Abstract
The three deleted in liver cancer genes (DLC1–3) encode Rho-GTPase-activating proteins (RhoGAPs) whose expression is frequently down-regulated or silenced in a variety of human malignancies. The RhoGAP activity is required for full DLC-dependent tumor suppressor activity. Here we report that DLC1 and DLC3 bind to human tensin1 and its chicken homolog. The binding has been mapped to the tensin Src homology 2 (SH2) and phosphotyrosine binding (PTB) domains at the C terminus of tensin proteins. Distinct DLC1 sequences are required for SH2 and PTB binding. DCL binding to both domains is constitutive under basal conditions. The SH2 binding depends on a tyrosine in DCL1 (Y442) but is phosphotyrosine-independent, a highly unusual feature for SH2 binding. DLC1 competed with the binding of other proteins to the tensin C terminus, including β3-integrin binding to the PTB domain. Point mutation of a critical tyrosine residue (Y442F) in DLC1 rendered the protein deficient for binding the tensin SH2 domain and binding full-length tensin. The Y442F protein was diffusely cytoplasmic, in contrast to the localization of wild-type DLC1 to focal adhesions, but it retained the ability to reduce the intracellular levels of Rho-GTP. The Y442F mutant displayed markedly reduced biological activity, as did a mutant that was RhoGAP-deficient. The results suggest that DLC1 is a multifunctional protein whose biological activity depends on cooperation between its tensin binding and RhoGAP activities, although neither activity depends on the other.
Keywords: DLC1, Src homology 2 and phosphotyrosine binding domains, tumor suppressor gene
It is widely recognized that a combination of genetic and epigenetic changes in the target cell contribute to cancer development (1). These changes may include the activation of oncogenes or antiapoptotic genes and the inactivation of tumor suppressor or proapoptotic genes. One feature of many advanced tumors is the activation of the Rho-GTPases RhoA and RhoC. These small GTPases, which are Ras-related proteins, may contribute to various parameters of abnormal cell growth, including viability, migration, invasion, proliferation, and anchorage-independent growth (2–4). Although point mutation in Ras genes frequently accounts for their activation in cancer, such mutations have not been reported for RhoA and RhoC, which appear to be wild type in cancer. As with most other small GTPases, the Rho-GTPases are active when bound to GTP and inactive when bound to GDP. They can be activated by stimulation of Rho-specific guanine nucleotide exchange factors, which increase the level of Rho-GTP and reduce the level of Rho-GDP, or inactivated by Rho-specific GTPase-activating proteins (RhoGAPs), which reduce the level of Rho-GTP and increase the level of Rho-GDP. By far the most common alteration reported for Rho regulators in cancer is the inactivation of RhoGAPs that are members of the deleted in liver cancer (DLC) family.
The prototypic member, designated DLC1, is localized to chromosome 8p21–22 in a region that is commonly deleted in hepatocellular carcinoma (5). Its expression is frequently down-regulated or silenced in various solid tumors and hematologic malignancies, predominantly by promoter methylation (6–13). Ectopic reexpression in DLC1-deficient cancer cell lines can suppress cell proliferation, induce apoptosis, and reduce tumorigenicity. The RhoGAP activity appears to be required for these antioncogenic activities because RhoGAP-deficient mutants have reduced biological activity. The DLC1 protein is reported to have a peripheral cellular location, and the rat homolog has been localized to caveolae and focal adhesions (6, 14, 15).
There are two other closely related genes, DLC2 (or STARD13), located on chromosome 13q12 (16), and DLC3 (or KIAA0189 or STARD8), located on the X chromosome at q13 (17). They have been less intensively examined, but their expression is also down-regulated in a spectrum of tumors (11, 17). All three DLC genes encode proteins that are ≈1,100 aa, with three recognized motifs: (i) a sterile α motif (SAM; other SAM motifs are often implicated in protein–protein interactions), (ii) a RhoGAP catalytic domain, and (iii) a START (STAR-related lipid transfer) domain. Other START motifs are implicated in the association with lipid-rich molecules.
Although these studies establish the DLC family as tumor suppressor genes, there is relatively little insight into their mechanism of action beyond the requirement for the RapGAP activity. It seems unlikely that this activity by itself accounts for the frequent inactivation of DLC members in tumors because other RhoGAPs do not appear to be commonly down-regulated in tumors. Furthermore, the three recognized motifs in DLC, namely, SAM, RhoGAP, and START, together account for ≈500 of the 1,100 aa, which suggests that other putative key functions may be carried out by these uncharacterized regions. To examine this possibility, we have carried out a yeast two-hybrid screen with sequences located between the SAM and RhoGAP domains. One interacting protein identified in this screen was tensin1, which is a peripheral protein implicated in several biological processes, including cell migration. Here we demonstrate that this interaction is biologically significant and show that two distinct regions of DLC1 bind two distinct domains of tensin.
Results
DLC1 and Tensin Form a Complex in Mammalian Cells.
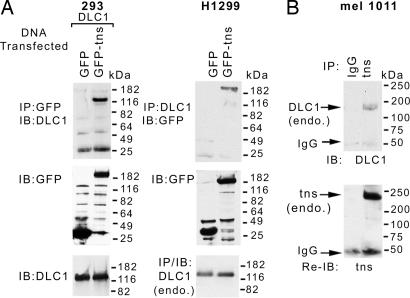
In a yeast two-hybrid screen with fragments of DLC1 and a human lung cDNA library, an interaction was identified between DLC1 and tensin [supporting information (SI) Fig. 7]. The positive yeast two-hybrid result involved a DLC1 fragment comprising amino acids 210–460, which are located between the SAM and RhoGAP domains, whereas the human tensin1 sequences comprised amino acids 1393–1736, which represent its C terminus. To confirm that this interaction can occur in mammalian cells, we cotransfected full-length (FL) DLC1 and GFP-tagged FL chicken tensin in 293 cells, which do not detectably express either gene, and carried out immunoprecipitation and Western blotting, which revealed DLC1 and tensin bands (Fig. 1A Left Top and Left Middle, respectively). Chicken tensin is homologous to human tensin1 and has been widely used to study tensin in mammalian cells (18). Transfection of GFP-tensin into H1299 cells, a lung cancer cell line that expresses endogenous DLC1 but very low levels of endogenous tensin, also gave positive results (Fig. 1A Right). To verify that endogenous human DLC1 and tensin1 form a complex with each other, we performed immunoprecipitation and Western blotting of extracts from a human melanoma cell line (mel 1011) that expressed both proteins (Fig. 1B).
Fig. 1.
DLC1 and tensin form a complex in mammalian cells. (A) DLC1 was cotransfected with GFP or GFP-tensin into 293 cells, and specifically detected DLC1 was coimmunoprecipitated in the anti-GFP immunopellet (Left). Reciprocally, GFP-tensin transfected into H1299 cells was specifically detected by coimmunoprecipitation of endogenous DLC1 with a DLC1 antibody (Right). (B) Extracts from mel 1011 cells, which contain endogenous DLC1 and tensin, were immunoprecipitated with control IgG or a tensin antibody. Coprecipitated DLC1 was detected by a DLC1 antibody (Upper). The blot was reprobed with the tensin antibody (Lower).
DLC1 and DLC3 Bind to both the Tensin–Src Homology 2 (SH2) and Tensin–Phosphotyrosine Binding (PTB) Domains.
The C terminus of tensin proteins contains an SH2 domain followed by a PTB domain, which together occupy the C-terminal ≈270 aa. Preliminary mapping with internal deletion mutants of DLC1 and a GST-tagged C-terminal fragment of chicken tensin that encodes both the SH2 and PTB domains (amino acids 1508–1787, a C-terminal polypeptide that lacks the last 5 aa of the protein) indicated that DLC1 mutants from which amino acids 415–430 and 451–470 had been deleted had reduced binding to the tensin C terminus, whereas those lacking amino acids 468–476 or 476–490 had binding activity similar to wild-type DLC1 (SI Fig. 8A).
SH2 and PTB domains are docking sites for protein–protein interaction (19, 20). Although PTB stands for phosphotyrosine (pY) binding, many PTB domains can bind their ligands in a pY-independent manner, and the PTB domain in tensin is phylogenetically grouped with this latter class of PTB domains (20). By contrast, almost all SH2 domains require pY (19). We therefore initially assumed that the PTB domain of tensin would be primarily responsible for the interaction with DLC1 because the binding to tensin appeared to be pY-independent, given the positive yeast two-hybrid results, where pY is low, and data showing that DLC1 efficiently bound the tensin SH2-PTB region in nontransformed mammalian cells, bound with similar efficiently in v-Src-transformed cells, and did not contain detectable levels of pY even in v-Src-transformed cells (SI Fig. 9).
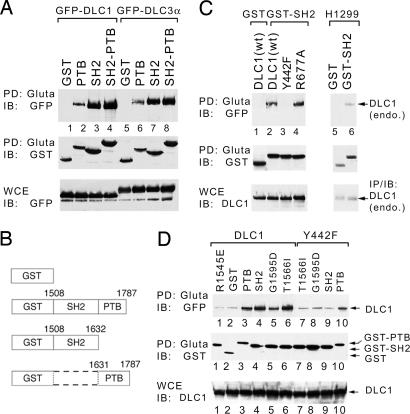
However, when we analyzed DLC1 binding to premature termination mutants of the C-terminal tensin fragment that includes both domains, we unexpectedly found that a mutant containing the tensin SH2 domain but lacking the PTB domain bound DLC1 with an efficiency that was only partially reduced compared with the starting fragment that contained both domains (data not shown). We therefore inferred that DLC1 predominantly binds the SH2 domain, while the reduced binding suggested that DLC1 might also bind the PTB domain. To verify these possibilities directly, we constructed plasmids encoding isolated tensin-SH2 and tensin-PTB domains, fused to GST (Fig. 2B), and determined that both were bound FL DLC1 and DLC3 (Fig. 2A). The SH2 domain also bound endogenous DLC1 in H1299 cells (Fig. 2C). Furthermore, the DLC1 sequences that bind the tensin PTB domain are located N terminal to those required for binding the tensin SH2 domain because a DLC1 fragment composed of amino acids 1–400 bound the PTB domain as efficiently as FL DLC1 but did not bind the SH2 domain (SI Fig. 10; data shown only for PTB binding).
Fig. 2.
DLC1 and DLC3α can be pulled down by tensin SH2 and PTB domains. (A) 293T cells were cotransfected with GFP-DLC1 or GFP-DLC3α and GST-tensin (chicken) fusion proteins as shown in B; amino acid numbers refer to chicken tensin. Cell extracts were pulled down by using glutathione Sepharose-4B (Gluta) and immunoblotted (IB) as indicated. The loading controls are also shown. (C) DLC1 and a RhoGAP-dead mutant R677A, but not a Y442F mutant, bind to the tensin SH2 domain in transfected 293T cells (Left). The endogenous DLC1 in H1299 cells was pulled down in association with the tensin SH2 domain (Right). (D) Characterization of tensin SH2 mutants for DLC1 binding. DLC1 and Y442F were cotransfected with GST and GST fusion proteins as indicated, and the pull-down was followed by immunoblotting.
A Point Mutation in DLC1 Renders the Protein Deficient for Binding the Tensin SH2 Domain.
The best studied SH2 domain known to bind a ligand in a pY-independent manner is the one in SAP, a 126-aa protein that consists mainly of a 93-aa SH2 domain that binds the SLAM receptors and is mutated in X-linked lymphoproliferative disorder (19, 21). Scanning the region of DLC1 we had implicated in tensin SH2 binding (DLC1 amino acids 415–470), we identified a sequence with remarkable homology to the motif in the SLAM receptors that binds SAP in a pY-independent manner. SAP-binding ligands prefer T/S-I-Y-X-X-V/I, and all three DLC proteins encode S-I-Y-D-N-V (amino acids 440–445 in DLC1). We therefore used the SLAM/SAP model to construct mutants in DLC1 (22). For SAP ligands, there are three key amino acids: the N-terminal T/S, the Y, and the C-terminal V/I, which in DLC1 correspond to amino acids 440, 442, and 445, respectively. Although substitution of F for Y impairs SH2 binding in other SH2 binding motifs that contain the critical Y residue, this mutation by itself does not render the SLAM peptide deficient for SAP binding (reviewed in ref. 19). However, a double mutation, of either the T/S or the V/I in conjunction with the F for Y mutation, does render the peptide deficient for SAP binding. In contrast to SAP, the substitution of F for Y in DLC1 was sufficient to render the protein defective for binding the SH2 domain of tensin, with double mutants, or deletion of amino acids 440–448 not appearing to be more deficient than the Y442F mutant (Fig. 2C and SI Fig. 8).
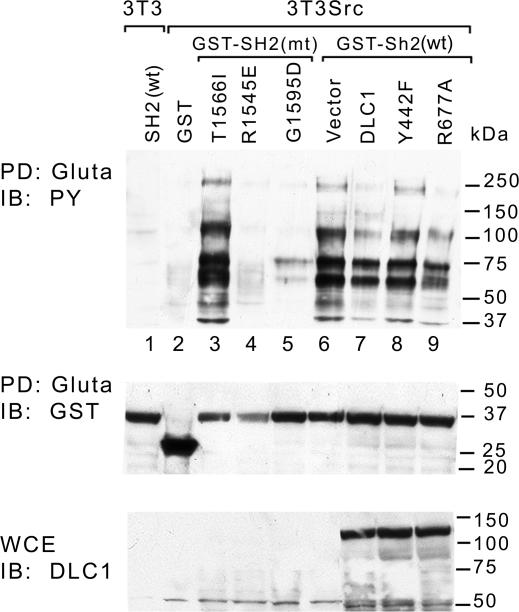
Sequence comparison between the tensin and SAP SH2 domains indicated that they share ≈25% sequence identity. The conserved residues include three that have SAP point mutations in patients with XLD [R32Q, T53I, and G93D (22, 23)], each of which results in a drastic reduction in SLAM binding. We therefore engineered the analogous mutations in the tensin SH2 domain to determine whether each might similarly reduce binding to DLC1: R1545E (the naturally occurring R32Q SAP mutant protein is very unstable, which led us to use a different mutant amino acid), T1566I, and G1595D, respectively. However, the T1566I tensin mutant bound DLC1 with an efficiency similar to that of wild type, and the G1595D mutant displayed ≈4-fold reduction in binding, whereas the analogous mutants in SAP are reported to reduce binding efficiency at least 20-fold (Fig. 2D). By contrast, the tensin SH2 domain with the R1545E mutation did not detectably bind DLC1 (Fig. 2D), analogous to the R32Q mutation in SAP. Because the tensin SH2 domain is known to bind several pY-containing proteins in transformed cells and this binding is apparently required for tensin to induce cell migration (24), we also studied the ability of the tensin SH2 mutants to bind pY-containing proteins in v-Src-transformed NIH 3T3 cells. The results paralleled those seen for DLC1 binding (Fig. 3Top), implying that the mutations in the tensin SH2 domain impaired the binding of these molecules and DLC1 to a similar degree. We also determined whether DLC1 expression might reduce the binding of the pY-containing proteins to the wild-type tensin SH2 domain. When v-Src-transformed 3T3 cells were cotransfected with constructs encoding DLC1 and the tensin SH2 domain, wild-type DLC1 and a RhoGAP-deficient mutant (R677A) reduced the binding of several protein bands, whereas the Y442F mutant did not interfere with binding (Fig. 3).
Fig. 3.
DLC1 interferes with proteins that bind the tensin SH2 domain in Src-transformed cells. v-Src-transformed 3T3 cells were cotransfected with the wild-type GST-SH2 domain with or without wild-type or mutant DLC1 as indicated. Transformed cells were also transfected with mutant GST-SH2 domains as indicated. Untransformed 3T3 cells were transfected with wild-type GST-SH2 domain. After Gluta pull-down, the proteins bound to the SH2 domain were detected by anti-pY blot (PY; Top). The expression of transfected GST fusion proteins and DLC1 is shown in Middle and Bottom, respectively.
Binding to the Tensin SH2 Domain Is Required for Colocalization of DLC1 with Tensin.
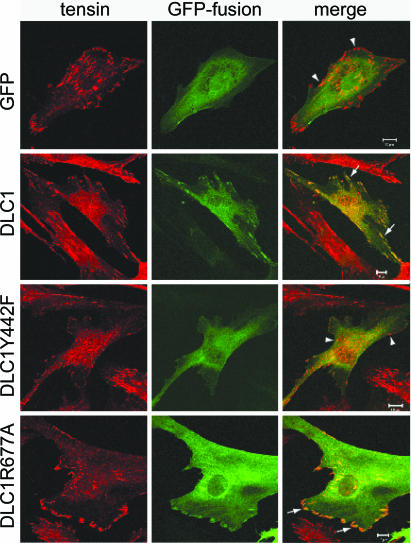
The above results suggested that the interaction between DLC1 and tensin might be impaired with the Y442F DLC1 mutant. Cotransfection of NIH 3T3 cells with a construct encoding the N-terminal 550 aa of DLC1 carrying a GST tag and GFP-tagged chicken tensin indicated that it colocalized with GFP-tensin, but the fragment carrying the Y442F mutation did not (SI Fig. 11). GFP-tagged FL DLC1 also colocalized with endogenous human tensin1 and vinculin (a marker for focal adhesions) in a human fibroblast cell line (1634), whereas the Y442F mutant did so less frequently (Fig. 4 and SI Fig. 11). As expected, a DLC1 mutant that was deficient for RhoGAP activity (R677A), which bound the SH2 domain similarly to wild type (Fig. 2C), colocalized with tensin.
Fig. 4.
Colocalization of DLC1 with endogenous tensin. Human fibroblast line 1634 grown on coverslips was transfected with GFP or GFP-tagged DLC1 (wild type, Y442F, and R677A) as indicated. Endogenous tensin was stained with a tensin antibody and rhodamine-conjugated secondary antibody. Arrows (but not arrowheads) indicate some colocalization of DLC1 and tensin. Wild-type DLC1 and R667A contain peripheral spots that colocalize with tensin in the merged image, whereas Y442F and GFP have diffuse staining and rarely colocalize. By using vinculin as a marker, it was confirmed that these spots represent typical focal adhesions (see SI Fig. 10). At least 100 cells of each type were viewed. The images represent 85–90% of total viewed cells, except that ≈15% of cells expressing the Y442F mutant had a nuclear as well as a cytoplasmic localization. (Scale bars: 10 μm.) Expression of the transfected genes was confirmed by immunoblotting (data not shown).
The Y442F Mutant Negatively Regulates Rho-GTP in Vivo.
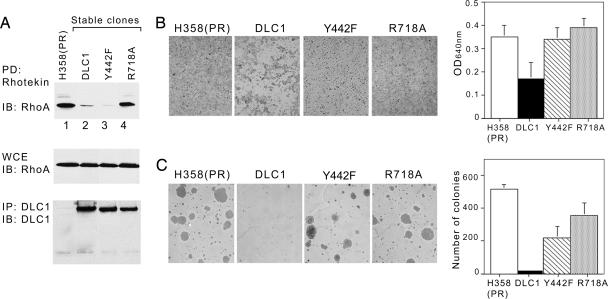
One important activity of DLC is the ability to negatively regulate Rho-GTP via its catalytic RhoGAP domain. To determine whether the impaired ability of the Y442F mutant to colocalize with tensin affected this activity, HEK293T cells, which have low endogenous levels of Rho-GTP, were transfected with wild-type or mutant DLC1 and treated with lysophosphatidic acid (LPA), which increases Rho-GTP (25). As expected, cells that received the vector control displayed an LPA-dependent increase in Rho-GTP, LPA-treated cells expressing wild-type DLC1 (or DLC3) had lower levels of Rho-GTP, and LPA-treated cells expressing either of two RhoGAP mutants (R677A and R718A) had higher Rho-GTP levels (SI Fig. 12). By contrast, the Y442F mutant reduced the LPA-stimulated levels of RhoGAP to a similar degree as wild-type DLC1 (Fig. 5). Similar results were seen when wild-type DLC1, Y442F, or R677A were stably transfected into the H358 lung cancer cell line, which lacks endogenous DLC1 (Fig. 5A). Thus, the failure to colocalize with tensin did not abrogate the ability of the Y442F mutant to carry out its RapGAP function in vivo.
Fig. 5.
The Y442F mutant reduces Rho-GTP but is deficient biologically. (A) Rho-GTP in H358 cells and stable clones expressing DLC1, Y442F, or R718A were analyzed by Rhotekin pull-down assay followed by anti-RhoA blotting (Top). The expression of DLC1 in the stable clones and total RhoA loading controls were confirmed by immunoblotting. These H358-derived cells were used for the biological experiments in the figure. (B) Transwell migration assay. After 3 days, migrated cells that had come through the transwell filter were photographed after staining and then solubilized and quantified colorimetrically. The data are the mean ± SD of triplicate well measurements from one representative experiment. (C) Anchorage-independent growth assay. Cells were grown in soft agar (0.4%) and photographed after 4 weeks. The quantitative data are the mean number of colonies (± SD) >400 μm in diameter from one representative experiment.
The Y442F DLC1 Mutant Is Less Active Biologically than Wild Type.
To evaluate the effect of the Y442F mutant on the biological activity of DLC1, the stable DLC1 transfectants of the H358 line were examined in cell migration and agar growth assays (Fig. 5 B and C, respectively). Cells expressing wild-type DLC1 migrated and formed colonies more slowly than the negative control, whereas those expressing the Y442F mutant or the R718A RhoGAP mutant migrated and formed colonies at a rate closer to the control cells. Thus, efficient inhibition of these biological activities by DLC1 requires cooperation between its RhoGAP activity and its tensin binding activity.
DLC1 Competes with β3-Integrin for Binding to the Tensin PTB Domain.
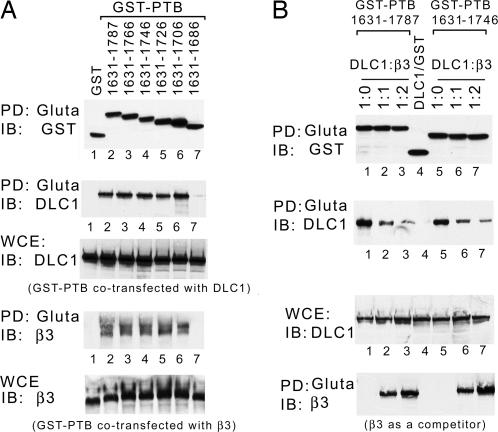
The tensin PTB domain binds the intracellular portion of several β-integrins, including β3-integrin (26), and is required for tensin-dependent cell migration (24). Given that DLC1 binds the tensin PTB domain, we speculated that DLC1 might compete with β3-integrin for binding and that similar sequences in the PTB domain might be required for binding each protein. To address this possibility, a series of premature termination mutants of the tensin PTB domain (amino acids 1631–1787) were analyzed for their ability to bind DLC1 or β3-integrin when cotransfected into HEK293 cells. Strikingly, although the PTB mutants containing amino acids 1631–1706 or additional C-terminal residues bound both DLC1 and β3-integrin with an efficiency similar to that of the FL PTB domain, a mutant that was 20 aa shorter (residues 1631–1686) was severely deficient for binding both proteins (Fig. 6A). These results suggested that DLC1 and β3-integrins were likely to compete with each other for binding this domain, analogous to its competition for the binding of pY-containing proteins to the tensin SH2 domain (Fig. 3). To confirm this hypothesis, β3-integrin was shown to compete with DLC1 for binding the PTB domain (Fig. 6B).
Fig. 6.
DLC1 can compete with β3-integrin for binding the tensin PTB domain. (A) Mapping the PTB domain of chicken tensin for DLC1 and β3-integrin binding. GST-PTB and premature termination mutants were cotransfected with equal amounts of DLC1 or β3-integrin in 293T cells. After GST-PTB pull-down, associated protein binding was assayed by anti-DLC1 and anti-β3 immunoblotting as indicated. Cell extracts were used as loading controls. (B) Competition assay. GST-PTB fragments and the combination of DLC1 vs. β3 were cotransfected at the indicated ratios in 293T cells. After Gluta pull-down, the DLC1 signal was reduced when using β3 as a competitor. The expression of transfected proteins and loading controls is shown.
Discussion
We have found that DLC1 binds human tensin1 and its chicken homolog, that this binding involves both the SH2 and PTB domains at the C terminus of tensin, and that distinct DCL1 amino acids mediate SH2 and PTB binding. The overall homology between chicken and human tensin proteins is 74%, and their 270 C-terminal amino acids, which contain the SH2 and PTB domains, are >98% homologous (87% identical). Oncogenic inhibition by DLC1 requires cooperation of its tensin binding and RhoGAP activities, but neither activity depends on the other.
The vast majority of SH2 ligands contain a critical Y that must be phosphorylated to bind the SH2 domain (19). However, although DLC1 contains such a Y (residue 442) in its SH2 binding region, its binding to the SH2 domain of tensin appears to be constitutive and not to depend on pY. The SLAM receptors, which bind the SH2 domain of SAP, are the best known prior example of SH2 ligands that bind in a pY-independent manner (21, 22), and we found remarkable amino acid sequence homology between DLC1 residues and the key residues of SLAM required for binding SAP. We verified that this region of homology is essential for DLC1 to bind the tensin SH2 region by showing that Y442F renders DLC1 deficient for binding wild-type tensin in vivo, although the ability to bind the isolated PTB domain remained intact. This degree of stringency for SH2 binding is greater than reported for SLAM, which requires mutation of a second residue, in addition to the Y-to-F mutation, to render it deficient for SAP binding. Whereas the T53I mutation in SAP is reported to markedly reduce SLAM/SAP binding, the analogous T1566I mutation in the tensin SH2 domain did not impair its binding to DLC1 or other ligands. Therefore, although the interaction between DLC1 and the tensin SH2 domain can occur in a pY-independent manner, as is also true of the interaction between SLAM and SAP, there are differences in the details of the requirements for binding.
The ability of DLC1 to bind the tensin family, which in mammalian cells contains four genes, tensin1 (TNS1), tensin2 (TENC1), tensin3 (TNS3), and cten (TNS4), has recently been examined. As with tensin1, the C terminus of all family members contains an SH2 domain and a PTB domain. However, they are less conserved than the homology between tensin1 and its chicken homolog. Compared with tensin1, the C-terminal 270 aa of tensin2 and tensin3 are each 89% homologous (64% and 72% identical, respectively), and those of cten are 78% homologous (51% identical). Yam et al. (27) have reported an interaction with tensin2, and Liao et al. (28) have reported an interaction with cten, which lacks the N-terminal sequences common to the other tensin genes. In a yeast two-hybrid assay, DLC1 interacted with the SH2 domain of cten, but not with its PTB domain. In mammalian cells, the DLC1 interaction was pY-independent, and both the Y442F and a S440A mutant of a DLC1 fragment (residues 1–535) gave a diffuse localization and were deficient for inhibiting growth in monolayer cultures. For the interaction with tensin2, it was concluded that DLC1 interacted with the PTB domain of tensin2 but not with the SH2 domain, amino acids 375–509 were sufficient for binding tensin2, a DLC1 mutant lacking amino acids 375–509 (which has deleted Y442 and many surrounding residues) was deficient for binding tensin2, and this mutant lacked the ability to cooperate with tensin2 in suppressing Ras-dependent activation of a serum response element in HEK293 cells transiently transfected with four different plasmids. Aspects of our independent results are in line with those reported for cten because we find that DLC1 binds tensin in a pY-independent manner, it predominantly binds the SH2 domain of tensin1 and chicken tensin, and the Y442F DLC1 mutant is deficient for interaction with tensin and displays reduced biological activity. Although we have not examined tensin2, the cten study reported, in their yeast two-hybrid assay, that DLC1 interacted with the SH2 domain of tensin1, tensin2, and tensin3. These data suggest that in mammalian cells DLC1 may bind the SH2 domain of tensin2 in addition to its PTB domain.
Our results demonstrate for the first time that DLC1 and DLC3 bind both the SH2 and PTB domains of the same tensin protein and that binding to these domains is mediated by different sequences of DLC1. Unlike the previous studies, we have evaluated the relationship between tensin binding and RhoGAP activity in cells. This analysis has shown that the Y442F mutant, despite its diffuse localization, reduces the overall level of Rho-GTP as efficiently as wild-type DLC. In contrast, the earlier studies (27, 28) had speculated that the principal role of tensin binding was to reduce the level of Rho-GTP. Our data therefore strongly suggest that tensin binding has a RhoGAP-independent role that contributes to the biological activity of DLC. Our biochemical analyses provide a potential alternate mechanism because we observe that DLC1 can compete with β3-integrin for binding the PTB domain and with several proteins for binding the SH2 domain; a previous mutant analysis of tensin1 reported that both of these tensin domains are required for tensin-dependent migration (24). Thus, DLC1, and presumably the other DLC family members, is a multifunctional protein whose tensin binding and RhoGAP activities do not depend on each other, but both activities are required for biological activity. The multifunctional nature of DLC1 may account for its frequent inactivation in tumors, in contrast to other RhoGAPs.
Materials and Methods
DNA Constructs, Cell Culture, and Transfection.
For DNA constructs, see SI Text. NIH 3T3 (3T3), HEK293 (American Type Culture Collection; 293), HEK293H (Invitrogen; 293H), HEK293T (ref. 29; 293T), and human fibroblasts 1634 were maintained in DMEM supplemented with 10% FBS. Human melanoma cell line mel 1011 (from S. Topalian, National Cancer Institute) and human NSCLC cell lines H1299 and H358 (from C. Harris, National Cancer Institute) were cultured in RPMI medium 1640 supplemented with 10% FBS. Cells were cultured at 37°C in a humidified 5% CO2 atmosphere. Transient transfections were carried out with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Stable 3T3v-Src clones and H358 clones expressing DLC1, Y442F, or R718A were generated by transfection with Lipofectamine followed by a selection with G418 (0.9 mg/ml).
Immunofluorescent Staining and Microscopy.
Cells were seeded on glass coverslips, transfected with GFP-tagged chicken tensin or DLC1, and incubated for 24 h. Cells were fixed in 2% formaldehyde at room temperature. After rinsing with PBS, cells were incubated with 1:25 anti-tensin rabbit polyclonal antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) for 2 h at 4°C, then 1:200 rhodamine-conjugated secondary antibody (Jackson ImmunoResearch) for 1 h, and visualized with a LSM510 confocal microscope.
Cell Migration and Soft Agar Assays.
Transwell assays were performed with 6.5-mm-diameter Falcon cell culture inserts (8-μm pore size; Becton Dickinson) precoated with 0.01% gelatin in 24-well cell culture plates. H358 cells and stable clones were trypsinized, resuspended in serum-free RPMI medium 1640, and transferred to the upper chamber (5 × 104 cells in 500 μl); 800 μl of medium containing 10% FBS was added to the lower chamber. After incubation for 3 days, cells remaining on the upper surface of the filter were removed with a cotton swab; cells that had migrated to the lower surface were fixed, stained (methylene blue:methanol:carbol fuchsin = 3:2:1) for 10 min, destained, visualized microscopically, and photographed. The migrated cells were then solubilized overnight with 1% Triton X-100. The collected lysates were quantified colorometrically in a spectrophotomer. For soft agar colony assays, H358 and stable clones (1 × 105 cells) were mixed with RPMI medium 1640 complete medium containing 0.4% agar (Difco) and placed over 0.6% of basal agar in 60-mm dishes. Cells were grown for 2 to 3 weeks, and colonies were visualized microscopically and photographed.
In Vivo Pull-Down Assay, Coimmunoprecipitation, and Immunoblotting.
Cells were cotransfected with plasmids expressing GST or GST fusion proteins and DLC1 or its derivatives. Two days after transfection, cells were lysed with golden lysis buffer (GLB: 20 mM Tris, pH 7.9/137 mM NaCl/10% glycerol/1% Triton X-100/5 mM EDTA/1 mM EGTA/1 mM Na3VO4/10 mM NaF/1 mM sodium pyrophosphate/1 mM β-glycerophosphate/protease inhibitor mixture tablet). The cleared supernatants were collected, and the amount of protein was estimated by using a BCA kit (Pierce). Equal amounts of protein from cell extracts were used for pull-down assays by adding 30 μl of glutathione Sepharose-4B slurry (Amersham) and rotating 3 h at 4°C. The pellets were washed once with GLB, once with high-salt HNTG (20 mM Hepes/500 mM NaCl/0.1% Triton X-100/10% glycerol), and twice with low-salt HNTG (20 mM Hepes/150 mM NaCl/0.1% Triton X-100/10% glycerol), and incubated with Laemmli sample buffer. Fifteen percent of each sample was used for detecting GST fusion protein, and 85% was used for detecting DLC1 and its mutant derivatives that had been pulled down in association with the GST fusion protein. For coimmunoprecipitation experiments, equal amounts of protein lysates were incubated with control IgG or specific antibodies. Thirty microliters of protein A/G slurry (Pierce) was added to each immune reaction and rotated overnight at 4°C. The immunopellets were washed four times as above. After separating protein samples by SDS/PAGE, immunoblotting was used to detect protein signals with anti-GST (Santa Cruz Biotechnology), DLC1 (BD Biosciences), β3-integrin (BD Biosciences), tensin (Santa Cruz Biotechnology), GFP (Covance), or anti-pY mAb (4G10; Upstate Biotechnology). For each blot, horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG (Amersham) was used for the second reaction at 1:10,000 dilution. Immunocomplexes were visualized by ECL using an ECL kit (Amersham).
Supplementary Material
Acknowledgments
We thank Curt Harris, Suzanne Topalian, and Marian Durkin for providing reagents and Paul Randazzo and Jeffrey Rubin for helpful discussions. This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Abbreviations
- SH2
Src homology 2
- pY
phosphotyrosine
- PTB
pY binding
- RhoGAP
Rho-specific GTPase-activating protein
- FL
full-length
- LPA
lysophosphatidic acid
- SAM
sterile α motif
- DLC
deleted in liver cancer.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703033104/DC1.
References
- 1.Vogelstein B, Kinzler KW. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 2.Karnoub AE, Symons M, Campbell SL, Der CJ. Breast Cancer Res Treat. 2004;84:61–71. doi: 10.1023/B:BREA.0000018427.84929.5c. [DOI] [PubMed] [Google Scholar]
- 3.Ridley AJ. Breast Cancer Res Treat. 2004;84:13–19. doi: 10.1023/B:BREA.0000018423.47497.c6. [DOI] [PubMed] [Google Scholar]
- 4.Gomez del Pulgar T, Benitah SA, Valeron PF, Espina C, Lacal JC. BioEssays. 2005;27:602–613. doi: 10.1002/bies.20238. [DOI] [PubMed] [Google Scholar]
- 5.Yuan BZ, Miller MJ, Keck CL, Zimonjic DB, Thorgeirsson SS, Popescu NC. Cancer Res. 1998;58:2196–2199. [PubMed] [Google Scholar]
- 6.Zhou X, Thorgeirsson SS, Popescu NC. Oncogene. 2004;23:1308–1313. doi: 10.1038/sj.onc.1207246. [DOI] [PubMed] [Google Scholar]
- 7.Goodison S, Yuan J, Sloan D, Kim R, Li C, Popescu NC, Urquidi V. Cancer Res. 2005;65:6042–6053. doi: 10.1158/0008-5472.CAN-04-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong CM, Yam JW, Ching YP, Yau TO, Leung TH, Jin DY, Ng IO. Cancer Res. 2005;65:8861–8868. doi: 10.1158/0008-5472.CAN-05-1318. [DOI] [PubMed] [Google Scholar]
- 9.Guan M, Zhou X, Soulitzis N, Spandidos DA, Popescu NC. Clin Cancer Res. 2006;12:1412–1419. doi: 10.1158/1078-0432.CCR-05-1906. [DOI] [PubMed] [Google Scholar]
- 10.Seng TJ, Low JS, Li H, Cui Y, Goh HK, Wong ML, Srivastava G, Sidransky D, Califano J, Steenbergen RD, et al. Oncogene. 2007;26:934–944. doi: 10.1038/sj.onc.1209839. [DOI] [PubMed] [Google Scholar]
- 11.Ullmannova V, Popescu NC. Int J Oncol. 2006;29:1127–1132. [PubMed] [Google Scholar]
- 12.Song YF, Xu R, Zhang XH, Chen BB, Chen Q, Chen YM, Xie Y. J Clin Pathol. 2006;59:947–951. doi: 10.1136/jcp.2005.031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi H, Guo J, Duff DJ, Rahmatpanah F, Chitima-Matsiga R, Al-Kuhlani M, Taylor KH, Sjahputera O, Andreski M, Wooldridge JE, Caldwell CW. Carcinogenesis. 2007;28:60–70. doi: 10.1093/carcin/bgl092. [DOI] [PubMed] [Google Scholar]
- 14.Kawai K, Yamaga M, Iwamae Y, Kiyota M, Kamata H, Hirata H, Homma Y, Yagisawa H. Biochem Soc Trans. 2004;32:1107–1109. doi: 10.1042/BST0321107. [DOI] [PubMed] [Google Scholar]
- 15.Yamaga M, Sekimata M, Fujii M, Kawai K, Kamata H, Hirata H, Homma Y, Yagisawa H. Genes Cells. 2004;9:25–37. doi: 10.1111/j.1356-9597.2004.00698.x. [DOI] [PubMed] [Google Scholar]
- 16.Ching YP, Wong CM, Chan SF, Leung TH, Ng DC, Jin DY, Ng IO. J Biol Chem. 2003;278:10824–10830. doi: 10.1074/jbc.M208310200. [DOI] [PubMed] [Google Scholar]
- 17.Durkin ME, Ulmannova V, Guan M, Popescu NC. Oncogene. 2007 doi: 10.1038/sj.onc.1210244. in press. [DOI] [PubMed] [Google Scholar]
- 18.Zamir E, Katz M, Posen Y, Erez N, Yamada KM, Katz BZ, Lin S, Lin DC, Bershadsky A, Kam Z, Geiger B. Nat Cell Biol. 2000;2:191–196. doi: 10.1038/35008607. [DOI] [PubMed] [Google Scholar]
- 19.Machida K, Mayer BJ. Biochim Biophys Acta. 2005;1747:1–25. doi: 10.1016/j.bbapap.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 20.Uhlik MT, Temple B, Bencharit S, Kimple AJ, Siderovski DP, Johnson GL. J Mol Biol. 2005;345:1–20. doi: 10.1016/j.jmb.2004.10.038. [DOI] [PubMed] [Google Scholar]
- 21.Veillette A. Nat Rev Immunol. 2006;6:56–66. doi: 10.1038/nri1761. [DOI] [PubMed] [Google Scholar]
- 22.Hwang PM, Li C, Morra M, Lillywhite J, Muhandiram DR, Gertler F, Terhorst C, Kay LE, Pawson T, Forman-Kay JD, Li SC. EMBO J. 2002;21:314–323. doi: 10.1093/emboj/21.3.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li C, Iosef C, Jia CY, Gkourasas T, Han VK, Shun-Cheng Li S. Biochemistry. 2003;42:14885–14892. doi: 10.1021/bi034798l. [DOI] [PubMed] [Google Scholar]
- 24.Chen H, Lo SH. Biochem J. 2003;370:1039–1045. doi: 10.1042/BJ20021308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moolenaar WH, van Meeteren LA, Giepmans BN. BioEssays. 2004;26:870–881. doi: 10.1002/bies.20081. [DOI] [PubMed] [Google Scholar]
- 26.Calderwood DA, Fujioka Y, de Pereda JM, Garcia-Alvarez B, Nakamoto T, Margolis B, McGlade CJ, Liddington RC, Ginsberg MH. Proc Natl Acad Sci USA. 2003;100:2272–2277. doi: 10.1073/pnas.262791999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yam JW, Ko FC, Chan CY, Jin DY, Ng IO. Cancer Res. 2006;66:8367–8372. doi: 10.1158/0008-5472.CAN-05-2850. [DOI] [PubMed] [Google Scholar]
- 28.Liao YC, Si L, Devere White RW, Lo SH. J Cell Biol. 2007;176:43–49. doi: 10.1083/jcb.200608015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buck CB, Pastrana DV, Lowy DR, Schiller JT. J Virol. 2004;78:751–757. doi: 10.1128/JVI.78.2.751-757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.