Abstract
PPARγ is required for fat cell development and is the molecular target of antidiabetic thiazolidinediones (TZDs), which exert insulin-sensitizing effects in adipose tissue, skeletal muscle, and liver. Unexpectedly, we found that inactivation of PPARγ in macrophages results in the development of significant glucose intolerance plus skeletal muscle and hepatic insulin resistance in lean mice fed a normal diet. This phenotype was associated with increased expression of inflammatory markers and impaired insulin signaling in adipose tissue, muscle, and liver. PPARγ-deficient macrophages secreted elevated levels of factors that impair insulin responsiveness in muscle cells in a manner that was enhanced by exposure to FFAs. Consistent with this, the relative degree of insulin resistance became more severe in mice lacking macrophage PPARγ following high-fat feeding, and these mice were only partially responsive to TZD treatment. These findings reveal an essential role of PPARγ in macrophages for the maintenance of whole-body insulin action and in mediating the antidiabetic actions of TZDs.
Introduction
Insulin resistance is a primary defect underlying the development of type 2 diabetes and is a central component defining the metabolic syndrome, a constellation of abnormalities including obesity, hypertension, glucose intolerance, and dyslipidemia (1–3). Recent findings indicate a close link between chronic inflammation and insulin resistance (4–7). Pharmacologic or genetic inhibition of pathways that underlie inflammatory responses protect experimental animals from diet-induced insulin resistance, suggesting a direct role of inflammation in promoting insulin-resistant states (4–8). Interestingly, macrophages have been found to accumulate in obese adipose tissue, where they produce inflammatory mediators that may contribute to the development of insulin resistance (9, 10). Furthermore, disabling the inflammatory pathway within macrophages by creating a myeloid cell–specific knockout of IκB kinase β (IKK-β) protected mice from diet-induced insulin resistance, with hyperinsulinemic-euglycemic clamp studies clearly showing protection of skeletal muscle and liver insulin action (4). Recent studies also suggest that physiological levels of FFAs may induce inflammatory responses in macrophages and adipocytes through TLR4, providing a potential link among diet, lipid metabolism, and function of the innate immune system (11, 12).
PPARγ, a member of the nuclear receptor superfamily of ligand-dependent transcription factors, plays an essential role in fat cell development and glucose homeostasis (13–15). PPARγ is also the molecular target of thiazolidinediones (TZDs), which are insulin-sensitizing drugs used clinically in the treatment of type 2 diabetes (16–18). TZDs are likely to exert insulin-sensitizing effects by both positively and negatively regulating gene expression in several cell types and tissues, with adipose tissue, skeletal muscle, and liver being primary targets (19–25). Positive regulation of gene expression in adipocytes drives the adipogenic programs of gene expression, but the relative contributions of PPARγ target genes such as adiponectin to the insulin-sensitizing activities of TZDs remain incompletely understood.
TZDs also act through PPARγ to negatively regulate the stimulus-dependent production of numerous inflammatory mediators that promote an insulin-resistant state (26–28). Within adipocytes, TZDs inhibit the expression and/or biological effects of IL-6, TNF-α, plasminogen activator inhibitor–1 (PAI-1), monocyte chemoattractant protein–1 (MCP-1), and angiotensinogen (29). PPARγ is also expressed in macrophages, where it has been shown to inhibit TLR- and IFN-γ–initiated inflammatory responses in a gene-specific manner (28, 30). TZDs also inhibit the expression of inflammatory mediators in atherosclerotic lesions that are characterized by the accumulation of macrophage-derived foam cells (31, 32).
The observations that macrophages are recruited to adipose tissue and are a source of mediators that promote insulin resistance coupled with the known antiinflammatory activities of PPARγ in macrophages raised the question of whether macrophage PPARγ is a therapeutic target of the insulin-sensitizing activities of TZDs. Here we present evidence supporting this hypothesis and further demonstrate that macrophage PPARγ plays a role in normal glucose homeostasis in mice.
Results
PPARγ deletion in myeloid cells.
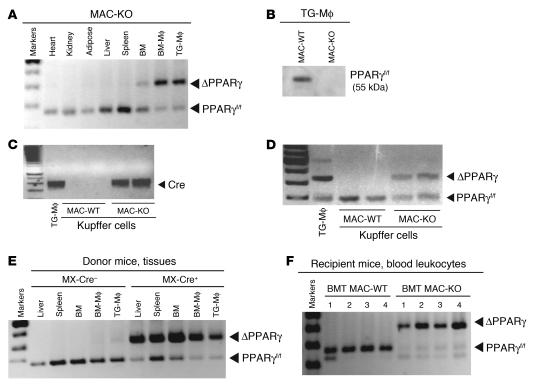
To investigate the role of macrophage PPARγ expression in the regulation of whole-body glucose metabolism and in mediating the insulin-sensitizing effects of TZDs, we used 2 independent strategies to inactivate PPARγ in macrophages in 2 different strains of mice, mixed white background and C57BL/6. In the first approach, mixed-background mice carrying 2 floxed alleles of PPARγ (PPARγf/f) were mated to mice expressing Cre recombinase under the control of the myeloid-specific lysozyme M (LysM) promoter (33). LysMCre+PPARγf/f mice exhibited more than 90% recombination of the floxed PPARγ locus in peritoneal macrophages and a corresponding decrease in PPARγ protein levels as compared with control (LysMCre–PPARγf/f) WT mice (Figure 1, A and B). Significant recombination was also observed in partially purified Kupffer cells from liver (Figure 1, C and D). In contrast, recombination of the PPARγ locus was not observed in other tissues, such as heart, kidney, adipose tissue, and spleen (Figure 1A). Throughout, we refer to LysMCre-negative littermates (LysMCre–PPARγf/f) as MAC-WT and LysMCre-positive littermates (LysMCre+PPARγf/f) as MAC-KO.
Figure 1. Efficiency of LysCre- and MXCre-mediated loxP recombination in peritoneal macrophages, BM-derived macrophages, and hepatic Kupffer cells.
(A) PCR analysis of genomic DNA isolated from the indicated tissues, BM, BM-Mφ, and TG-Mφ of conditionally targeted LysMCre+PPARγf/f (MAC-KO) mice. (B) Western blot analysis of protein isolated from peritoneal macrophages harvested from MAC-WT and MAC-KO mice. (C and D) PCR analysis of genomic DNA from purified Kupffer cells harvested from MAC-WT and MAC-KO mice. (E) PCR analysis of genomic DNA isolated from hematopoietic tissues (liver, spleen, BM, BM-Mφ, and TG-Mφ) from MXCre–PPARγf/f control and conditionally targeted MXCre+PPARγf/f donor mice after poly-I:C induction. (F) PCR analysis of genomic DNA isolated from circulating blood leukocytes from BMT MAC-WT and BMT MAC-KO mice 4–6 weeks after BMT.
As a second strategy, mice carrying the floxed PPARγ alleles were also bred to a C57BL/6 mouse line expressing Cre recombinase under the control of the inducible MX promoter (34, 35). Induction of MXCre by injection of double-stranded RNA (poly-I:C) results in nearly quantitative deletion of the floxed region of the PPARγ floxed allele in hepatocytes and hematopoietic cells (ref. 33 and Figure 1E). Following poly-I:C–induced recombination of the floxed PPARγ locus, BM progenitor cells were isolated and used for BM transplantation (BMT) in lethally irradiated 6-month-old male C57BL/6 mice. We refer to mice reconstituted with BM progenitor cells obtained from poly-I:C–treated MXCre+PPARγf/f mice as BMT MAC-KO. Control BMT mice were reconstituted with BM progenitor cells obtained from poly-I:C–treated MXCre–PPARγf/f mice and are referred to as BMT MAC-WT. By 6 weeks following transplantation, peripheral blood mononuclear cells from BMT Mac-KO mice exhibited more than 90% recombination of the PPARγ locus, while MAC-WT mice exhibited no recombination (Figure 1F).
Myeloid-specific PPARγ deletion causes glucose intolerance and insulin resistance.
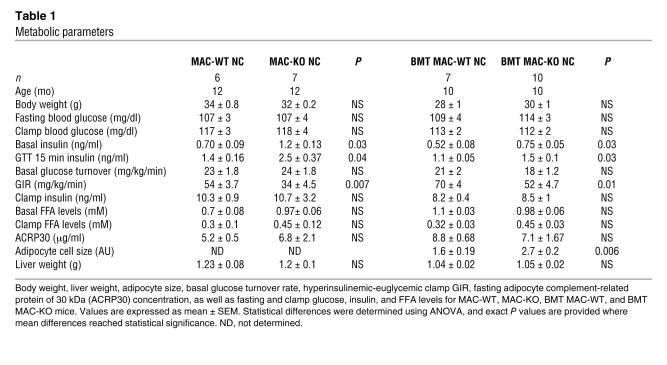
Despite some subtle baseline differences observed between the MAC-WT and BMT MAC-WT models, most likely due to background strain, age, and body weight differences in the animals used for these studies, within-model differences between WT and KO mice were quite consistent and not strain dependent. Thus, basal fasting insulin levels were elevated significantly in MAC-KO and BMT MAC-KO compared with respective WT control mice (Table 1). Macrophage-specific disruption of PPARγ caused glucose intolerance in both the 12-month-old mixed white background MAC-KOs as well as the 10-month-old C57BL/6 background BMT MAC-KO mice, as shown by the exaggerated glucose excursion curves during glucose tolerance tests (GTTs) (Figure 2, A and B). The standard glucose challenge also elicited a significantly elevated insulin response at 15 minutes (Table 1) in MAC-KO and BMT MAC-KO mice, suggestive of peripheral insulin resistance.
Table 1 .
Metabolic parameters
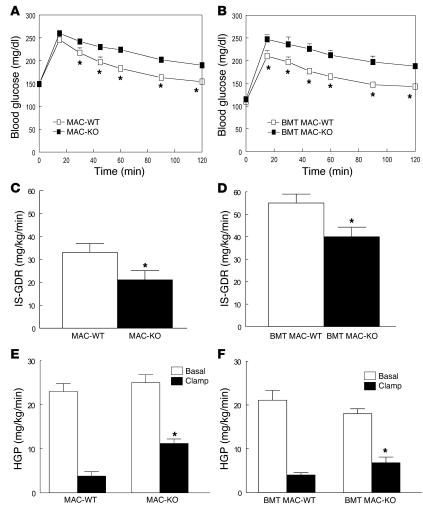
Figure 2. Macrophage-specific PPARγ gene deletion causes glucose intolerance and skeletal muscle and hepatic insulin resistance.
GTTs were performed following a 6-hour fast in 12-month-old MAC-WT (n = 6) versus MAC-KO (n = 7) (A) and 10-month-old BMT MAC-WT (n = 7) versus BMT MAC-KO (n = 10) mice (B). *P < 0.05, mean values for WT (open squares) versus KO (filled squares); repeated-measures ANOVA with Tukey’s post-hoc procedure. Insulin’s ability to stimulate glucose disposal (IS-GDR) into skeletal muscle of 12-month-old MAC-WT (n = 6) versus MAC-KO (n = 7) (C) and 10-month-old BMT MAC-WT (n = 7) versus BMT MAC-KO (n = 10) mice (D). IS-GDR values are expressed as mean ± SEM. Mean differences were detected using 1-way ANOVA. *P < 0.05 between genotypes. Insulin’s ability to suppress HGP was determined in MAC-WT versus MAC-KO (E) and BMT MAC-WT versus BMT MAC-KO (F) mice. HGP values are expressed as mean ± SEM for basal conditions versus clamp. Mean differences between genotypes within condition were detected using 1-way ANOVA. *P < 0.05 between genotypes, within condition.
Hyperinsulinemic-euglycemic clamp studies were performed in normal chow–fed (NC-fed) animals to more accurately quantify whole-body alterations in insulin sensitivity and delineate the tissue-specific site(s) responsible for diminished glucose tolerance. While there were no differences in basal glucose turnover between WT and KO mice for either genetic strategy (Table 1), during insulin stimulation, the amount of exogenous glucose required to maintain euglycemia during the clamp was significantly diminished in both strains of macrophage-specific PPARγ-KO versus WT mice (Table 1). The insulin-stimulated glucose disposal rate (IS-GDR), primarily reflecting skeletal muscle glucose disposal, was significantly diminished in MAC-KO and BMT MAC-KO versus WT mice (Figure 2, C and D). This finding is quite striking for the BMT MAC-KO mice, given that the mice were exposed to PPARγ-devoid myeloid cells for only 16 weeks. Insulin’s ability to suppress hepatic glucose production (HGP) was also impaired in both KO models (Figure 2, E and F), as evidenced by the increased rate of glucose production during insulin stimulation (clamp). In a separate set of studies, we performed transplantation of marrow from WT mice into MAC-WT and MAC-KO mice (Supplemental Figure 1, A–D; supplemental material available online with this article; doi:10.1172/JCI31561DS1). The replacement of PPARγ-competent myeloid cells in the MAC-KO mice led to complete normalization of glucose tolerance (Supplemental Figure 1, A and B) as well as skeletal muscle (Supplemental Figure 1C) and hepatic (Supplemental Figure 1D) insulin sensitivity. Taken together, these data show that loss of macrophage PPARγ expression causes systemic glucose intolerance as well as skeletal muscle and liver insulin resistance.
Impaired insulin signaling in skeletal muscle from BMT MAC-KO mice.
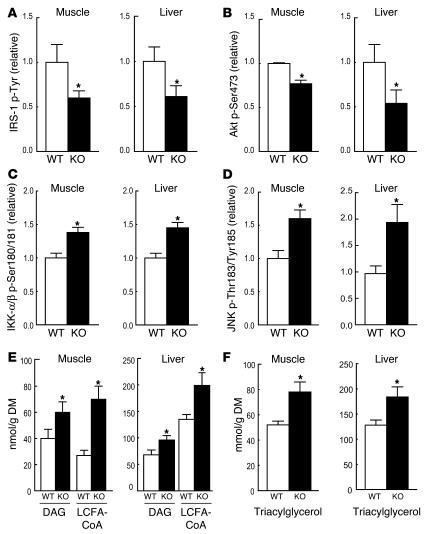
We assessed the effect of macrophage-specific PPARγ KO on insulin signaling in skeletal muscle and liver and found a significant reduction in insulin-stimulated tyrosine phosphorylation of insulin receptor substrate–1 (IRS-1) (Figure 3A) along with a corresponding decrease in serine phosphorylation of Akt in both tissues (Figure 3B). The changes in phosphorylation were independent of changes in total protein (data not shown). Reductions in IRS-1 and Akt phosphorylation in muscle and liver were paralleled by significant increases in the activation of IKK-α/β (phospho-IKK) (Figure 3C) and JNK (phospho-JNK) (Figure 3D), serine kinases previously shown to cause inhibition of IRS-1 function and insulin signaling (36–38). In addition, loss of macrophage-specific PPARγ led to a significant increase in skeletal muscle and liver diacylglycerol (DAG), long-chain fatty acyl-CoA (LCFACoA; Figure 3E), and triacylglycerol (Figure 3F), which are lipid metabolites implicated in insulin resistance (39–41). Together, these findings indicate that loss of PPARγ expression in macrophages results in impaired insulin signaling and accumulation of lipid metabolites in liver and muscle, which is fully consistent with the results of the glucose clamp studies and increased F4/80 staining in quadriceps muscle (Supplemental Figure 2A).
Figure 3. Macrophage-specific PPARγ gene deletion causes impaired insulin signaling and lipid accumulation in skeletal muscle and liver.
Skeletal muscle (quadriceps) and liver samples were harvested from BMT MAC-WT (n = 6; white bars) and BMT MAC-KO (n = 6; black bars) mice following the glucose clamp, and homogenates were immunoprecipitated and immunoblotted for the quantification of IRS-1 tyrosine phosphorylation (A) as well as for Akt1/2 (Ser473), (C) IKK-α/β (Ser180/181), and (D) JNK (Thr183/Tyr185) phosphorylation relative to protein content (B). Muscle and liver samples were also dehydrated and analyzed for the accumulation of lipid intermediates including diacylglycerol (DAG), long-chain fatty acyl-CoA (LCFACoA) (E), and triacylglycerol (F). All values are expressed as a mean ± SEM. Mean differences were detected using 1-way ANOVA; *P < 0.05.
Gene expression profiles in BMT MAC-KO mice.
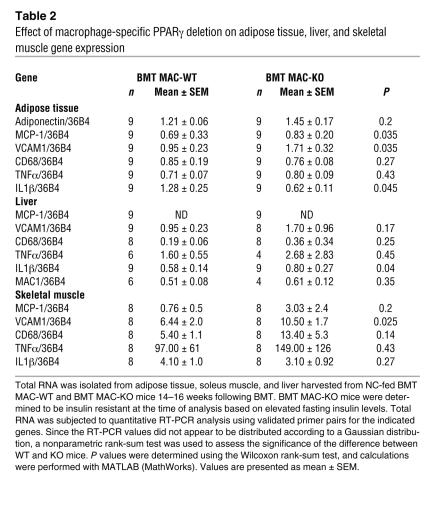
In adipose tissue, MCP-1, VCAM1, and IL-1β levels were significantly elevated in BMT MAC-KO mice (Table 2). Histological analyses of adipose tissue indicated scattered, focal increases in F4/80-positive cells and a significant increase in adipocyte size in NC-fed BMT MAC-KO animals as compared with WT (Table 1 and Supplemental Figure 2, B and C). Skeletal muscle harvested from BMT MAC-KO mice exhibited a significant increase in VCAM1 expression, while IL-1β expression was significantly elevated in liver of BMT MAC-KO mice (Table 2). These changes in gene expression are consistent with the observed increase in the active forms of IKK-α/β and JNK in these tissues (Figure 3, C and D).
Table 2 .
Effect of macrophage-specific PPARγ deletion on adipose tissue, liver, and skeletal muscle gene expression
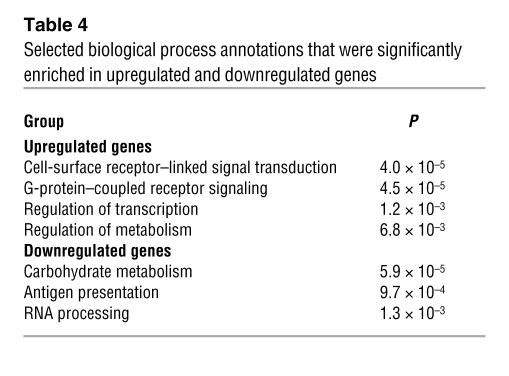
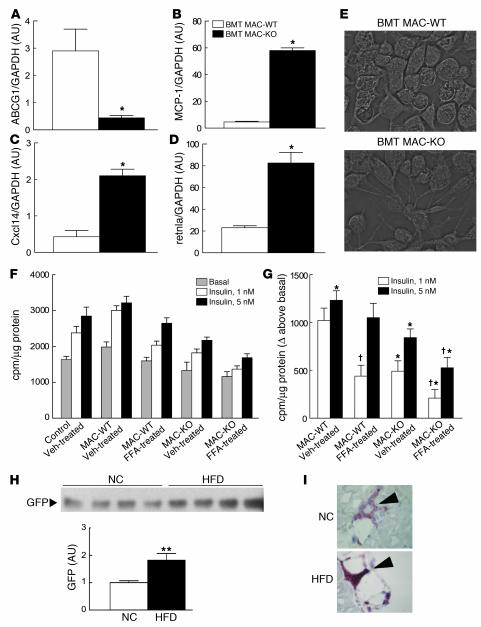
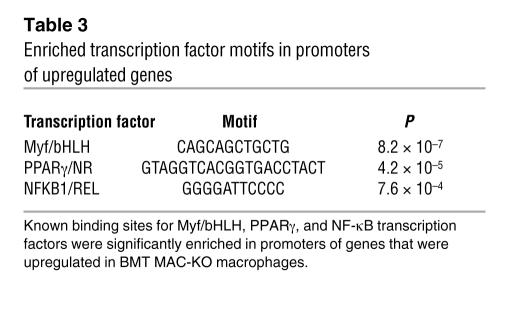
To assess the effects of loss of PPARγ on patterns of macrophage gene expression in vivo, we analyzed approximately 10,000 transcript levels in elicited peritoneal macrophages from BMT MAC-WT and BMT MAC-KO mice. Expression profiling experiments were performed 12 weeks after BM reconstitution, at which time insulin resistance had developed in BMT MAC-KO mice. More than 220 genes were upregulated greater than 2-fold (P < 0.01), and more than 170 genes were downregulated greater than 2-fold (P < 0.01) (Supplemental Table 1). Computational motif discovery analysis identified a sequence motif corresponding to the upstream half-site and 5ι conserved region of the consensus PPARγ response element as being significantly enriched in the promoters of the genes that were downregulated in PPARγ-deficient macrophages, suggesting that many of these genes are direct targets of PPARγ (Figure 4A). Conversely, binding sites for Myf/bHLH factors, PPARγ, and NF-κB/REL were overrepresented in upregulated genes (Figure 4B). Gene ontology analysis suggested complex effects of PPARγ deletion on lipid metabolism and immune function (Table 4). Enrichments for biological process annotations linked to signal transduction, transcription, and metabolism were observed in upregulated genes, while annotations linked to carbohydrate metabolism and antigen processing were enriched in downregulated genes. Approximately 15% of the differentially regulated genes lacked functional annotations. 3-Hydroxy-3-methylglutaryl-coenzyme A lyase (Hmgcl), Abcg1, aldehyde dehydrogenase 3B1 (Aldh3a1), glutaryl-coenzyme A dehydrogenase (Gcdh), peroxisomal enoyl coenzyme A hydratase 1 (Ech1), and uncoupling protein 2 (Ucp2) were among the significantly downregulated genes in BMT MAC-KO cells, while Cxcl1, Cxcl14, Ccl24, Ccr1, and resistin-like molecule–α (retnla/RELMa/FIZZ1) were among the significantly upregulated genes. Expression changes were confirmed by RT-PCR for a representative subset of these genes (Figure 5, A–D, and data not shown). Intriguingly, MCP-1 and Cxcl14 (42), chemokines involved in macrophage/dendritic cell recruitment into tissues, and retnla (43), a resistin-related molecule that is expressed in the stromal fraction of adipose tissue, were elevated 2- to 10-fold in thioglycollate-elicited peritoneal macrophages (TG-Mφ) from BMT MAC-KO mice. At a morphological level, PPARγ-deficient macrophages (BMT MAC-KO) exhibited an activated phenotype, as evidenced by the extensive stress fiber development compared with BMT MAC-WT (Figure 5E).
Figure 4. Enriched PPARγ motif in promoters of downregulated genes.
Computational motif discovery identified a motif with a significant match to the known PPARγ consensus recognition element in promoters of genes downregulated in macrophages from BMT MAC-KO compared with BMT MAC-WT.
Table 4 .
Selected biological process annotations that were significantly enriched in upregulated and downregulated genes
Figure 5. Loss of PPARγ causes macrophage activation and secretion of insulin resistance producing molecules.
Altered expression of ABCG1 (A), MCP-1 (B), Cxcl14 (C), and retnla (D) in TG-Mφ from NC-fed BMT MAC-KO (n = 9; black bars) versus BMT MAC-WT (n = 9; white bars) mice. (E) TG-Mφ showing advanced stress fiber formation in the BMT MAC-KOs (NC-fed). Impaired insulin-stimulated 2-deoxyglucose uptake into L6 myocytes incubated with CM from vehicle- (Veh-) or FFA-treated TG-Mφ harvested from BMT MAC-KO (n = 6) versus BMT MAC-WT (n = 6) mice (F and G) and increased GFP-labeled BM-derived cells in skeletal muscle from mice fed a HFD (black bar) versus NC (white bar) (H). Values for RT-PCR are expressed as mean ± SEM. *P < 0.05 between genotypes. Insulin-stimulated 2-DOG assays represent 6 wells per condition and are expressed as mean ± SEM in cpm/μg of protein for absolute tracer uptake values (F) and uptake expressed above basal (G). *P < 0.05, BMT MAC-WT versus BMT MAC-KO, within incubation condition; †P < 0.05, vehicle versus FFA, within genotype. (H) GFP immunoblots were quantified using densitometric analysis, and mean expression values ± SEM are presented. n = 4 per group. **P < 0.05 between dietary groups. (I) Immunohistochemical detection shows increased CD11c- and F4/80-expressing cells in skeletal muscle following 8 weeks of high-fat feeding.
PPARγ-deficient macrophages cause impaired insulin action in myocytes.
These observations raised the question of whether PPARγ functions in macrophages to stimulate the production of insulin-sensitizing factors and/or inhibit the secretion of factors that cause insulin resistance. To distinguish between these possibilities, we directly tested the effect of macrophage-specific PPARγ deletion on muscle insulin action in vitro. Elicited peritoneal macrophages were obtained from BMT MAC-WT and BMT MAC-KO mice and cultured in the presence or absence of FFAs that are known to activate macrophage inflammatory pathways (11). Conditioned medium (CM) from these macrophages was then tested for its effect on insulin-stimulated glucose uptake in L6 myocytes. When L6 myocytes were incubated with CM harvested from the WT and KO macrophages, we found that CM from BMT MAC-KOs caused a decrease in insulin-stimulated glucose transport into myocytes compared with CM from BMT MAC-WT macrophages (Figure 5, F and G). Specifically, CM from BMT MAC-WT macrophages was without effect on insulin-stimulated glucose transport, whereas, CM harvested from the BMT MAC-KOs inhibited insulin-mediated glucose transport by 52% (P = 0.0002; Figure 5G), indicating marked cellular insulin resistance. After macrophage FFA treatment, CM from WT cells was now able to inhibit glucose transport, but the effects of BMT MAC-KO CM were much greater (P = 0.0004; Figure 5G). These in vitro observations recapitulated the in vivo findings from the glucose clamp studies, suggesting that PPARγ deletion from macrophages results in increased production of factors that can cause insulin resistance through a paracrine or endocrine mechanism.
BM-derived cells track to skeletal muscle.
While the relationship between macrophage tissue infiltration and insulin resistance is well studied in mouse and human adipose tissue, relatively little attention has been paid to the role of macrophage infiltration in skeletal muscle. To test the potential of macrophages to influence skeletal muscle insulin signaling in a paracrine manner, we transplanted BM from transgenic mice with constitutive expression of GFP into irradiated C57BL/6 recipients (age-matched to BMT MAC-WT and BMT MAC-KO mice) fed NC or a high-fat diet (HFD). GFP expression was easily detected in skeletal muscle from mice fed NC and was significantly elevated (~2-fold; P = 0.025; Figure 5H) in skeletal muscle harvested from mice fed an HFD. Western blot analyses of GFP was further supported by immunohistochemistry studies showing increased abundance of CD11c- and F4/80-expressing cells in muscle from mice fed an HFD (Figure 5I and data not shown), suggesting that an increased number of BM-derived macrophages and dendritic cells are recruited to skeletal muscle during high-fat feeding.
Macrophage PPARγ is required for the full antidiabetic effects of TZDs.
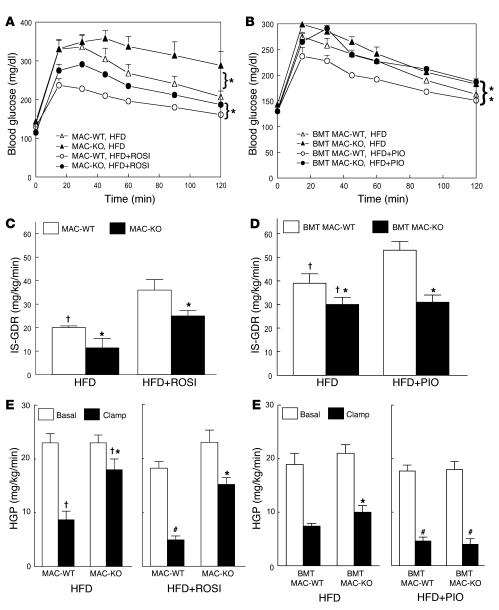
To assess the role of macrophage PPARγ in TZD-induced insulin sensitization, we treated WT and KO animals with a TZD in current clinical use, rosiglitazone or pioglitazone. Since TZDs are ineffective in animals with normal insulin sensitivity, for these studies we placed all animal groups (MAC-WT, MAC-KO, BMT MAC-WT, BMT MAC-KO) on an HFD so that both WT and KO mice would be rendered insulin resistant. As shown in Figure 6, both MAC-KO (Figure 6A) and BMT MAC-KO (Figure 6B) mice became more glucose intolerant and hyperinsulinemic (basal insulin levels 1.9-fold higher than those in WT, P = 0.04; 15-minute insulin response to GTT 2.3-fold higher than in WT, P = 0.04) on an HFD than WT controls. As expected, the HFD produced skeletal muscle (decreased IS-GDR) and hepatic (blunted HGP suppression) insulin resistance in both groups of WT animals (Figure 6, C–F), with more severe insulin resistance in the KO animals, particularly the MAC-KOs (Figure 6, C and E).
Figure 6. Macrophage-specific PPARγ deletion causes susceptibility to HFD-induced insulin resistance and diminished TZD effectiveness.
GTTs were performed on 12-month-old male MAC-KO versus MAC-WT mice fed a HFD with or without rosiglitazone (ROSI) (A) and 10-month-old male BMT MAC-WT versus BMT MAC-KO mice fed a HFD with or without pioglitazone (PIO) (B) for 8 weeks. Mean blood glucose concentrations ± SEM are shown for both groups of WT mice fed a HFD (open triangles) or HFD plus TZD (open circles) and for both groups of KO mice fed a HFD (filled triangles) or HFD plus TZD (filled circles). *P < 0.05; statistical differences were determined using repeated-measures ANOVA. (C and D) IS-GDR was determined in both groups of HFD-fed TZD-treated and untreated WT (MAC-WT and BMT MAC-WT; white bars) and KO (MAC-KO and BMT MAC-KO; black bars) mice. HGP at basal conditions (white bars) and during insulin stimulation (black bars) was determined for MAC-WT and MAC-KO (E) and for BMT MAC-WT and BMT MAC-KO (F) mice. Values are expressed as mean ± SEM (n = 5–7 mice/group). Significant differences were detected using 1-way ANOVA. †P < 0.05, NC (Figure 2, C–F) versus HFD, within genotype; *P < 0.05 between genotypes within diet and condition; #P < 0.05, HFD versus HFD plus TZD, within genotype and condition.
More importantly, while TZD treatment returned to normal values glucose tolerance in both groups of control WT mice, it did not restore normal glucose tolerance in KO mice (Figure 6, A and B). These results were confirmed and extended by the glucose clamp studies. The IS-GDR, reflecting skeletal muscle insulin sensitivity, was completely returned to normal values in both groups of control WT mice by either rosiglitazone (Figure 6C) or pioglitazone (Figure 6D) treatment. In sharp contrast, rosiglitazone had only a partial effect (Figure 6C), while pioglitazone had no effect in improving IS-GDR in BMT MAC-KO mice (Figure 6D). Similar to these observations in muscle, hepatic insulin sensitivity was restored to normal in the HFD-fed control WT animals following either rosiglitazone (Figure 6E, right panel) or pioglitazone (Figure 6F, right panel) treatment. While pioglitazone was effective at improving hepatic insulin sensitivity in the BMT MAC-KOs (Figure 6F, right panel), rosiglitazone was only partially effective in MAC-KO mice (Figure 6E, right panel). This reduced Kupffer cell depletion of PPARγ is the most likely explanation for the greater hepatic insulin sensitivity in BMT MAC-KOs (Figure 6F) compared with MAC-KOs (Figure 6E) fed a TZD-enriched HFD.
Discussion
The present studies, using 2 different strategies for deleting PPARγ from macrophages, reveal an unexpected role for macrophage PPARγ in regulating insulin action in muscle and liver. The finding that deletion of PPARγ in macrophages leads to systemic insulin resistance, even in lean animals, suggests that the macrophage may be an initiator cell type in insulin-resistant states. Furthermore, the observation that the insulin-sensitizing effects of TZDs were attenuated in the KO animals is of particular interest with respect to the use of TZDs in the treatment of insulin resistance associated with type 2 diabetes. While TZDs clearly exert insulin-sensitizing effects in adipocytes (19), liver (24), and skeletal muscle (21, 22), the present studies establish a significant role for PPARγ in macrophages as a critical therapeutic target of these compounds.
Although the 2 models exhibited qualitatively similar metabolic defects in most respects, there were some subtle differences between the MAC-KO and BMT MAC-KO mice with respect to baseline metabolic parameters and TZD responsiveness in liver. The MAC-WT and MAC-KO mice were older and slightly heavier than the those in the BMT groups, and differences in baseline values for both groups of WT mice were most likely due to effects of age, weight, and strain background. In addition, the different genetic methods used for PPARγ gene deletion in the 2 models resulted in different efficiencies of recombination/replacement in specific populations of tissue macrophages. For example, the greater hepatic TZD responsiveness in BMT MAC-KO mice as compared with MAC-KOs was most likely due to incomplete turnover of Kupffer cells (BM-derived liver macrophages) following BMT. Full replacement of these cells by BM-derived cells takes several months following transplantation, whereas Kupffer cells from MAC-KO mice are devoid of PPARγ essentially from birth. This reduced Kupffer cell depletion of PPARγ is the most likely explanation for the greater hepatic insulin sensitivity in BMT MAC-KOs compared with MAC-KO mice fed a TZD-enriched HFD.
The findings of increased activity of IKKs and JNK, impaired insulin receptor signaling, and altered levels of lipid metabolites in both muscle and liver of BMT MAC-KO mice suggests that by repressing the inflammatory response, macrophage PPARγ can inhibit the macrophage from promoting an insulin-resistant state in these tissues. Although cause-effect relationships remain to be established, altered levels of DAG, LCFACoA, and triacylglycerol are most likely secondary to activation of inflammatory pathways in our mouse models.
Gene expression and CM experiments are consistent with the notion that PPARγ negatively regulates macrophage genes that encode proteins such as released cytokines/chemokines that contribute to insulin resistance. These results are congruent with previous findings showing that PPARγ-deficient macrophages are not only resistant to antiinflammatory effects of TZDs, but are hyperresponsive to inflammatory signals (26, 28). Thus, physiologic levels of metabolites that can promote inflammation, such as FFAs, are predicted to lead to exaggerated secretion of additional inflammatory mediators in PPARγ-deficient macrophages. The findings from the gene expression analyses and studies of insulin action in liver and muscle are consistent with this notion. In addition, the results from our CM studies directly support our model, as media harvested from FFA-treated BMT MAC-KO macrophages caused a much higher degree of cellular insulin resistance (decreased insulin-stimulated glucose transport) in L6 myocytes compared with CM from FFA-treated BMT MAC-WT macrophages.
The identities of the key secreted factors that promote insulin resistance remain to be established. We performed a number of assays of CM for candidate proteins, and while some factors are measurable and change as expected (e.g., MCP-1, KC, and G-CSF), many of the proteins of greatest interest were expressed below the levels required for reliable detection (e.g., TNF-α) but still sufficient for biologic activity. Blocking antibody against TNF-α reduces, but does not abolish, the inhibitory effects of CM on insulin-dependent glucose uptake in L6 cells. Thus, it is likely that multiple factors are involved, possibly including lipid metabolites (e.g., prostaglandins; ref. 42). Of interest, expression profiling experiments revealed increased expression of retnla, a resistin-like molecule, in BMT MAC-KO macrophages, and increased expression of retnla was observed in adipose tissue of BMT MAC-KO mice following high- fat feeding. In humans, resistin itself is primarily expressed in macrophages and is negatively regulated by TZDs (43), raising the possibility that retnla and resistin have conserved functions in macrophages in mice and humans (44), respectively, and are among the significant targets of the insulin-sensitizing effects of TZDs in macrophages. Recent studies have also demonstrated that macrophages can themselves become insulin resistant (45), and it will be of interest to directly study insulin signaling in PPARγ-deficient macrophages.
It remains unclear whether the factors secreted by macrophages that promote insulin resistance function in a paracrine or endocrine manner. Since macrophages are resident in all tissues, it is possible that loss of PPARγ in macrophages results in altered paracrine communication between macrophages and other cells types in adipose tissue, skeletal muscle, and liver. The increased adipose tissue macrophage content (Supplemental Figure 2, B and D) and elevated levels of Cxcl14 (46), retnla (44), IL-1β, and JNK in adipose tissue (Supplemental Figure 2, E–H) following high-fat feeding are consistent with this possibility. Furthermore, transplantation of BM from mice with constitutive expression of GFP into irradiated C57BL/6 recipients (age-matched to BMT MAC-WT and BMT MAC-KO mice) resulted in significant GFP expression in muscle. Following an inflammatory stimulus known to cause insulin resistance, i.e., high-fat feeding, GFP expression representing the tracking of BM-derived cells to skeletal muscle was significantly increased. This increase in infiltration of BM-derived cells into skeletal muscle following 8 weeks of high-fat feeding also coincided with increased markers of antigen-presenting cells known to be activated by elevated FFAs. Our findings in mouse muscle are consistent with findings in human diabetic muscle showing a strong relationship among macrophage abundance, tissue inflammation, and insulin resistance (47).
The current studies have potential therapeutic implications. Although currently available TZDs are effective insulin sensitizers, they are also associated with side effects including weight gain, increased adiposity, and edema. It would therefore be desirable to develop selective PPARγ modulators that retain antidiabetic and antiatherogenic activities but do not have undesirable side effects. In this regard, it has recently been demonstrated that a selective PPARγ modulator that is relatively ineffective in promoting transcriptional activation is fully active as an inducer of PPARγ-mediated repression of inflammatory response genes by promoting ligand-dependent sumoylation of PPARγ (48, 49), suggesting the possibility of new avenues for development of safer, more effective drugs.
In summary, the current studies demonstrate that macrophage-specific PPARγ deletion leads to glucose intolerance, hyperinsulinemia, and systemic insulin resistance, with decreased insulin sensitivity manifested in both skeletal muscle and liver. In addition, the insulin-sensitizing effects of TZDs are attenuated in macrophage PPARγ-KO animals, indicating that macrophage PPARγ is an important therapeutic target of these drugs. In future studies, it will be important to more clearly define the key macrophage-specific PPARγ target genes responsible for these effects in liver and muscle.
Methods
Animals.
Mice carrying floxed alleles of PPARγ were created as described previously (35). PPARγf/f mice were crossed with either MXCre (34) or LysMCre (33) mice to generate mice with a macrophage-specific PPARγ gene deletion. The heterozygous offspring of both the loxP-targeted PPARγ gene (PPARγf/+) and the Cre transgene (PPARγf/+ MXCre+ or LysMCre+) were then crossed with PPARγf/f mice to generate mice homozygous for the Cre transgene and for the PPARγ floxed allele (MxCre+PPARγf/f or LysMCre+PPARγf/f). Mice, MXCre+PPARγf/f or LysMCre+PPARγf/f (MAC-KO) and PPARγf/f (MAC-WT), were interbred to generate experimental and control animals. All mice were bred on a mixed background of 129/SV and C57BL/6. Mice were genotyped by PCR using the primers 2F, 1R, and H5 as described previously (50). To induce MXCre expression, MXCre+PPARγf/f mice and PPARγf/f littermate controls were injected with poly-I:C as described previously (28). Male mice were fed either standard rodent chow or HFD (45% calories from fat; no. TD96132; Harlan Teklad) with or without rosiglitazone (3 mg/kg/d; 8 weeks) or with or without pioglitazone (15 mg/kg/d; 8 weeks). All experimental procedures were approved by the Animal Subjects Committee at UCSF according to NIH guidelines.
BMT.
BM progenitor cells were isolated by flushing the femurs and tibiae of donor animals of MXCre–PPARγf/f or MXCre+PPARγf/f mice (8–10 weeks of age) with PBS-T. T cells were depleted from BM cells by magnetic separation using anti-CD90 according to the manufacturer’s instructions (Miltenyi Biotec). Approximately 3 × 106 donor cells were transferred into γ-irradiated (10 Gy) male C57BL/6 mice (6 months old) via tail vein injection, as previously described (51). Four to 6 weeks after the BMT, mice were bled to monitor repopulation. Genomic DNA was extracted from the circulating blood leukocytes and genotyped by PCR. After 6–8 weeks recovery on the standard chow diet, animals were then randomly assigned to 1 of 3 feeding protocols for an additional 8 weeks: (a) NC; (b) HFD (no. TD96132; Harlan Teklad); (c) HFD plus pioglitazone (15 mg/kg/d).
Kupffer cell isolation.
Kupffer cells were isolated from MAC-WT and MAC-KO mice by collagenase digestion and differential centrifugation using Percoll (GE HealthCare) as described elsewhere (52) with slight modifications. Briefly, the liver was perfused in situ through the portal vein with HEPES-buffered saline at 37°C for 5 minutes followed by perfusion with HEPES containing 0.3% collagenase IV (Sigma-Aldrich) for 3 minutes. After digestion, the liver was excised, and the suspension was filtered. The filtrate was centrifuged twice at 50 g at 4°C for 2 minutes. The supernatant was collected and centrifuged at 350 g for 5 minutes, and the pellet was resuspended with buffer. The cell suspension was then layered on top of a density cushion of 20%/50% discontinuous Percoll (GE HealthCare). The Kupffer cell fraction was obtained and washed with buffer after centrifugation at 800 g for 20 minutes. The cells were sorted by FACS analysis using anti-Cd11b antibody (Dako).
Peritoneal macrophage harvesting and preparation.
Macrophages were harvested from C57BL/6 BMT mice, MAC-WT and MAC-KO, and RNA isolated for analysis of alterations in gene expression as previously described (28). Briefly, thioglycollate solution was injected into the peritoneum to elicit the accumulation of macrophages. On day 3 following injection, animals were asphyxiated with isoflurane while the peritoneum was washed 3 times with 3 ml of sterile PBS. Once the TG-Mφ were obtained, they were immediately spun down (300 g for 10 minutes at 4°C), collected into a pellet, and retained at –80°C for subsequent RNA analysis. In a separate set of experiments, TG-Mφ were harvested from both genotypes of BMT mice and used for CM studies.
CM preparation from TG-Mφ.
TG-Mφ were harvested from BMT MAC-KO (n = 6) and BMT MAC-WT (n = 6) mice as previously described (28). Cells were plated in DMEM low-glucose medium supplemented with 10% low-endotoxin FBS, penicillin/streptomycin, and glutamine (Invitrogen). TG-Mφ were treated 24 hours later with a 500 μM FFA cocktail (100 μM arachidonic acid, 100 μM lauric acid, 100 μM linoleic acid, 100 μM myristic acid, 100 μM oleic acid; Sigma-Aldrich) or vehicle control in medium containing 0.68% BSA and 0.5 % FBS for 3 hours. After treatment, cells were rinsed twice in low-glucose DMEM to remove the FFA cocktail and then were allowed to recover for 12 hours in low-glucose DMEM medium containing 2% FBS. Following the 12-hour recovery period, CM was harvested and stored in single-use aliquots at –80°C, and TG-Mφ from both genotypes of mice were lysed and RNA isolated for subsequent RT-PCR gene expression analyses.
2-Deoxyglucose uptake assay in conditioned L6 myotubes.
L6 myoblast clones overexpressing Glut4 were obtained as a gift from Amira Clip (The Hospital for Sick Children, University of Toronto, Toronto, Ontario, Canada). Cells were cultured in an MEM medium supplemented with 10% FBS under 5% CO2 at 37°C. To induce differentiation, at 80%–90% confluence L6 cells were switched to an MEM medium containing 2% FBS (medium changed every 2–3 days). Eight to 10 days after differentiation, mature L6 myotubes were incubated for 48 hours with CM (diluted 1:4 in an MEM medium plus 2% FBS) obtained from TG-Mφ harvested from BMT MAC-WT and BMT MAC-KO mice. Cellular insulin-stimulated glucose uptake was subsequently assayed. Briefly, myotubes were serum starved for 3 hours. Medium was then replaced with HEPES salt buffer, and cells were stimulated with or without insulin (1 and 5 nM) for 20 minutes. 2-Deoxy-[3H] d-glucose (0.4 μCi/ml final) was added for 10 minutes at 37°C. Reactions were terminated by washing the myotubes twice in ice-cold PBS, which was followed by cell lysis for radioactivity determination.
Assessment of glucose tolerance and insulin sensitivity in vivo.
Two weeks prior to the GTT, blood was drawn from the tail and plasma was immediately analyzed for insulin (Linco), FFA (Wako), and ACRP30 (Linco) levels. To assess glucose tolerance, dextrose (1,000 mg/kg) was injected into the peritoneum, and the glucose excursion following injection was monitored over time and compared between the genotypes. Blood samples were drawn at time 0 and at 15, 30, 45, 60, 90, and 120 minutes following dextrose injection. Blood samples were analyzed for glucose and insulin.
Glucose clamp studies were performed in chronically cannulated mice 3 days after surgery as previously described (22). A basal blood sample was taken at –90 minutes and measured for glucose, insulin, and FFA levels. Following this, a primed constant infusion of 5.0 μCi/h, 0.12 ml/h of [3-3H] d-glucose (NEN Life Science Products) was initiated. At time 0, a basal blood sample was drawn for determination of glucose-specific activity. Following basal sampling, glucose (50% dextrose; Abbott) and insulin (12 mU/kg/min; Novo Nordisk) plus tracer (5.0 μCi/h) infusions were initiated simultaneously, and glucose levels clamped at euglycemia using a variable glucose infusion rate (GIR). Steady state was achieved when blood glucose was successfully clamped and the GIR fixed for a minimum of 30 minutes. At the end of the clamp, a blood sample was taken at 100 and 120 minutes for the determination of tracer-specific activity. No difference in specific activity between these 2 time points was detected (P = 0.4). At steady state, the rate of glucose disappearance or the total GDR is equal to the sum of the rate of endogenous or HGP plus the exogenous (cold) GIR (53, 54). The IS-GDR is equal to the total GDR minus the basal glucose turnover rate.
RNA isolation and quantitative real-time-PCR.
Total RNA was isolated from skeletal muscle (soleus), liver, adipose tissue, and macrophages using TRI zol (Invitrogen) and DNase I digested and then were column purified (RNeasy MinElute Cleanup; QIAGEN). First-strand cDNA was synthesized using SuperScript III and random hexamers (Invitrogen). Samples were run in 20-μl reactions using an ABI 7300 (Applied Biosystems). Samples were incubated at 95°C for 15 minutes, followed by 40 cycles at 95°C for 10 seconds, 56°C for 20 seconds, and 72°C for 30 seconds. SYBR green oligonucleotides were used for detection and quantification of a given gene, expressed as mRNA level normalized to a standard housekeeping gene (GAPDH or 36B4) using the ΔΔCT method as described by the manufacturer (Invitrogen). We performed separate control experiments to ensure that the efficiencies of target and reference amplification were equal. The specificity of the PCR amplification was verified by melting curve analysis of the final products using Opticon 3 software (Bio-Rad).
Primer sequences.
Primer sequences were: PPARγ (forward, 5′-GCCCTTTGGTGACTTTATGG-3′; reverse, 5′-CAGCAGGTTGTCTTGGATGT-3′); ACRP30 (forward, 5′-GCTCCTGCTTTGGTCCCTCCAC-3′; reverse, 5′-GCCCTTCAGCTCCTGTCATTCC-3′); ABCG1 (forward, 5′-AAGGCCTACTACCTGGCAAAGA-3′; reverse, 5′-GCAGTAGGCCACAGGGAACA-3′); TNF-α (forward, 5′-CCAGACCCTCACACTCAGATC-3′); reverse, 5′-CACTTGGTGGTTTGCTACGAC-3′); CD-68 (forward, 5′-CTTCCCACAGGCAGCACAG-3′; reverse, 5′-AATGATGAGAGGCAGCAAGAGG-3′); F4/80 (forward, 5′-CTTGGCTATGGGCTTCCAGTC-3′; reverse, 5′-GCAAGGAGGACAGAGTTTATCGTG-3′); VCAM1 (forward, 5′-CCATTGAAGATACCGGGAAAT-3′; reverse, 5′-TAGCTGTCTGCTCCACAGGAT-3′); MCP-1 (forward, 5′-AGGTCCCTGTCATGCTTCTG-3′; reverse, 5′-GCTGCTGGTGATCCTCTTGT-3′); Cxcl14 (forward, 5′-ACAATGCCTGGAACGAGAAG-3′; reverse, 5′-TCTCTCAACTGGCCTGGAGT-3′); IL-1β (forward, 5′-TGGGCCTCAAAGGAAAGAAT-3′; reverse, 5′-CAGGCTTGTGCTCTGCTTGT-3′); retnla (forward, 5′-TCGTGGAGAATAAGGTCAAGG-3′; reverse, 5′-GGAGGCCCATCTGTTCATAG-3′); MAC-1 (forward, 5′-TGTGAGCAGCACTGAGATCC-3′; reverse, 5′-ATGAGAGCCAAGAGCACCAG-3′); JNK1 (forward, 5′-AGCAGAAGCAAACGTGACAAC-3′; reverse, 5′-GCTGCACACACTATTCCTTGAG-3′).
Western blot analysis.
Nuclear extracts from TG-Mφ were isolated according to the protocols provided in the NE-PER Nuclear and Cytoplasmic Extraction Reagents Kit (Pierce). To detect PPAR-γ by immunoblot analysis, a rabbit polyclonal antibody antibody against PPARγ (E-8; Santa Cruz Biotechnology Inc.) was used. The secondary antibody was obtained from Dako. Immunoreactive proteins were detected using chemiluminescence (Pierce).
Skeletal muscle and liver samples were pulverized in liquid nitrogen and homogenized in RIPA lysis buffer containing protease and phosphatase inhibitors. Lysates were clarified, centrifuged, and resolved by SDS-PAGE. Samples were transferred overnight to nitrocellulose membranes that were subsequently probed with the following antibodies for protein and phosphorprotein detection: actin (Sigma-Aldrich), IRS-1 (Santa Cruz Biotechnology Inc.), AKT/p-Ser473 (Santa Cruz Biotechnology Inc.), IKK-β/p-Ser180/181 (Cell Signaling Technology), and JNK/p-Thr183/Tyr185 (Cell Signaling Technology). To analyze tyrosine phosphorylation of IRS-1, lysates were immunoprecipitated overnight with IRS-1 antibody (Santa Cruz Biotechnology Inc.) and immobilized on protein A agarose beads (Upstate). The beads were collected, washed, and separated from the protein. Tyrosine phosphorylation of IRS-1 was detected by immunoblotting using an anti-PY20 antibody (BD Biosciences). Muscle lysates obtained from GFP BMT mice were probed with a GFP antibody obtained from Santa Cruz Biotechnology Inc. and compared with positive (GFP-labeled adipocytes) and negative (muscle lysates from BMT MAC-WT mice) control lysates. Densitometric analysis was performed using NIH ImageReady software, version 1.6 (http://www.scioncorp.com).
Immunohistochemistry and adipocyte cell sizing.
Following the glucose clamp, a portion of the epididymal fat pad not used for RNA analysis was placed in 8% formalin for 24 hours. Tissue samples were then placed in individual cassettes and kept in 10% ethanol for subsequent analyses. Next samples were paraffin embedded and serial sections taken from each sample. Samples were deparaffinized, washed 4 times, then stained with either H&E or an antibody against the macrophage-specific glycoprotein F4/80 (Serotec) or against dendritic cell CD11c (Biosource). Samples stained with an anti-F4/80 antibody were viewed under a microscope at ×40, and pictures were taken using a Motic B1 series camera to determine the presence of macrophages in the epididymal adipose tissue. Surprisingly, only small, isolated clusters of macrophages were identified in MAC-KO insulin-resistant animals or mice rendered insulin resistant following an 8-week HFD. Using a micrometer, adipocyte (n = 100 adipocytes) diameter was measured, and an average diameter was recorded for each animal. Values were recorded in arbitrary units.
Muscle and liver lipid analyses.
Triacylglycerol content was analyzed as previously described (55). Tissues were dissected free of visible connective tissue and blood. Lipid was extracted by a Folch extraction, the triacylglycerol was saponified in an ethanol/KOH solution at 60°C, and glycerol content was determined fluorometrically. Diacylglycerol was extracted and quantified according to the methods of Preiss et al. (56). Lipids were extracted from freeze-dried, powdered muscle using chloroform/methanol/PBS plus 0.2% SDS (1:2:0.8). Diacylglycerol kinase and [32P]ATP (15 mCi/mmol cold ATP) were added to lysates preincubated with cardiolipin/octylglucoside, and the reaction was terminated after 2 hours by the addition of chloroform/methanol (2:1). Samples were spotted onto thin-layer chromatography plates and developed. 32P-labeled phosphatidic acid was identified, dried, scraped from the TLC plate, and counted in a liquid scintillation counter (Tri-Carb 2500TR; Packard). Total long-chain acyl-CoAs were analyzed by a sensitive enzymatic recycling assay as described previously (57).
Statistics.
Proximal promoter regions were extracted for each gene represented on the microarray using the May 2006 Mouse Genome Assembly (NCB136) with the method as described previously (58). Analysis was restricted to the region 2 kb upstream and 500 bp downstream of the transcription start site. In cases where several possible alternative promoters may be present, analysis was focused on the most 5′ transcription start site. Motif discovery was performed using a comparative algorithm similar to those previously described (59–61). Promoters were initially divided into 2 sets: those that were upregulated by PPARγ deletion and those that were present on the array but did not change in response to PPARγ deletion. An exhaustive search for all n-mers (6 < n < 12) was performed, and each n-mer with up to 2 mismatches was scored for its enrichment in the promoters up regulated by PPARγ deletion using the hypergeometric distribution. The top 50 n-mers with the lowest P values where then converted into probability matrices and optimized in a manner similar to that used by Liu and Liu (61), except that we used the hypergeometric distribution as the scoring function instead of the MAP score. Statistical differences in mRNA levels between the genotypes were detected using ANOVA and Wilcoxon rank-sum analyses as indicated in the figure legends. Differences in blood glucose values between the genotypes during the GTT were detected using ANOVA repeated measures. Differences in circulating parameters, tissue size, GIR, HGP, IS-GDR, insulin signaling molecules, lipid intermediate levels, muscle 2-DOG uptake, and muscle GFP expression between groups were detected using ANOVA with Tukey’s post-hoc analysis where appropriate. Significance was set at P < 0.05.
Supplementary Material
Table 3 .
Enriched transcription factor motifs in promoters of upregulated genes
Acknowledgments
MXCre transgenic mice were obtained from R. Kuhn and K. Rajewsky (University of Cologne, Cologne, Germany). LysMCre mice were provided by I. Förster (University of Düsseldorf, Düsseldorf, Germany). Immunohistochemical analyses were performed by the Center for Histology Shared Resources at the UCSD Cancer Center, directed by N. Varki. We would also like to thank K. Saijo and J. Neels for assistance with the marrow transplantation procedure and Vanessa Nuñez for technical assistance. This study was supported by an American Heart Association Grant-in-Aid, the Spanish Ministry of Education and Science (SAF2006-01010), a Marie Curie International Reintegration Grant (IRG-016187), and Fundación “Mutua Madrileña” (to M. Ricote) and by NIH grants to C.K. Glass (GM069338, DK063491, HL56989, and DK074868), J.M. Olefsky (DK33651), and A.L. Hevener (DK60484 and DK73227). M. Ricote is supported by the Ramón y Cajal Program (MEC). This study was also funded in part by a University of California Discovery Program BioStar grant (J.M. Olefsky) with matching funds from Pfizer Inc. and from a grant provided by Takeda Pharmaceutical Co. Ltd. (J.M. Olefsky).
Footnotes
Nonstandard abbreviations used: BMT, BM transplantation; CM, conditioned medium; GDR, glucose disposal rate; GIR, glucose infusion rate; GTT, glucose tolerance test; HFD, high-fat diet; HGP, hepatic glucose production; IKK-β, IκB kinase β; IRS-1, insulin receptor substrate–1; IS-GDR, insulin-stimulated GDR; LysM, lysozyme M; MCP-1, monocyte chemoattractant protein–1; retnla, resistin-like molecule–α; TG-Mφ, thioglycollate-elicited peritoneal macrophages; TZD, thiazolidinedione.
Conflict of interest: J.M. Olefsky is a consultant for Pfizer Inc.
Citation for this article: J. Clin. Invest. 117:1658–1669 (2007). doi:10.1172/JCI31561
References
- 1.DeFronzo R.A., Bonadonna R.C., Ferrannini E. Pathogenesis of NIDDM. A balanced overview. Diabetes Care. 1992;15:318–368. doi: 10.2337/diacare.15.3.318. [DOI] [PubMed] [Google Scholar]
- 2.Reaven G.M. Role of insulin resistance in human disease (syndrome X): an expanded definition. Annu. Rev. Med. 1993;44:121–131. doi: 10.1146/annurev.me.44.020193.001005. [DOI] [PubMed] [Google Scholar]
- 3.Miranda P.J., DeFronzo R.A., Califf R.M., Guyton J.R. Metabolic syndrome: definition, pathophysiology, and mechanisms. Am. Heart J. 2005;149:33–45. doi: 10.1016/j.ahj.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 4.Arkan M.C., et al. IKK-β links inflammation to obesity-induced insulin resistance. Nat. Med. 2005;11:191–198. doi: 10.1038/nm1185. [DOI] [PubMed] [Google Scholar]
- 5.Cai D., et al. Local and systemic insulin resistance due to hepatic activation of IKK-β and NF-κB. Nat. Med. 2005;11:183–190. doi: 10.1038/nm1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan M., et al. Reversal of obesity- and diet-induced insulin resistance with salicylates or targeted disruption of Ikkbeta. Science. 2001;293:1673–1677. doi: 10.1126/science.1061620. [DOI] [PubMed] [Google Scholar]
- 7.Hirosumi J., et al. A central role for JNK in obesity and insulin resistance. Nature. 2002;420:333–336. doi: 10.1038/nature01137. [DOI] [PubMed] [Google Scholar]
- 8.Hotamisligil G.S. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 9.Weisberg S.P., McCann D., Rosenbaum D.M., Leibel R.L., Ferrante A.W. Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Invest. 2003;112:1796–1808. doi: 10.1172/JCI200319246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu H., et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J. Clin. Invest. 2003;112:1821–1830. doi: 10.1172/JCI200319451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi H., Kokoeva M.V., Inouye K., Tzameli I., Yin H., Flier J.S. TLR4 links innate immunity and fatty acid-induced insulin resistance. J. Clin. Invest. 2006;116:3015–3025. doi: 10.1172/JCI28898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suganami T., et al. Role of the toll-like receptor 4/NF-κB pathway in saturated fatty acid-induced inflammatory changes in the interaction between adipocytes and macrophages. Arterioscler. Thromb. Vasc. Biol. . 2007;27:84–91. doi: 10.1161/01.ATV.0000251608.09329.9a. [DOI] [PubMed] [Google Scholar]
- 13.Spiegelman B.M. PPARγ: adipogenic regulator and thiazolidinedione receptor. Diabetes. . 1998;47:507–514. doi: 10.2337/diabetes.47.4.507. [DOI] [PubMed] [Google Scholar]
- 14.Willson T.M., Lambert M.H., Kliewer S.A. Peroxisome proliferator-activated receptor gamma and metabolic disease. Annu. Rev. Biochem. 2001;70:341–367. doi: 10.1146/annurev.biochem.70.1.341. [DOI] [PubMed] [Google Scholar]
- 15.Picard F., Auwerx J. PPAR (gamma) and glucose homeostasis. Annu. Rev. Nutr. 2002;22:167–197. doi: 10.1146/annurev.nutr.22.010402.102808. [DOI] [PubMed] [Google Scholar]
- 16.Lehmann J.M., et al. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor gamma (PPAR gamma). J. Biol. Chem. . 1995;270:12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- 17.Suter S.L., Nolan J.J., Wallace P., Gumbiner B., Olefsky J.M. Metabolic effects of new oral hypoglycemic agent CS-045 in NIDDM subjects. Diabetes Care. . 1992;15:193–203. doi: 10.2337/diacare.15.2.193. [DOI] [PubMed] [Google Scholar]
- 18.Nolan J.J., Ludvik B., Beerdsen P., Joyce M., Olefsky J. Improvements in glucose tolerance and insulin resistance in obese subjects treated with troglitazone. N. Engl. J. Med. 1994;331:1188–1193. doi: 10.1056/NEJM199411033311803. [DOI] [PubMed] [Google Scholar]
- 19.Ibrahimi A., et al. Evidence for a common mechanism of action for fatty acids and thiazolidinedione antidiabetic agents on gene expression in preadipose cells. Mol. Pharmacol. . 1994;46:1070–1076. [PubMed] [Google Scholar]
- 20.He W., et al. 2003Adipose-specific peroxisome proliferator-activated receptor g knockout causes insulin resistance in fat and liver but not in muscle. Proc. Natl. Acad. Sci. U. S. A. 10015712–15717. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verma N.K., Singh J.H., Dey C.S. PPARγ expression modulates insulin sensitivity in C2C12 skeletal muscle cells. Br. J. Pharmacol. 2004;143:1006–1013. doi: 10.1038/sj.bjp.0706002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hevener A.L., et al. Muscle-specific Pparg deletion causes insulin resistance. Nat. Med. 2003;9:1491–1497. doi: 10.1038/nm956. [DOI] [PubMed] [Google Scholar]
- 23.Norris A.W., et al. Muscle-specific PPARγ-deficient mice develop increased adiposity and insulin resistance but respond to thiazolidinediones. J. Clin. Invest. 2003;112:608–618. doi: 10.1172/JCI200317305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujiwara T., Okuno A., Yoshioka S., Horikoshi H. Suppression of hepatic gluconeogenesis in long-term troglitazone treated diabetic KK and C57BL/KsJ-db/db mice. Metabolism. 1995;44:486–490. doi: 10.1016/0026-0495(95)90056-x. [DOI] [PubMed] [Google Scholar]
- 25.Gavrilova O., et al. Liver peroxisome proliferator-activated receptor gamma contributes to hepatic steatosis, triglyceride clearance, and regulation of body fat mass. J. Biol. Chem. 2003;278:34268–34276. doi: 10.1074/jbc.M300043200. [DOI] [PubMed] [Google Scholar]
- 26.Ricote M., Li A.C., Willson T.M., Kelly C.J., Glass C.K. The peroxisome proliferator-activated receptor-γ is a negative regulator of macrophage activation. Nature. . 1998;391:79–82. doi: 10.1038/34178. [DOI] [PubMed] [Google Scholar]
- 27.Jiang C., Ting A.T., Seed B. PPARg agonists inhibit production of monocyte inflammatory cytokines. Nature. 1998;391:82–86. doi: 10.1038/34184. [DOI] [PubMed] [Google Scholar]
- 28.Welch J.S., Ricote M., Akiyama T.E., Gonzalez F.J., Glass C.K. PPARg and PPARd negatively regulate specific subsets of lipopolysaccharide and IFNg target genes in macrophages. Proc. Natl. Acad. Sci. U. S. A. 2003;100:6712–6717. doi: 10.1073/pnas.1031789100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma A.M., Staels B. Peroxisome proliferator-activated receptor γ (PPARγ) and adipose tissue — understanding obesity-related changes in regulation of lipid and glucose metabolism. J. Clin. Endocrinol. Metab. 2007;92:386–395. doi: 10.1210/jc.2006-1268. [DOI] [PubMed] [Google Scholar]
- 30.Ogawa S., et al. Molecular determinants of crosstalk between nuclear receptors and Toll-like receptors mediating counter-regulation of inflammatory responses. Cell. 2005;122:707–721. doi: 10.1016/j.cell.2005.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li A., et al. Peroxisome proliferator-activated receptor γ ligands inhibit development of atherosclerosis in LDL receptor-deficient mice. J. Clin. Invest. 2000;106:523–531. doi: 10.1172/JCI10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li A.C., et al. Differential inhibition of macrophage foam cell formation and atherosclerosis in mice by PPAR α, β/δ, and γ. J. Clin. Invest. 2004;114:1564–1576. doi: 10.1172/JCI200418730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clausen B.E., Burkhardt C., Reith W., Renkawitz R., Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 34.Kuhn R., Schwenk F., Aguet M., Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 35.Akiyama T.E., et al. Conditional disruption of the peroxisome proliferator-activated receptor γ gene in mice results in lowered expression of ABCA1, ABCG1, and apoE in macrophages and reduced cholesterol efflux. Mol. Cell. Biol. 2002;22:2607–2619. doi: 10.1128/MCB.22.8.2607-2619.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee Y.H., Giraud J., Davis R.J., White M.F. C-Jun N-terminal kinase (JNK) mediates feedback inhibition of the insulin signaling cascade. J. Biol. Chem. 2003;278:2896–2902. doi: 10.1074/jbc.M208359200. [DOI] [PubMed] [Google Scholar]
- 37.Liu Y.-F., et al. Serine phosphorylation proximal to its phosphotyrosine binding domain inhibits insulin receptor substrate 1 function and promotes insulin resistance. Mol. Cell. Biol. 2004;24:9668–9681. doi: 10.1128/MCB.24.21.9668-9681.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wellen K.E., Hotamisligil G.S. Inflammation, stress, and diabetes. J. Clin. Invest. 2005;115:1111–1119. doi: 10.1172/JCI200525102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Itani S.I., Ruderman N.B., Schmieder F., Boden G. Lipid-induced insulin resistance in human muscle is associated with changes in diacylglycerol, protein kinase C, and IkappaB-alpha. Diabetes. . 2002;51:2005–2011. doi: 10.2337/diabetes.51.7.2005. [DOI] [PubMed] [Google Scholar]
- 40.Chavez J.A., Summers S.A. Characterizing the effects of saturated fatty acids on insulin signaling and ceramide and diacylglycerol accumulation in 3T3-L1 adipocytes and C2C12 myotubes. Arch. Biochem. Biophys. 2003;419:101–109. doi: 10.1016/j.abb.2003.08.020. [DOI] [PubMed] [Google Scholar]
- 41.Pan D.A., et al. Skeletal muscle triglyceride levels are inversely related to insulin action. Diabetes. 1997;46:983–988. doi: 10.2337/diab.46.6.983. [DOI] [PubMed] [Google Scholar]
- 42.Casteleijn E., et al. Hormonal control of glycogenolysis in paranchymal liver cells by kupffer and endothelial liver cells. J. Biol. Chem. 1988;263:2699–2703. [PubMed] [Google Scholar]
- 43.Patel L., et al. Resistin is expressed in human macrophages and is directly regulated by PPAR gamma activators. Biochem. Biophys. Res. Comm. 2003;300:472–476. doi: 10.1016/s0006-291x(02)02841-3. [DOI] [PubMed] [Google Scholar]
- 44.Rajala M.W., et al. Cell type-specific expression and coregulation of murine resistin and resistin-like molecule-α in adipose tissue. Mol. Endocrinol. 2002;16:1920–1930. doi: 10.1210/me.2002-0048. [DOI] [PubMed] [Google Scholar]
- 45.Liang C.P., et al. Increased CD36 protein as a response to defective insulin signaling in macrophages. J. Clin. Invest. 2004;113:764–773. doi: 10.1172/JCI200419528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frederick M.J., et al. In vivo expression of the novel CXC chemokine BRAK in normal and cancerous human tissue. Am. J. Pathol. . 2000;156:1937–1950. doi: 10.1016/S0002-9440(10)65067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Torres S.H., De Sanctis J.B., de Briceno M., Hernandez N., Finol H.J. Inflammation and nitric oxide production in skeletal muscle of type 2 diabetic patients. J. Endocrinol. 2004;181:419–427. doi: 10.1677/joe.0.1810419. [DOI] [PubMed] [Google Scholar]
- 48.Li M., Pascual G., Glass C.K. Peroxisome proliferator-activated receptor dependent repression of the inducible nitric oxide synthase gene. Mol. Cell. Biol. 2000;20:4699–4707. doi: 10.1128/mcb.20.13.4699-4707.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pascual G., et al. A SUMOylation-dependent pathway mediates transpression of inflammatory response genes by PPAR-γ. Nature. 2005;437:759–763. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nicol C.J., et al. PPARgamma influences susceptibility to DMBA-induced mammary, ovarian and skin carcinogenesis. Carcinogenesis. . 2004;25:1747–1755. doi: 10.1093/carcin/bgh160. [DOI] [PubMed] [Google Scholar]
- 51.Schiller N.K., Kubo N., Boisvert W.A., Curtiss L.A. Effect of gamma-irradiation and bone marrow transplantation on atherolsclerosis in LDL receptor-deficient mice. Arterioscler. Thromb. Vasc. Biol. 2001;21:1674–1680. doi: 10.1161/hq1001.096724. [DOI] [PubMed] [Google Scholar]
- 52. Handel-Fernandez, M.E., and Lopez, D.M. 2000. Isolation of macrophages from tissues, fluids, and immune response sites. In Macrophages: practical approach series. D.M. Paulnock, editor. Oxford University Press. New York, New York, USA. 1–30. [Google Scholar]
- 53.Steele R. Influences of glucose loading and of injected insulin on hepatic glucose output. Ann. N. Y. Acad. Sci. 1959;82:420–430. doi: 10.1111/j.1749-6632.1959.tb44923.x. [DOI] [PubMed] [Google Scholar]
- 54.DeFronzo R.A., et al. The effect of insulin on the disposal of intravenous glucose. Diabetes. 1981;30:1000–1007. doi: 10.2337/diab.30.12.1000. [DOI] [PubMed] [Google Scholar]
- 55.Frayn K.N., Maycock P.F. Skeletal muscle triacylglycerol in the rat: methods for sampling and measurement, and studies of biological variability. J. Lipid Res. 1980;21:139–144. [PubMed] [Google Scholar]
- 56.Preiss J., et al. Quantitative measurement of sn-1,2-diacylglycerols present in platelets, hepatocytes, and ras and sis-transformed normal rat kidney cells. J. Biol. Chem. 1986;261:8597–8600. [PubMed] [Google Scholar]
- 57.Allred J.B., Guy D.G. Determination of coenzyme A and acetyl CoA in tissue extracts. Anal. Biochem. 1969;29:293–299. doi: 10.1016/0003-2697(69)90312-1. [DOI] [PubMed] [Google Scholar]
- 58.Halees A.S., Leyfer D., Weng Z. PromoSer: a large-scale mammalian promoter and transcription start site identification service. Nucleic Acids Res. . 2003;31:3554–3559. doi: 10.1093/nar/gkg549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Barash, Y., Bejerano, G., and Friedman, N. 2001. A simple hyper-geometric approach for discovering putative transcription factor binding sites. Algorithms in bioinformatics: First International Workshop, WABI 2001. August 28–31. Aarhus, Denmark. O. Gascuel and B.M.E. Moret, editors. Springer. Berlin, Germany/Heidelberg, Germany/New York, New York, USA. 278–293. [Google Scholar]
- 60. Segal, E., Barash, Y., Simon, I., Friedman, N., and Koller, D. 2002. From promoter sequence to expression: a probabilistic framework. Proceedings of the Sixth Annual International Conference on Computational Biology. April 18–21. Washington, DC, USA. G. Myers et al., editors. ACM Press. New York, New York, USA. 263–272. [Google Scholar]
- 61.Liu X., Brutlag D., Liu J.2002An algorithm for finding protein-DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat. Biotechnol. 20835 – 839. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.