Abstract
MDP (muramyl dipeptide), a component of peptidoglycan, interacts with NOD2 (nucleotide-binding oligomerization domain 2) stimulating the NOD2–RIP2 (receptor-interacting protein 2) complex to activate signalling pathways important for antibacterial defence. Here we demonstrate that the protein kinase activity of RIP2 has two functions, namely to limit the strength of downstream signalling and to stabilize the active enzyme. Thus pharmacological inhibition of RIP2 kinase with either SB 203580 [a p38 MAPK (mitogen-activated protein kinase) inhibitor] or the Src family kinase inhibitor PP2 induces a rapid and drastic decrease in the level of the RIP2 protein, which may explain why these RIP2 inhibitors block MDP-stimulated downstream signalling and the production of IL-1β (interleukin-1β) and TNFα (tumour necrosis factor-α). We also show that RIP2 induces the activation of the protein kinase TAK1 (transforming-growth-factor-β-activated kinase-1), that a dominant-negative mutant of TAK1 inhibits RIP2-induced activation of JNK (c-Jun N-terminal kinase) and p38α MAPK, and that signalling downstream of NOD2 or RIP2 is reduced by the TAK1 inhibitor (5Z)-7-oxozeaenol or in TAK1-deficient cells. We also show that MDP activates ERK1 (extracellular-signal-regulated kinase 1)/ERK2 and p38α MAPK in human peripheral-blood mononuclear cells and that the activity of both MAPKs and TAK1 are required for MDP-induced signalling and production of IL-1β and TNFα in these cells. Taken together, our results indicate that the MDP–NOD2/RIP2 and LPS (lipopolysaccharide)–TLR4 (Toll-like receptor 4) signalling pathways converge at the level of TAK1 and that many subsequent events that lead to the production of pro-inflammatory cytokines are common to both pathways.
Keywords: muramyl dipeptide (MDP), nucleotide-binding oligomerization domain 2 (NOD2), pro-inflammatory cytokines, receptor-interacting protein 2 (RIP2), transforming-growth-factor-β-activated kinase-1 (TAK1)
Abbreviations: CK1, ‘casein kinase 1’; ERK, extracellular-signal-regulated kinase; FCS, foetal-calf serum; GAK, cyclin G-associated kinase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GST, glutathione S-transferase; HA, haemagglutinin; HEK-293, human embryonic kidney 293 cells; IκB, inhibitor of NF-κB; IKKβ, inhibitor of nuclear factor κB kinase β; IL, interleukin; JNK, c-Jun N-terminal kinase; K63-pUb, Lys63-linked polyubiquitin; KI, kinase inactive; LPS, lipopolysaccharide; MAPK, mitogen-activated protein kinase; MDP, muramyl dipeptide; MKK1, MAPK kinase-1; M-TriDAP, muramyl-N-acetyl-L-Ala-D-Glu-m-diaminopimelic acid; NEMO, nuclear factor κB essential modifier; NF-κB, nuclear factor κB; NOD, nucleotide-binding oligomerization domain; PBMCs, peripheral-blood mononuclear cells; RIP2, receptor-interacting protein 2; shRNA, short hairpin RNA; TAB, TAK1-binding protein; TAK1, transforming-growth-factor-β-activated kinase-1; TLR, Toll-like receptor; TNF, tumour necrosis factor; Tpl2, tumour progression locus 2; TRAF6, TNF-receptor-associated factor 6; VSV-G, vesicular-stomatitis-virus envelope glycoprotein; WT, wild-type
INTRODUCTION
The NOD (nucleotide binding and oligomerization domain) proteins NOD1 and NOD2 are the intracellular receptors for peptidoglycan, a major component of bacterial cell walls. The minimal subfragments of peptidoglycan recognized by NOD1 and NOD2 are M-TriDAP (muramyl-N-acetyl-L-Ala-D-Glu-m-diaminopimelic acid) and MDP (muramyl dipeptide) respectively, which bind to the C-terminal LRR (leucine-rich repeat) domains of these proteins. NOD1 and NOD2 interact with a protein kinase called RIP2 (receptor-interacting protein 2; also called RICK, RIPK2 and CARDIAK), which is required for downstream signalling. NOD1 and NOD2 acting via RIP2, switch on the transcription factor NF-κB (nuclear factor-κB) and the MAPKs (mitogen-activated protein kinases) termed p38α MAPK, JNK (c-Jun N-terminal kinase) and ERK1 and ERK2 (extracellular signal-regulated kinases 1 and 2) [1–3], which stimulate production of the pro-inflammatory cytokines that mount innate immune responses to combat invading bacteria.
The NOD1/NOD2–RIP2 signalling pathway is thought to be used for host defence against intracellular bacteria that have evaded the TLR (Toll-like receptor)-mediated defence mechanisms in the plasma membrane, such TLR4, which responds to bacterial LPS (lipopolysaccharide). The NOD2 signalling pathway may also provide a defence against Gram-positive bacteria, the cell walls of which do not contain LPS [1]. The importance of NOD2 in protection against inflammatory bowel disease has been highlighted by the finding that particular mutations in NOD2 are associated with the susceptibility to Crohn's disease [1,3–7]. Significantly, PBMCs (peripheral-blood mononuclear cells) from these patients do not produce the cytokines IL-1β (interleukin 1β), IL-10 or TNFα (tumour necrosis factor α) in response to either MDP or M-TriDAP, demonstrating that NOD2 is also required for signalling by NOD1 in these cells [8]. However, NOD2 clearly plays additional roles in regulating systemic responses to pathogens, because NOD2-deficient mice are protected against LPS-induced septic shock, even though LPS-induced signalling pathways and pro-inflammatory cytokine production are apparently normal [3]. RIP2-deficient mice are also protected against LPS-induced septic shock and have impaired ability to defend against infection by Listeria monocytogenes [9].
How the MDP–NOD2–RIP2 signalling module actually switches on downstream signalling is unclear, because, surprisingly, when overexpressed in HEK-293 (human embryonic kidney 293) cells, a catalytically inactive [KI (‘kinase inactive’)] mutant of RIP2 was reported to be as effective as wild-type (WT) RIP2 in activating NF-κB-dependent gene transcription and JNK [10] or in triggering apoptosis [11]. Thus the protein kinase activity of RIP2 is thought not to be essential for the MDP-induced activation of these signalling pathways. These observations raised the question of how RIP2 switches on downstream signalling events and what function its associated kinase activity might have.
In the present paper we demonstrate that the protein kinase activity of RIP2 plays at least two roles in the MDP–NOD2 signalling system. First, we find that KI-RIP2 is even more effective than WT-RIP2 in activating NF-κB-dependent gene transcription, JNK1/JNK2 and p38α MAPK, suggesting that RIP2 kinase activity functions to limit the strength of downstream signalling. Secondly, we find that RIP2 kinase activity is required to maintain RIP2 expression levels in transfected HEK-293 cells, which may explain our finding that pharmacological inhibition of the endogenous RIP2 kinase activity suppresses the MDP-induced activation of NF-κB. We also find that, when overexpressed, RIP2 interacts with, and activates, TAK1 (transforming-growth-factor-β-activated kinase 1) and that MDP–NOD2- or RIP2-induced NF-κB gene transcription does not occur when TAK1 is inhibited or in TAK1-deficient cells. Finally, we find that the MDP-induced signalling and production of IL-1β and TNFα in human PBMCs is attenuated by pharmacological inhibition of p38α MAPK, MKK1 (MAPK kinase-1) or TAK1. Taken together, our results suggest that the signalling pathways by which MDP–NOD2 and LPS–TLR4 induce the production of IL-1β and TNFα converge at the level of TAK1.
EXPERIMENTAL
Materials
PD 184352, synthesized by an improved method [12], and BIRB 0796, synthesized from 4,4-dimethyl-3-exopentanenitrile [13], were provided by Dr Natalia Shpiro and Dr Rudolfo Marquez (both of the Division of Biological Chemistry and Molecular Microbiology, School of Life Sciences, University of Dundee, Dundee, Scotland, U.K.). SB 203580 was purchased from Promega, the Src family kinase inhibitors PP1 and PP2 from Calbiochem, the TAK1 inhibitor (5Z)-7-oxozeaenol from AnalytiCon Discovery GmbH, LPS and MDP free of LPS from InvivoGen and the MEF2 nucleofection kit from Amaxa.
DNA constructs
DNA encoding human RIP2 (NP_003812) was amplified from IMAGE clone 4026156 (Geneservice) with oligonucleotides MP1155 and MP1160 using the GC Rich PCR System (Roche) and cloned into a pCR2.1 vector. The construct was sequenced then ligated into the BamH1/Not1 sites in the pCMV HA-1 vector. A vector in which Asp164 was changed to alanine (pCMV HA-RIP2[D164A]) was created using the QuikChange® Mutagenesis kit (Stratagene) with oligonucleotides MP2345 and MP2346, and a vector in which Thr95 was changed to methionine (pCMV HA-RIP2[T95M] was created using oligonucleotides MP2847 and MP2848. Mouse NOD2 (BC044774) was amplified from IMAGE clone 5324763 as described above with oligonucleotides MP1879 and MP1880 and cloned into the BamH1 site of pCMV HA-1.
The oligonucleotide sequences were as follows: MP1155, GCGCGGCCGCCAACGGGGAGGCCATCTGCAGC; MP1160, GCGGCCGCTTACATGCTTTTATTTTGAAGTAAATTTAAAGATGGTGATCTAGAAA; MP1879, GCGGATCCATGTGCTCACAGGAAGAGTTCCAGGC; MP1880, GCGGATCCTCACAACAAGAGTCTGGCGTCCCTG; MP2345, CATGTTAAGATTGCAGCTTTTGGTTTATCAAAG; MP2346, CTTTGATAAACCAAAAGCTGCAATCTTAACATG; MP2847, TTTTTGGGAATAGTTATGGAATACATGCCAAAT; MP2848, ATTTGGCATGTATTCCATAACTATTCCCAAAAA.
DNA encoding a ConA basal promoter into which three copies of the NF-κB DNA response element had been inserted (termed ‘3× κB luciferase reporter construct’) and constructs encoding the HIV-1 gag-Pol polyprotein and VSV-G (vesicular-stomatitis-virus envelope glycoprotein) for the production of lentiviruses were provided by Dr Ron Hay, College of Life Sciences, University of Dundee, Dundee, Scotland, U.K. [14]. The TAK1 shRNA (short hairpin RNA) lentiviral plasmid was from Sigma and the pRL-TK vector driving Renilla luciferase from Promega.
Production of lentiviruses and infection
Lentiviruses carrying a TAK1 shRNA plasmid (TRCN0000001558; Sigma) were produced using a gag-pol construct and a VSV-G encoded plasmid by triple transfection as described in [15]. To create stable cell lines, 200 μl of viral supernatant was used to infect HEK-293 cells on a 10 cm2 dish. After 48 h, 3 μg/ml puromycin was added to the medium for selection. Stably transfected cells were used for experiments.
Cell culture, transfection and NF-κB reporter gene assay
HEK-293 cells that stably express the IL-1 receptor (termed ‘IL-1R cells’; a gift from Tularik, South San Francisco, CA, U.S.A.) and mouse embryonic fibroblasts from TAK1+/+ and TAK1−/− mice [16] were cultured in 10 cm2 dishes in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) FCS (foetal calf serum). The HEK-293 cells were transfected with DNA vectors mixed with polyethyleneimine [17], whereas mouse embryonic fibroblasts were transfected with the Amaxa MEF2 kit according to the manufacturer's instructions. RAW 264.7 cells were maintained in RPMI 1640 medium (Invitrogen) supplemented with 100 units/ml penicillin and 100 μg/ml streptomycin, 1 mM sodium pyruvate, 2 mM L-glutamine and 10% heat-inactivated FCS. Transient transfection of the RAW 264.7 cell line was achieved by electroporation. For this, cells were harvested, washed twice in Opti-MEM (Invitrogen) and resuspended at 4×107 cells/ml in Opti-MEM. A 0.5 ml portion of cell suspension was then mixed with plasmid DNA and the cell/DNA mixture added to a 0.4-cm-electrode-gap electroporation cuvette and shocked in a Bio-Rad GenePulser II (300 V, 950 μF) at 21 °C. Cells were resuspended immediately in 1 ml of pre-warmed growth medium and aliquots (1.6×106 cells) added to six-well plates and treated as described in the Results section.
For the measurement of NF-κB-dependent luciferase gene expression, cells were lysed in Passive Lysis Buffer (Promega) and luciferase activity measured using a Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. Firefly luciferase activity was normalized on the basis of Renilla luciferase activity.
Antibodies
Anti-TAK1, the antibody recognizing TAB1 (TAK1-binding protein 1) phosphorylated at Thr431 [18] and an antibody that recognizes TAK1 phosphorylated at Thr187 (raised against the peptide IQTHMT*NNKGS, where T* is phosphothreonine) were raised in sheep. The antibody that recognizes TAK1 phosphorylated at Thr187 was affinity-purified on a phosphopeptide antigen–agarose column and used for immunoblotting in the presence of 10 μg/ml of the unphosphorylated peptide immunogen. Antibodies recognizing the active phosphorylated forms of JNK, p38α MAPK and ERK1/ERK2 were from Cell Signalling Technologies, anti-TAK1 was from Santa Cruz, anti-HA (anti-haemagglutinin) and anti-Myc were from Sigma Chemical Co., and anti-(human IL-1β) was from R&D Systems.
Immunoblotting
Cell extracts were obtained using standard lysis buffer [50 mM Tris/HCl, pH 7.5, 1 mM EGTA, 1 mM EDTA, 1% (w/w) Triton X-100, 1 mM Na3VO4, 50 mM NaF, 5 mM sodium pyrophosphate, 0.27 M sucrose, 0.1% mercaptoethanol and ‘Complete’ proteinase inhibitor cocktail (Roche)]. After centrifugation for 15 min at 18000 g, the supernatant (termed the ‘cell extract’) was decanted and protein concentrations were measured by the Bradford method.
Isolation of PBMCs and stimulation with MDP and LPS
PBMCs were isolated from buffy coats (provided by the Ninewells Hospital Blood Transfusion Centre, Dundee, Scotland, U.K.) and diluted with an equal volume of RPMI 1640 with 25 mM Hepes and L-glutamine (Invitrogen). The suspensions were subjected to density-gradient centrifugation over Ficoll-Paque™ (GE Healthcare) and the PBMC fraction was washed three times with RPMI 1640/25 mM Hepes/L-glutamine before the cells were plated in the same medium. The cells were then treated as described in the Results section.
Expression, purification and assay of RIP2
RIP2 was expressed from a baculovirus vector into insect (Spodoptera frugiperda) Sf21 cells as a His6-tagged fusion protein and purified by chromatography on Ni2+-nitrilotriacetate–agarose. The purified RIP2 was diluted in 50 mM Tris/HCl (pH 7.5)/0.1% 2-mercaptoethanol/0.1 mM EGTA/1 mg/ml BSA and assayed for 10 min at 30 °C in a reaction volume of 50 μl containing 50 mM Tris/HCl, pH 7.5, 0.1% 2-mercaptoethanol, 0.1 mM EGTA, 1 mg/ml myelin basic protein, 10 mM magnesium acetate, 0.1 mM [γ-32P]ATP (50–1000 c.p.m./pmol). The reactions were initiated by the addition of ATP after incubating the other components for 5 min at room temperature. Reactions were stopped by spotting 40 μl of assay mixture on to 1.5 cm×1.5 cm squares of Whatman P81 paper, which were washed three times in 75 mM phosphoric acid and once in acetone before air-drying and measurement of 32P radioactivity incorporated by Cerenkov counting.
RESULTS
Catalytically inactive (KI-) RIP2 is more effective than WT-RIP2 in switching on downstream signalling pathways
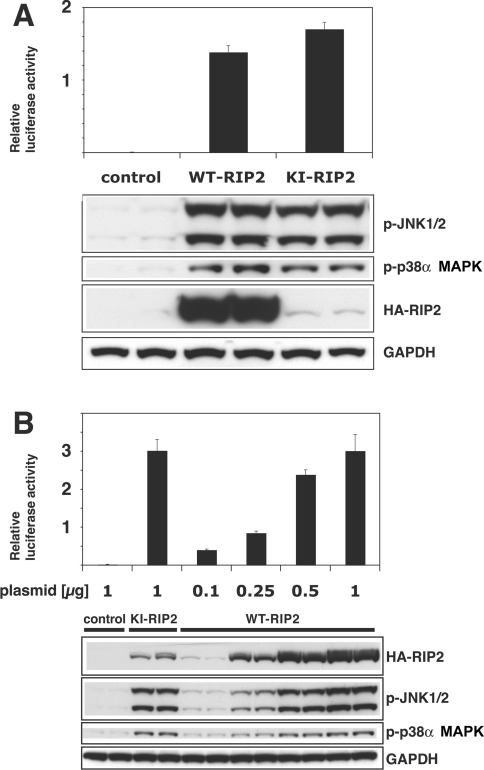
The overexpression of either WT-RIP2 or KI-RIP2 in HEK-293 cells induced NF-κB-dependent gene transcription and phosphorylation of the activation loops of p38α MAPK and JNK1/JNK2 (Figure 1A), as expected from previous studies. However, we noticed that KI-RIP2 was expressed at a far lower level than the WT protein (Figure 1A), suggesting that the KI-RIP2 might be even more effective in switching on downstream signalling pathways. In order to investigate this possibility, we therefore decreased the amount of DNA encoding WT-RIP2 that we transfected into the cells until the level of expression of the protein became similar to that of KI-RIP2. These experiments revealed that KI-RIP2 was indeed far more effective than the WT-RIP2 in switching on JNK and p38α MAPK activation and NF-κB-dependent gene transcription (Figure 1B). For example, transfection of 0.25 μg of DNA encoding WT-RIP2 induced a 3–4-fold lower activation of NF-κB-dependent gene transcription or phosphorylation of JNK and p38α MAPK than did transfection of 1 μg of DNA encoding KI-RIP2, even though the expression of the WT-RIP2 protein was higher than that of the KI-RIP2 protein under these conditions.
Figure 1. RIP2 activates JNK, p38α MAPK and NF-κB-dependent gene transcription in transfected HEK-293 cells.
(A) Cells were co-transfected with 1 μg of plasmid RIP2 DNA encoding either HA-tagged WT- or KI-RIP2 and 0.1 μg of DNA encoding the 3×κB luciferase reporter construct (see the Experimental section). After 48 h, the cells were extracted in Passive Lysis Buffer and NF-κB-dependent luciferase reporter gene expression was measured (uppermost panel). In identical experiments carried out in parallel, the cells were extracted with standard cell lysis buffer and the extract (20 μg of protein) was immunoblotted with anti-HA antibodies to detect the HA-tagged RIP2 and with phospho-specific antibodies that recognize the active phosphorylated forms of JNK1 and JNK2 (p-JNK1/2) and p38α MAPK (p-p38α MAPK). The extracts were also immunoblotted with antibodies that recognize GAPDH (glyceraldehyde-3-phosphate dehydrogenase) as a loading control. The error bars in the upper panel and results in the lower panel show the S.D. between duplicate experiments. Similar results were obtained in several other experiments. (B) The experiment was carried out as described in (A), except that the amounts of plasmid DNA encoding HA-tagged WT-RIP2 were varied as indicated.
The protein kinase activity of RIP2 controls RIP2 stability
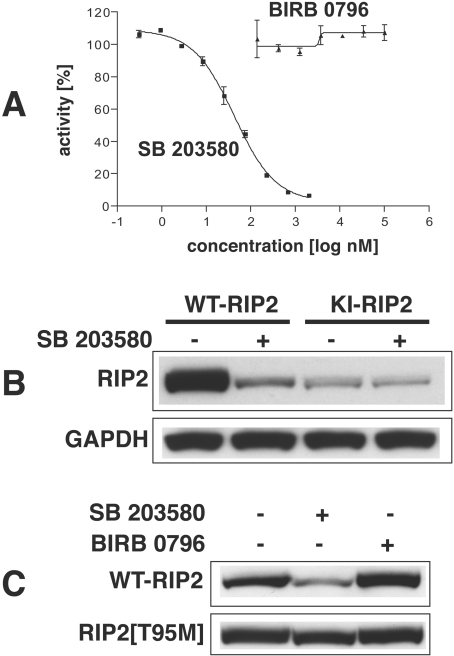
The observation that KI-RIP2 (RIP2[D164A]) (Figure 1A) and another KI mutant RIP2[K47A] (results not shown) were expressed at a greatly reduced level compared with WT-RIP2 (Figure 1A) suggested that RIP2 kinase activity might also control the level of expression of the RIP2 protein. However, it remained entirely possible that these amino acid substitutions had affected the translation, stability or degradation of the protein in a way that was unrelated to the loss of protein kinase activity caused by the mutations. In order to distinguish between these possibilities we therefore exploited the protein kinase inhibitor SB 203580. This compound is a relatively specific inhibitor of p38α MAPK [19,20], but was later found to inhibit RIP2 kinase activity in vitro with even greater potency ([21]; see Figure 2A). We therefore transfected HEK-293 cells with DNA encoding either WT-RIP2 or KI-RIP2 and then exposed the cells to SB 203580. Strikingly, SB 203580 reduced the level of expression of WT-RIP2 to that of the KI-RIP2. Moreover, SB 203580 did not affect the level of expression of the catalytically inactive mutant (Figure 2B).
Figure 2. RIP2 inhibitor SB 203580 decreases the protein expression levels of WT-RIP2, but not KI-RIP2, in transfected HEK-293 cells.
All experiments were carried out at least twice with similar results. (A) Effect of SB 203580 and BIRB 0796 on the activity of RIP2 in vitro. RIP2 was assayed as described in the Experimental section at the indicated concentrations of SB 203580 or BIRB 0796. The IC50 value for inhibition by SB 203580 was 43 nM. (B) Cells were transfected with DNA constructs encoding HA-tagged versions of either WT-RIP2 or KI-RIP2 (1 μg of plasmid RIP2 DNA). After 24 h the cells were incubated for a further 24 h with (+) or without (−) 10 μM SB 203580, then extracted with standard lysis buffer. The level of transfected RIP2 in the extract was analysed by immunoblotting with an anti-HA antibody. (C) Cells were transfected for 48 h with a construct encoding HA-tagged WT-RIP2 or RIP2[T95M] (1 μg of plasmid RIP2 DNA). The cells were incubated with 10 μM SB 203580 or 0.1 μM BIRB 0796 for 1 h before the end of transfection period, lysed, and 20 μg of extract protein was analysed by immunoblotting with an anti-HA antibody. The extracts were also immunoblotted with antibodies that recognize GAPDH as a loading control.
SB 203580 inhibits not only p38α MAPK and RIP2, but also two other protein kinases [CK1 (formerly termed ‘casein kinase I’) and GAK (cyclin G-associated protein kinase)] [21]. It could therefore be argued that the effect of SB 203580 to reduce the level of expression of the RIP2 protein was caused not by the inhibition of RIP2 kinase activity, but by the inhibition of p38α MAPK, CK1 or GAK. In order to address this possibility, we repeated the experiment using BIRB 0796, a much more potent inhibitor of p38α MAPK than is SB 203580 [22,23] and which does not inhibit RIP2 (Figure 2A), CK1 or GAK (L. Plater, H. McLaughlan and P. Cohen, unpublished work) in vitro, even at concentrations 1000-fold higher than those required to inhibit p38α MAPK. In contrast with SB 203580, BIRB 0796 did not affect the level of expression of WT-RIP2 (Figure 2C), implying that it was the inhibition of RIP2 kinase activity by SB 203580, and not the inhibition of p38α MAPK, that had caused the expression of RIP2 to decline.
The p38α MAPK can be largely desensitized to inhibition by SB 203580 by mutating Thr106 to methionine, the larger side chain of methionine preventing access of SB 203580 to a hydrophobic pocket near the ATP-binding site [24,25]. Since RIP2 possesses a threonine residue at the equivalent position, it seemed likely that the presence of this residue was critical for the sensitivity of RIP2 kinase activity to the drug. We therefore mutated Thr95 of RIP2 to methionine and examined whether SB 203580 was now able to reduce the level of expression of RIP2. These experiments demonstrated that SB 203580 decreased the level of WT-RIP2, but not RIP2[T95M] (Figure 2C). Taken together, these results establish that it is the protein kinase activity of RIP2 that maintains the level of expression of activated RIP2 kinase, at least when overexpressed in HEK-293 cells.
SB 203580 and PP2, but not BIRB 0796, suppresses MDP–NOD2-induced activation of NF-κB in HEK-293 and RAW 264.7 cells
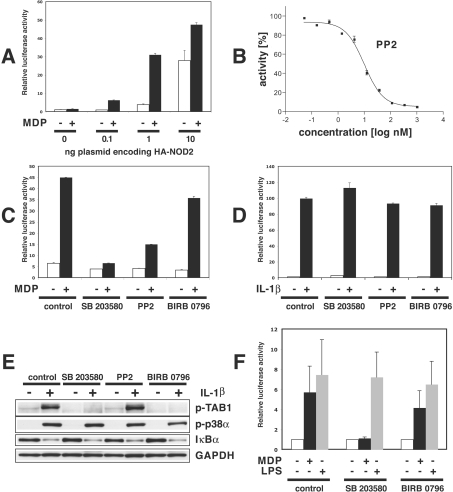
In order to investigate whether the endogenous RIP2 kinase activity is required for MDP-induced signalling, we initially performed experiments in HEK-293 cells. Although these cells do not express NOD2 and therefore do not respond to MDP, sensitivity to MDP can be conferred by transfection with DNA encoding NOD2 [26]. When relatively large amounts of NOD2 DNA (10 ng/10 cm2 dish) were transfected, NF-κB-dependent reporter-gene expression was activated, but little further stimulation by MDP could be detected (Figure 3A), presumably because NOD2 possesses some intrinsic ability to signal, even in the absence of MDP, which in overexpression experiments is sufficient to activate the pathway. However, when the amount of NOD2 DNA transfected was reduced to 1 ng, there was little activation of NF-κB transcription in the absence of MDP, but MDP could now induce a 5–10-fold increase in NF-κB-dependent gene transcription (Figure 3A).
Figure 3. RIP2 kinase inhibitors SB 203580 and PP2 suppress MDP–NOD2-induced activation of NF-κB.
All experiments were performed in duplicate, and similar results were obtained in at least two independent experiments. (A) HEK-293 cells were co-transfected with 0.1 μg of DNA encoding 3×κB luciferase reporter construct and 0.1 μg of pTK-RL plasmid DNA (encoding Renilla luciferase) plus 0.8 μg of empty pCMV5 vector (to bring the final amount of vector to 1.0 μg as used for all experiments) and the indicated amounts of plasmid DNA encoding HA-tagged NOD2. After 24 h, cells were incubated without (white bars) or with (black bars) 1 μM MDP. After 24 h the cells were lysed and NF-κB-dependent luciferase reporter gene expression was analysed. The error bars show the S.D. between duplicate experiments. (B) Effect of PP2 on the activity of RIP2 in vitro. RIP2 was assayed as described in the Experimental section at the indicated concentrations of PP2. The IC50 value for inhibition by PP2 was 10 nM. (C) Experiments were performed as described for (A), using 1 ng of DNA encoding HA-NOD2 for transfection. At 1 h before stimulation with (black bars) or without (white bars) MDP the cells were incubated without (control) or with 10 μM SB 203580, 0.1 μM BIRB 0796 or 1 μM PP2. The error bars show the S.D. between duplicate experiments. (D) HEK-293 IL-1R cells were transfected with the 3× κB luciferase reporter construct and pTK-RL plasmid DNA as described in (A), treated with the indicated inhibitors as in (C) and stimulated with 5 ng/ml IL-1β instead of MDP. The error bars show the S.D. between duplicate experiments. (E) HEK-293 IL-1R cells were incubated without (control) or with 10 μM SB 203580, 0.1 μM BIRB 0796 or 1 μM PP2 1 h before stimulation with 5 ng/ml IL-1β for 20 min. Cells were lysed and 20 μg of extract protein was analysed by immunoblotting with antibodies recognizing phosphorylated TAB1 (phosphothreonine-431) and p38α MAPK as well as IκBα and GAPDH as a loading control. (F) RAW 264.7 cells were transiently co-transfected via electroporation with 50 μg of DNA encoding 3× κB luciferase reporter construct and 5 μg of pTK-RL plasmid DNA (encoding Renilla luciferase) (see the legend to Figure 1A). Cells were left to recover for 4 h in RPMI 1640 supplemented with 10% heat-inactivated FCS, and the medium was then replaced with RPMI 1640 without FCS. The cells were incubated for 60 min with (+) or without (−) 5 μM SB 203580 or 0.1 μM BIRB 0796, then stimulated with (+) or without (−) 1 μM MDP or 100 ng/ml LPS or left untreated. After 24 h, the cells were extracted in 0.5 ml of Passive Lysis Buffer and 20 μl aliquots of the extract were analysed for NF-κB-dependent luciferase reporter-gene expression. The results are given as means±S.D. for three different experiments, each performed in duplicate.
We found that PP1 and PP2, which are potent inhibitors of Src family protein kinases, are even more potent inhibitors of RIP2. The IC50 value for inhibition of RIP2 in vitro was 10–20 nM for PP2 (Figure 3B) and PP1 (results not shown), about 2-fold lower than the IC50 for inhibition of Src kinase and the related Lck kinase (results not shown). These observations led us to study the effect of PP2, as well as SB 203580, on MDP-induced signalling. Both of these compounds, but not BIRB 0796, suppressed MDP-stimulated NF-κB gene transcription (Figure 3C), indicating that this signalling pathway requires RIP2 kinase activity. By contrast, neither SB 203580 nor PP2 inhibited IL-1β-stimulated NF-κB gene transcription in HEK-293 cells that stably express the IL-1 receptor (termed ‘IL-1R cells’) (Figure 3D) or IκB (inhibitor of NF-κB) degradation (Figure 3E), indicating that the effects of SB 203580 and PP2 are specific for MDP signalling and that RIP2 kinase activity is not rate-limiting for IL-1-stimulated NF-κB gene transcription or IκB degradation. Moreover, at the concentrations used in these experiments, SB 203580 and BIRB 0796, but not PP2, attenuated the phosphorylation of TAB1 (Figure 3E), a well-authenticated physiological substrate of p38α MAPK [18]. Thus the effects of SB 203580 and PP2 are not mediated by the inhibition of p38α MAPK.
To investigate whether SB 203580 inhibits MDP signalling in a cell that normally expresses NOD2 and RIP2, we carried out further experiments in RAW 264.7 cells, a transformed mouse macrophage cell line. In these cells, MDP (presumably acting through the endogenous NOD2 and RIP2) stimulated NF-κB-dependent reporter gene transcription, which was suppressed by SB 203580, but not by BIRB 0796 (Figure 3F). In contrast, LPS-stimulated NF-κB-dependent gene transcription was unaffected by either SB 203580 or BIRB 0796 (Figure 3F).
TAK1 is required for signalling downstream of NOD2 and RIP2
The protein kinase TAK1 is known to play an essential role in mediating the LPS-induced activation of NF-κB, JNK and p38α MAPK and the production of pro-inflammatory cytokines in vivo [16,27–29], raising the question of whether TAK1 was also essential for MDP–NOD2/RIP2 signalling to NF-κB and MAP kinases. Consistent with the potential involvement of TAK1, this protein kinase has been reported to interact with NOD2 in co-transfection experiments [30]. These investigators also reported that the MDP-induced activation of NF-κB-dependent gene expression was suppressed by the overexpression of a catalytically inactive dominant-negative form of TAK1. Moreover, they also observed that the overexpression of NOD2 inhibited TAK1-induced NF-κB activation in RIP2-deficient fibroblasts. On the basis of these experiments they concluded that the NOD2 signalling pathway antagonizes TAK1-mediated signalling and vice versa [30].
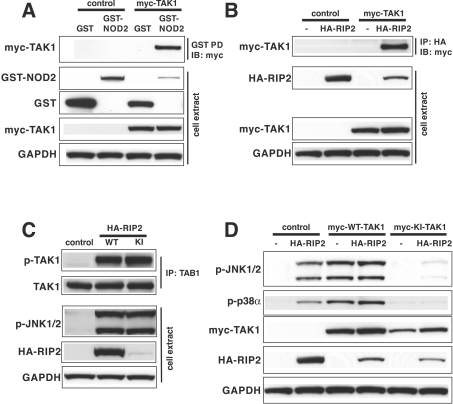
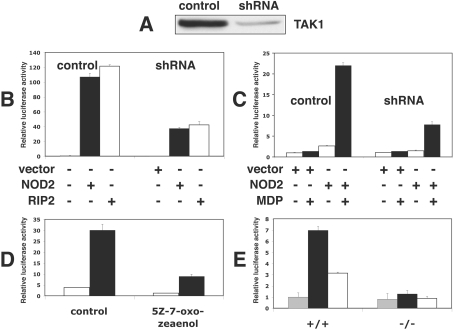
We have confirmed the previous report that NOD2 interacts with TAK1 (Figure 4A) and, in addition, found that RIP2 interacts with TAK1 in co-transfection experiments (Figure 4B). This interaction between RIP2 and TAK1 is likely to be direct and not indirect via NOD2, because HEK-293 cells do not express NOD2. Importantly, we also found that the endogenous TAK1 (measured by the phosphorylation of Thr187 [31]) and JNK were activated by transfection with DNA encoding either WT-RIP2 or KI-RIP (Figure 4C) and that a catalytically inactive TAK1 mutant suppressed the RIP2-induced phosphorylation (activation) of endogenous JNK and p38α MAPK (Figure 4D). Therefore, in contrast with the conclusions reached previously [30], our experiments suggested that TAK1 was not a negative regulator of NOD2–RIP2 signalling, but instead might be essential for signalling through this pathway. In order to investigate whether this was the case, we first reduced the level of expression of TAK1 by 80–90% in HEK-293 cells by introducing a vector expressing an shRNA (Figure 5A). This suppressed by 65–70% the activation of NF-κB induced either by the overexpression NOD2 or RIP2 (Figure 5B) or by MDP in the presence of low levels of transfected NOD2 (Figure 5C). In parallel experiments RIP2-induced JNK activation was similarly reduced by shRNA knockdown of TAK1 (results not shown).
Figure 4. RIP2 and NOD2 interact with, and activate, TAK1.
(A) HEK-293 cells in 10-cm-diameter dishes were co-transfected for 48 h with 3.75 μg of plasmid DNA encoding GST (glutathione S-transferase)-tagged NOD2 (or GST as a control) and 3.75 μg of DNA encoding Myc-tagged WT-TAK1 or an empty vector (as a control) and then lysed. An aliquot of the cell extract (0.5 mg of protein) was added to 20 μl of glutathione–Sepharose beads and, after 1 h incubation at 4 °C, the beads to which GST-NOD2 or GST were now bound were collected [GST PD (GST-pull down)], washed once with 1 ml of lysis buffer, twice with 1 ml of lysis buffer containing 500 mM NaCl and once with 1 ml of 10 mM Tris/HCl, pH 8. After denaturation in 1% (w/v) SDS, the proteins released were subjected to SDS/PAGE and immunoblotted with anti-Myc antibodies (IB: myc) to detect TAK1. Further aliquots of the cell extract (20 μg of protein) were also immunoblotted with the antibodies indicated. (B) Same as (A), except that HA–RIP2 replaced GST–NOD2 and, after cell lysis, an aliquot of the cell extract (0.5 mg of protein) was added to 5 μg of anti-HA antibody. Following incubation for 45 min at 4 °C, 20 μl of Protein G–Sepharose was added and, after another 45 min at 4 °C, the HA immunoprecipitates (IP: HA) were collected, washed as described in (A) and immunoblotted with anti-Myc antibodies (IB: myc) to detect TAK1. (C) Same as (B), except that 7.5 μg of DNA encoding HA–WT-RIP2 or HA–KI-RIP2 was transfected into HEK-293 cells and, after immunoprecipitation of the TAK1 complex (using antibodies raised against the TAB1 regulatory subunit), followed by washing as in (A), immunoblotting was performed with antibodies that recognize TAK1 phosphorylated at Thr187 (p-TAK1) and antibodies that recognize the phosphorylated and unphosphorylated forms of TAK1 equally well (TAK1). Further aliquots of the cell extract (20 μg of protein) were immunoblotted with antibodies that recognize the active phosphorylated forms of JNK1 and JNK2, anti-HA antibodies (to detect RIP2) and anti-GAPDH antibodies as a loading control. (D) HEK-293 cells were co-transfected with vectors expressing HA–RIP2 and Myc-tagged WT-TAK1 or KI-TAK1. After cell lysis, the extracts were then immunoblotted with the antibodies indicated.
Figure 5. TAK1 is required for MDP–NOD2/RIP2 signalling.
All experiments were carried at least twice with similar results. (A) Control HEK-293 cells or cells depleted of TAK1 by transfection with an shRNA were generated as described in the Experimental section. The cells were lysed, TAK1 immunoprecipitated as described in the legend to Figure 4, and the pellet extracted in SDS and immunoblotted with anti-TAK1 antibodies. (B) HEK-293 cells were co-transfected with 0.1 μg of DNA encoding 3× κB luciferase reporter construct (see the Experimental section), 0.1 μg of pTK-RL plasmid DNA (encoding Renilla luciferase) plus 0.8 μg of empty pCMV5 vector (black bars) or plasmids encoding GST-NOD2 (black bars) or HA-tagged WT-RIP2 (white bars). After 48 h, the cells were lysed and NF-κB-dependent luciferase reporter-gene expression was analysed. The error bars show the S.D. between duplicate experiments. (C) Experiments were performed as described in the legend to Figure 3(A), using 1 ng of DNA encoding HA–NOD2 or empty-vector DNA for transfection of control HEK-293 cells or cells depleted of TAK1. After 24 h, cells were incubated without (white bars) or with (black bars) 1 μM MDP. After 24 h the cells were lysed and NF-κB-dependent luciferase reporter-gene expression was analysed. The error bars show the S.D. between duplicate experiments. (D) Experiments were performed as in (C). At 1 h before stimulation with (black bars) or without (white bars) MDP, the cells were incubated without (control) or with 1 μM (5Z)-7-oxozeaenol, a TAK1 inhibitor. The error bars show the S.D. between duplicate experiments. (E) WT (+/+) or TAK1−/− (−/−) mouse embryonic fibroblasts were co-transfected with 0.5 μg of DNA encoding 3× κB luciferase reporter construct (see the Experimental section), 0.5 μg of pTK-RL plasmid DNA (encoding Renilla luciferase), plus 4 μg of empty pCMV5 vector (grey bars) or plasmids encoding GST–NOD2 (black bars) or HA-tagged WT-RIP2 (white bars) using the Amaxa Nucleofection MEF2 kit according to the manufacturer's instructions. After 48 h the cells were lysed and NF-κB-dependent luciferase reporter-gene expression was analysed. The error bars show the S.D. between duplicate experiments.
In order to confirm these results by an independent method, we studied the effect of (5Z)-7-oxozeaenol, which has been reported to be a potent and relatively specific inhibitor of TAK1 [32]. These experiments showed that this compound suppressed MDP-stimulated NF-κB gene transcription by about 70% when included in the cell culture medium at 1 μM (Figure 5D). We found that this concentration was needed to suppress completely IL-1-stimulated activated of JNK, p38α MAPK and ERK1/ERK2 in IL-1R cells (results not shown).
We then studied the effects of NOD2 and RIP2 on NF-κB gene transcription in immortalized embryonic fibroblasts from WT and TAK1−/− mice. These experiments showed that transfection with DNA expressing either NOD2 or RIP2 stimulated NF-κB-dependent gene transcription in the WT cells, but no stimulation occurred in the TAK1-deficient cells (Figure 5E).
MDP-induced production of IL-1β in PBMCs is prevented by specific inhibition of TAK1, p38α MAPK or MKK1/MKK2
The experiments described above indicated that TAK1 represents an important point of convergence of the LPS–TLR4 and MDP–NOD2 signalling pathways, suggesting that many subsequent downstream events might be common to both pathways. We therefore investigated the signalling pathways required for the MDP-induced production of the pro-inflammatory cytokines IL-1β and TNFα in human PBMCs a more physiological system in which the cells are known to respond particularly well to this agonist [8].
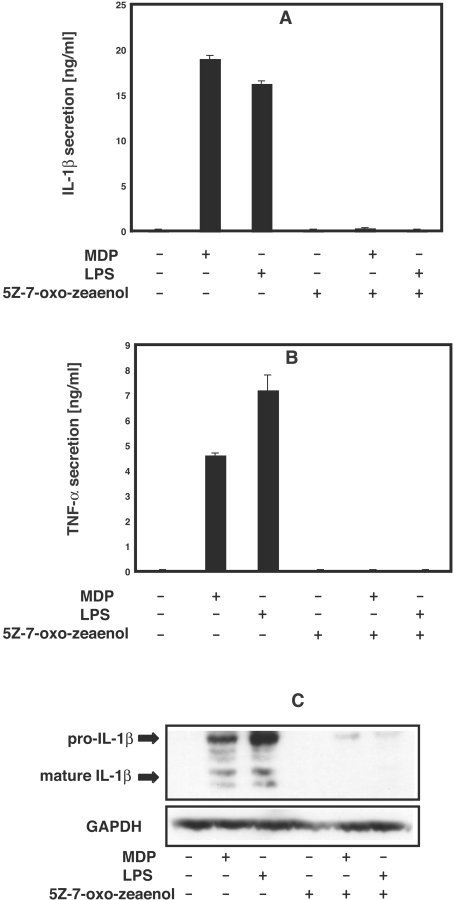
To examine whether TAK1 activity is required for signalling by MDP in human PBMCs, we studied the effect of the TAK1 inhibitor (5Z)-7-oxozeaenol. When added to the culture medium at 1 μM, this compound prevented both the MDP-stimulated and LPS-stimulated secretion of IL-1β (Figure 6A) and TNFα (Figure 6B) and the MDP- or LPS-induced production of pro-IL-1β (Figure 6C) in human PBMCs.
Figure 6. The TAK1 inhibitor (5Z)-7-oxozeaenol suppresses MDP-stimulated production of IL-1β and TNFα.
Human PBMCs were resuspended in RPMI 1640 medium without FCS. Cells were grown in 24-well plates (3×107 cells/well) and incubated for 60 min with (+) or without (−) 1 μM (5Z)-7-oxozeaenol, then stimulated with 50 nM MDP or 100 ng/ml LPS (+) or left unstimulated (−). After 16 h the cells were pelleted by centrifugation for 2 min at 3000 g and the supernatants collected. (A, B) Aliquots of the supernatant were diluted appropriately and the concentration of IL-1β (A) or TNFα (B) determined by ELISA (R&D systems) as described by the manufacturer. The results are presented as means±S.D. for duplicate determinations on one donor. Similar results were obtained in experiments with a second donor. (C) The cell pellet from (A) was lysed, and levels of pro-IL-1β and mature IL-1β in the extracts analysed by immunoblotting with an IL-1β-specific antibody. The results shown are representative of experiments with cells derived from two independent donors.
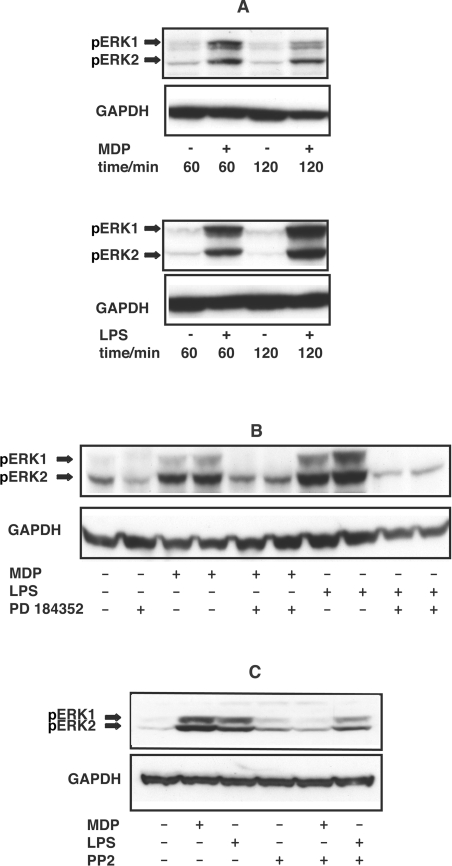
Two protein kinases activated downstream of TAK1 and, required for the LPS-induced production of a number of pro-inflammatory cytokines, are p38α MAPK [20] and Tpl2 [tumour progression locus 2; also called COT (cancer osaka thyroid)] [33]. The p38α MAPK activates MAPKAP-K2 (MAPK-activated protein kinase 2), which is required for the LPS-induced production of several pro-inflammatory cytokines [34], while Tpl2 activates the MAPK kinases MKK1 and MKK2 and hence ERK1/ERK2 [33]. We found that, as for LPS, MDP induced the activation of ERK1/ERK2 within 60 min in human PBMCs (Figure 7A) and that this was prevented by PD 184352 (Figure 7B), a potent and specific inhibitor of MKK1/MKK2 [35,36]. PP2, a potent inhibitor of RIP2 (Figure 3B), suppressed the MDP-, but not LPS-induced, activation of ERK1/ERK2, indicating that RIP2 kinase activity is required for MDP signalling to ERK1/ERK2 in human PBMCs (Figure 7C).
Figure 7. MDP induces the phosphorylation of ERK1 and ERK2 by activating MKK1.
Each of the results shown is representative of experiments with cells derived from at least two independent donors. PBMCs resuspended in RPMI 1640 without FCS were kept in 10-cm-diameter dishes and left to recover for 3 h. (A) PBMC were stimulated with (+) or without (−) 50 nM MDP or 100 ng/ml LPS, then lysed at the times indicated. Levels of phosphorylation of ERK1 and ERK2 (pERK1, pERK2) were analysed by immunoblotting with a phosphospecific antibody. (B) The experiment was carried out as in (A), except that the PBMCs were incubated for 60 min with (+) or without (−) 2 μM PD 184352 before stimulation with (+) or without (−) 50 nM MDP or 100 ng/ml LPS. After 60 min, cells were lysed and the levels of phosphorylated ERK1/ERK2 were analysed as in (A). (C) The experiment was carried out as in (B), except that PBMCs were incubated for 60 min with (+) or without (−) 1 μM PP2 instead of 2 μM PD 184352, prior to stimulation with MDP or LPS.
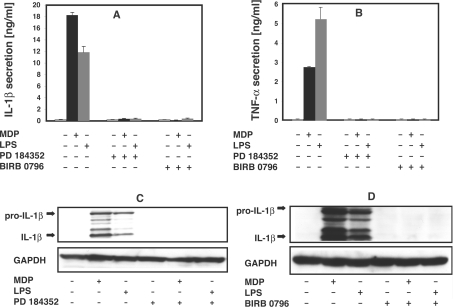
Like LPS, MDP stimulated the secretion IL-1β and TNFα in human PBMCs, and this was suppressed by PD 184352 or the p38α MAPK inhibitor BIRB 0796 (Figures 8A and 8B). The BIRB 0796 was used at 0.1 μM, a concentration at which it inhibits p38α/β MAPKs specifically, but does not inhibit RIP2 (Figure 2A) or many other protein kinases tested [22]. PD 184352 (Figure 8C) or BIRB 0796 (Figure 8D) also prevented the MDP- or LPS-induced expression of pro-IL-1β, demonstrating that these drugs exert their effects by blocking pro-IL-1β expression, rather than just IL-1β secretion.
Figure 8. MDP-induced secretion of IL-1β and TNF-α and expression of pro-IL-1β are prevented by inhibition of MKK1 or p38α MAPK.
Human PBMCs were resuspended in RPMI 1640 medium without FCS. (A, B) Cells were grown in 24-well plates (3×107 cells/well) and incubated for 60 min with (+) or without (−) 2 μM PD 184352 or 0.1 μM BIRB 0796, then stimulated with 50 nM MDP or 100 ng/ml LPS (+) or left unstimulated (−). After 16 h the cells were pelleted by centrifugation for 2 min at 3000 g and the supernatants collected. Aliquots were diluted appropriately and the concentration of IL-1β (A) and TNFα (B) determined by ELISA (R&D systems) as described by the manufacturer. The results are presented with the S.D. for duplicate determinations on one donor. Similar results were obtained in experiments with a second donor. (C, D) Cells were grown in 10-cm-diameter dishes (1×108 cells/dish) and incubated for 60 min with (+) or without (−) 2 μM PD 184352 (C) or 0.1 μM BIRB 0796 (D), and then stimulated with MDP or LPS as described in (A, B). After 16 h, cells were lysed, and levels of pro-IL-1β and mature IL-1β in the extracts analysed by immunoblotting with an IL-1β-specific antibody. The results shown are representative of experiments with cells derived from two independent donors.
It should be mentioned that PBMC from different human donors showed large quantitative differences in the amount of IL-1β or TNFα produced in response to MDP or LPS, although sensitivity to BIRB 0796 and PD 184352 was unaltered. However, in two donors, MDP-stimulated production of IL-1β in PBMC was only inhibited 50 and 70% respectively by BIRB 0796, and 90 and 70% respectively by PD 184352, whereas in other donors these inhibitors caused almost complete suppression (C. Lang, unpublished work).
DISCUSSION
At the time when RIP2 was first identified, it was noticed that, when overexpressed in HEK-293 cells, a catalytically inactive [KI (kinase inactive)] mutant was as effective as the WT protein in activating JNK or NF-κB [10] or inducing the apoptosis of MCF7 breast sarcoma cells [11]. These observations raised the question of what role the protein kinase activity of RIP2 might play. Here, we demonstrate that KI-RIP2 is much more effective than WT-RIP2 in activating JNK and NF-κB in transfected HEK-293 cells (Figures 1 and 2), indicating that the protein kinase activity functions to limit the strength of signalling under these conditions. The molecular events that underlie this action are unknown, but since HEK-293 cells do not express NOD2, it cannot be explained by RIP2 exerting a negative-feedback control on NOD2. Thus mechanisms involving a RIP2-kinase-catalysed enhancement of the interaction of NOD2 with an inhibitory protein, such as Erbin (Erbb2 interacting protein) [37,38], appear to be excluded. RIP2 is known to phosphorylate itself at multiple sites [39], raising the possibility that the autophosphorylated protein kinase is less effective than the unphosphorylated form of the enzyme in activating TAK1 and/or other proteins that drive downstream signalling events.
We have also shown that the protein kinase activity of RIP2 is required to maintain its level of expression in transfected HEK-293 cells. Thus KI-RIP2 was expressed at much lower levels than WT-RIP2, whereas pharmacological inhibition with SB 203580 (Figures 2B and 2C) or PP2 (results not shown) reduced the expression of WT-RIP2 to a level similar to that of KI-RIP2, without affecting the level of expression of KI-RIP2. Moreover, SB 203580 had a much smaller effect on the expression of a RIP2 mutant that is less sensitive to SB 203580 (Figure 2C). Interestingly, the SB 203580-induced decrease in expression of RIP2 is quite fast, being complete within 30–60 min, but preliminary experiments indicate that this cannot be prevented by either the proteasome inhibitor MG 132 or a specific inhibitor of caspase 1 (M. Windheim and P. Cohen, unpublished work), a proteinase that interacts with, and is activated by, NOD2 [40]. Thus the mechanism that leads to the disappearance of activated RIP2 has still to be clarified.
Importantly, we found that the MDP-stimulated activation of NF-κB gene transcription in NOD2-transfected HEK-293 cells or in untransfected RAW 264.7 cells was prevented by SB 203580 or PP2, but not by the much more potent p38α MAPK inhibitor BIRB 0796, which does not inhibit RIP2 (Figure 3). Our working hypothesis is that MDP induces the active conformation of RIP2, which then phosphorylates itself or another regulatory protein, leading to stabilization of the activated enzyme. Pharmacological inhibition of RIP2 kinase prevents this stabilization, and the MDP-induced downstream signalling does not occur because the level of expression of RIP2 falls below the threshold needed for signalling to occur. We assume that KI-RIP2 is able to trigger MDP-dependent NF-κB gene transcription in transfected cells, because it is expressed at supraphysiological levels under these conditions, levels well in excess of those needed to support signalling. To our knowledge, the results of the present study provide the first evidence that the protein kinase activity of RIP2 is required for signalling downstream of MDP–NOD2, albeit indirectly. Interestingly, knock-in mice that express the catalytically inactive RIP2[K47A] mutant instead of WT-RIP2 also exhibit decreased expression of the mutant enzyme [41]. It would clearly be of considerable interest to know whether MDP is capable of stimulating NF-κB-dependent gene transcription and pro-inflammatory cytokine production in these animals, but this was not investigated in the published study [41].
In contrast with the inhibition of MDP-stimulated NF-κB gene transcription by SB 203580 and PP2, LPS-stimulated and IL-1β stimulated NF-κB reporter-gene transcription in RAW 264.7 (Figure 3F) and IL-1R cells (Figure 3D) respectively were unaffected by SB 203580 and PP2. Moreover, PP2 suppressed MDP-, but not LPS-, induced activation of ERK1/ERK2 in human PBMCs (Figure 7C). Thus, although LPS stimulates RIP2 kinase activity [39,41], RIP2 kinase activity is not rate-limiting for LPS-induced activation of NF-κB and ERK1/ERK2. While the present paper being revised, other investigators [42] also reported that RIP2 was not required for signalling through LPS–TLR4 or other TLRs.
TAK1 is thought to play an essential role in the LPS-induced activation of signalling pathways and the production of pro-inflammatory cytokines [16,27–29], and in the present paper we demonstrate that TAK1 is also required for MDP–NOD2/RIP2 signalling (Figures 4 and 5) and proinflammatory cytokine production (Figure 6). These results imply that TAK1 represents one of the key points at which the LPS–TLR4 and MDP–NOD2 signalling pathways converge (Figure 9), explaining why downstream events, such as the activation of NF-κB, JNK, p38α MAPK and ERK1/ERK2, and the production of IL-1β and TNFα, are common to both pathways (Figures 4–8). Consistent with a key role for TAK1, it has been reported that the overexpression of NOD2 or RIP2 activates IKKβ (inhibitor of NF-κB kinase β) by inducing the phosphorylation of Ser177 and Ser181 [43], the sites phosphorylated by TAK1. Moreover, as for LPS, the activation of p38α MAPK and ERK1/ERK2 are both required for the MDP-induced production of Pro-IL-1β, secreted IL-1β or secreted TNFα in human PBMCs (Figure 8), which express endogenous levels of NOD2 and RIP2.
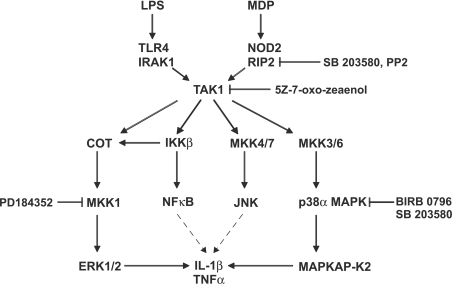
Figure 9. Signalling pathways required for the MDP- and LPS-induced production of pro-inflammatory cytokines.
The results described in the present paper indicate that MDP- and LPS-stimulated signalling converges at the level of TAK1 and that the activation of MKK1 and p38α MAPK is required for the production of IL-1β and TNF-α. The targets for the drugs used in the present study [SB 203580, PP2, BIRB 0796, (5Z)-7-oxozeaenol and PD 184352] are indicated. NF-κB and JNK may promote IL-1β and TNFα production by stimulating transcription of their genes, as indicated by the broken-line arrows.
RIP2 is also known to be required for the signalling by M-TriDAP–NOD1 (see the Introduction). While the present paper was being revised, other investigators [44] reported that dominant-negative TAK1 and siRNA (small interfering RNA) knockdown of TAK1 reduced M-TriDAP–NOD1-induced activation of JNK and IL-8 secretion. That study, together with ours, indicates that TAK1 lies downstream of RIP2 in the NOD1 as well as the NOD2 signalling pathway.
A major outstanding question concerns the mechanism by which RIP2 triggers the activation of TAK1 and other downstream signalling events. The activation of TAK1 and IKKβ by TNFα, IL-1β or LPS is thought to be initiated by the formation of K63-pUb (Lys63-linked polyubiquitin) chains that become linked covalently to the protein kinase RIP ([45,45a,46]) and the ubiquitin E3 ligase TRAF-6 (tumour-necrosis-factor-receptor-associated factor 6) [47]. These K63-pUb scaffolds interact with the NEMO (NF-κB essential modifier) regulatory subunit of the IKK complex and the TAB2/TAB3 regulatory subunits of the TAK1 complex, which may co-localize TAK1 and IKKβ and so facilitate the TAK1-catalysed phosphorylation and activation of IKKβ. The catalytic domain of RIP is most similar to RIP2, and, like RIP2, the catalytic activity of RIP does not appear to be required for signalling by TNFα [48], raising the question of whether RIP2 might also trigger downstream signalling by a mechanism that requires the Lys-63-linked polyubiquitination of one or more proteins. Indeed, it has been reported that NEMO undergoes Lys-63-linked polyubiquitination when co-transfected with either NOD2 or RIP2 in HEK-293 cells [43]. Moreover, these effects were suppressed by co-transfection with vectors expressing the Lys-63-specific deubiquitinating enzyme CYLD [cylindromatosis (turban tumor syndrome)] [43]. These observations have provided the first indication that Lys-63-linked polyubiquitination of NEMO may be critical for the NOD2/RIP2-induced activation of IKKβ, which can lead to the activation of ERK1/ERK2 [49,50], as well as the activation of NF-κB. However, as the NEMO–IKKβ complex does not participate in the activation of p38α MAPK or JNK (Figure 9), it seems likely that additional devices are needed to mediate the NOD2/RIP2-induced activation of these MAPKs.
Acknowledgments
We thank Professor Shizuo Akira (Department of Host Defense, Research Institute for Microbial Diseases, Osaka University, Osaka, Japan) for providing TAK1−/− cells, Dr Natalia Shpiro and Dr Rudolfo Marquez for synthesizing PD 184352 and BIRB 0796, Dr Ron Hay for providing several constructs, Marjan Ford (MRC Laboratory for Molecular Biology, Cambridge, U.K.) for GAK, and the Ninewells Hospital Blood Transfusion Centre, Dundee, Scotland, U.K., for providing buffy coats. We are grateful to the protein and antibody production teams of the Division of Signal Transduction Therapy, University of Dundee (co-ordinated by Dr Hilary McLaughlan and Dr James Hastie) for His6-RIP2 purified from insect Sf21 cells and for antibodies, and the DNA Sequencing Service, University of Dundee, Dundee, Scotland, U.K. (www.dnaseq.co.uk). M.W. acknowledges a postdoctoral position from EU Research Training Network Framework 5. This work was supported by the UK Medical Research Council, The Royal Society, AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Merck and Co, Merck KGaA and Pfizer.
References
- 1.Inohara N., Chamaillard M., McDonald C., Nuñez G. NOD–LRR proteins: role in host–microbial interactions and inflammatory disease. Annu. Rev. Biochem. 2005;74:355–383. doi: 10.1146/annurev.biochem.74.082803.133347. [DOI] [PubMed] [Google Scholar]
- 2.Kobayashi K. S., Chamaillard M., Ogura Y., Henegariu O., Inohara N., Nunez G., Flavell R. A. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 3.Pauleau A. L., Murray P. J. Role of nod2 in the response of macrophages to toll-like receptor agonists. Mol. Cell. Biol. 2003;23:7531–7539. doi: 10.1128/MCB.23.21.7531-7539.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brant S. R., Shugart Y. Y. Inflammatory bowel disease gene hunting by linkage analysis: rationale, methodology, and present status of the field. Inflamm. Bowel Dis. 2004;10:300–311. doi: 10.1097/00054725-200405000-00019. [DOI] [PubMed] [Google Scholar]
- 5.Hampe J., Cuthbert A., Croucher P. J., Mirza M. M., Mascheretti S., Fisher S., Frenzel H., King K., Hasselmeyer A., MacPherson A. J., et al. Association between insertion mutation in NOD2 gene and Crohn's disease in German and British populations. Lancet. 2001;357:1925–1928. doi: 10.1016/S0140-6736(00)05063-7. [DOI] [PubMed] [Google Scholar]
- 6.Hugot J. P., Chamaillard M., Zouali H., Lesage S., Cezard J. P., Belaiche J., Almer S., Tysk C., O'Morain C. A., Gassull M., et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 7.Ogura Y., Bonen D. K., Inohara N., Nicolae D. L., Chen F. F., Ramos R., Britton H., Moran T., Karaliuskas R., Duerr R. H., et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 8.Netea M. G., Ferwerda G., de Jong D. J., Werts C., Boneca I. G., Jehanno M., Van Der Meer J. W., Mengin-Lecreulx D., Sansonetti P. J., Philpott D. J., et al. The frameshift mutation in Nod2 results in unresponsiveness not only to Nod2- but also Nod1-activating peptidoglycan agonists. J. Biol. Chem. 2005;280:35859–35867. doi: 10.1074/jbc.M504924200. [DOI] [PubMed] [Google Scholar]
- 9.Chin A. I., Dempsey P. W., Bruhn K., Miller J. F., Xu Y., Cheng G. Involvement of receptor-interacting protein 2 in innate and adaptive immune responses. Nature. 2002;416:190–194. doi: 10.1038/416190a. [DOI] [PubMed] [Google Scholar]
- 10.Thome M., Hofmann K., Burns K., Martinon F., Bodmer J. L., Mattmann C., Tschopp J. Identification of CARDIAK, a RIP-like kinase that associates with caspase-1. Curr. Biol. 1998;8:885–888. doi: 10.1016/s0960-9822(07)00352-1. [DOI] [PubMed] [Google Scholar]
- 11.McCarthy J. V., Ni J., Dixit V. M. RIP2 is a novel NF-κB-activating and cell death-inducing kinase. J. Biol. Chem. 1998;273:16968–16975. doi: 10.1074/jbc.273.27.16968. [DOI] [PubMed] [Google Scholar]
- 12.Shpiro N., Marquez R. An improved synthesis of the potent MEK inhibitor PD184352. Synth. Commun. 2005;35:2265. [Google Scholar]
- 13.Regan J., Breitfelder S., Cirillo P., Gilmore T., Graham A. G., Hickey E., Klaus B., Madwed J., Moriak M., Moss N., et al. Pyrazole urea-based inhibitors of p38 MAP kinase: from lead compound to clinical candidate. J. Med. Chem. 2002;45:2994–3008. doi: 10.1021/jm020057r. [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez M. S., Thompson J., Hay R. T., Dargemont C. Nuclear retention of IκBα protects it from signal-induced degradation and inhibits nuclear factor κB transcriptional activation. J. Biol. Chem. 1999;274:9108–9115. doi: 10.1074/jbc.274.13.9108. [DOI] [PubMed] [Google Scholar]
- 15.Zufferey R., Nagy D., Mandel R. J., Naldini L., Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
- 16.Sato S., Sanjo H., Takeda K., Ninomiya-Tsuji J., Yamamoto M., Kawai T., Matsumoto K., Takeuchi O., Akira S. Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat. Immunol. 2005;6:1087–1095. doi: 10.1038/ni1255. [DOI] [PubMed] [Google Scholar]
- 17.Durocher Y., Perret S., Kamen A. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 2002;30:E9. doi: 10.1093/nar/30.2.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheung P. C., Campbell D. G., Nebreda A. R., Cohen P. Feedback control of the protein kinase TAK1 by SAPK2a/p38α. EMBO J. 2003;22:5793–5805. doi: 10.1093/emboj/cdg552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cuenda A., Rouse J., Doza Y. N., Meier R., Cohen P., Gallagher T. F., Young P. R., Lee J. C. SB 203580 is a specific inhibitor of a MAP kinase homologue which is stimulated by cellular stresses and interleukin-1. FEBS Lett. 1995;364:229–233. doi: 10.1016/0014-5793(95)00357-f. [DOI] [PubMed] [Google Scholar]
- 20.Lee J. C., Laydon J. T., McDonnell P. C., Gallagher T. F., Kumar S., Green D., McNulty D., Blumenthal M. J., Heys J. R., Landvatter S. W., et al. A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature. 1994;372:739–746. doi: 10.1038/372739a0. [DOI] [PubMed] [Google Scholar]
- 21.Godl K., Wissing J., Kurtenbach A., Habenberger P., Blencke S., Gutbrod H., Salassidis K., Stein-Gerlach M., Missio A., Cotten M., Daub H. An efficient proteomics method to identify the cellular targets of protein kinase inhibitors. Proc. Natl. Acad. Sci. U.S.A. 2003;100:15434–15439. doi: 10.1073/pnas.2535024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuma Y., Sabio G., Bain J., Shpiro N., Marquez R., Cuenda A. BIRB796 inhibits all p38 MAPK isoforms in vitro and in vivo. J. Biol. Chem. 2005;280:19472–19479. doi: 10.1074/jbc.M414221200. [DOI] [PubMed] [Google Scholar]
- 23.Pargellis C., Tong L., Churchill L., Cirillo P. F., Gilmore T., Graham A. G., Grob P. M., Hickey E. R., Moss N., Pav S., Regan J. Inhibition of p38 MAP kinase by utilizing a novel allosteric binding site. Nat. Struct. Biol. 2002;9:268–272. doi: 10.1038/nsb770. [DOI] [PubMed] [Google Scholar]
- 24.Eyers P. A., Craxton M., Morrice N., Cohen P., Goedert M. Conversion of SB 203580-insensitive MAP kinase family members to drug-sensitive forms by a single amino-acid substitution. Chem. Biol. 1998;5:321–328. doi: 10.1016/s1074-5521(98)90170-3. [DOI] [PubMed] [Google Scholar]
- 25.Gum R. J., McLaughlin M. M., Kumar S., Wang Z., Bower M. J., Lee J. C., Adams J. L., Livi G. P., Goldsmith E. J., Young P. R. Acquisition of sensitivity of stress-activated protein kinases to the p38 inhibitor, SB 203580, by alteration of one or more amino acids within the ATP binding pocket. J. Biol. Chem. 1998;273:15605–15610. doi: 10.1074/jbc.273.25.15605. [DOI] [PubMed] [Google Scholar]
- 26.Inohara N., Ogura Y., Fontalba A., Gutierrez O., Pons F., Crespo J., Fukase K., Inamura S., Kusumoto S., Hashimoto M., et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn's disease. J. Biol. Chem. 2003;278:5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 27.Liu H. H., Xie M., Schneider M. D., Chen Z. J. Essential role of TAK1 in thymocyte development and activation. Proc. Natl. Acad. Sci. U.S.A. 2006;103:11677–11682. doi: 10.1073/pnas.0603089103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Omori E., Matsumoto K., Sanjo H., Sato S., Akira S., Smart R. C., Ninomiya-Tsuji J. TAK1 is a master regulator of epidermal homeostasis involving skin inflammation and apoptosis. J. Biol. Chem. 2006;281:19610–19617. doi: 10.1074/jbc.M603384200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shim J. H., Xiao C., Paschal A. E., Bailey S. T., Rao P., Hayden M. S., Lee K. Y., Bussey C., Steckel M., Tanaka N., et al. TAK1, but not TAB1 or TAB2, plays an essential role in multiple signalling pathways in vivo. Genes Dev. 2005;19:2668–2681. doi: 10.1101/gad.1360605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen C. M., Gong Y., Zhang M., Chen J. J. Reciprocal cross-talk between Nod2 and TAK1 signalling pathways. J. Biol. Chem. 2004;279:25876–25882. doi: 10.1074/jbc.M400682200. [DOI] [PubMed] [Google Scholar]
- 31.Singhirunnusorn P., Suzuki S., Kawasaki N., Saiki I., Sakurai H. Critical roles of threonine 187 phosphorylation in cellular stress-induced rapid and transient activation of transforming growth factor-β-activated kinase 1 (TAK1) in a signalling complex containing TAK1-binding protein TAB1 and TAB2. J. Biol. Chem. 2005;280:7359–7368. doi: 10.1074/jbc.M407537200. [DOI] [PubMed] [Google Scholar]
- 32.Ninomiya-Tsuji J., Kajino T., Ono K., Ohtomo T., Matsumoto M., Shiina M., Mihara M., Tsuchiya M., Matsumoto K. A resorcylic acid lactone, 5Z-7-oxozeaenol, prevents inflammation by inhibiting the catalytic activity of TAK1 MAPK kinase kinase. J. Biol. Chem. 2003;278:18485–18490. doi: 10.1074/jbc.M207453200. [DOI] [PubMed] [Google Scholar]
- 33.Dumitru C. D., Ceci J. D., Tsatsanis C., Kontoyiannis D., Stamatakis K., Lin J. H., Patriotis C., Jenkins N. A., Copeland N. G., Kollias G., Tsichlis P. N. TNF-α induction by LPS is regulated posttranscriptionally via a Tpl2/ERK-dependent pathway. Cell. 2000;103:1071–1083. doi: 10.1016/s0092-8674(00)00210-5. [DOI] [PubMed] [Google Scholar]
- 34.Kotlyarov A., Neininger A., Schubert C., Eckert R., Birchmeier C., Volk H. D., Gaestel M. MAPKAP kinase 2 is essential for LPS-induced TNF-α biosynthesis. Nat. Cell Biol. 1999;1:94–97. doi: 10.1038/10061. [DOI] [PubMed] [Google Scholar]
- 35.Davies S. P., Reddy H., Caivano M., Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sebolt-Leopold J. S., Dudley D. T., Herrera R., Van Becelaere K., Wiland A., Gowan R. C., Tecle H., Barrett S. D., Bridges A., Przybranowski S., et al. Blockade of the MAP kinase pathway suppresses growth of colon tumors in vivo. Nat. Med. 1999;5:810–816. doi: 10.1038/10533. [DOI] [PubMed] [Google Scholar]
- 37.Kufer T. A., Kremmer E., Banks D. J., Philpott D. J. Role for Erbin in bacterial activation of Nod2. Infect. Immun. 2006;74:3115–3124. doi: 10.1128/IAI.00035-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDonald C., Chen F. F., Ollendorff V., Ogura Y., Marchetto S., Lecine P., Borg J. P., Nunez G. A role for Erbin in the regulation of Nod2-dependent NF-κB signalling. J. Biol. Chem. 2005;280:40301–40309. doi: 10.1074/jbc.M508538200. [DOI] [PubMed] [Google Scholar]
- 39.Dorsch M., Wang A., Cheng H., Lu C., Bielecki A., Charron K., Clauser K., Ren H., Polakiewicz R. D., Parsons T., et al. Identification of a regulatory autophosphorylation site in the serine-threonine kinase RIP2. Cell Signalling. 2006;18:2223–2229. doi: 10.1016/j.cellsig.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 40.Netea M. G., Azam T., Ferwerda G., Girardin S. E., Walsh M., Park J. S., Abraham E., Kim J. M., Yoon D. Y., Dinarello C. A., Kim S. H. IL-32 synergizes with nucleotide oligomerization domain (NOD) 1 and NOD2 ligands for IL-1beta and IL-6 production through a caspase 1-dependent mechanism. Proc. Natl. Acad. Sci. U.S.A. 2005;102:16309–16314. doi: 10.1073/pnas.0508237102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu C., Wang A., Dorsch M., Tian J., Nagashima K., Coyle A. J., Jaffee B., Ocain T. D., Xu Y. Participation of Rip2 in lipopolysaccharide signalling is independent of its kinase activity. J. Biol. Chem. 2005;280:16278–16283. doi: 10.1074/jbc.M410114200. [DOI] [PubMed] [Google Scholar]
- 42.Park J. H., Kim Y. G., McDonald C., Kanneganti T. D., Hasegawa M., Body-Malapel M., Inohara N., Nunez G. RICK/RIP2 mediates innate immune responses induced through Nod1 and Nod2 but not TLRs. J. Immunol. 2007;178:2380–2386. doi: 10.4049/jimmunol.178.4.2380. [DOI] [PubMed] [Google Scholar]
- 43.Abbott D. W., Wilkins A., Asara J. M., Cantley L. C. The Crohn's disease protein, NOD2, requires RIP2 in order to induce ubiquitinylation of a novel site on NEMO. Curr. Biol. 2004;14:2217–2227. doi: 10.1016/j.cub.2004.12.032. [DOI] [PubMed] [Google Scholar]
- 44.da Silva Correia J., Miranda Y., Leonard N., Hsu J., Ulevitch R. J. Regulation of Nod1-mediated signalling pathways. Cell Death Differ. 2006;14:830–839. doi: 10.1038/sj.cdd.4402070. [DOI] [PubMed] [Google Scholar]
- 45.Wu C. J., Conze D. B., Li T., Srinivasula S. M., Ashwell J. D. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-κB activation. Nat. Cell Biol. 2006;8:398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- 45a.Erratum. Nat. Cell Biol. 2006;8:424. [Google Scholar]
- 46.Ea C. K., Deng L., Xia Z. P., Pineda G., Chen Z. J. Activation of IKK by TNFα requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol. Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 47.Deng L., Wang C., Spencer E., Yang L., Braun A., You J., Slaughter C., Pickart C., Chen Z. J. Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 48.Ting A. T., Pimentel-Muinos F. X., Seed B. RIP mediates tumor necrosis factor receptor 1 activation of NFκB but not Fas/APO-1-initiated apoptosis. EMBO J. 1996;15:6189–6196. [PMC free article] [PubMed] [Google Scholar]
- 49.Beinke S., Robinson M. J., Hugunin M., Ley S. C. Lipopolysaccharide activation of the TPL-2/MEK/extracellular signal-regulated kinase mitogen-activated protein kinase cascade is regulated by IκB kinase-induced proteolysis of NF-κB1 p105. Mol. Cell. Biol. 2004;24:9658–9667. doi: 10.1128/MCB.24.21.9658-9667.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waterfield M., Jin W., Reiley W., Zhang M., Sun S. C. IκB kinase is an essential component of the Tpl2 signalling pathway. Mol. Cell. Biol. 2004;24:6040–6048. doi: 10.1128/MCB.24.13.6040-6048.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]