Abstract
Background
The endosymbiont Wolbachia pipientis infects a broad range of arthropod and filarial nematode hosts. These diverse associations form an attractive model for understanding host:symbiont coevolution. Wolbachia's ubiquity and ability to dramatically alter host reproductive biology also form the foundation of research strategies aimed at controlling insect pests and vector-borne disease. The Wolbachia strains that infect nematodes are phylogenetically distinct, strictly vertically transmitted, and required by their hosts for growth and reproduction. Insects in contrast form more fluid associations with Wolbachia. In these taxa, host populations are most often polymorphic for infection, horizontal transmission occurs between distantly related hosts, and direct fitness effects on hosts are mild. Despite extensive interest in the Wolbachia system for many years, relatively little is known about the molecular mechanisms that mediate its varied interactions with different hosts. We have compared the genomes of the Wolbachia that infect Drosophila melanogaster, wMel and the nematode Brugia malayi, wBm to that of an outgroup Anaplasma marginale to identify genes that have experienced diversifying selection in the Wolbachia lineages. The goal of the study was to identify likely molecular mechanisms of the symbiosis and to understand the nature of the diverse association across different hosts.
Results
The prevalence of selection was far greater in wMel than wBm. Genes contributing to DNA metabolism, cofactor biosynthesis, and secretion were positively selected in both lineages. In wMel there was a greater emphasis on DNA repair, cell division, protein stability, and cell envelope synthesis.
Conclusion
Secretion pathways and outer surface protein encoding genes are highly affected by selection in keeping with host:parasite theory. If evidence of selection on various cofactor molecules reflects possible provisioning, then both insect as well as nematode Wolbachia may be providing substances to hosts. Selection on cell envelope synthesis, DNA replication and repair machinery, heat shock, and two component switching suggest strategies insect Wolbachia may employ to cope with diverse host and intra-host environments.
Background
Intracellular bacterial symbiont associations are extremely common in invertebrates. The capacity for these symbionts to shape host biology is immense and includes documented effects on host reproduction [1], food preference [2], locomotion [3], and interspecific competition [4]. Teasing apart the contributions of insect and symbiont genomes to such multi-organism determined phenotypes is necessary if the evolution and ecology of both partners are to be understood. This can be challenging, because the complex biotic interaction also makes these systems less tractable experimentally. Comparative study of sequenced symbiont genomes and their relatives is offering new means to direct empirical study of symbiosis [5].
The endosymbiont Wolbachia pipientis infects a wide range of arthropod and filarial nematode hosts. Across its host range the microbe is associated with diverse phenotypic outcomes. The Wolbachia-nematode associations are mutualistic while all other associations could be described as commensal or parasitic in nature. In nematodes the infection is confined to the nematode reproductive tract and the hypodermal tissue where the microbe plays an integral role in host viability and reproduction [6,7]. Phylogenies of Wolbachia and their nematode hosts are congruent, reflecting a long history of strict vertical transmission [8]. Tight associations like these are predicted to generate genome reduction [9], as host support of symbiont requirements leads to degradation and loss of the genes in these redundant pathways. Consistent with this prediction, the genome of the Wolbachia strain that infects Brugia malayi (wBm) is much smaller and highly streamlined relative to the genomes of free-living bacteria and other Wolbachia [10,11].
The Wolbachia-arthropod association, in contrast, is more fluid in nature. Infections are not fixed in populations and most appear to be mild in their effects on host fitness [12,13]. Horizontal transmission among host lineages is common on a phylogenetic time scale, meaning closely related Wolbachia can be found in taxonomically diverse hosts [14]. Infections can be found in numerous somatic tissues as well as the gonads [15]. The presence of Wolbachia in insect hemolymph in combination with recent experimental work also suggests that the bacteria may be exposed to extracellular environments for sustained periods [16]. Across the arthropods Wolbachia also induces a broad range of reproductive manipulations including feminization, male killing, cytoplasmic incompatibility, and parthenogenesis [1,17,18]. The pattern of Wolbachia tissue distribution, infection densities, induced fitness effects, and reproductive manipulations vary greatly within the arthropods and are the result of host and bacterial genotype interactions [19-22].
Here we report the results of genome wide screens for the presence of diversifying selection in the Wolbachia that infect the filarial nematode, Brugia malayi wBm [10] and the insect Drosophila melanogaster, wMel [11]. Per gene estimates of nonsynonyous substitution per nonsynonymous site versus synonymous substitution per synonymous site (dN/dS) in the Wolbachia relative to the outgroup species, Anaplasma marginale [23] were used to infer past history of positive selection [24]. This approach has been utilized previously to explore the genetic basis of complex phenotypes in a diverse range of taxa [24-29]. By identifying key molecular adaptations in each of the two Wolbachia lineages, we sought to shed light the mechanistic basis of the Wolbachia symbiosis and how it might vary with respect to different hosts. We hypothesized that genes whose encoded proteins were involved with secretion or were localized to the Wolbachia cell surface would show evidence of strong selection due to their interaction with the host. We also expected to find evidence of selection on pathways that could be used for host provisioning in wBm. The screen confirmed both these hypotheses. The genomic comparisons also revealed possible points of host provisioning in wMel and strategies Wolbachia may have evolved for coping with diverse hosts and intra host environments.
Results & Discussion
Summary
Of the 591 loci examined, 60 in wBm and 101 in wMel bore signatures of positive selection (see Additional file 1). The proportion of genes affected by diversifying selection in Wolbachia was higher than reports from other screens in bacteria [26] and may reflect the well-documented phenomenon of rapid evolution in endosymbionts [30-32]. The small effective population sizes of these bacteria would predict more rapid fixation of nonsynonymous mutations due to drift and hence generate higher average ratios of dN to dS [33]. The distribution of significant genes was not clumped with respect to genome position (data not shown) with the exception of the ribosomal protein encoding genes, which are members of an operon. Excluding hypothetical and unknown groups, the significant genes represent 13 functional classes (GenomeAtlas annotation) in wBm and 15 in wMel (TIGR annotation) (Fig. 1 and see Additional file 1). Genes comprising the biological role categories nucleotide biosynthesis, amino acid biosynthesis, and transport/secretion were similarly affected by diversifying selection in both genomes. In general, evidence of selection was more common in the wMel genome. Larger numbers of genes in the role categories of DNA metabolism, energy metabolism, protein synthesis, cell envelope synthesis, cofactor biosynthesis, and protein fate were disproportionately affected in wMel (Fig. 1). We have reviewed the gene composition for several of these functional categories and speculate on their role in the evolution of insect vs. nematodee symbiont associations.
Figure 1.
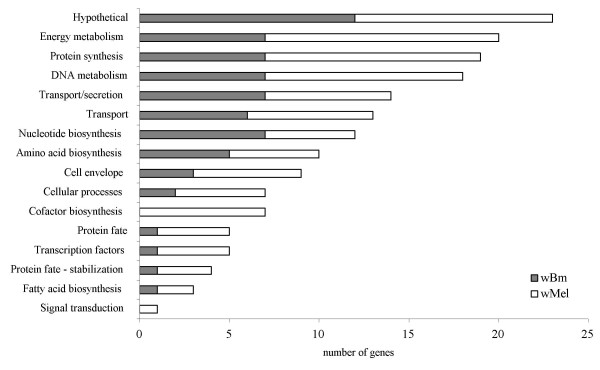
Positive selection by functional role. Total number of genes exhibiting significant positive selection by functional role in the wBm and wMel genomes.
Purifying selection
An examination of the dN/dS ratios also highlighted those genes experiencing extreme levels purifying selection in either of the Wolbachia lineages. A total of 323 genes in the wBm lineage and 250 genes in wMel had a ratio < 1.0. We then examined only the most severely affected genes (dN/dS < 0.2) in each lineage and asked whether genes from any of the functional categories were over-represented. Most of the major functional classes were represented by only a small number of genes. The exceptions were the categories of synthesis and modification of ribosomal proteins in both genomes and the biosynthesis and degradation of cell envelope in wBm only. The former represented ~15% of genes with dN/dS < 0.2 and the latter 8% of the genes for wBm. The extreme conservation in ribosomal protein evolution is not surprising given their essential and conserved cellular functions for all kingdoms of life. Purifying selection on cell envelope component genes in wBm is interesting given that these same genes are experiencing diversifying selection in wMel (see Additional file 1). The Wolbachia cell envelope may be exposed to vastly different environments in the insect versus nematode hosts. Differences in how selection is operating on the genes encoding membrane proteins may reflect adaptation to lineage specific ecological niches (see Direct contact with the host below).
Evidence of provisioning
The basis of Wolbachia's dependence on its host and the nature of any benefits provided to hosts are two fundamental unknown aspects of this symbiotic association. The completed genome sequences of wBm and wMel [10,11] have only recently advanced our understanding of what Wolbachia can and cannot synthesize and what it may be transporting across its membrane. Symbiont provisioning of insect hosts is hypothesized for many associations and has been documented in numerous insects including; aphids [34], tsetse flies [35], rice weevils [36], and ants [37]. Evidence of provisioning would not be surprising in the nematode relationships as Wolbachia is clearly acting as a mutualist. Arthropod Wolbachia have traditionally been thought of as parasitic and therefore the presence of diversifying selection on a number of cofactor biosynthesis genes is particularly exciting (Fig. 2 & see Additional file 1).
Figure 2.

Cofactor biosynthesis. Specific genes and dN/dS exhibiting selection (BH p = 0.001 model and Fisher's exact test) by select functional role for each genome. Each box corresponds to a significant gene in the sub role and degree of shading indicates magnitude of dN/dS. "Unrep" indicates the ratio was at the reportable limit of codeml (see methods). Names of gene products are listed in order to the right of boxes. See Additional file 1 for individual p values, gene ids/descriptions, WD#s, and dN & dS values.
Both Wolbachia genomes lack complete pathways for de novo synthesis of coenzyme A, NAD, biotin, lipoate, ubiquinone, and folate; presumably the host supplements these compounds [10,11]. Several genes that encode for components of these disrupted biosynthetic pathways show evidence of positive selection in wMel and may reflect the molecular evolutionary process of integrating host and symbiont systems (Fig. 2 & see Additional file 1). Selection on genes in these same pathways was also detected in wBm, but under less stringent rejection criteria (see Additional file 1). Unlike the above listed cofactors, riboflavin biosynthesis pathways are complete in both Wolbachia strains. Evidence for positive selection on riboflavin synthesis was present in wMel (Fig. 2, Model p < 0.001, & Fisher's p < 0.001) and again in wBm under slightly less stringent criteria (see Additional file 1). Symbiont provisioning of riboflavin has been documented in both weevil-SOPE and aphid-Buchnera associations [36,38]. Two members of the heme biosynthetic pathway (of seven genes in total) were affected by selection in wMel. Additional genes in the heme biosynthesis pathway were also identified in both wBm and wMel when less stringent rejection criteria were applied (see Additional file 1). An examination of the Brugia malayi genome [10] suggests that the nematode may be incapable of synthesizing its own heme and therefore it is possible that wBm Wolbachia may be provisioning its host with heme intermediates. Although insect hosts are not dependent on Wolbachia for heme biosynthesis, the microbe may supplement host stores or play an additional role in iron homeostasis.
In addition to the provision of metabolic cofactors, invertebrate hosts may also benefit from an additional source of nucleotides provided by Wolbachia. Multiple genes in this functional category (seven in wBm and five in wMel, Fig. 1) were affected by positive selection (see Additional file 1). Other endosymbionts, including the parasitic Rickettsia or beneficial Buchnera, scavenge nucleotides from the host environment via ATP/ADP translocases. Wolbachia, however encodes complete purine and pyrimidine biosynthetic pathways, and lacks the nucleotide translocase found in the closely related Rickettsia [10,11]. The provision of nucleotides by wBm and wMel could benefit their hosts during periods of rapid DNA replication and cellular division, such as during oogenesis and embryogenesis [10]. Lastly, there is widespread evidence of diversifying selection in both genomes on amino acid biosynthetic pathway genes (Fig 1 and see Additional file 1). Wolbachia lack many genes in the biosynthetic pathways for amino acids and therefore it is less likely they are provisioning hosts in this regard [10,11].
Coordination of cell division with the host
The coordination of symbiont replication with host cell division is required to prevent either loss of the symbiont within the host or over replication leading to pathology within the host [1], such as that occurring with wMelPop. The mechanisms underlying this balancing act in Wolbachia-host associations are unknown. Filarial Wolbachia densities increase when the infection passes from the insect vector into the mammalian host [39,40]. Arthropod Wolbachia are also present at different densities depending on host species [20], host developmental phase [41], and tissue distribution [15,42]. For a number of insect species, Wolbachia has the additional challenge of dealing with host diapause where the microbe's replication would have to be slowed or stopped temporarily to maintain synchrony with host cell division [43].
Several genes associated with cell division particularly with regulation of growth rates, appear to be positively selected in wMel (Fig. 3), including the cell division genes ftsA [44], ftsK [45], and rne [46]. Also affected in wMel, is surE [47] whose expression is associated with adaptation to stressful conditions and survival of stationary phase in E. coli. Another rate limiting step in terms of growth and cell division that may be targeted by selection is protein synthesis. The processes of synthesis and modification of ribosomal proteins, translation factors, and base modification were heavily affected in both genomes (included in Protein synthesis, Fig. 1 and see Additional file 1). These molecular adaptations may affect rates of cell cycling indirectly by regulating rates of protein synthesis.
Figure 3.
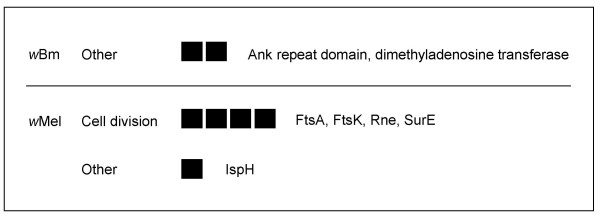
Cellular processes. Specific genes and dN/dS exhibiting selection (BH p = 0.001 model and Fisher's exact test) by select functional role for each genome. Each box corresponds to a significant gene in the sub role and degree of shading indicates magnitude of dN/dS. "Unrep" indicates the ratio was at the reportable limit of codeml (see methods). Names of gene products are listed in order to the right of boxes. See Additional file 1 for individual p values, gene ids/descriptions, WD#s, and dN & dS values.
In both genomes, diversifying selection on genes involved with DNA replication was surprisingly common given the fundamental conserved nature of the DNA replication process (Fig. 4). A recent screen of uropathogenic E. coli relative to non-pathogenic strains, also revealed diversifying selection on cell division & DNA metabolism genes [26]. These pathogenic strains in their shift from commensal origins have gained the ability to invade and live inside host cells. Heightened evolutionary change in cell division and DNA replication genes may affect efficiency of growth and underpin coordination with host cell activities. This is a particular challenge for wMel given the need to a adapt to a broader range of host cell types, host cell division rates, extracellular/intracellular environments, and ambient temperatures.
Figure 4.
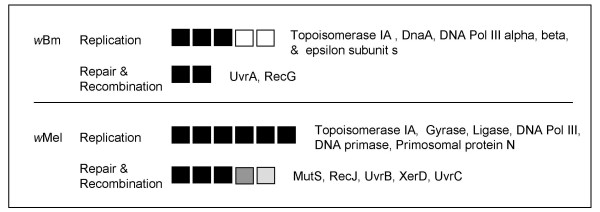
DNA metabolism. Specific genes and dN/dS exhibiting selection (BH p = 0.001 model and Fisher's exact test) by select functional role for each genome. Each box corresponds to a significant gene in the sub role and degree of shading indicates magnitude of dN/dS. "Unrep" indicates the ratio was at the reportable limit of codeml (see methods). Names of gene products are listed in order to the right of boxes. See Additional file 1 for individual p values, gene ids/descriptions, WD#s, and dN & dS values.
Coping with Muller's ratchet
The accumulation of mildly deleterious mutations in symbionts due to repeated bottlenecking during transmission between hosts has been used to predict the irreversible degradation of symbiont genomes via the process of Muller's ratchet [48]. Selection for more effective repair or recombination systems may mitigate the effects of the ratchet upstream in the process. Both Wolbachia genomes appear to contain a functional set of DNA repair enzymes. Two genes in wBm and five genes in wMel encoding recombination and/or repair proteins were affected by positive selection. Muller's ratchet could be mitigated by genetic recombination among divergent strains of Wolbachia that infect a single host. However, this is not likely to occur for wBm where multiple divergent strains of Wolbachia do not coexist within a single host. Multiple genes involved with aminoacylation of tRNAs were also affected by positive selection (see Additional file 1). These proteins ensure fidelity of translation by providing error correction [49]. The prevalence of selection was roughly equal in wBm and wMel (six vs. nine genes, respectively) and could represent another strategy for minimizing effects of other sources of error on protein performance.
Variable environments
The accumulation of slightly deleterious mutants in Buchnera [48] by the process of Muller's ratchet has predicted the importance of chaperones in maintaining the integrity of proteins in symbionts. Evidence for positive selection on groEL in Buchnera has been interpreted as support for the action of the ratchet [50]. The protein, GroEL compensates for mildly deleterious mutations by permitting proper structures to form. This screen has identified selection on other heat shock genes and regulators of the heat shock process in including dnaK [50], htpG [51]hscA [52], several clp genes [53], and multiple proteases (Fig. 5). The prevalence of selection on genes encoding heat shock proteins is higher in wMel than in wBm. Several heat shock encoding genes do display evidence of diversifying in wBm under less stringent rejection criteria (see Additional file 1). It is possible that the heat shock system forms part of a strategy for dealing with variable environments that may include, exposure to changing temperatures, different intracellular and extracellular environments. While the superhosts of filarial Wolbachia include the insect vector, the vast proportion of the nematode lifecycle is spent within a mammalian host. Shifts between mammalian and insect hosts would expose the Wolbachia to different temperatures and though required for long-term survival would be temporally infrequent. Wolbachia that infect arthropods on the other hand may be exposed to more fluctuating temperature regimes on shorter time scales as insects cannot thermoregulate and thus body temperature is more likely to vary over a 24-hour period. These Wolbachia are also likely to be exposed to both extracellular as well as diverse intracellular environments [15,16].
Figure 5.
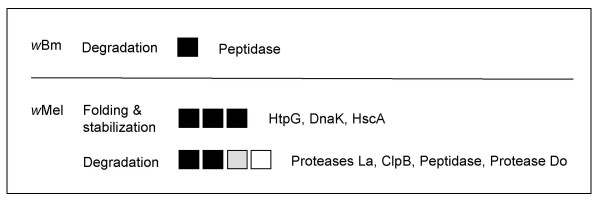
Protein fate. Specific genes and dN/dS exhibiting selection (BH p = 0.001 model and Fisher's exact test) by select functional role for each genome. Each box corresponds to a significant gene in the sub role and degree of shading indicates magnitude of dN/dS. "Unrep" indicates the ratio was at the reportable limit of codeml (see methods). Names of gene products are listed in order to the right of boxes. See Additional file 1 for individual p values, gene ids/descriptions, WD#s, and dN & dS values.
In wMel one of the genes encoding part of the two-component system also exhibited evidence of positive selection (see Additional file 1, signal transduction). The two-component system forms the basis of a small-molecule signaling pathway and is thought to play a role in quorum sensing [54]. In other bacteria these pathways affect exopolysaccharide synthesis, biofilm formation, motility, cell differentiation, and virulence. Genes comprising quorum-sensing systems have previously been shown to be targets of selection [55]. Selection on this pathway in wMel may indicate a mechanism for rapidly inducing widespread transcriptional changes in response to shifting habitats.
Direct contact with the host
The cell envelope and surface proteins represent the most obvious candidates for host interaction. Strong diversifying selection on genes encoding surface proteins in parasites, including Wolbachia [27] has been well documented [56]. Five genes in wMel and one gene in wBm involved with biosynthesis of peptidoglycan or cell envelope assembly were positively selected (Fig. 6). Peptidoglycan serves as one of the primary recognition molecules for the insect innate immune response and host immune systems exploit variation in the structure and metabolism of peptidoglycan for the recognition of invading bacteria [57]. It is possible that the diversifying selection in cell envelope synthesis genes has been driven by immune evasion pressure. This may be especially important in wMel in times of extracellularity such as infection of hemolymph [15,16]. An alternate explanation is that because production of peptidoglycan is tightly linked with DNA replication, cell growth, and cell division, selection pressure on aspects of bacterial growth including growth phase and growth rate may have driven selection in peptidoglycan synthesis [58]. Several other genes encoding outer membrane proteins also exhibited evidence of selection in both genomes (Fig. 6 other and see Additional file 1, Hypotheticals).
Figure 6.
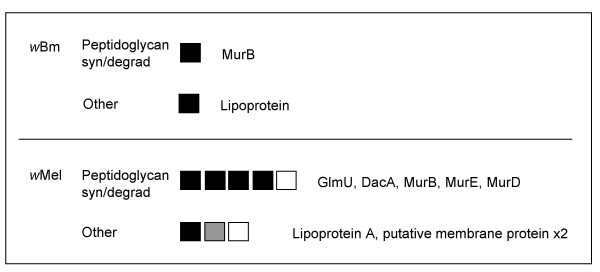
Cell envelope. Specific genes and dN/dS exhibiting selection (BH p = 0.001 model and Fisher's exact test) by select functional role for each genome. Each box corresponds to a significant gene in the sub role and degree of shading indicates magnitude of dN/dS. "Unrep" indicates the ratio was at the reportable limit of codeml (see methods). Names of gene products are listed in order to the right of boxes. See Additional file 1 for individual p values, gene ids/descriptions, WD#s, and dN & dS values.
Communication with the host & extracellular environment
For an intracellular microbe, secretion and import represent the main route of communication with the host and the extracellular environment. While both wMel and wBm must communicate with their primary hosts, filarial Wolbachia may also play a role in communication with the mammalian or insect super hosts via their occupation of the hypodermal cells. These cells form channels and are involved with secretion between nematode and super hosts [59,60]. A large number of genes encoding proteins underlying secretion pathways were under selection, three in wBm and four in wMel (Fig. 7). These genes represent the Type I secretion system (ABC transporter), Type IV secretion system (vir genes), and SRP (signal recognition protein) and Sec pathways (secY, yidC, and yajC). The Type I system is widespread in bacteria and aids in the secretion of diverse proteins [61]. Type IV secretion facilitates host-endosymbiont communication in a broad range of intracellular bacteria [62]. The Sec pathway comprises chaperones, transport machinery, and a system of pores that carry proteins from the ribosome to the extracellular space. The SRP pathway targets proteins from the ribosome to Sec pathway pores [63].
Figure 7.
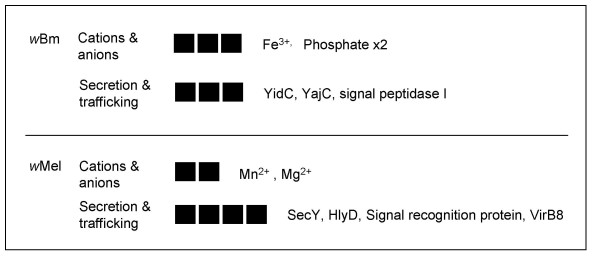
Transport. Specific genes and dN/dS exhibiting selection (BH p = 0.001 model and Fisher's exact test) by select functional role for each genome. Each box corresponds to a significant gene in the sub role and degree of shading indicates magnitude of dN/dS. "Unrep" indicates the ratio was at the reportable limit of codeml (see methods). Names of gene products are listed in order to the right of boxes. See Additional file 1 for individual p values, gene ids/descriptions, WD#s, and dN & dS values.
Ankyrin repeat domain-containing proteins are common in eukaryotes and viruses and are thought to mediate protein-protein interactions. ANK encoding genes are unusually common in the Wolbachia genome relative to other bacteria. The ANK containing proteins are especially interesting in the Wolbachia system given their possible involvement in determining reproductive phenotypes or host specificity [64,65]. In Anaplasma phagocytophilum [66], one of these proteins is secreted into the host cell where it binds host chromatin and may affect host gene expression. Only one gene encoding an ANK protein exhibited diversifying selection in our screen (Fig. 3). The functional role of this protein in Wolbachia is not known.
Conclusion
There are a number of caveats associated with the interpretation of genome wide screens for selection [67]. The methods employed here should be fairly conservative given, the use of per gene measures of dN/dS that are more likely to detect only dominant features of a gene, the statistical tests of difference between dN & dS, and use of multiple test correction procedures. We cannot completely exclude issues of saturation and increased fixation of nonsynonymous mutations in populations with small Ne [33]. The results are also highly defined by the choice of outgroup. As more genome sequences become available future screens between strains within the Wolbachia genus may provide finer scale comparison among lineages. The trends identified here in terms of biological process, while not proof of adaptation, highlight the most likely points of interaction between hosts and symbionts. These areas may be targeted for empirical study in hopes of better understanding the mechanistic basis of Wolbachia symbiosis.
From this screen we would suggest the following hypotheses for further empirical testing. Both wMel and wBm may provision hosts with the following compounds or their intermediates; heme, riboflavin, ubiquinone, folate, and nucleotides. Rearing the hosts under restricted diets or more natural field conditions could reveal yet undescribed Wolbachia associated fitness benefits, particularly in insects. Regulating DNA replication and cell division may not only be a requirement for successful intracellularity, but also the key to adaptation to diverse cellular environments, temperatures, and host ranges in insect hosts. Enhanced DNA repair, improved translation fidelity, and the heat shock response may be adaptive responses to the action of Muller's ratchet in these small bottlenecked populations. The heat shock response in combination and two-component switching may be employed by insect Wolbachia to cope with variable host environments. Communication with the host is fundamental for both Wolbachia and evidence of diversifying selection is present in multiple secretion pathways. Insect Wolbachia are uniquely experiencing selection on cell envelope synthesis genes. This may reflect a greater exposure to effector molecules of the host immune system.
Methods
Selection and alignment of gene orthologs
Anaplasma marginale (St. marie's strain) [68] was selected as the outgroup as it is the closest known relative to Wolbachia [23]. A member of the α-proteobacteria, A. marginale is a pathogen of cattle that is vectored primarily by ticks [68]. Sequences of wMel, wBm and A. marginale protein encoding genes – 1195, 805 and 949 respectively – were obtained from the RefSeq database. Proteins were considered orthologous if each combination of Blast searches (six in this three-way comparison) identified the same gene as the best scoring match [25,69]. Ambiguous matches with little sequence similarity and very short alignments were eliminated by accepting only Blast hits with e-values less than or equal to 1 × 10-6. All known pseudogenes and phage sequences were excluded. The amino acid sequences for the 591 orthologs selected by the above procedure were then aligned with ClustalW ver. 1.83 [70] using default parameters and the resulting alignments back-translated into their DNA sequence, preserving patterns of indels from protein alignments.
Inference of positive selection
The likelihood ratio test of the null hypothesis of constant rates of nonsynonymous substitutions per nonsynonymous site over synonymous substitutions per synonymous site (dN/dS) among all three lineages was performed on each triplet of genes using codon-based maximum likelihood models. The models were implemented using codeml – a program for codon-based substitution models from PAML package ver. 3.14 [24]. All models were implemented to utilize one dN/dS ratio among all amino acid sites [71]. The likelihood test was performed as a one-sided chi-square test of the null hypothesis H0 assuming one dN/dS ratio among all three lineages versus alternative hypotheses HA and HB allowing for two dN/dS ratios – one for wBm or wMel respectively, and a second for the remaining two lineages (branch-specific model).
Obtained log likelihood ratios were tested for significance using the upper critical value of chi-square distribution for one degree of freedom. The null hypothesis of constant dN/dS ratio among all three lineages was rejected when two times the log likelihood was greater than 3.84. A Benjamini & Hochberg multiple test correction [72] was employed in combination with a critical rejection value, α = 0.001. As random numbers are used to start the maximum likelihood iterations, we repeated the above analysis five times to check for convergence of the models. Average value and standard deviation of the focal lineages dN/dS ratios were used to assess model convergence. The supplemental tables report mean dN and dS values across the five replicate analyses. A number of genes with very small mean dS produced artificially inflated ratios at the reportable limit of codeml (999). In these cases the ratios themselves are not particularly informative (Fig. 2, dN/dS = 999, unreportable). We therefore have used Fisher's exact test [73] (p ≤ 0.001 & Benjamini & Hochberg multiple test correction) [72] for all loci to identify genes where dN was significantly different (and larger) from dS. All genes of interest reported here have therefore met both the significance criteria under the appropriate model of selection and possess report a mean dN that is significantly different and greater from the mean dS.
Genome characteristics and assumptions of Codeml
The assumptions of codeml include similarity of base composition and codon usage patterns as well as calculable genetic distances across the sequences being compared. The wBm and wMel genomes have very similar base compositions, 34.1 [10] and 35.2% GC [11]. Anaplasma marginale is 48.9% GC [68]. A comparison of codon usage patterns between the three genomes by paired t-tests revealed no statistical differences (data not shown). Mean dN and dS values were 0.056 ± 0.002 and 0.14 ± 0.008 for wBm/HA and 0.049 ± 0.001 and 0.10 ± 0.007 for wMel/HB, respectively. Genetic distances are large enough that dN/dS [74] should not suffer from a time lag. Alternatively, genes experiencing a high degree of divergence and more specifically saturation could lead to overestimates of dN/dS. Anismova et al modeled the effects of various parameters including divergence on both power and accuracy of the likelihood ratio test [75]. Our datasets (three taxa, mean gene length in codons ≈ 343 Transition/transversion ratio ≈ 4.0, and median dN/dS ≈ 0.3 for both HA and HB) are most similar to the reported results of experiment C. These simulations identified no type I error at α = 0.01. This study relies on a more stringent α and inspection of the data indicates that most significant genes possess high dN values relative to dS (see Additional file 1) and are therefore not likely to be artifacts of saturation.
Authors' contributions
JB and MA carried out the genome analysis, participated in the design of the study, contributed to data interpretation, and assisted with drafting the manuscript. BS assisted with data analysis & interpretation and contributed to drafts of the manuscript. EM conceived the study, participated in the design, assisted with data interpretation, and drafted the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Codeml output & Fisher's exact test for all 591 orthologs under each evolutionary model. The following information is listed for all orthologs under model HA or HB; A. marginale, wBm, and wMel gene ids, wMel WD locus id, uncorrected and corrected p-value for fit to model, annotated functional roles both main and sub, N (number of nonsynonymous sites), S (number of synonymous sites), dN, dS, dN/dS, Fisher's exact test uncorrected and corrected p-values. Two additional sheets list only the subset of genes demonstrating significant evidence of positive selection.
Acknowledgments
Acknowledgements
The authors wish to thank Gavin Huttley, Cynthia Riginos, and five anonymous reviewers for advice regarding data analysis and Scott O'Neill for his ongoing collaboration. The research was supported by The Australian Research Council through Discovery Project Grant DP557987 to CI's Brownlie and McGraw.
Contributor Information
Jeremy C Brownlie, Email: j.brownlie@uq.edu.au.
Marcin Adamski, Email: m.adamski@uq.edu.au.
Barton Slatko, Email: dnaseq@neb.com.
Elizabeth A McGraw, Email: e.mcgraw@uq.edu.au.
References
- O'Neill SL, Hoffmann AA, Werren JH. Influential passengers : inherited microorganisms and arthropod reproduction. Oxford ; New York, Oxford University Press; 1997. p. xi, 214. [Google Scholar]
- Tsuchida T, Koga R, Fukatsu T. Host plant specialization governed by facultative symbiont. Science. 2004;303:1989. doi: 10.1126/science.1094611. [DOI] [PubMed] [Google Scholar]
- Fleury F, Vavre F, Ris N, Fouillet P, Bouletreau M. Physiological cost induced by the maternally-transmitted endosymbiont Wolbachia in the Drosophila parasitoid Leptopilina heterotoma. Parasitology. 2000;121 Pt 5:493–500. doi: 10.1017/s0031182099006599. [DOI] [PubMed] [Google Scholar]
- Sicard M, Hinsinger J, Le Brun N, Pages S, Boemare N, Moulia C. Interspecific competition between entomopathogenic nematodes (Steinernema) is modified by their bacterial symbionts (Xenorhabdus) BMC Evol Biol. 2006;6:68. doi: 10.1186/1471-2148-6-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale C, Moran NA. Molecular interactions between bacterial symbionts and their hosts. Cell. 2006;126:453–465. doi: 10.1016/j.cell.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Taylor MJ, Hoerauf A. Wolbachia bacteria of filarial nematodes. Parasitology Today. 1999;15:437–442. doi: 10.1016/s0169-4758(99)01533-1. [DOI] [PubMed] [Google Scholar]
- Taylor MJ, Bandi C, Hoerauf A. Wolbachia bacterial endosymbionts of filarial nematodes. Adv Parasitol. 2005;60:245–284. doi: 10.1016/S0065-308X(05)60004-8. [DOI] [PubMed] [Google Scholar]
- Casiraghi M, Anderson TJ, Bandi C, Bazzocchi C, Genchi C. A phylogenetic analysis of filarial nematodes: comparison with the phylogeny of Wolbachia endosymbionts. Parasitology. 2001;122 doi: 10.1017/s0031182000007149. [DOI] [PubMed] [Google Scholar]
- Moran NA. Tracing the evolution of gene loss in obligate bacterial symbionts. Curr Opin Microbiol. 2003;6:512–518. doi: 10.1016/j.mib.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Foster J, Ganatra M, Kamal I, Ware J, Makarova K, Ivanova N, Bhattacharyya A, Kapatral V, Kumar S, Posfai J, Vincze T, Ingram J, Moran L, Lapidus A, Omelchenko M, Kyrpides N, Ghedin E, Wang S, Goltsman E, Joukov V, Ostrovskaya O, Tsukerman K, Mazur M, Comb D, Koonin E, Slatko B. The Wolbachia genome of Brugia malayi: endosymbiont evolution within a human pathogenic nematode. PLoS Biol. 2005;3:e121. doi: 10.1371/journal.pbio.0030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Sun LV, Vamathevan J, Riegler M, Deboy R, Brownlie JC, McGraw EA, Martin W, Esser C, Ahmadinejad N, Wiegand C, Madupu R, Beanan MJ, Brinkac LM, Daugherty SC, Durkin AS, Kolonay JF, Nelson WC, Mohamoud Y, Lee P, Berry K, Young MB, Utterback T, Weidman J, Nierman WC, Paulsen IT, Nelson KE, Tettelin H, O'Neill SL, Eisen JA. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2004;2:E69. doi: 10.1371/journal.pbio.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry AJ, Palmer MR, Rand DM. Variable fitness effects of Wolbachia infection in Drosophila melanogaster. Heredity. 2004;93:379–389. doi: 10.1038/sj.hdy.6800514. [DOI] [PubMed] [Google Scholar]
- Harcombe W, Hoffmann AA. Wolbachia effects in Drosophila melanogaster: in search of fitness benefits. J Invertebr Pathol. 2004;87:45–50. doi: 10.1016/j.jip.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Werren JH, Zhang W, Guo LR. Evolution and phylogeny of Wolbachia: reproductive parasites of arthropods. Proc Biol Sci. 1995;261:55–63. doi: 10.1098/rspb.1995.0117. [DOI] [PubMed] [Google Scholar]
- Dobson SL, Bourtzis K, Braig HR, Jones BF, Zhou W, Rousset F, O'Neill SL. Wolbachia infections are distributed throughout insect somatic and germ line tissues. Insect Biochemistry and Molecular Biology. 1999;29:153–160. doi: 10.1016/s0965-1748(98)00119-2. [DOI] [PubMed] [Google Scholar]
- Rasgon JL, Gamston CE, Ren X. Survival of Wolbachia pipientis in cell-free medium. Appl Environ Microbiol. 2006;72:6934–6937. doi: 10.1128/AEM.01673-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH, O'Neill SL. The evolution of heritable symbionts. In: O'Neill SL, Hoffmann AA and Werren JH, editor. Influential Passengers: inherited microorganisms and arthropod reproduction. Oxford, Oxford University Press; 1997. pp. 1–41. [Google Scholar]
- Stouthamer R, Breeuwer JA, Hurst GD. Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annu Rev Microbiol. 1999;53:71–102. doi: 10.1146/annurev.micro.53.1.71. [DOI] [PubMed] [Google Scholar]
- McGraw EA, Merritt DJ, Droller JN, O'Neill SL. Wolbachia-mediated sperm modification is dependent on the host genotype in Drosophila. Proc R Soc Lond B Biol Sci. 2001;268:2565–2570. doi: 10.1098/rspb.2001.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw EA, Merritt DJ, Droller JN, O'Neill SL. Wolbachia density and virulence attenuation after transfer into a novel host. Proc Natl Acad Sci U S A. 2002;99:2918–2923. doi: 10.1073/pnas.052466499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds KT, Hoffmann AA. Male age, host effects and the weak expression or non-expression of cytoplasmic incompatibility in Drosophila strains infected by maternally transmitted Wolbachia. Genet Res. 2002;80:79–87. doi: 10.1017/s0016672302005827. [DOI] [PubMed] [Google Scholar]
- Mouton L, Henri H, Bouletreau M, Vavre F. Effect of temperature on Wolbachia density and impact on cytoplasmic incompatibility. Parasitology. 2006;132:49–56. doi: 10.1017/S0031182005008723. [DOI] [PubMed] [Google Scholar]
- Dumler JS, Barbet AF, Bekker CP, Dasch GA, Palmer GH, Ray SC, Rikihisa Y, Rurangirwa FR. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and 'HGE agent' as subjective synonyms of Ehrlichia phagocytophila. Int J Syst Evol Microbiol. 2001;51:2145–2165. doi: 10.1099/00207713-51-6-2145. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Clark AG, Glanowski S, Nielsen R, Thomas PD, Kejariwal A, Todd MA, Tanenbaum DM, Civello D, Lu F, Murphy B, Ferriera S, Wang G, Zheng X, White TJ, Sninsky JJ, Adams MD, Cargill M. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science. 2003;302:1960–1963. doi: 10.1126/science.1088821. [DOI] [PubMed] [Google Scholar]
- Chen SL, Hung CS, Xu J, Reigstad CS, Magrini V, Sabo A, Blasiar D, Bieri T, Meyer RR, Ozersky P, Armstrong JR, Fulton RS, Latreille JP, Spieth J, Hooton TM, Mardis ER, Hultgren SJ, Gordon JI. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: A comparative genomics approach. Proc Natl Acad Sci U S A. 2006;103:5977–5982. doi: 10.1073/pnas.0600938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins FM, Hurst GD, Yang Z. Host-symbiont conflicts: positive selection on an outer membrane protein of parasitic but not mutualistic Rickettsiaceae. Mol Biol Evol. 2002;19:1341–1349. doi: 10.1093/oxfordjournals.molbev.a004195. [DOI] [PubMed] [Google Scholar]
- Civetta A. Positive selection within sperm-egg adhesion domains of fertilin: an ADAM gene with a potential role in fertilization. Mol Biol Evol. 2003;20:21–29. doi: 10.1093/molbev/msg002. [DOI] [PubMed] [Google Scholar]
- Travers SA, O'Connell MJ, McCormack GP, McInerney JO. Evidence for heterogeneous selective pressures in the evolution of the env gene in different human immunodeficiency virus type 1 subtypes. J Virol. 2005;79:1836–1841. doi: 10.1128/JVI.79.3.1836-1841.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MA, Moran NA, Baumann P. Sequence evolution in bacterial endosymbionts having extreme base compositions. Mol Biol Evol. 1999;16:1586–1598. doi: 10.1093/oxfordjournals.molbev.a026071. [DOI] [PubMed] [Google Scholar]
- Canback B, Tamas I, Andersson SG. A phylogenomic study of endosymbiotic bacteria. Mol Biol Evol. 2004;21:1110–1122. doi: 10.1093/molbev/msh122. [DOI] [PubMed] [Google Scholar]
- Fry AJ, Wernegreen JJ. The roles of positive and negative selection in the molecular evolution of insect endosymbionts. Gene. 2005;355:1–10. doi: 10.1016/j.gene.2005.05.021. [DOI] [PubMed] [Google Scholar]
- Woolfit M, Bromham L. Increased rates of sequence evolution in endosymbiotic bacteria and fungi with small effective population sizes. Mol Biol Evol. 2003;20:1545–1555. doi: 10.1093/molbev/msg167. [DOI] [PubMed] [Google Scholar]
- Baumann P, Baumann L, Lai CY, Rouhbakhsh D, Moran NA, Clark MA. Genetics, physiology, and evolutionary relationships of the genus Buchnera: intracellular symbionts of aphids. Annu Rev Microbiol. 1995;49:55–94. doi: 10.1146/annurev.mi.49.100195.000415. [DOI] [PubMed] [Google Scholar]
- Rio RV, Lefevre C, Heddi A, Aksoy S. Comparative genomics of insect-symbiotic bacteria: influence of host environment on microbial genome composition. Appl Environ Microbiol. 2003;69:6825–6832. doi: 10.1128/AEM.69.11.6825-6832.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heddi A, Grenier AM, Khatchadourian C, Charles H, Nardon P. Four intracellular genomes direct weevil biology: nuclear, mitochondrial, principal endosymbiont, and Wolbachia. Proc Natl Acad Sci U S A. 1999;96:6814–6819. doi: 10.1073/pnas.96.12.6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil R, Silva FJ, Zientz E, Delmotte F, Gonzalez-Candelas F, Latorre A, Rausell C, Kamerbeek J, Gadau J, Holldobler B, van Ham RC, Gross R, Moya A. The genome sequence of Blochmannia floridanus: comparative analysis of reduced genomes. Proc Natl Acad Sci U S A. 2003;100:9388–9393. doi: 10.1073/pnas.1533499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabachi A, Ishikawa H. Provision of riboflavin to the host aphid, Acyrthosiphon pisum, by endosymbiotic bacteria, Buchnera. J Insect Physiol. 1999;45:1–6. doi: 10.1016/s0022-1910(98)00104-8. [DOI] [PubMed] [Google Scholar]
- McGarry HF, Egerton GL, Taylor MJ. Population dynamics of Wolbachia bacterial endosymbionts in Brugia malayi. Mol Biochem Parasitol. 2004;135:57–67. doi: 10.1016/j.molbiopara.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Fenn K, Blaxter M. Quantification of Wolbachia bacteria in Brugia malayi through the nematode lifecycle. Mol Biochem Parasitol. 2004;137:361–364. doi: 10.1016/j.molbiopara.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Ikeda T, Ishikawa H, Sasaki T. Regulation of Wolbachia density in the Mediterranean flour moth, Ephestia kuehniella, and the almond moth, Cadra cautella. Zoolog Sci. 2003;20:153–157. doi: 10.2108/zsj.20.153. [DOI] [PubMed] [Google Scholar]
- Ijichi N, Kondo N, Matsumoto R, Shimada M, Ishikawa H, Fukatsu T. Internal spatiotemporal population dynamics of infection with three Wolbachia strains in the adzuki bean beetle, Callosobruchus chinensis (Coleoptera: Bruchidae) Appl Environ Microbiol. 2002;68:4074–4080. doi: 10.1128/AEM.68.8.4074-4080.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruang-areerate T, Kittayapong P, McGraw EA, Baimai V, O'Neill SL. Wolbachia replication and host cell division in Aedes albopictus. Curr Microbiol. 2004;49:10–12. doi: 10.1007/s00284-003-4245-8. [DOI] [PubMed] [Google Scholar]
- Dai K, Lutkenhaus J. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J Bacteriol. 1992;174:6145–6151. doi: 10.1128/jb.174.19.6145-6151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez AA, Farewell A, Nannmark U, Nystrom T. A mutation in the ftsK gene of Escherichia coli affects cell-cell separation, stationary-phase survival, stress adaptation, and expression of the gene encoding the stress protein UspA. J Bacteriol. 1997;179:5878–5883. doi: 10.1128/jb.179.18.5878-5883.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada A, Nagai K, Wachi M. A decreased level of FtsZ is responsible for inviability of RNase E-deficient cells. Genes Cells. 2005;10:733–741. doi: 10.1111/j.1365-2443.2005.00872.x. [DOI] [PubMed] [Google Scholar]
- Li C, Ichikawa JK, Ravetto JJ, Kuo HC, Fu JC, Clarke S. A new gene involved in stationary-phase survival located at 59 minutes on the Escherichia coli chromosome. J Bacteriol. 1994;176:6015–6022. doi: 10.1128/jb.176.19.6015-6022.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA. Accelerated evolution and Muller's rachet in endosymbiotic bacteria. Proc Natl Acad Sci U S A. 1996;93:2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy H, Ling J, Alfonzo J, Ibba M. Loss of editing activity during the evolution of mitochondrial phenylalanyl-tRNA synthetase. J Biol Chem. 2005;280:38186–38192. doi: 10.1074/jbc.M508281200. [DOI] [PubMed] [Google Scholar]
- Fares MA, Moya A, Barrio E. Adaptive evolution in GroEL from distantly related endosymbiotic bacteria of insects. J Evol Biol. 2005;18:651–660. doi: 10.1111/j.1420-9101.2004.00861.x. [DOI] [PubMed] [Google Scholar]
- Thomas JG, Baneyx F. Roles of the Escherichia coli small heat shock proteins IbpA and IbpB in thermal stress management: comparison with ClpA, ClpB, and HtpG In vivo. J Bacteriol. 1998;180:5165–5172. doi: 10.1128/jb.180.19.5165-5172.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickery LE, Silberg JJ, Ta DT. Hsc66 and Hsc20, a new heat shock cognate molecular chaperone system from Escherichia coli. Protein Sci. 1997;6:1047–1056. doi: 10.1002/pro.5560060511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer EC, Glover JR, Singer MA, Lindquist S. HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem Sci. 1996;21:289–296. [PubMed] [Google Scholar]
- Camilli A, Bassler BL. Bacterial small-molecule signaling pathways. Science. 2006;311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansaldi M, Dubnau D. Diversifying selection at the Bacillus quorum-sensing locus and determinants of modification specificity during synthesis of the ComX pheromone. J Bacteriol. 2004;186:15–21. doi: 10.1128/JB.186.1.15-21.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T, Ikeo K, Gojobori T. Large-scale search for genes on which positive selection my operate. Mol Biol Evol. 1996;13:685–690. doi: 10.1093/oxfordjournals.molbev.a025629. [DOI] [PubMed] [Google Scholar]
- Mengin-Lecreulx D, Lemaitre B. Structure and metabolism of peptidoglycan and molecular requirements allowing its detection by the Drosophila innate immune system. J Endotoxin Res. 2005;11:105–111. doi: 10.1179/096805105X35233. [DOI] [PubMed] [Google Scholar]
- Mengin-Lecreulx D, van Heijenoort J. Effect of growth conditions on peptidoglycan content and cytoplasmic steps of its biosynthesis in Escherichia coli. J Bacteriol. 1985;163:208–212. doi: 10.1128/jb.163.1.208-212.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair KL, Barsuhn CL, Day JS, Ho NF, Geary TG, Thompson DP. Biophysical model for organic acid excretion in Ascaris suum. Mol Biochem Parasitol. 1998;93:179–190. doi: 10.1016/s0166-6851(98)00036-x. [DOI] [PubMed] [Google Scholar]
- Blair KL, Geary TG, Mensch SK, Vidmar TJ, Li SK, Ho NF, Thompson DP. Biophysical characterization of a large conductance anion channel in hypodermal membranes of the gastrointestinal nematode, Ascaris suum. Comp Biochem Physiol A Mol Integr Physiol. 2003;134:805–818. doi: 10.1016/s1095-6433(03)00014-x. [DOI] [PubMed] [Google Scholar]
- Delepelaire P. Type I secretion in gram-negative bacteria. Biochim Biophys Acta. 2004;1694:149–161. doi: 10.1016/j.bbamcr.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol. 2005;59:451–485. doi: 10.1146/annurev.micro.58.030603.123630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M, Koch HG, Beck K, Schafer U. Protein traffic in bacteria: multiple routes from the ribosome to and across the membrane. Prog Nucleic Acid Res Mol Biol. 2001;66:107–157. doi: 10.1016/s0079-6603(00)66028-2. [DOI] [PubMed] [Google Scholar]
- Iturbe-Ormaetxe I, Burke GR, Riegler M, O'Neill SL. Distribution, expression, and motif variability of ankyrin domain genes in Wolbachia pipientis. J Bacteriol. 2005;187:5136–5145. doi: 10.1128/JB.187.15.5136-5145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkins SP, Walker T, Lynd AR, Steven AR, Makepeace BL, Godfray HC, Parkhill J. Wolbachia variability and host effects on crossing type in Culex mosquitoes. Nature. 2005;436:257–260. doi: 10.1038/nature03629. [DOI] [PubMed] [Google Scholar]
- Caturegli P, Asanovich KM, Walls JJ, Bakken JS, Madigan JE, Popov VL, Dumler JS. ankA: an Ehrlichia phagocytophila group gene encoding a cytoplasmic protein antigen with ankyrin repeats. Infect Immun. 2000;68:5277–5283. doi: 10.1128/iai.68.9.5277-5283.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum C, Hill E. Being positive about selection. PLoS Biol. 2006;4:e87. doi: 10.1371/journal.pbio.0040087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brayton KA, Kappmeyer LS, Herndon DR, Dark MJ, Tibbals DL, Palmer GH, McGuire TC, Knowles DP., Jr. Complete genome sequencing of Anaplasma marginale reveals that the surface is skewed to two superfamilies of outer membrane proteins. Proc Natl Acad Sci U S A. 2005;102:844–849. doi: 10.1073/pnas.0406656102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mural RJ, Adams MD, Myers EW, Smith HO, Miklos GL, Wides R, Halpern A, Li PW, Sutton GG, Nadeau J, Salzberg SL, Holt RA, Kodira CD, Lu F, Chen L, Deng Z, Evangelista CC, Gan W, Heiman TJ, Li J, Li Z, Merkulov GV, Milshina NV, Naik AK, Qi R, Shue BC, Wang A, Wang J, Wang X, Yan X, Ye J, Yooseph S, Zhao Q, Zheng L, Zhu SC, Biddick K, Bolanos R, Delcher AL, Dew IM, Fasulo D, Flanigan MJ, Huson DH, Kravitz SA, Miller JR, Mobarry CM, Reinert K, Remington KA, Zhang Q, Zheng XH, Nusskern DR, Lai Z, Lei Y, Zhong W, Yao A, Guan P, Ji RR, Gu Z, Wang ZY, Zhong F, Xiao C, Chiang CC, Yandell M, Wortman JR, Amanatides PG, Hladun SL, Pratts EC, Johnson JE, Dodson KL, Woodford KJ, Evans CA, Gropman B, Rusch DB, Venter E, Wang M, Smith TJ, Houck JT, Tompkins DE, Haynes C, Jacob D, Chin SH, Allen DR, Dahlke CE, Sanders R, Li K, Liu X, Levitsky AA, Majoros WH, Chen Q, Xia AC, Lopez JR, Donnelly MT, Newman MH, Glodek A, Kraft CL, Nodell M, Ali F, An HJ, Baldwin-Pitts D, Beeson KY, Cai S, Carnes M, Carver A, Caulk PM, Center A, Chen YH, Cheng ML, Coyne MD, Crowder M, Danaher S, Davenport LB, Desilets R, Dietz SM, Doup L, Dullaghan P, Ferriera S, Fosler CR, Gire HC, Gluecksmann A, Gocayne JD, Gray J, Hart B, Haynes J, Hoover J, Howland T, Ibegwam C, Jalali M, Johns D, Kline L, Ma DS, MacCawley S, Magoon A, Mann F, May D, McIntosh TC, Mehta S, Moy L, Moy MC, Murphy BJ, Murphy SD, Nelson KA, Nuri Z, Parker KA, Prudhomme AC, Puri VN, Qureshi H, Raley JC, Reardon MS, Regier MA, Rogers YH, Romblad DL, Schutz J, Scott JL, Scott R, Sitter CD, Smallwood M, Sprague AC, Stewart E, Strong RV, Suh E, Sylvester K, Thomas R, Tint NN, Tsonis C, Wang G, Williams MS, Williams SM, Windsor SM, Wolfe K, Wu MM, Zaveri J, Chaturvedi K, Gabrielian AE, Ke Z, Sun J, Subramanian G, Venter JC, Pfannkoch CM, Barnstead M, Stephenson LD. A comparison of whole-genome shotgun-derived mouse chromosome 16 and the human genome. Science. 2002;296:1661–1671. doi: 10.1126/science.1069193. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Hasegawa M. Models of amino acid substitution and applications. Mol Biol Evol. 1998;15:1600–1611. doi: 10.1093/oxfordjournals.molbev.a025888. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- Zhang J, Kumar S, Nei M. Small-sample tests of episodic adaptive evolution: a case study of primate lysozymes. Mol Biol Evol. 1997;14:1335–1338. doi: 10.1093/oxfordjournals.molbev.a025743. [DOI] [PubMed] [Google Scholar]
- Rocha EP, Smith JM, Hurst LD, Holden MT, Cooper JE, Smith NH, Feil EJ. Comparisons of dN/dS are time dependent for closely related bacterial genomes. J Theor Biol. 2006;239:226–235. doi: 10.1016/j.jtbi.2005.08.037. [DOI] [PubMed] [Google Scholar]
- Anisimova M, Bielawski JP, Yang Z. Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol Biol Evol. 2001;18:1585–1592. doi: 10.1093/oxfordjournals.molbev.a003945. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Codeml output & Fisher's exact test for all 591 orthologs under each evolutionary model. The following information is listed for all orthologs under model HA or HB; A. marginale, wBm, and wMel gene ids, wMel WD locus id, uncorrected and corrected p-value for fit to model, annotated functional roles both main and sub, N (number of nonsynonymous sites), S (number of synonymous sites), dN, dS, dN/dS, Fisher's exact test uncorrected and corrected p-values. Two additional sheets list only the subset of genes demonstrating significant evidence of positive selection.


