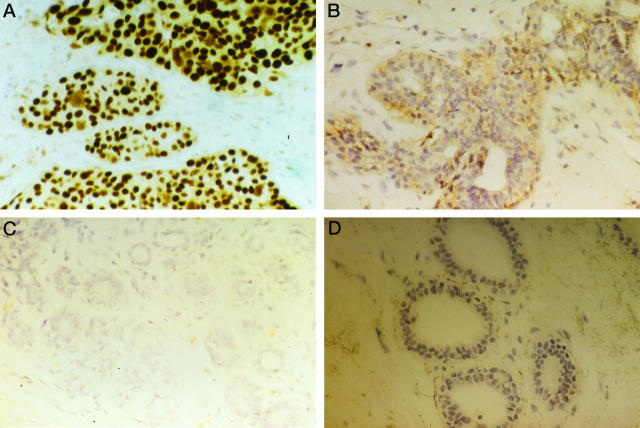
In order to determine if CDK4 gene amplification also resulted in overexpression of the gene and increased levels of the gene product, immunohistochemical analysis of the tumor tissue specimens including the 15 breast cancers with CDK4 gene amplification was performed by using an antibody directed against CDK4 protein. Moderate and intense nuclear and/or cytoplasmic stainings were observed in breast cancer cells, and distribution of positive cells was heterogeneous in a cancer nest and between cancer nests (Figure 3, A and B) ▶ . All 15 tumors with CDK4 gene amplification showed overexpression of the CDK4 protein. Overexpression of the CDK4 protein was also observed in three tumors without gene amplification. The other breast cancer samples were negative for CDK4 protein, although scattered CDK4-positive tumor cells were found in some instances (Figure 3C) ▶ .
Figure 3.
Immunohistochemical demonstration of CDK4 protein expression in breast carcinomas. Representative examples of invasive ductal adenocarcinomas with CDK4 amplification showing strong nuclear staining (A) or moderate cytoplasmic staining (B). Negative staining results in an invasive ductal breast carcinoma without CDK4 amplification (C) and in normal mammary epithelial cells (D). Original magnification, ×400.
Relationships between CDK4 Gene Amplification and Clinicopathological Parameters
The association between CDK4 gene amplification and clinicopathological characteristics was determined. No significant associations were found between CDK4 gene amplification and patient’s age, tumor size, and lymph-node status. Although statistically not significant, CDK4 gene amplification was observed more frequently in tumors of higher histological grade (Table 1) ▶ . The mean Ki-67 labeling index was significantly higher in breast cancers with CDK4 gene amplification and overexpression than in tumors with normal copy number of CDK4 gene (P < 0.05; Table 2 ▶ ).
Table 1.
Correlation between CDK4 Amplification and Clinicopathological Characteristics of 95 Breast Cancer Patients
| Characteristics | n = 95 | CDK4 amplification | P value |
|---|---|---|---|
| Age (years) | |||
| <50 | 25 | 4 | P > 0.05 |
| ≥50 | 70 | 11 | |
| Tumor size | |||
| pT1 | 50 | 6 | |
| pT2 | 33 | 6 | P > 0.05 |
| pT3/4 | 12 | 3 | |
| Lymph node status | |||
| N− | 47 | 6 | P > 0.05 |
| N+ | 48 | 9 | |
| Grade | |||
| G1 | 12 | 1 | |
| G2 | 58 | 8 | P > 0.05 |
| G3 | 25 | 6 | |
| ER | |||
| Negative | 43 | 7 | P > 0.05 |
| Positive | 52 | 8 | |
| PgR | |||
| Negative | 39 | 6 | P > 0.05 |
| Positive | 56 | 9 |
Table 2.
Correlation between CDK4 Amplification and Nuclear Antigen Ki-67 Labeling Index of 95 Breast Cancers
| CDK4 | n | Index of Ki-67 (%) mean ± standard |
|---|---|---|
| Overexpression with gene amplification | 15 | 21.3 ± 4.9* |
| Overexpression without gene amplification | 3 | 14.8 |
| No amplification | 77 | 4.7 ± 3.2 |
*Significant difference compared with tumors without CDK4 gene amplification (P < 0.05, t-test).
Discussion
Oncogene amplification is a common mechanism of proto-oncogene activation and contributes to neoplastic cell transformation and tumor progression. Detection of oncogene amplification using fluorescent differential PCR as well as application of automated fluorescent DNA technology for LOH analysis have been described by various groups. 18,23,24 In this study, differential PCR and automated fluorescent DNA technology were applied for quantitative determination of CDK4 amplification in breast cancers. This approach offers several advantages compared with other detection and staining methods: 1) Separation and quantitative determination of PCR products can be performed automatically without the delay inherent in autoradiography, requiring approximately 3 hours only. 2) The enhanced sensitivity of the fluorescent detection method requires 25 PCR cycles only to achieve detectable results and allows to analyze PCR products in the exponential phase of the PCR reaction. Furthermore, linearity of fluorescence detection covers a much wider range than scanning of autoradiograms or ethidium bromide or silver-stained gels, resulting in an improved quality of data. Therefore, this approach is particularly suitable for the routine clinical analysis of genetic alterations in large series of samples.
By cytogenetic analysis, translocations and amplifications on 12q13 have been detected in karyotypes of breast carcinomas. 15,16 Furthermore, increased expression of CDK4 occurs frequently in carcinogen-induced rat mammary tumors. 17 These findings have raised the hypothesis that amplification of CDK4 might be one of the genetic alterations in breast cancer, which contribute to disruption of cell cycle control in the pathogenesis of breast cancer. In the present study, we provide the first evidence of amplification of the CDK4 gene in breast carcinomas. The frequency of CDK4 amplification in breast carcinomas (15.8%) determined here is similar to that observed in malignant gliomas (13 to 15%) but lower than the frequencies reported for human sarcomas (27 to 50%). Two previous studies on expression of CDK4 protein in breast cancer reported that immunoreactivity of CDK4 protein was found in between 80 and 95% of breast carcinomas analyzed by immunohistochemical staining. 25,26 In contrast, overexpression of CDK4 protein was detected in 19% of our series of breast cancers. This discrepancy should be attributed to the difference in applied methods, the criteria for scoring of immunoreactivity, and characteristics between the various primary carcinoma tissues used for different studies. All breast tumors of our series with CDK4 gene amplification showed overexpression of CDK4 protein determined by immunostaining. These findings are in line with recent studies on other cancers and cancer cell lines revealing increased levels of CDK4 protein in association with gene amplification. 27,28 In addition, overexpression of CDK4 without gene amplification in three breast cancers may represent increased transcription or post-translational modification of the protein in cancer cells. The recent report of Zhang et al 29 has demonstrated that overexpression of CDK4 protein is accompanied by a significant increase in the number of proliferating cells in colon adenomas as shown by BrdU incorporation and immunostaining of proliferating cell nuclear antigen (PCNA). Ito et al 25 have found that overexpression of CDK4 determined by immunostaining was significantly more heterogeneous in breast tumors of larger sizes and/or at higher stages. In agreement with these observations, a significant relationship between CDK4 amplification and Ki-67 labeling index has been found in breast cancers in this study. The results presented here as well as the finding of Ito et al thus indicate an important role of CDK4 amplification and overexpression in human breast carcinomas.
Alterations in the cell cycle regulatory pathway involving p16, pRB, cyclin D1, and CDK4 have been implicated in tumorigenesis. Inactivation of RB and CDKN2A or activation of cyclin D1 or CDK4 have been demonstrated in various types of human cancers. 1-3,5 Furthermore, it has been hypothesized that alterations of any of the components in the growth-regulatory pathway (p16, pRB, cyclin D1, and CDK4) may have a similar effect leading to disturbance of the cell cycle regulatory system in favor of cell proliferation. 2,30 This hypothesis is supported by the following observations: 1) CDK4 amplification and alteration of CDKN2A are not found concomitant in human gliomas and sarcomas 27,31,32 ; 2) loss of expression of p16 is restricted to tumors with wild-type RB in lung cancers and glioblastomas 33,34 ; and 3) amplification of cyclin D1 is associated inversely with mutation of RB in esophageal cancers. 35 Previous studies in breast cancer have reported that amplification of cyclin D1 10,11,35 and inactivation of RB 12,13 are present in a significant fraction of breast carcinomas. Hypermethylation of the 5′-CpG island of the CDKN2A gene associated with loss of CDKN2A transcription is also frequently found in primary breast cancers, 14 although homozygous deletions and point mutations of this gene are rare in breast cancers. 36 As presented here, CDK4 gene amplification may be an additional genetic alteration in breast cancer. Additional functional studies of CDK4 protein kinase activity and the interactions of cell cycle regulatory proteins in breast cancer with CDK4 amplification and overexpression may clarify and extend the importance of the role of this regulatory pathway in the pathogenesis of breast cancers.
Acknowledgments
The authors thank the staff from the operating room and the technicians from the Laboratory of the Pathomorphology, Frauenklinik, Heinrich-Heine Universität, Düsseldorf, Germany, for the recruitment of tumors and their persistent support. Thanks to Mrs. Dorotheé Larbig for expert technical assistance.
Footnotes
Address reprint requests to Dr. D. Niederacher, Molekulargenetisches Labor, Frauenklinik, Heinrich-Heine Universität, Moorenstrasse 5, D-40225 Düsseldorf, Germany. E-mail: niederac@uni-duesseldorf.de.
Supported in part by Grant Be 1215/6-3 from the Deutsche Forschungsgemeinschaft, Bonn, Germany.
Han-Xiang An’s current address is Division of Cytogenetics (H400), German Cancer Research Center, INF 280, 69120 Heidelberg, Germany.
References
- 1.Sherr CJ: Mammalian G1 cyclins. Cell 1993, 73:1059-1065 [DOI] [PubMed] [Google Scholar]
- 2.Hartwell LH, Kastan MB: Cell cycle control and cancer. Science 1994, 266:1821-1828 [DOI] [PubMed] [Google Scholar]
- 3.Hunter T, Pines J: Cyclins and cancer: II. Cyclin D and CDK inhibitors come of age. Cell 1994, 79:573-582 [DOI] [PubMed] [Google Scholar]
- 4.Matsushime H, Ewen ME, Strom DK, Kato JY, Hanks SK, Roussel MF, Sherr CJ: Identification and properties of an atypical catalytic subunit (p34PSK-J3/cdk4) for mammalian D type G1 cyclins. Cell 1992, 71:323-334 [DOI] [PubMed] [Google Scholar]
- 5.Xiong Y, Zhang H, Beach D: Subunit rearrangement of the cyclin-dependent kinases is associated with cellular transformation. Genes Dev 1993, 7:1572-1583 [DOI] [PubMed] [Google Scholar]
- 6.Khatib ZA, Matsushime H, Valentine M, Shapiro DN, Sherr CJ, Look AT: Coamplification of the CDK4 gene with MDM2 and GLI in human sarcomas. Cancer Res 1993, 53:5535-5541 [PubMed] [Google Scholar]
- 7.Collins VP: Gene amplification in human gliomas. Glia 1995, 15:289-296 [DOI] [PubMed] [Google Scholar]
- 8.Reifenberger G, Reifenberger J, Ichimura K, Meltzer PS, Collins VP: Amplification of multiple genes from chromosomal region 12q13–14 in human malignant gliomas: preliminary mapping of the amplicons shows preferential involvement of CDK4, SAS, and MDM2. Cancer Res 1994, 54:4299-4303 [PubMed] [Google Scholar]
- 9.Wölfel T, Hauer M, Schneider J, Serrano M, Wàlfel C, Klehmann-Hieb E, De Plaen E, Hankeln T, Meyer zum Buschenfelde KH, Beach DA: p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science 1995, 269:1281-1284 [DOI] [PubMed] [Google Scholar]
- 10.Buckley MF, Sweeney KJ, Hamilton JA, Sini RL, Manning DL, Nicholson RI, deFazio A, Watts CK, Musgrove EA, Sutherland RL: Expression and amplification of cyclin genes in human breast cancer. Oncogene 1993, 8:2127-2133 [PubMed] [Google Scholar]
- 11.Gillett C, Fantl V, Smith R, Fisher C, Bartek J, Dickson C, Barnes D, Peters G: Amplification and overexpression of cyclin D1 in breast cancer detected by immunohistochemical staining. Cancer Res 1994, 54:1812-1817 [PubMed] [Google Scholar]
- 12.Lee EY, To H, Shew JY, Bookstein R, Scully P, Lee WH: Inactivation of the retinoblastoma susceptibility gene in human breast cancers. Science 1988, 241:214-221 [DOI] [PubMed] [Google Scholar]
- 13.Lungberg C, Skoog L, Cavenee WK, Nordenskjöld M: Loss of heterozygosity in human ductal breast cancer indicates a recessive mutation on chromosome 13. Proc Natl Acad Sci USA 1987, 84:2372-2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herman JG, Merlo A, Mao L, Lapidus RG, Issa JP, Davidson NE, Sidransky D, Baylin SB: Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res 1995, 55:4525-4530 [PubMed] [Google Scholar]
- 15.Geleick D, Müller H, Matter A, Torhorst J, Regenass U: Cytogenetics of breast cancer. Cancer Genet Cytogenet 1990, 46:217-229 [DOI] [PubMed] [Google Scholar]
- 16.Courjal F, Cuny M, Simony-Lafontaine J, Louason G, Speiser P, Zeillinger R, Rodriguez C, Theillet C: Mapping of DNA amplifications at 15 chromosomal localizations in 1875 breast tumors: definition of phenotypic groups. Cancer Res 1997, 57:4360-4367 [PubMed] [Google Scholar]
- 17.Sgambato A, Han EK, Zhang YJ, Moon RC, Santella RM, Weinstein IB: Deregulated expression of cyclin D1 and other cell cycle-related genes in carcinogen-induced rat mammary tumors. Carcinogenesis 1995, 16:2193-2198 [DOI] [PubMed] [Google Scholar]
- 18.An HX, Niederacher D, Beckmann MW, Göhring UJ, Scharl A, Picard F, van Roeyen C, Schnürch H-G, Bender HG: ERBB2 gene amplification detected by fluorescent differential polymerase chain reaction in paraffin-embedded breast carcinoma tissues. Int J Cancer 1995, 64:291-297 [DOI] [PubMed] [Google Scholar]
- 19.Elston CW, Ellis IO: Pathological prognostic factors in breast cancer: I. the value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 1991, 19:403-410 [DOI] [PubMed] [Google Scholar]
- 20.Beckmann MW, Niederacher D, Massenkeil G, Tutschek B, Beckmann A, Schenko G, Schnürch H-G, Bender HG: Expression analyses of epidermal growth factor receptor and HER-2/neu: no advantage of prediction of recurrence or survival in breast cancer patients. Oncology 1996, 53:441-447 [DOI] [PubMed] [Google Scholar]
- 21.Hsu SM, Raine L, Fanger H: Use of avidin-biotin-peroxidase complex (ABC) in immunoperoxidase techniques: a comparison between ABC and unlabeled antibody (PAP) procedures. J Histochem Cytochem 1981, 29:577-580 [DOI] [PubMed] [Google Scholar]
- 22.Volm M, Koomägi R, Rittgen W: Clinical implications of cyclins, cyclin-dependent kinase, RB and E2F1 in squamous-cell lung carcinoma. Int J Cancer 1998, 79:294-299 [DOI] [PubMed] [Google Scholar]
- 23.Cawkwell L, Lewis FA, Quirke P: Frequency of allele loss of DCC, p53, RB1, WT1, NF1, NM23 and APC/MCC in colorectal cancer assayed by fluorescent multiplex polymerase chain reaction. Br J Cancer 1993, 70:813-818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niederacher D, Picard F, van Roeyen C, An HX, Bender HG, Beckmann MW: Pattern of allelic loss on chromosome 17 in sporadic breast carcinomas detected by fluorescent labelled microsatellite analysis. Genes Chromosome Cancer 1997, 18:181-192 [DOI] [PubMed] [Google Scholar]
- 25.Ito Y, Kobayashi T, Takeda T, Nakano Y, Tamaki Y, Komoike Y, Wakasugi E, Shin E, Takatsuka Y, Kikkawa N, Matsuura N, Monden M: Expression of p16 and cyclin-dependent kinase 4 proteins in primary breast carcinomas. Oncology 1997, 54:508-515 [DOI] [PubMed] [Google Scholar]
- 26.Sasano H, Frost AR, Saitoh R, Taniyama Y, Nagura H, Matsunaga G, Takehana K, Kimura M, Silverberg SG: Immunolocalisation of cyclin D and E and cyclin dependent kinase (cdk) 2 and 4 in human breast carcinoma. Anticancer Res 1997, 17:3685-3690 [PubMed] [Google Scholar]
- 27.He J, Allen JR, Collins VP, Allalunis-Turner MJ, Godbout R, Day RS, III, James CD: CDK4 amplification is an alternative mechanism to p16 gene homozygous deletion in glioma cell lines. Cancer Res 1994, 54:5804-5807 [PubMed] [Google Scholar]
- 28.Rollbrocker B, Waha A, Louis DN, Wiestler OD, von Deimling A: Amplification of the cyclin-dependent kinase 4 (CDK4) gene is associated with high cdk4 protein levels in glioblastoma multiforme. Acta Neuropathol 1996, 92:70-74 [DOI] [PubMed] [Google Scholar]
- 29.Zhang T, Nanney LB, Luongo C, Lamps L, Heppner KJ, DuBois RN, Beauchamp RD: Concurrent overexpression of cyclin D1 and cyclin-dependent kinase 4 (Cdk4) in intestinal adenomas from multiple intestinal neoplasia (Min) mice and human familial adenomatous polyposis patients. Cancer Res 1997, 57:169-175 [PubMed] [Google Scholar]
- 30.Serrano M, Hannon GJ, Beach DA: A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D1/CDK4. Nature 1993, 366:704-707 [DOI] [PubMed] [Google Scholar]
- 31.Maelandsmo GM, Florenes VA, Hovig E, Oyjord T, Engebraaten O, Holm R, Borresen AL, Fodstad O: Involvement of the pRb/p16/cdk4/cyclin D1 pathway in the tumorigenesis of sporadic malignant melanomas. Br J Cancer 1996, 73:909-916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmidt EE, Ichimura K, Reifenberger G, Collins VP: CDKN2 (p16/MTS1) gene deletion or CDK4 amplification occurs in the majority of glioblastomas. Cancer Res 1994, 54:6321-6324 [PubMed] [Google Scholar]
- 33.Otterson GA, Kratzke RA, Coxon A, Kim YW, Kaye FJ: Absence of p16INK4 protein is restricted to the subset of lung cancer lines that retains wildtype RB. Oncogene 1994, 9:3375-3378 [PubMed] [Google Scholar]
- 34.Ueki K, Ono Y, Henson JW, Efird JT, von Deimling A, Louis DN: CDKN2/p16 or RB alterations occur in the majority of glioblastomas and are inversely correlated. Cancer Res 1996, 56:150-153 [PubMed] [Google Scholar]
- 35.Jiang W, Zhang YJ, Kahn SM, Hollstein MC, Santella RM, Lu SH, Harris CC, Montesano R, Weinstein IB: Altered expression of the cyclin D1 and retinoblastoma genes in human esophageal cancer. Proc Natl Acad Sci USA 1993, 90:9026-9030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu L, Sgroi D, Sterner CJ, Beauchamp RL, Pinney DM, Keel S, Ueki K, Rutter JL, Buckler AJ, Louis DN, Gusella JF, Ramesh V: Mutational analysis of CDKN2 (MTS1/p16ink4) in human breast carcinomas. Cancer Res 1994, 54:5262-5264 [PubMed] [Google Scholar]