Abstract
We have developed a simple and highly efficient method to disrupt chromosomal genes in Escherichia coli in which PCR primers provide the homology to the targeted gene(s). In this procedure, recombination requires the phage λ Red recombinase, which is synthesized under the control of an inducible promoter on an easily curable, low copy number plasmid. To demonstrate the utility of this approach, we generated PCR products by using primers with 36- to 50-nt extensions that are homologous to regions adjacent to the gene to be inactivated and template plasmids carrying antibiotic resistance genes that are flanked by FRT (FLP recognition target) sites. By using the respective PCR products, we made 13 different disruptions of chromosomal genes. Mutants of the arcB, cyaA, lacZYA, ompR-envZ, phnR, pstB, pstCA, pstS, pstSCAB-phoU, recA, and torSTRCAD genes or operons were isolated as antibiotic-resistant colonies after the introduction into bacteria carrying a Red expression plasmid of synthetic (PCR-generated) DNA. The resistance genes were then eliminated by using a helper plasmid encoding the FLP recombinase which is also easily curable. This procedure should be widely useful, especially in genome analysis of E. coli and other bacteria because the procedure can be done in wild-type cells.
Keywords: bacterial genomics, FLP recombinase, FRT sites, Red recombinase
The availability of complete bacterial genome sequences has provided a wealth of information on the molecular structure and organization of a myriad of genes and ORFs whose functions are poorly understood. A systematic mutational analysis of genes in their normal location can provide significant insight into their function. Although a number of general allele replacement methods (1–7) can be used to inactivate bacterial chromosomal genes, these all require creating the gene disruption on a suitable plasmid before recombining it onto the chromosome. In contrast, genes can be directly disrupted in Saccharomyces cerevisiae by transformation with PCR fragments encoding a selectable marker and having only 35 nt of flanking DNA homologous to the chromosome (8). This PCR-mediated gene replacement method has greatly facilitated the generation of specific mutants in the functional analysis of the yeast genome; it relies on the high efficiency of mitotic recombination in yeast (9). Directed disruption of chromosomal genes can also be done in Candida albicans by using similar PCR fragments with 50- to 60-nt homology extensions (10).
In contrast to yeast and a few naturally competent bacteria, most bacteria are not readily transformable with linear DNA. One reason Escherichia coli is not so transformable is because of the presence of intracellular exonucleases that degrade linear DNA (11). However, recombination-proficient mutants lacking exonuclease V of the RecBCD recombination complex are transformable with linear DNA (12). Recombination can occur in recB or recC mutants carrying a suppressor (sbcA or sbcB) mutation that activates an alternative recombination pathway; sbcA activates the RecET recombinase of the Rac prophage, whereas sbcB enhances recombination by the RecF pathway (13). Such recBC sbcB mutants have been especially useful for recombining in vitro constructed mutations onto the E. coli chromosome by using linear DNA (14). The discovery that recD mutants are recombinase proficient but lack exonuclease V (15, 16) has led to using singly mutated recD derivatives of E. coli (1) in similar gene disruption experiments.
It has been known for a long time that many bacteriophages encode their own homologous recombination systems (17). It has also recently been shown that the λ Red (γ, β, exo) function promotes a greatly enhanced rate of recombination over that exhibited by recBC sbcB or recD mutants when using linear DNA (18). Yet this system has produced no chromosomal gene disruptions when using PCR fragments with short homology extensions (unpublished data). A system has been developed that uses the RecET recombinase to disrupt plasmid-borne genes with such fragments (19); it has also been used to make a single chromosomal deletion, but in that instance very long (138-nt) primers were used.
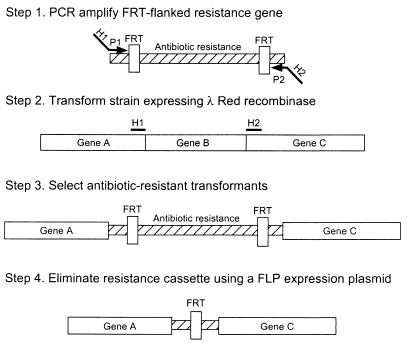
Here we describe a procedure based on the Red system that has allowed us to make more than 40 different disruptions on the E. coli chromosome without a single failure. The basic strategy is to replace a chromosomal sequence (e.g., gene B in Fig. 1) with a selectable antibiotic resistance gene that is generated by PCR by using primers with 36-nt homology extensions (H1 and H2). This is accomplished by Red-mediated recombination in these flanking homologies. After selection, the resistance gene can also be eliminated by using a helper plasmid expressing the FLP recombinase, which acts on the directly repeated FRT (FLP recognition target) sites flanking the resistance gene. The Red and FLP helper plasmids can be simply cured by growth at 37°C because they are temperature-sensitive replicons.
Figure 1.
A simple gene disruption strategy. H1 and H2 refer to the homology extensions or regions. P1 and P2 refer to priming sites.
Materials and Methods
Media, Chemicals and Other Reagents.
Ampicillin-, chloramphenicol- (CmR), and kanamycin-resistant (KmR) transformants were selected on tryptone-yeast extract agar medium (20) containing the respective antibiotic at 100, 25, and 25 μg/ml. 5-Bromo-4-chloro-3-indolyl β-d-galactopyranoside (Bachem) or 5-bromo-4-chloro-3-indolyl-phosphate-p-toluidine (Bachem) were used at 40 μg/ml to detect β-galactosidase or bacterial alkaline phosphatase (Bap) activity, respectively. Bap constitutive mutants were streaked along with controls on media without an indicator dye and verified by dripping onto the colonies a solution of 0.4% p-nitrophenyl-phosphate (Sigma) in 1 M Tris⋅HCl, pH 8 (21). A total of 1 mM l-arabinose or isopropyl-β-d-galactopyranoside (Sigma) was used for induction. SOB and SOC media were prepared as described elsewhere (22). Oligonucleotides were from IDT (Coralville, IA). Enzymes were from New England Biolabs unless indicated otherwise. Taq polymerase was used in all PCR tests. Taq and Pfu (Stratagene) polymerases were mixed 10:1 and used per Taq instructions to generate DNAs for cloning and mutagenesis. Qiagen products (Hilden, Germany) were used to isolate plasmid DNAs, gel-purify fragments, or purify PCR products.
Bacteria.
BW25113 (lacIq rrnBT14 ΔlacZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78), BW25993 (lacIq hsdR514 ΔaraBADAH33 ΔrhaBADLD78), and BW25141 (lacIq rrnBT14 ΔlacZWJ16 ΔphoBR580 hsdR514 ΔaraBADAH33 ΔrhaBADLD78 galU95 endABT333 uidA(ΔMluI)∷pir+ recA1) are derivatives of the F-, λ-, E. coli K-12 strain BD792 [CGSC6159 (23)] and have no other known mutations. The ΔaraBADAH33, ΔphoBR580, ΔrhaBADLD78, uidA(ΔMluI)∷pir+, and linked rrnBT14 ΔlacZWJ16 mutations were recombined onto the chromosome by allele replacement (3, 24, 25). Conditional replicative oriRγ plasmids were maintained in the pir+ host BW25141 or similar ones (25). Several mutations were transferred by using P1kc transduction (24). BT340 [DH5α carrying pCP20 (26)], MG1655 [CGSC6300 (27)], and the endABT333, galU95, and hsdR514 alleles have been described (25).
Plasmids.
pANTSγ (28) and pINT-ts (29) were from M. Koob (University of Wisconsin, Madison), pBAD18 (30) from L. Guzman (Harvard Medical School, Boston), pCP15 (26) from W. Wackernagel (Universitat Oldenburg, Oldenburg, Germany), pSC140 from S. Chiang (Harvard Medical School) and S. Chiang and J. J. Mekalanos, personal communication; and pTP223 from K. Murphy (18). The Red helper plasmids (see Fig. 2) are derivatives of pINT-ts which contain araC-ParaB and γ β exo (without or with tL3) DNA fragments that were PCR-generated by using pBAD18 and λ DNA as template, respectively. The template plasmids are derivatives of pANTSγ that contain an FRT-flanked kanamycin resistance (kan) or chloramphenicol resistance (cat) gene from pCP15 or pSC140, respectively. Synthetic DNAs containing FRT sites without or with a juxtaposed ribosome-binding site were generated by PCR. Details will be reported elsewhere. All relevant segments generated by PCR were sequenced on both strands in the Microbiology and Molecular Genetics Core Facility at Harvard Medical School.
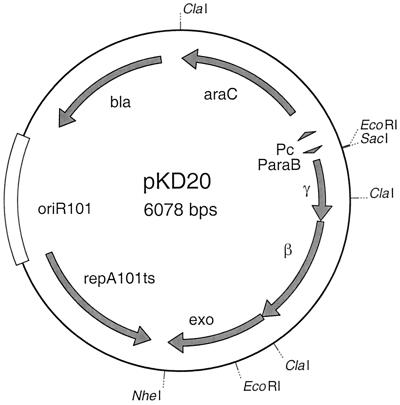
Figure 2.
Red recombinase expression plasmids. pKD20 and pKD46 (not shown) include 1,894 nt (31348–33241) and 2,154 nt (31088–33241) of phage λ (GenBank accession no. J02459), respectively.
Gene Disruption.
Transformants carrying a Red helper plasmid were grown in 5-ml SOB cultures with ampicillin and l-arabinose at 30°C to an OD600 of ≈0.6 and then made electrocompetent by concentrating 100-fold and washing three times with ice-cold 10% glycerol. PCR products were gel-purified, digested with DpnI, repurified, and suspended in elution buffer (10 mM Tris, pH 8.0). Electroporation was done by using a Cell-Porator with a voltage booster and 0.15-cm chambers according to the manufacturer's instructions (GIBCO/BRL) by using 25 μl of cells and 10–100 ng of PCR product. Shocked cells were added to 1-ml SOC, incubated 1 h at 37°C, and then one-half was spread onto agar to select CmR or KmR transformants. If none grew within 24 h, the remainder was spread after standing overnight at room temperature. After primary selection, mutants were maintained on medium without an antibiotic. They were colony-purified once nonselectively at 37°C and then tested for ampicillin sensitivity to test for loss of the helper plasmid. If it was not lost, then a few were colony-purified once at 43°C and similarly tested.
PCR Verification.
Three PCRs were used to show that all mutants have the correct structure. A freshly isolated colony was suspended in 20-μl water with a plastic tip from which 5-μl portions were used in separate 20-μl PCRs following a 2-min preincubation, “hot start,” at 95°C. Common test primers included: c1 (TTATACGCAAGGCGACAAGG) and c2 (GATCTTCCGTCACAGGTAGG) for cat, and k1 (CAGTCATAGCCGAATAGCCT), k2 (CGGTGCCCTGAATGAACTGC), and kt (CGGCCACAGTCGATGAATCC) for kan. Two reactions were done by using nearby locus-specific primers with the respective common test primer (c1, c2, k1, or k2) to test for both new junction fragments. A third reaction was carried out with the flanking locus-specific primers to verify simultaneous loss of the parental (nonmutant) fragment and gain of the new mutant-specific fragment. The latter was repeated after elimination of the resistance gene. A fourth reaction was sometimes done with primers k2 and kt to test for a 471-nt kan fragment. Control colonies were always tested side-by-side.
Eliminating Antibiotic Resistance Gene.
pCP20 is an ampicillin and CmR plasmid that shows temperature-sensitive replication and thermal induction of FLP synthesis (26). CmR and KmR mutants were transformed with pCP20, and ampicillin-resistant transformants were selected at 30°C, after which a few were colony-purified once nonselectively at 43°C and then tested for loss of all antibiotic resistances. The majority lost the FRT-flanked resistance gene and the FLP helper plasmid simultaneously.
Nomenclature.
One allele number signifies all mutations generated with a particular primer pair, as these are identical except for the inserted DNA. An allele number followed by a double colon and a descriptor for the inserted sequence indicates those with an insertion. For example, DE(lacZYA)514∷cat and DE(lacZYA)514∷kan refer to mutations made with the same primers and pKD3 (cat) or pKD4 (kan) as template, respectively. After eviction of the resistance gene, the mutation(s) is simply called DE(lacZYA)514, as these are identical regardless of history.
Results
Description of the Red Disruption System.
The Red system includes three genes: γ, β, and exo, whose products are called Gam, Bet, and Exo, respectively (18). Gam inhibits the host RecBCD exonuclease V so that Bet and Exo can gain access to DNA ends to promote recombination. In preliminary studies we attempted to make chromosomal mutations by using the multicopy Red plasmid pTP223 and PCR products with short homology extensions, but were unsuccessful. We therefore made several low copy plasmids such as pKD20 (Fig. 2) encoding the Red recombinase. pKD20 has an optimized ribosome-binding site for efficient translation of γ and expresses γ, β, and exo from the arabinose-inducible ParaB promoter. It is also a temperature-sensitive replicon to allow for its easy elimination.
We carried out several preliminary gene disruption experiments by using pKD20 and similar plasmids, essentially as described in Materials and Methods, except with different template plasmids. In these experiments, we found a high proportion of antibiotic-resistant transformants without gene disruptions. Instead, many carried what appeared to be new plasmids that apparently escaped DpnI digestion [which was done to eliminate methylated (unamplified) template DNA (19)] by being synthesized in an aberrant PCR. These apparently resulted from our use of plasmids as templates (data not shown). To circumvent their occurrence, we constructed new template plasmids that are conditional (oriRγ) replicons that require the trans-acting Π protein (the pir gene product) for replication. They also have resistance genes that are flanked by directly repeated FRT sites. By using pKD20 and these template plasmids (Fig. 3), we made several chromosomal gene disruptions as described below.
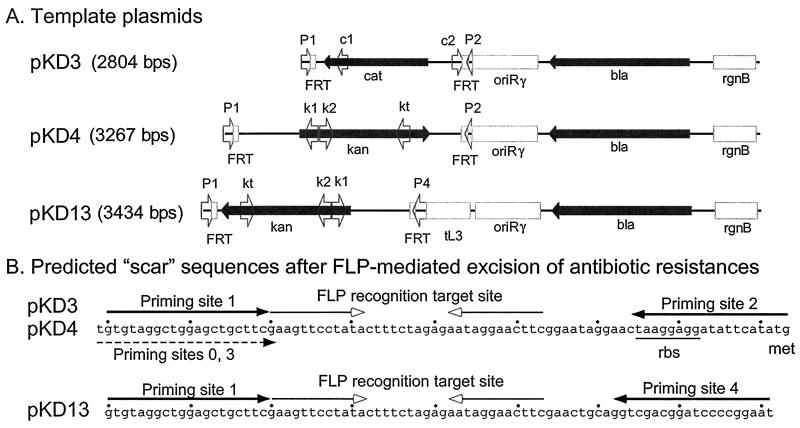
Figure 3.
Template plasmids. (A) Linear representations of the template plasmids. Arrowheads show locations and orientations of priming sites. P1: priming sites 0, 1, and 3; P2: priming site 2; P4: priming site 4. c1, c2, k1, k2, and kt: common test primers. (B) Sequences remaining after FLP-mediated excision of the antibiotic resistance genes. Priming sites 0, 1, and 3 begin at nucleotides 1, 2, and 3, respectively. Priming sites 2 and 4 begin at the left or right ends, as shown. Arrows with open arrowheads show the nearly perfect FRT site inverted repeats. The ribosome binding site (rbs) and methionine (met) start codon are marked.
Our standard protocol is illustrated in Fig. 1. PCR products were generated by using several pairs of 56- to 70-nt-long primers that included 36- to 50-nt homology extensions and 20-nt priming sequences for pKD3, pKD4, or pKD13 as template (Table 1). The respective 1.1-, 1.6-, or 1.4-kbp PCR products were purified, treated with DpnI, and then transformed into bacteria carrying the Red helper plasmid as described in Materials and Methods. We routinely obtained tens or hundreds of CmR or KmR transformants of BW25113 (or similar strains) carrying pKD20. None was found in absence of arabinose, thus showing a requirement for the Red recombinase. Fewer were usually found for MG1655 carrying pKD20. Differences are likely due to BW25113, unlike MG1655, being ΔaraBAD, hsdR, or both. Because similar numbers of transformants were found when MG1655 carrying pKD20 was grown with 10 mM arabinose, hsdR+ is not a major problem despite the presence of E. coli K12 restriction recognition sites (31) within the FRT-flanked resistance cassettes. Restriction would have a lesser effect if single-stranded DNA were the primary recombination substrate under these conditions. We found similar numbers of recombinants when using 36- to 50-nt homology extensions; however, this may have resulted from our use of unpurified (and unmodified) primers in these experiments as longer primers are expected to be less pure. Importantly, when using PCR products targeted to the lac and pst genes, all transformants displayed the expected Lac− or Bap constitutive phenotype, respectively. The resistance genes were eliminated by using a FLP helper plasmid.
Table 1.
Allele designations of chromosomal gene disruptions
| Mutation* | Template(s) | Homology extensions† | Priming sites‡ |
|---|---|---|---|
| DE(lacZYA)514 | pKD3, pKD4 | 36 nt; H1: 365662C; H2: 360842 | P0; P2 |
| DE(ompR-envZ)516 | pKD13 | 50 nt; H1: 3534269C; H2: 3532141 | P1; P4 |
| DE(torSTRCAD)517 | pKD13 | 50 nt; H1: 1052654; H2: 1061625C | P1; P4 |
| ΔarcB40 | pKD3 | 50 nt; H1: 3350706C; H2: 3348269 | P1; P2 |
| ΔcyaA1403 | pKD13 | 38 nt; H1: 3988674; H2: 3991330C | P1; P4 |
| ΔphnR171 | pKD4 | 36 nt; H1: 2242; H2: 3016C | P3; P2 |
| ΔpstB608 | pKD4 | 44 nt; H1: 3906010C; H2: 3905260 | P1; P2 |
| ΔpstCA607 | pKD4 | 44 nt; H1: 3908039C; H2: 3905991 | P1; P2 |
| ΔpstS605 | pKD3, pKD4 | 36 nt; H1: 3909143C; H2: 3908059 | P3; P2 |
| ΔpstSCAB-phoU606 | pKD13 | 36 nt; H1: 3909339C; H2: 3904418 | P1; P4 |
| ΔrecA635 | pKD13 | 50 nt; H1: 2821868C; H2: 2820743 | P1; P4 |
All except two were made in pKD20 transformants of BW25113. DE(lacZYA)514 mutants were made in the isogenic Lac+ strain BW25993; phnR was disrupted in three similar strains carrying the Salmonella typhimurium LT2 phosphonatase gene cluster (32) on the chromosome. Some were also made in MG1655 transformants.
Extension lengths are given first. Numerals identify the 3′ nucleotide of the extension in the E. coli genome sequence [(ref. 33; GenBank accession no. U00096) or the S. typhimurium phnR entry (GenBank accession no. U69493)]. C, complement; H1, homology 1; H2, homology 2.
One primer had the H1 extension and the 3′ sequence for priming site 0 (P0), 1 (P1), or 3 (P3). The other had the H2 extension and the 3′ sequence for the complement of priming site 2 (P2) or 4 (P4).
Disruptions of the lac Operon.
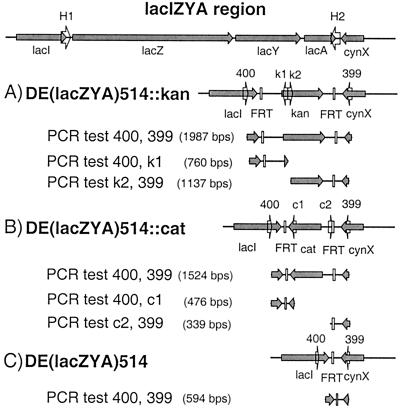
We used both pKD3 and pKD4 as templates to delete precisely a 5,178-nt segment of the lacZYA operon with the same 56-nt primers (Table 1). DE(lacZYA)514 leaves lacI intact, removes the lac promoter, lacZ, lacY, and lacA entirely, and leaves intact the terminator for the downstream, oppositely oriented, cynX (Fig. 4). Two or three representative mutants were characterized from each of several experiments in which CmR or KmR colonies were isolated after transformation of different hosts with PCR fragments that were generated independently. PCR tests using locus-specific primers and cat- or kan-specific primers revealed that all had new junction and locus-specific fragments of the expected sizes. On elimination of the resistance gene, the resultant mutants also gave the expected size new fragment in a PCR test with locus-specific primers. We also showed that, as expected, the gene disruptions can be transferred into new strains by P1 transduction. The resultant CmR and KmR transductants as well as their antibiotic-sensitive derivatives gave the expected fragments in similar PCR tests. Thus, these mutants have the correct structures.
Figure 4.
Structures and PCR verification of DE(lacZYA)514 mutations. Top line shows the region near the wild-type lac operon. (A—C) Structures of the DE(lacZYA)514 alleles generated using pKD4 as template, pKD3 as template, and after elimination of the resistance genes, respectively. Numerals above the structures refer to locus-specific primers. The predicted PCR test products are shown. See Fig. 3 for other notations.
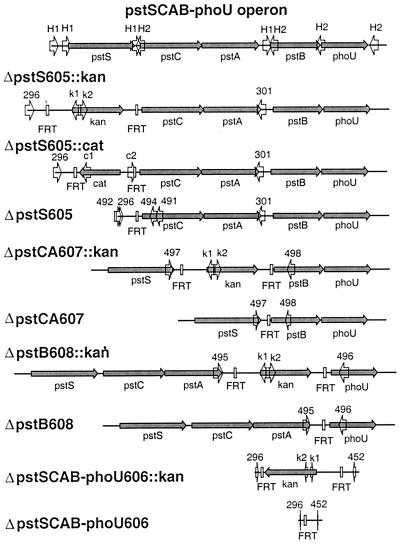
Disruptions of the pstSCAB-phoU Operon.
We made four different disruptions of the pstSCAB-phoU operon (Table 1). One precisely removes pstS, one removes pstCA, one removes pstB, and a fourth removes the entire operon including its promoter (Fig. 5). Mutations of this operon result in constitutive expression of the phosphate (Pho) regulon (34). After elimination of the resistance genes, we showed the pstS and pstB mutations are nonpolar, as such mutants were complemented by plasmids carrying pstS+ or pstB+ alone, respectively. The pstCA mutant was not tested because of the lack of an appropriate complementing plasmid. All mutations were verified by using locus-specific and common test primers (Fig. 4) as described above. The new junction fragments from representative ΔpstS605, ΔpstCA607, and ΔpstB608 mutants were also directly sequenced following PCR amplification. Although most had the exact predicted sequence, occasional mutants lacked 1 nt within the region for one of the PCR primers. The same synthetic DNA also always gave recombinants with the correct sequence.
Figure 5.
Structures of pstSCAB-phoU operon mutations. Top line shows the wild-type pstSCAB-phoU operon. Lower lines show the gene disruptions. The new junction fragments for the ΔpstS605, ΔpstCA607, and ΔpstB608 mutations were amplified by using primer 492 with 491, 497 with 498, and 495 with 496, respectively, and sequenced. See Fig. 4 for other notations.
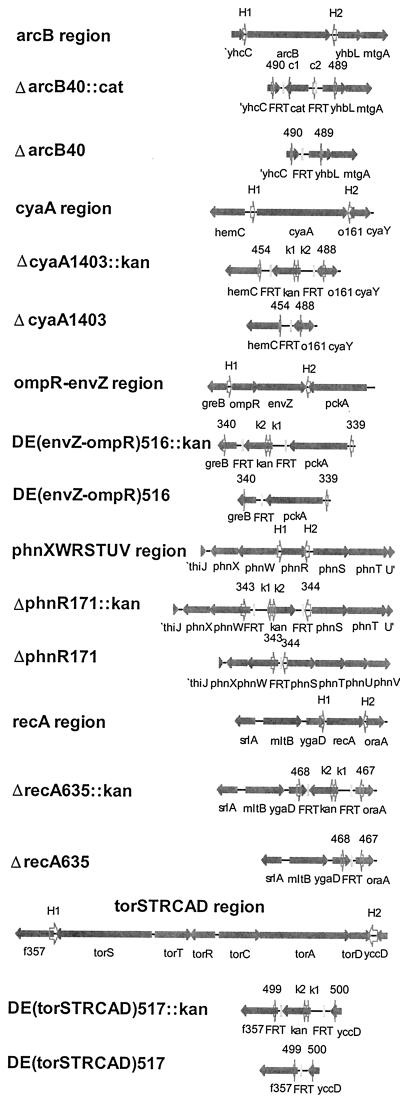
More Gene Disruptions.
We targeted disruptions to six additional chromosomal loci (Fig. 6; Table 1). In all but one case, all of the CmR or KmR transformants tested had the predicted structure using similar PCR tests. The one exception concerned the cya (adenylate cyclase) locus. In this case, all KmR transformants grew poorly, as expected for cya mutants. Yet subsequent tests revealed many to be spontaneous KmR mutants, which can also arise and often grow slowly (35, 36). Importantly, all transformants shown to carry kan were also shown to be correct by PCR as well as by phenotype.
Figure 6.
Structures of other gene disruptions. See Fig. 4 for notations.
Discussion
Our method for disrupting E. coli chromosomal genes is analogous to one that has been used for many years in yeast (8). It is based on results of K. Murphy (18) who provided us with his materials before publication. Because multicopy plasmids might interfere with recombination by acting as competitive inhibitors (18), we cloned the Red genes (γ, β, and exo) into a low copy number plasmid. We used a vector which shows temperature-sensitive replication (37) to permit its easy curing from the resultant mutants. The plasmids pKD20 and pKD46 (Fig. 2) express the Red system under control of a well-regulated promoter to avoid unwanted recombinational events under noninducing conditions. They differ in that the latter has the native tL3 terminator downstream of exo. Although all recombinants described here were made by using pKD20, we now use pKD46 instead because we recently discovered that pKD46 yields a greatly enhanced number of recombinants. The reason is unknown. Curiously, tL3 encodes a small ORF that may be responsible for a host inhibition (Hin) phenotype, for which the basis is unknown (38).
We also constructed special template plasmids. These have the conditional oriRγ origin to reduce a background number of resistant colonies carrying template-like plasmids that can predominate (at least when small circular plasmids are used as templates). When making gene disruptions using the templates and priming sites in Fig. 3, elimination of the antibiotic resistance gene leaves behind an 82- to 85-nt scar in place of the disrupted gene(s). pKD3 and pKD4 are identical except for the region between the FRT sites, so they create an identical scar that has stop codons in all six reading frames. As drawn, this scar has an idealized ribosome binding site and start codon for downstream (rightward) gene expression (Fig. 3). When using these as templates, gene disruptions within the pst operon were also nonpolar. They can therefore be used to create nonpolar gene deletions within operons or deletions that remove only an N-terminal coding region and express a C-terminal protein domain. In contrast, the pKD13 scar has no translation signals. Therefore, we have used it primarily to disrupt single genes or entire operons. Its scar has stop codons in all three forward but in only one reverse reading frame. Because of the presence of two ORFs in the orientation opposite that of Fig. 3, pKD13 might also be useful for creating in-frame deletions in which the scar encodes a new 27-residue internal peptide(s).
These scars could be problematic under certain conditions. Because of the limited homology that is required for gene disruption when using the Red system, a new PCR fragment can recombine at the new targeted gene or at the scar of an earlier gene disruption. To avoid such occurrences, we have made single gene disruptions in wild-type hosts and constructed multiple mutants in standard P1 crosses. However, other chromosomal rearrangements might result from FLP-promoted recombination events between FRT sites at different loci. Although we have seen no such events, we routinely rechecked all gene disruption sites by PCR on elimination of a resistance gene from a new locus in such multiple mutants.
All gene disruption mutants were verified by a PCR strategy which tested for the presence of new locus- and junction-specific fragments of predicted sizes. As further verification, the locus-specific fragments from selected mutants were PCR-amplified and sequenced after elimination of the resistance gene. Although this revealed that the majority were correct, about 10% had 1-nt deletions. The incorrect ones probably resulted from PCR products generated from a primer lacking an internal base. Oligonucleotides with internal 1-nt deletions arise from chemical synthesis and are difficult to remove by conventional purification methods (39, 40), especially when using 60-nt or longer primers. All 1-nt deletions occurred at or very near the junction of a priming site and homology extension. This is expected as PCR primers with 1-nt deletions elsewhere are likely to prime less well or be incorporated into PCR products that recombine less efficiently. Accordingly, the junction fragments of all mutants whose actual sequence is critical for their further study are routinely sequenced to avoid mutants with 1-nt deletions.
In summary, we have isolated chromosomal mutants with 13 different gene disruptions by direct transformation of E. coli carrying a Red helper plasmid with PCR products having short homology extensions for the targeted locus. This method should be widely useful. It should also be rather straightforward to extend its use to other bacteria. To adapt it to more distantly related bacteria, it may be necessary to express the Red system under different control or from another low copy number vector. In some cases, it may be advantageous to substitute an analogous recombinase from a phage(s) specific for a particular group of bacteria.
Acknowledgments
This manuscript is dedicated to the memory of H. E. Umbarger who died on November 15, 1999. We thank individuals cited in the text for samples; Don Court, Jean-Marc Ghigo, and Kenan Murphy for communicating unpublished results; Jill Hutchcroft and Irwin Tessman for critically reading the manuscript; and lab members for helpful discussions. This research was supported by Award MCB-9730034 from the National Science Foundation.
Abbreviations
- Bap
bacterial alkaline phosphatase
- CmR
chloramphenicol-resistant
- FRT
FLP recognition target
- KmR
kanamycin-resistant
- kan
kanamycin resistance gene
- cat
chloramphenicol resistance gene
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.120163297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.120163297
References
- 1.Russell C B, Thaler D S, Dahlquist F W. J Bacteriol. 1989;171:2609–2613. doi: 10.1128/jb.171.5.2609-2613.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamilton C M, Aldea M, Washburn B K, Babitzke P, Kushner S R. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Metcalf W W, Jiang W, Daniels L L, Kim S-K, Haldimann A, Wanner B L. Plasmid. 1996;35:1–13. doi: 10.1006/plas.1996.0001. [DOI] [PubMed] [Google Scholar]
- 4.Link A J, Phillips D, Church G M. J Bacteriol. 1997;179:6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dabert P, Smith G R. Genetics. 1997;145:877–889. doi: 10.1093/genetics/145.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kato C, Ohmiya R, Mizuno T. Biosci Biotechnol Biochem. 1998;62:1826–1829. doi: 10.1271/bbb.62.1826. [DOI] [PubMed] [Google Scholar]
- 7.Pósfai G, Kolisnychenko V, Bereczki Z, Blattner F R. Nucleic Acids Res. 1999;27:4409–4415. doi: 10.1093/nar/27.22.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oliver S G, Winson M K, Kell D B, Baganz F. Trends Biotechnol. 1998;16:373–378. doi: 10.1016/s0167-7799(98)01214-1. [DOI] [PubMed] [Google Scholar]
- 10.Wilson R B, Davis D, Mitchell A P. J Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lorenz M G, Wackernagel W. Microbiol Rev. 1994;58:563–602. doi: 10.1128/mr.58.3.563-602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cosloy S D, Oishi M. Proc Natl Acad Sci USA. 1973;70:84–87. doi: 10.1073/pnas.70.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark A J, Sandler S J. Crit Rev Microbiol. 1994;20:125–142. doi: 10.3109/10408419409113552. [DOI] [PubMed] [Google Scholar]
- 14.Winans S C, Elledge S J, Krueger J H, Walker G C. J Bacteriol. 1985;161:1219–1221. doi: 10.1128/jb.161.3.1219-1221.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amundsen S K, Taylor A F, Chaudhury A M, Smith G R. Proc Natl Acad Sci USA. 1986;83:5558–5562. doi: 10.1073/pnas.83.15.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biek D P, Cohen S N. J Bacteriol. 1986;167:594–603. doi: 10.1128/jb.167.2.594-603.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith G R. Microbiol Rev. 1988;52:1–28. doi: 10.1128/mr.52.1.1-28.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy K C. J Bacteriol. 1998;180:2063–2071. doi: 10.1128/jb.180.8.2063-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y M, Buchholz F, Muyrers J P P, Stewart A F. Nat Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 20.Wanner B L. In: Methods in Molecular Genetics. Adolph K W, editor. Vol. 3. Orlando, FL: Academic; 1994. pp. 291–310. [Google Scholar]
- 21.Wanner B L, Latterell P. Genetics. 1980;96:242–266. doi: 10.1093/genetics/96.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanahan D. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 23.Wanner B L. J Mol Biol. 1983;166:283–308. doi: 10.1016/s0022-2836(83)80086-2. [DOI] [PubMed] [Google Scholar]
- 24.Haldimann A, Fisher S L, Daniels L L, Walsh C T, Wanner B L. J Bacteriol. 1997;179:5903–5913. doi: 10.1128/jb.179.18.5903-5913.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haldimann A, Daniels L L, Wanner B L. J Bacteriol. 1998;180:1277–1286. doi: 10.1128/jb.180.5.1277-1286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cherepanov P P, Wackernagel W. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 27.Metcalf W W, Steed P M, Wanner B L. J Bacteriol. 1990;172:3191–3200. doi: 10.1128/jb.172.6.3191-3200.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pósfai G, Koob M, Hradecná Z, Hasan N, Filutowicz M, Szybalski W. Nucleic Acids Res. 1994;22:2392–2398. doi: 10.1093/nar/22.12.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hasan N, Koob M, Szybalski W. Gene. 1994;150:51–56. doi: 10.1016/0378-1119(94)90856-7. [DOI] [PubMed] [Google Scholar]
- 30.Guzman L-M, Belin D, Carson M J, Beckwith J. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kan N C, Lautenberger J A, Edgell M H, Hutchison C A., III J Mol Biol. 1979;130:191–209. doi: 10.1016/0022-2836(79)90426-1. [DOI] [PubMed] [Google Scholar]
- 32.Jiang W, Metcalf W W, Lee K-S, Wanner B L. J Bacteriol. 1995;177:6411–6421. doi: 10.1128/jb.177.22.6411-6421.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blattner F R, Plunkett G, III, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 34.Wanner B L. In: Escherichia coli and Salmonella typhimurium Cellular and Molecular Biology. Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B Jr, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1357–1381. [Google Scholar]
- 35.Thorbjarnardottir S H, Magnusdottir R A, Eggertsson G. Mol Gen Genet. 1978;161:89–98. doi: 10.1007/BF00266619. [DOI] [PubMed] [Google Scholar]
- 36.Sâsârman A, Horodniceanu T. J Bacteriol. 1967;94:1268–1269. doi: 10.1128/jb.94.4.1268-1269.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hashimoto-Gotoh T, Franklin F C H, Nordheim A, Timmis K N. Gene. 1981;16:227–235. doi: 10.1016/0378-1119(81)90079-2. [DOI] [PubMed] [Google Scholar]
- 38.Court D, Oppenheim A. In: Lambda II. Hendrix R W, Roberts J W, Stahl F W, Weisberg R A, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1983. pp. 251–277. [Google Scholar]
- 39.Temsamani J, Kubert M, Agrawal S. Nucleic Acids Res. 1995;23:1841–1844. doi: 10.1093/nar/23.11.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen D, Yan Z, Cole D L, Srivatsa G S. Nucleic Acids Res. 1999;27:389–395. doi: 10.1093/nar/27.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]