Abstract
Tumour-specific chromosomal rearrangements are known to create chimaeric products with the ability to generate many human cancers. hTAFII68-TEC (where hTAFII68 is human TATA-binding protein-associated factor II 68 and TEC is translocated in extraskeletal chondrosarcoma) is such a fusion product, resulting from a t(9;17) chromosomal translocation found in extraskeletal myxoid chondrosarcomas, where the hTAFII68 NTD (N-terminal domain) is fused to TEC protein. To identify proteins that control hTAFII68-TEC function, we used affinity chromatography on immobilized hTAFII68 (NTD) and MALDI-TOF (matrix-assisted laser-desorption ionization–time-of-flight) MS and isolated a novel hTAFII68-TEC-interacting protein, GAPDH (glyceraldehyde-3-phosphate dehydrogenase). GAPDH is a glycolytic enzyme that is also involved in the early steps of apoptosis, nuclear tRNA export, DNA replication, DNA repair and transcription. hTAFII68-TEC and GAPDH were co-immunoprecipitated from cell extracts, and glutathione S-transferase pull-down assays revealed that the C-terminus of hTAFII68 (NTD) was required for interaction with GAPDH. In addition, three independent regions of GAPDH (amino acids 1–66, 67–160 and 160–248) were involved in binding to hTAFII68 (NTD). hTAFII68-TEC-dependent transcription was enhanced by GAPDH, but not by a GAPDH mutant defective in hTAFII68-TEC binding. Moreover, a fusion of GAPDH with the GAL4 DNA-binding domain increased the promoter activity of a reporter containing GAL4 DNA-binding sites, demonstrating the presence of a transactivation domain(s) in GAPDH. The results of the present study suggest that the transactivation potential of the hTAFII68-TEC oncogene product is positively modulated by GAPDH.
Keywords: chromosome translocation, co-activator, extraskeletal myxoid chondrosarcoma, hTAFII68-TEC, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), protein–protein interaction, transactivation
Abbreviations: EMC, extraskeletal myxoid chondrosarcoma; EWS, Ewing's sarcoma; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GST, glutathione S-transferase; HEK, human embryonic kidney; hTAFII68, human TATA-binding protein-associated factor II 68; MALDI-TOF MS, matrix-assisted laser-desorption ionization–time-of-flight MS; NBRE, nerve growth factor inducible factor B response element; NTD, N-terminal domain; OCA-S, Oct-1 co-activator in S-phase; PML, promyelocytic leukaemia protein; TEC, translocated in extraskeletal chondrosarcoma; TLS, translocated in liposarcoma
INTRODUCTION
Human EMCs (extraskeletal myxoid chondrosarcomas) are soft tissue tumours of uncertain histogenetic origin arising primarily in the musculature, most commonly the thigh and knee [1–3]. A proportion of EMCs have been shown to harbour a characteristic translocation t(9;17)(q22;q11.2) involving the hTAFII68 (human TATA-binding protein-associated factor II 68) gene at 17q11.2 and the TEC (translocated in extraskeletal chondrosarcoma) gene at 9q22 [4,5].
hTAFII68 encodes a putative RNA/ss (single-stranded) DNA-binding protein, and was originally identified by its homology to the proto-oncogenes EWS (Ewing's sarcoma) and TLS (translocated in liposarcoma; another member of the EWS gene family) [6,7]. The two latter genes were cloned as the 5′-components of translocation-generated fusion genes in Ewing's sarcomas and myxoid liposarcomas [8,9]. The EWS and TLS genes are involved in several tumour-related chromosomal translocations that produce fusions with genes postulated to function as transcription factors [10,11]. In each case, the translocation produces chimaeric molecules containing the NTD (N-terminal domain) of EWS or TLS fused to the DNA-binding domain of the partner. TEC (also known as CHN and MINOR) is the human homologue of the rat NOR-1 receptor [12], and encodes a novel orphan nuclear receptor belonging to the steroid/thyroid receptor gene superfamily [1,2].
GAPDH (glyceraldehyde-3-phosphate dehydrogenase) is a multi-functional nuclear and cytoplasmic protein with glycolytic and non-glycolytic functions. It is present in several cellular compartments, including the cytoplasm, nucleus and plasma membrane [13–16]. In those subcellular locales, it functions in the catalysis of membrane fusion and transport [17–20], microtubule bundling [21,22], phosphate group transfer [23,24], nuclear RNA export [25,26], DNA repair [27–30] and RNA binding [31–37]. In addition, it plays an important role in stress responses leading to apoptosis, and in such cases it is translocated to the nucleus prior to the onset of apoptosis [38–41]. Serum withdrawal, aging of cultures, treatment with anticancer agents and potassium depolarization cause nuclear accumulation of GAPDH [30,39,40,42–44]. Consistent with this, depletion of GAPDH mRNA inhibits apoptosis, whereas overexpression of the GAPDH gene induces programmed cell death [41,43,45,46].
Previously GAPDH was identified as a component of the eukaryotic transcription machinery [47]. OCA-S is a multicomponent Oct-1 co-activator that is essential for S-phase-dependent histone H2B transcription [47]. Using an in vitro assay involving stimulation of Oct-1 transcription, OCA-S was chromatographically purified from a HeLa cell nuclear extract, and subsequent analysis demonstrated that GAPDH was part of the OCA-S complex implicated in regulating histone gene expression. Interestingly, GAPDH binds directly to Oct-1, is selectively recruited to the H2B promoter in S-phase, and has an intrinsic activation domain, indicating that it interacts with an as-yet-unidentified component of the basal RNA polymerase II transcription machinery [47]. GAPDH also interacts with eukaryotic RNA polymerase II [48,49] and with PML (promyelocytic leukaemia protein) [50]. It has been reported that the PML nuclear bodies associate with transcriptionally active genomic regions [51].
hTAFII68 (NTD) is believed to act as a transactivation domain for hTAFII68-TEC oncoprotein. To search for binding partners that control hTAFII68-TEC function in vivo, we performed GST (glutathione S-transferase) pull-downs in HEK (human embryonic kidney)-293T cell lysates, followed by MALDI-TOF (matrix-assisted laser-desorption ionization–time-of-flight) MS, and identified GAPDH as a binding partner. We confirmed the interaction between hTAFII68-TEC and GAPDH in vitro using bacterially expressed fusion proteins and in vivo using immunoprecipitation and Western blot analysis. In transient transfection assays, the transcriptional activity of hTAFII68-TEC was stimulated by GAPDH, but not by a GAPDH mutant defective in hTAFII68-TEC binding. In addition, fusion of GAPDH to the GAL4 DNA-binding domain produced a chimaera capable of transactivating a reporter gene containing GAL4-binding sites, indicating that GAPDH is a potential transcriptional co-activator. These data demonstrate that hTAFII68-TEC-mediated transcriptional activation is positively regulated by GAPDH.
MATERIALS AND METHODS
Materials and general methods
Restriction endonucleases, polynucleotide kinase, calf intestinal alkaline phosphatase, the Klenow fragment of DNA polymerase I and T4 DNA ligase were purchased from New England Biolabs. Plasmid DNA preparation, restriction enzyme digestion, agarose gel electrophoresis of DNA, DNA ligation, bacterial transformation and SDS/PAGE of proteins were carried out using standard methods [52]. PCR amplification products were sequenced by the chain termination method, using double-stranded DNA templates, to ensure the absence of mutations.
Isolation of hTAFII68-TEC complexes and MS
For affinity purification of hTAFII68-TEC-associated proteins, a GST–hTAFII68 (NTD) was constructed as described below. Pull-downs were carried out in binding buffer [50 mM Tris/HCl (pH 8.0), 1% Nonidet P40, 2 mM EDTA, 150 mM NaCl, 1 mM DTT (dithiothreitol) and 1× Complete Protease Inhibitor Cocktail (Roche Diagnostics)] as described previously [53], using HEK-293T lysates. Beads were collected by brief centrifugation (12000 g for 10 s at 4 °C) and washed four times with wash buffer (same composition as the binding buffer minus the 1× Complete Protease Inhibitor Cocktail). Bound proteins were eluted with SDS/PAGE sample buffer and subjected to SDS/PAGE. Gels were stained with Coomassie Blue for MALDI-TOF MS [54].
Plasmid construction
The construction of GST–hTAFII68 (NTD) [55] and GST–EWS (NTD) has been previously described [56]. For Tag2A/Flag-hTAFII68-TEC, plasmid pSKII/hTAFII68-TEC was digested with BamHI and cloned into the same site of pCMV-Tag2A (Invitrogen). Details on the construction of GST–TLS (NTD), pcDNA3/GST and pcDNA3/GST–hTAFII68 (NTD) have been given previously [55,56].
GST-fusion plasmids GST–hTAFII68 (1–72) and GST–hTAFII68 (29–105) have been previously described [55]. For GST-hTAFII68 (106–159), plasmid pGEX(4T-1)-hTAFII68 (NTD) was amplified by PCR using primers 5′-hTAFII68-106 (5′-GATCGAATTCGACTATGGTCAACAAGAT-3′, EcoRI site underlined) and 3′-hTAFII68-159 (5′-GATCCTCGAGTCTTGTGTGTGGTGGCTG-3′, XhoI site underlined), digested with EcoRI and XhoI, and cloned into the same sites of pGEX (4T-1).
The six histidine-containing fusion plasmids (His)6-GAPDH, (His)6-GAPDH (1–66), (His)6-GAPDH (67–160), (His)6-GAPDH (160–248) and (His)6-GAPDH (217–335) were generated using the following steps. (i) For (His)6-GAPDH, plasmid pcDNA3/GAPDH was digested with BamHI and EcoRI and cloned into the same sites of pTag3B (Stratagene). This in turn was digested with SacI and KpnI and cloned into the same sites of pTRC-HisB (Invitrogen). (ii) For (His)6-GAPDH (1–66), pTRC-HisB/GAPDH was digested with HindIII and self-ligated with DNA ligase. (iii) For (His)6-GAPDH (67–160), GAPDH (67–160) was amplified from pcDNA3/GAPDH by PCR using primers 5′-GAPDH-67 (5′-GATCGGATCCAAGCTTGTCATCAATGGA-3′, BamHI site underlined) and 3′-GAPDH-160 (5′-GATCGAATTCCAGGGGTGCTAAGCAGTT-3′, EcoRI site underlined), digested with BamHI and EcoRI, and cloned into the same sites of pTrc-HisA. (iv) For (His)6-GAPDH (160–248), GAPDH (160–335) was amplified from pcDNA3/GAPDH by PCR using primers 5′-GAPDH-161 (5′-GATCGGATCCGCCAAGGTCATCCATGAC-3′, BamHI site underlined) and 3′-GAPDH-335 (5′-GATCGAATTCTTACTCCTTGGAGGCCAT-3′ EcoRI site underlined), digested with BamHI and EcoRI, and cloned into the same sites of pTrc-HisA to generate pTrc-HisA/GAPDH (160–335). This in turn was digested with XbaI and EcoRI, blunted with Klenow, and self-ligated with DNA ligase. (v) For (His)6-GAPDH (217–335), GAPDH (217–335) was amplified from pcDNA3/GAPDH by PCR using primers 5′-GAPDH-217 (5′-GATCGGATCCGTGGGCAAGGTCATCCCT-3′, BamHI site underlined) and 3′-GAPDH-335 (5′-GATCGAATTCTTACTCCTTGGAGGCCAT-3′, EcoRI site underlined), digested with BamHI and EcoRI, and cloned into the same sites of pTRC-HisA.
The reporter plasmid p(B1a)8-Luc has been previously described [55]. For Tag3B/GAPDH, pGEX (4T-1)/GAPDH was digested with BamHI and XhoI and cloned into the same sites of Tag3B. For Tag3B/GAPDH (217–335), pTRC-HisA/GAPDH (217–335) was digested with BamHI and EcoRI and cloned into the same sites of Tag3B.
Subcellular localization of GAPDH
Subcellular fractions were prepared essentially as described by Dignam [57]. Nuclear and cytoplasmic fractions were boiled in 2% SDS for 10 min and the insoluble material was removed by centrifugation in a microcentrifuge at 12000 g for 10 min at 4 °C. Equivalent amounts of each supernatant on a cell basis were fractionated by SDS/PAGE, transferred to PVDF membranes and probed with the indicated antibodies. Immunocytochemical analyses were performed as previously described [55]. Briefly, HeLa, COS-7 and C28/I2 human juvenile costal chondrocyte cells (provided by Dr M. Goldring, Hospital for Special Surgery, Cornell Medical College in New York City, NY, U.S.A.) were plated on glass coverslips and transfected with pTag2A/Flag-hTAFII68-TEC plasmid. After transfection (48 h), the cells were washed in PBS and fixed for 10 min at −20 °C with acetone/methanol (1:1, v/v). To detect hTAFII68-TEC and GAPDH, we used primary antibodies for Flag-tagged hTAFII68-TEC antibody (M2; Sigma) and GAPDH (V-18; Santa Cruz Biotechnology) respectively. The subcellular distribution of hTAFII68-TEC or GAPDH was examined using a confocal laser scanning microscope (LSM5 Pascal, Carl Zeiss Co).
Purification of six histidine-tagged GAPDH mutant proteins
Recombinant six histidine-tagged GAPDH proteins were purified as described in [53]. The elution profiles of GAPDH mutant proteins were monitored by Western blot analysis using the monoclonal Xpress antibody (Invitrogen).
GST pull-down assays, co-affinity precipitations and co-immunoprecipitations
GST pull-down assays were performed as described previously [53]. Co-affinity precipitations and co-immunoprecipitation assays were also performed as described in [53,56] and bound proteins were detected by immunoblotting [58].
Cell culture, electroporation and reporter assays
HeLa, COS-7 or HEK-293T cells were maintained in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% heat-inactivated fetal calf serum (Hyclone), penicillin and streptomycin. Cells were transiently transfected with plasmids by electroporation using the Gene Pulser II RF module system according to the manufacturer's instructions (Bio-Rad). Luciferase assays were performed using the Promega Luciferase Assay System or Dual-luciferase Assay System (Promega). Renilla luciferase activities were used to normalize transfection efficiencies.
RESULTS
Identification of GAPDH as an hTAFII68-TEC-interacting protein
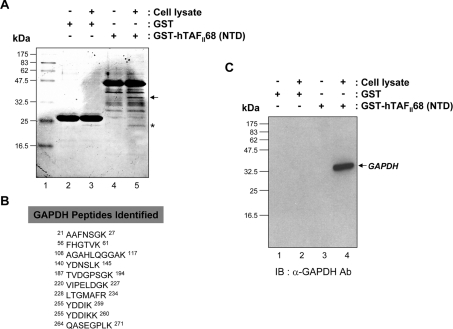
To isolate potential regulators of the hTAFII68-TEC oncoprotein, we used MS to identify proteins associating with it. Bacterially expressed GST or GST–hTAFII68 NTD-fusion proteins immobilized on glutathione–Sepharose beads were mixed with HEK-293T cell extracts. After extensive washing, bound proteins were eluted with sample buffer and analysed by SDS/PAGE and Coomassie Blue staining. Two proteins, of ∼34 kDa (denoted by an arrow) and ∼24 kDa (denoted by an asterisk), were visible on the Coomassie Blue-stained gel (Figure 1A). The 24 kDa species was present in eluates from both the GST and GST–hTAFII68 (NTD) affinity matrices (Figure 1A, lanes 3 and 5) and was not observed if the HEK-293T cell extract was omitted from the GST or GST–hTAFII68 (NTD) beads (Figure 1A, lanes 2 and 4). This suggested that the 24 kDa protein species was not specifically retained by hTAFII68 (NTD), and this species was not further characterized. The 34 kDa protein species was observed in eluates from the GST–hTAFII68 (NTD) beads (Figure 1A, lane 5), not from the column that had GST coupled to it and had been exposed to HEK-293T cell extracts (Figure 1A, lane 3), and not in eluates from GST–hTAFII68 (NTD) beads that had not been exposed to HEK-293T cell extracts (Figure 1A, lane 4). These results indicated that the ∼34 kDa protein species was specifically retained by and eluted from the GST–hTAFII68 (NTD) affinity matrix. The band was extracted from the gel matrix by trypsin digestion and analysed by MALDI-TOF MS, revealing peptides derived from GAPDH (Figure 1B). Binding to GAPDH was confirmed by Western blot analysis using a monoclonal anti-GAPDH antibody (Figure 1C).
Figure 1. Identification of GAPDH as an hTAFII68-TEC-associated protein.
(A) SDS/PAGE of hTAFII68 (NTD) complexes isolated from HEK-293T cells. Total HEK-293T cell lysates were incubated with either GST or a GST-fusion protein containing the NTD of hTAFII68 (indicated above the panel). Complexes were resolved by SDS/PAGE (15% gels) and stained with Coomassie Blue. Molecular mass markers are shown on the left; they are derived from prestained protein standards (broad range; New England Biolabs). The band investigated in this analysis is indicated by the arrow to the right. Lane 1, molecular mass marker; lane 2, GST; lane 3, GST plus cell lysate; lane 4, GST–hTAFII68 (NTD); lane 5, GST–hTAFII68 (NTD) plus cell lysate. (B) Peptide sequences of GAPDH identified by MALDI-TOF analysis. The GAPDH peptides matched by MALDI-TOF are shown with amino acid numbers displayed at both ends. (C) Association of hTAFII68 (NTD) with GAPDH. HEK-293T cell lysate was incubated with either GST–hTAFII68 (NTD) or GST. After affinity-selection, the pellets from GST pull-down assays were analysed by SDS/PAGE (15% gels), and bound GAPDH was detected with anti-GAPDH antibody (MAB374; Chemicon) and chemiluminescence (Perkin Elmer Life Science). The positions of the molecular mass markers are indicated on the left-hand side, and the GAPDH band is indicated by the arrow on the right-hand side. Lane 1, GST; lane 2, GST plus cell lysate; lane 3, GST–hTAFII68 (NTD); lane 4, GST–hTAFII68 (NTD) plus cell lysate. IB, immunoblotting; Ab, antibody.
hTAFII68-TEC interacts with GAPDH in vitro and in vivo
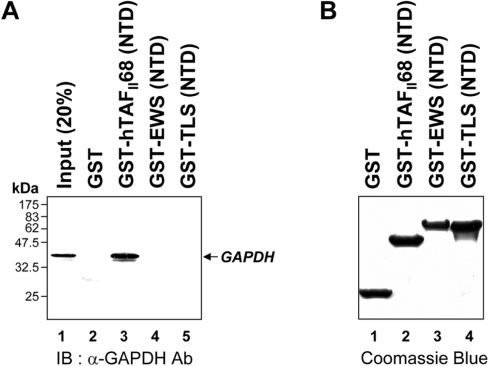
The in vitro interaction between hTAFII68-TEC and GAPDH was further investigated using GST-fusion proteins containing the NTDs of hTAFII68 and its homologues, such as EWS and TLS. As shown in Figure 2(A), approximately 30% of input GAPDH protein was retained on hTAFII68 (NTD)-conjugated Sepharose beads. However, neither EWS (NTD) nor TLS (NTD) was able to bind GAPDH (Figure 2A, lanes 4 and 5) suggesting that GAPDH binds differentially to the NTDs of TET (hTAFII68, EWS and TLS) family members. This provides a molecular criterion for distinguishing between them. As GAPDH did not bind to GST alone (Figure 2A, lane 2), the interaction was considered to be specific. The amounts of GST-fusion proteins utilized in this assay were fractionated on SDS/PAGE (15% gels) and visualized by Coomassie Blue staining which revealed that similar amounts of protein had been used in the pull-down assay (Figure 2B).
Figure 2. Binding of GAPDH to hTAFII68 (NTD) in vitro.
(A) Specific association of hTAFII68 (NTD) with GAPDH. GST-fusion proteins containing the NTDs of hTAFII68, EWS or TLS were incubated with cell lysates. An aliquot of the inputs (20%) and the pellets from the various pull-downs were analysed on SDS/PAGE (15% gels) and bound GAPDH protein was detected using an anti-GAPDH antibody (MAB374). The identities of the GST-fusion proteins are indicated above the panel. The positions of the molecular mass markers are indicated on the left-hand side. GAPDH is indicated by an arrow on the right-hand side. Three independent experiments were performed, all of which gave similar results. Lane 1, 20% input; lane 2, GST alone; lane 3, GST–hTAFII68 (NTD); lane 4, GST–EWS (NTD); lane 5, GST–TLS (NTD). IB, immunoblotting; Ab, antibody. (B) Quantitation of the GST-fusion proteins used in the GST pull-down assays. The GST-fusion proteins utilized in the pull-down assays were fractionated by SDS/PAGE (15% gels) and visualized by Coomassie Blue staining. Three independent experiments were performed, all of which gave similar results. Lane 1, GST alone; lane 2, GST–hTAFII68 (NTD); lane 3, GST–EWS (NTD); lane 4, GST–TLS (NTD).
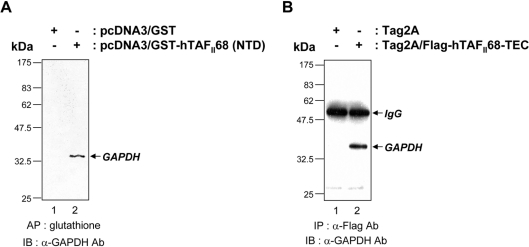
To determine whether the interaction between hTAFII68 (NTD) and GAPDH occurred in vivo, we performed co-affinity precipitations following transfection of COS-7 cells with an expression vector driving the synthesis of hTAFII68 (NTD). For these assays, mammalian expression vectors containing the NTD of hTAFII68 fused to GST [pcDNA3/GST–hTAFII68 (NTD)] and the GST domain alone (pcDNA3/GST) were transfected into COS-7 cells. The cells were lysed 48 h after transfection and the fusions were incubated with glutathione beads. Immunoblotting of the eluates with an anti-GAPDH antibody revealed that endogenous GAPDH was co-precipitated with GST–hTAFII68 (NTD), but not with GST alone (Figure 3A). These results demonstrate that hTAFII68 (NTD) and GAPDH can associate in vivo.
Figure 3. Association of hTAFII68-TEC with GAPDH in vivo.
(A) Co-affinity purification of hTAFII68 (NTD) with GAPDH from COS-7 cells. After transfection (48 h) of COS-7 cells with 10 μg of either pcDNA3/GST or pcDNA3/GST–hTAFII68 (NTD), cell extracts were prepared as described in the Materials and methods section and affinity-precipitated with glutathione–Sepharose beads. After fractionation by SDS/PAGE (15% gels), the proteins were analysed by Western blot with an anti-GAPDH antibody (MAB374). The identities of the transfected DNAs are indicated above the panel. The positions of the molecular mass markers are indicated on the left-hand side and the position of GAPDH is indicated by the arrow on the right-hand side. Three independent experiments were performed, all of which gave similar results. AP, affinity precipitation; IB, immunoblotting; Ab, antibody. (B) Co-immunoprecipitation of hTAFII68-TEC and GAPDH in vivo. After transfection (48 h) of HeLa cells with 10 μg of either Tag2A or Tag2A/Flag-hTAFII68-TEC, cell extracts were immunoprecipitated with an anti-Flag antibody (M2), resolved by SDS/PAGE (15% gels), and probed with an anti-GAPDH antibody (MAB374). The identities of the transfected DNAs are indicated above the panel. The positions of the molecular mass markers are indicated on the left-hand side, and the positions of IgG and GAPDH are indicated by arrows on the right-hand side. Three independent experiments were performed, all of which gave similar results. IP, immunoprecipitation; IB, immunoblotting; Ab, antibody.
To further examine whether the hTAFII68-TEC chimaeric protein and GAPDH associate in mammalian cells, we performed immunoprecipitation experiments with cell extracts of transiently transfected HeLa cells. The cells were lysed 48 h after transfection with expression vectors Tag2A/Flag-hTAFII68-TEC or Tag2A, and immunoprecipitated with an α-Flag antibody (M2). The eluates were immunoblotted with an anti-GAPDH antibody (MAB374; Chemicon) and GAPDH was found to co-precipitate specifically with hTAFII68-TEC (Figure 3B). Similar results were obtained with COS-7 cells (results not shown). These results suggest that hTAFII68-TEC and GAPDH also associate in vivo.
The C-terminal region of hTAFII68 (NTD) is involved in the GAPDH interaction
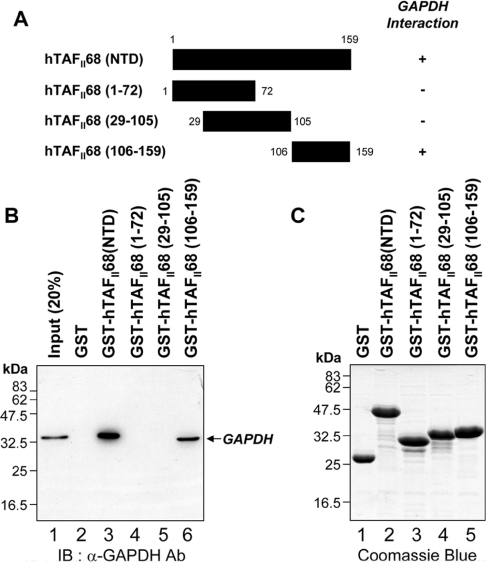
To define the domain within hTAFII68 (NTD) required for the interaction with GAPDH, in vitro GST pull-down assays were performed with a series of hTAFII68 (NTD) deletion mutants. The structures of the mutants are shown schematically in Figure 4(A). As shown in Figure 4(B), GAPDH strongly bound to GST–hTAFII68 (NTD) (Figure 4B, lane 3) and GST–hTAFII68 (106–159) (Figure 4B, lane 6). However, the GST-fusions with hTAFII68 (1–72) (Figure 4B, lane 4) and hTAFII68 (29–105) (Figure 4B, lane 5) failed to bind, indicating that amino acids 106–159 of the hTAFII68 (NTD) are necessary and sufficient for the interaction with GAPDH. The amounts of GST-fusion proteins utilized in this assay were fractionated on SDS/PAGE (15% gels) and visualized by Coomassie Blue staining which revealed that similar amounts of protein had been used in the pull-down assay (Figure 4C).
Figure 4. Mapping the hTAFII68 (NTD) region that interacts with GAPDH.
(A) Schematic representation of the GST–hTAFII68 (NTD) fusion proteins and their ability to bind to GAPDH. Numbers refer to amino acid residues, and binding ability is indicated by + or −. (B) Strong binding of GAPDH to GST–hTAFII68 (106–159). Recombinant GST–hTAFII68 (NTD) deletion mutants were incubated with HEK-293T cell lysates. Following GST pull-down assays, the bound proteins were eluted with SDS loading buffer and analysed by Western blotting with an anti-GAPDH antibody. The positions of molecular mass markers and of GAPDH are indicated. Three independent experiments were performed, all of which gave similar results. Lane 1, 20% input; lane 2, GST alone; lane 3, GST–hTAFII68 (NTD); lane 4, GST–hTAFII68 (1–72); lane 5, GST–hTAFII68 (29–105); lane 6, GST–hTAFII68 (106–159). IB, immunoblotting; Ab, antibody. (C) Coomassie Blue staining of the GST–hTAFII68 deletions. The amounts of GST-fusion proteins utilized in these assays were fractionated on SDS/PAGE (15% gels) and visualized by Coomassie Blue staining. Lane 1, GST alone; lane 2, GST–hTAFII68 (NTD); lane 3, GST–hTAFII68 (1–72); lane 4, GST–hTAFII68 (29–105); lane 5, GST–hTAFII68 (106–159).
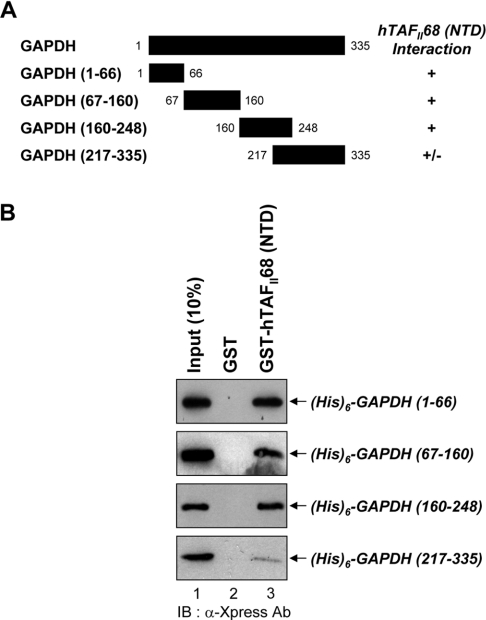
GAPDH contains at least three independent hTAFII68 (NTD)-interacting motifs
To delineate the regions of GAPDH responsible for the interaction with hTAFII68 (NTD), GAPDH (1–66), GAPDH (67–160), GAPDH (160–248) or GAPDH (217–335) were expressed as six histidine proteins in Escherichia coli and purified by Ni2+-NTA (Ni2+-nitrilotriacetate)–agarose resin (Figure 5A). In GST pull-down assays with GST or the GST–hTAFII68 (NTD) fusion protein, deletions of GAPDH containing amino acids 1–66, 67–160 and 160–248 interacted strongly with hTAFII68 (NTD), whereas GAPDH (217–335) interacted only weakly (Figure 5B). These result suggested that at least three sites (amino acids 1–66, 67–160 and 160–248) of GAPDH can bind to hTAFII68 (NTD) independently. The amounts of GST-fusion proteins utilized in this assay were fractionated on SDS/PAGE (15% gels) and visualized by Coomassie Blue staining, which revealed that similar amounts of protein had been used in the pull-down assay (Figure 2B).
Figure 5. Involvement of at least three independent domains of GAPDH in the interaction with hTAFII68 (NTD).
(A) Schematic representation of the deletion mutants of GAPDH and their ability to bind to hTAFII68 (NTD). Numbers refer to amino acid residues, and binding ability is indicated by + or −. (B) Binding of hTAFII68 (NTD) to GAPDH (1–66), GAPDH (67–160), GAPDH (160–248) and GAPDH (217–335). Deletion mutants of GAPDH containing six histidines were expressed in E. coli, purified with Ni2+-NTA–agarose resin, and incubated with either GST or GST-fusion hTAFII68 (NTD) bound to glutathione–Sepharose beads. An aliquot of the input and the pellets from the GST pull-down assays were analysed by SDS/PAGE (15% gels), and bound GAPDH was detected by Western blotting. The positions of the GAPDH deletion mutants are indicated with arrows on the right-hand side. Three independent experiments were performed, all of which gave similar results. Lane 1, 10% input; lane 2, GST alone; lane 3, GST–hTAFII68 (NTD). IB, immunoblotting; Ab, antibody.
hTAFII68-TEC co-localizes with a nuclear form of GAPDH
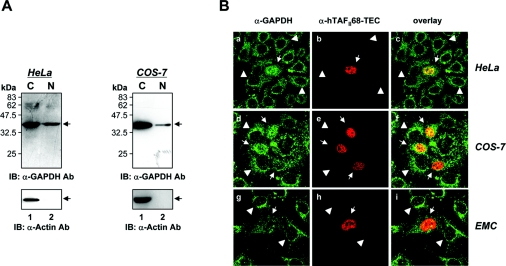
Since hTAFII68-TEC is a nuclear protein [55], we wanted to establish the subcellular location of endogenous GAPDH. HeLa (Figure 6A, left panels) and COS-7 (Figure 6A, right panels) cells were lysed, and cytosolic and nuclear protein fractions were prepared and subjected to SDS/PAGE and Western blot analysis. As shown in Figure 6(A), GAPDH was present in both the cytoplasm and nucleus in HeLa and COS-7 cells. As a further control for the quality of the fractionation procedure, a cytoplasmic protein, actin [59], was detected only in the cytoplasmic fraction (Figure 6A, lower panel).
Figure 6. Co-localization of hTAFII68-TEC and GAPDH.
(A) Subcellular localization of GAPDH in HeLa and COS-7 cells. To determine the subcellular location of GAPDH, HeLa and COS-7 cells were fractionated into nuclear and cytoplasmic preparations. Proteins were separated by SDS/PAGE (15% gels) and detected by Western blotting using anti-GAPDH (MAB 374, upper panels) or anti-actin (I-19, lower panels) antibodies. Three independent experiments were performed, all of which gave similar results. IB, immunoblotting; Ab, antibody. (B) Subcellular distribution of hTAFII68-TEC and GAPDH. HeLa, COS-7, and human chondrocyte cells (C28/I2) grown on coverslips were transfected with a mammalian expression vector encoding Flag-tagged hTAFII68-TEC protein. The transiently transfected cells were fixed with an acetone/methanol mixture and incubated with primary antibodies for GAPDH (V-18) or Flag tag (M2). The subcellular distribution of GAPDH or hTAFII68-TEC was examined using a confocal laser scanning microscope (LSM5 Pascal, Carl Zeiss Co., Ltd.). The merged image (overlay) shows the co-localization. Three independent experiments were performed, all of which gave similar results.
To confirm the hTAFII68-TEC–GAPDH interaction in vivo (Figure 3), we also performed immunofluorescence staining of the transfected cells. To do this we constructed a pTag2A/Flag-hTAFII68-TEC construct that expressed Flag-tagged hTAFII68-TEC protein. In control HeLa cells not expressing hTAFII68-TEC, the majority of GAPDH protein was localized to the cytoplasm [Figure 6B(a), arrowheads]. A significant amount of GAPDH was also detected in the nucleus, as previously reported [16,47,50]. However, in HeLa cells expressing hTAFII68-TEC, the amount of nuclear GAPDH was as high as that of cytoplasmic GAPDH, suggesting that hTAFII68-TEC promotes translocation of GAPDH to the nucleus [Figure 6B(a), arrows]. Consistent with a previous report [55], hTAFII68-TEC protein localized to the nucleus in HeLa cells [Figure 6B(b)]. The overlay images indicated that hTAFII68-TEC and GAPDH in the nucleoplasm partially overlapped [Figure 6B(c)]. Similar results were obtained in COS-7 and C28/I2 human chondrocyte cells [Figure 6B(d–i)]. Therefore we conclude that hTAFII68-TEC and a nuclear form of GAPDH co-localize or are in close proximity, and that the formation of hTAFII68-TEC–GAPDH complexes results in a redistribution of GAPDH.
GAPDH modulates hTAFII68-TEC-dependent transactivation
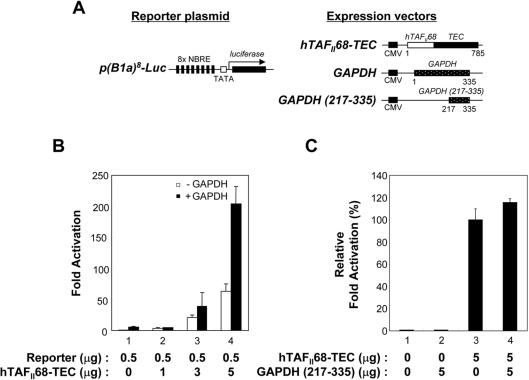
To investigate the consequences of the hTAFII68-TEC–GAPDH interaction, we measured the effect of GAPDH expression on transcriptional activation by hTAFII68-TEC. This was performed in HeLa cells by examining gene expression from a luciferase-based reporter plasmid transfected along with Tag2A/hTAFII68-TEC, with or without co-transfection of Tag3B/GAPDH. The structures of the reporter plasmid and the expression vectors used in this study are shown schematically in Figure 7(A). The introduction of hTAFII68-TEC into HeLa cells activated transcription (up to ∼60-fold) from p(B1a)8-Luc, a reporter plasmid containing the TATA minimal promoter with eight NBREs (nerve growth factor inducible factor B response elements) [60] driving synthesis of Renilla luciferase (Figure 7B, open bars). Interestingly, GAPDH augmented hTAFII68-TEC-mediated transactivation up to ∼210-fold (Figure 7B, solid bars). These results indicated that GAPDH can stimulate hTAFII68-TEC-mediated activation.
Figure 7. GAPDH enhances hTAFII68-TEC-mediated transcription.
(A) Schematic representation of the reporter and expression plasmids used in the present study. The p(B1a)8-Luc reporter plasmid contains eight copies of NBRE upstream of a basal promoter-luciferase gene construct. The eight copies of hTAFII68-TEC recognition sites are indicated by solid bars, the TATA box is represented by an open box, and the luciferase gene is indicated by a solid box. The expression vectors driving the production of hTAFII68-TEC, GAPDH or GAPDH (217–335) are also shown. The positions of the first and last amino acids are indicated below each construct. CMV, cytomegalovirus promoter. (B) Stimulation of hTAFII68-TEC-mediated transactivation by GAPDH. Aliquots of hTAFII68-TEC (bars 1–4) were co-transfected into HeLa cells with 2 μg of empty vector (open bars) or GAPDH expression plasmid (black bars). After 48 h the cells were harvested and luciferase assays performed. The experiments were repeated three times, and the averages of two independent experiments are presented and error bars are shown. (C) A GAPDH (217–335) mutant lacking the hTAFII68-TEC-interacting domain does not stimulate hTAFII68-TEC-mediated transcriptional activation. HeLa cells were transfected with 1 μg of p(B1a)8-Luc reporter plasmid, 5 μg of hTAFII68-TEC, and 5 μg of GAPDH (217–335) or empty vector, and luciferase activity was assayed. Relative transcriptional activation values were calculated with hTAFII68-TEC taken as 100%. The experiments were repeated three times, and the average of two independent experiments are presented with error bars.
To demonstrate that binding between GAPDH and hTAFII68-TEC is required for the observed effects on transcriptional activation, we constructed a GAPDH deletion mutant lacking the hTAFII68-TEC-binding domain, called pTag3B/GAPDH (217–335). This deletion mutant did not activate hTAFII68-TEC-dependent transactivation (Figure 7C), indicating that GAPDH modulated hTAFII68-TEC-mediated transactivation by physical association with hTAFII68-TEC.
GAPDH is a potential transcriptional co-activator
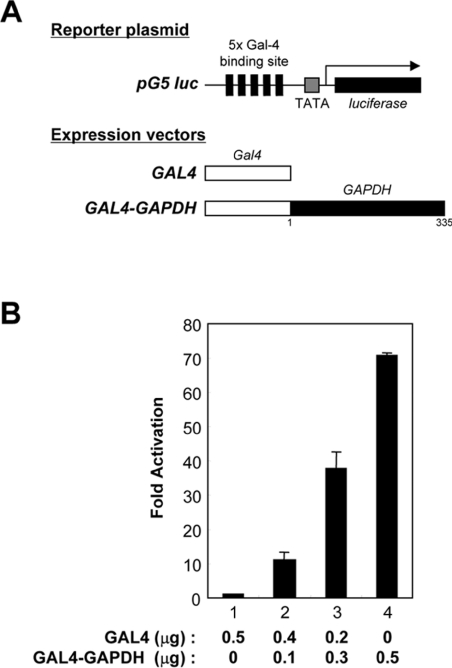
To assess how GAPDH activates hTAFII68-TEC-mediated transcription, it was fused to the GAL4 DNA-binding domain, generating GAL4–GAPDH (Figure 8A). The pG5 luc reporter contains five GAL4 DNA-binding sites upstream of the TATA box and was used as a reporter. An expression vector driving the synthesis of only the GAL4 DNA-binding domain, pGAL4, had no effect on the levels of luciferase produced from pG5 luc when transfected into COS-7 cells (Figure 8B, bar 1). In contrast, pGAL4–GAPDH activated luciferase production up to 70-fold (Figure 8B, bars 2–4), indicating that GAPDH contained a transactivation domain(s). These results demonstrate that GAPDH can act as a co-activator of transcription if it is recruited to the vicinity of the promoter.
Figure 8. Transactivation by a GAL4–GAPDH fusion protein.
(A) Schematic representation of the reporter plasmid and expression vectors. The pG5 luc reporter plasmid contains five GAL4-binding sites upstream of the luciferase gene. The GAL4 recognition sites are indicated with solid bars, the TATA box is represented by a shaded box, and the luciferase gene by a solid box. The expression vectors driving the production of GAL4 or GAL4–GAPDH are also presented. The positions of the first and last amino acids of the GAPDH construct are indicated below the construct. (B) Transactivation by GAPDH. COS-7 cells were co-transfected with 0.1 μg of pG5 luc reporter plasmid and 0, 0.1, 0.3 or 0.5 μg of GAL4–GAPDH (bars 1–4). For all reactions the total amount of transfected pGAL4–GAPDH DNA was adjusted to 0.5 μg with empty vector, pGAL4. After 48 h the cells were harvested and luciferase assays were performed. The experiments were repeated three times, and the average of two independent experiments is presented and the error bars are shown.
DISCUSSION
Aberrant expression or structural alteration of transcription factors is often a critical event in tumour development, and the characterization of specific chromosome abnormalities associated with human tumours has led to the discovery of new mechanisms involved in tumorigenic transformation. EMC is a malignant soft tissue tumour that occurs primarily in the extremities [61]. A portion of EMCs harbours a characteristic translocation t(9;17)(q22;q11) involving the hTAFII68 gene at 17q11 and the TEC gene at 9q22 [4,5]. This chimaeric hTAFII68-TEC encodes a potent transcriptional activator (S. Kim and J. Kim, unpublished work).
The results reported in the present study begin to reveal the regulation of the hTAFII68-TEC oncoprotein. Using affinity chromatography and MALDI-TOF MS, we showed that GAPDH interacts with hTAFII68-TEC and we confirmed this interaction using GST pull-down assays in vitro (Figure 2) and by co-immunoprecipitation experiments on extracts of cultured cells (Figure 3). Although it is unclear whether the interaction is direct, residues 106–159 of the hTAFII68 N-terminal domain were found to be required for the association with GAPDH (Figure 4).
GAPDH is an essential glycolytic enzyme in all prokaryotic and eukaryotic organisms, converting glyceraldehyde 3-phosphate into 1,3-diphosphoglycerate. However, it has a number of other seemingly independent activities, including a uracil DNA-glycosylase function, RNA binding, modulation of apoptosis, DNA replication and DNA repair [13], and we have now shown that it is able to stimulate hTAFII68-TEC-mediated transactivation (Figure 7). This effect was dose-dependent and required the region of GAPDH that interacts with hTAFII68-TEC. It seems possible that GAPDH also interacts with other transcriptional regulators.
Because of its abundance, GAPDH is often regarded as a stable marker protein in contrast with the dynamic changes of other proteins. However, GAPDH expression is also dynamic, being affected by hypoxic stress [62]. It also accumulates in the nucleus after cells are treated with genotoxic drugs such as mercaptopurine or thioguanine [16,63]. Because hTAFII68-TEC is a transcriptional activator and its activity is modulated by GAPDH (Figure 7), it would be interesting to see whether exposure to hypoxic stress or genotoxic drugs modulates its activity. In fact, treatment with 6-mercaptopurine stimulates the transcriptional activity of the orphan nuclear hormone receptor NOR-1 (the rat homologue of the human TEC receptor) and Nurr1 [64,65], but little is currently known of these effects. Our finding of the presence of some GAPDH in the nucleus (Figure 6) is consistent with other reports [16,66]. Since hTAFII68-TEC is nuclear [55], the interaction between hTAFII68-TEC and GAPDH may occur within the nucleoplasm. Interestingly, this interaction also appears to lead to the translocation of a fraction of GAPDH from the cytoplasm to the nucleus (Figure 6B). Using GST pull-down assays, we showed that the C-terminal part of hTAFII68 (NTD) harbours a binding site for GAPDH, and it is interesting that amino acids 106–159 of hTAFII68 (NTD) are required for the interaction with GAPDH, because we have shown that amino acids 75–159 are involved in the interaction with v-Src [55]. It is therefore possible that the C-terminal part of hTAFII68 (NTD) is specialized to participate in protein–protein interactions. Using recombinant proteins in GST pull-down assays, we showed that GAPDH harbours at least three independent binding sites for hTAFII68 (NTD) (Figure 5), suggesting that the association of GAPDH with hTAFII68 (NTD) can be mediated by three different domains of a single molecule or that the stoichiometry of the interaction is greater than one. Further experiments are required to distinguish these possibilities.
We showed that GAPDH can modulate hTAFII68-TEC-mediated transactivation (Figure 7). However, although GAPDH has intrinsic transactivation activity (Figure 8), the precise mechanism underlying this effect is unclear. One possibility is that GAPDH acts as a bridging factor linking an hTAFII68-TEC molecule to components of the transcription initiation machinery. Consistent with this speculation, GAPDH was recently identified as a cofactor of the Oct-1 coactivator complex, OCA-S (Oct-1 CoActivator in S-phase) [47], which is essential for S-phase-dependent histone H2B transcription; using stimulation of Oct-1 transcription in an in vitro assay, OCA-S was purified from a HeLa cell nuclear extract and found to contain seven proteins: nm23-H1, nm23-H2, uracil-DNA glycosylase, lactate dehydrogenase, Hsp70, Sti1 and GAPDH. The last also associates with PML in an interaction that is dependent on the presence of RNA [50], and the possible significance of this GAPDH–PML interaction was indicated by a recent study which revealed an association of PML nuclear bodies with transcriptionally active genomic regions [51].
In conclusion, the findings of the present study provide new evidence that hTAFII68-TEC function can be modulated by GAPDH. Unlike EWS-TEC which can transform CFK2 chondrogenic cells [67], the transforming activity of hTAFII68-TEC has yet to be assessed. Thus it would be of interest to determine whether GAPDH can collaborate with hTAFII68-TEC to transform cells into EMC tumours.
Acknowledgments
This research was supported by a Korea Research Foundation Grant funded by the Korean Government (MOEHRD, Basic Research Promotion Fund; KRF-2006-312-C00414), by the Seoul Research and Business Development Program (10816), and by the Advanced Basic Research Laboratory Program (R14-2002-012-01002-0) of the KOSEF. S.K. and J.L. were recipients of Seoul Science Fellowships and of a research fellowship BK21 from the Ministry of Education and Human Resources Development.
References
- 1.Clark J., Benjamin H., Gill S., Sidhar S., Goodwin G., Crew J., Gusterson B. A., Shipley J., Cooper C. S. Fusion of the EWS gene to CHN, a member of the steroid/thyroid receptor gene superfamily, in a human myxoid chondrosarcoma. Oncogene. 1996;12:229–235. [PubMed] [Google Scholar]
- 2.Labelle Y., Zucman J., Stenman G., Kindblom L. G., Knight J., Turc-Carel C., Dockhorn-Dworniczak B., Mandahl N., Desmaze C., Peter M., et al. Oncogenic conversion of a novel orphan nuclear receptor by chromosome translocation. Hum. Mol. Genet. 1995;4:2219–2226. doi: 10.1093/hmg/4.12.2219. [DOI] [PubMed] [Google Scholar]
- 3.Labelle Y., Bussieres J., Courjal F., Goldring M. B. The EWS/TEC fusion protein encoded by the t(9;22) chromosomal translocation in human chondrosarcomas is a highly potent transcriptional activator. Oncogene. 1999;18:3303–3308. doi: 10.1038/sj.onc.1202675. [DOI] [PubMed] [Google Scholar]
- 4.Sjogren H., Meis-Kindblom J., Kindblom L. G., Aman P., Stenman G. Fusion of the EWS-related gene TAF2N to TEC in extraskeletal myxoid chondrosarcoma. Cancer Res. 1999;59:5064–5067. [PubMed] [Google Scholar]
- 5.Attwooll C., Tariq M., Harris M., Coyne J. D., Telford N., Varley J. M. Identification of a novel fusion gene involving hTAFII68 and CHN from a t(9;17)(q22;q11.2) translocation in an extraskeletal myxoid chondrosarcoma. Oncogene. 1999;18:7599–7601. doi: 10.1038/sj.onc.1203156. [DOI] [PubMed] [Google Scholar]
- 6.Bertolotti A., Lutz Y., Heard D. J., Chambon P., Tora L. hTAF(II)68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. EMBO J. 1996;15:5022–5031. [PMC free article] [PubMed] [Google Scholar]
- 7.Morohoshi F., Arai K., Takahashi E. I., Tanigami A., Ohki M. Cloning and mapping of a human RBP56 gene encoding a putative RNA binding protein similar to FUS/TLS and EWS proteins. Genomics. 1996;38:51–57. doi: 10.1006/geno.1996.0591. [DOI] [PubMed] [Google Scholar]
- 8.Delattre O., Zucman J., Melot T., Garau X. S., Zucker J. M., Lenoir G. M., Ambros P. F., Sheer D., Turc-Carel C., Triche T. J., et al. The Ewing family of tumors – a subgroup of small-round-cell tumors defined by specific chimeric transcripts. N. Engl. J. Med. 1994;331:294–299. doi: 10.1056/NEJM199408043310503. [DOI] [PubMed] [Google Scholar]
- 9.Rabbitts T. H., Forster A., Larson R., Nathan P. Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignant liposarcoma. Nat. Genet. 1993;4:175–180. doi: 10.1038/ng0693-175. [DOI] [PubMed] [Google Scholar]
- 10.Ladanyi M. The emerging molecular genetics of sarcoma translocations. Diagn. Mol. Pathol. 1995;4:162–173. doi: 10.1097/00019606-199509000-00003. [DOI] [PubMed] [Google Scholar]
- 11.Kim J., Pelletier J. Molecular genetics of chromosome translocations involving EWS and related family members. Physiol. Genomics. 1999;1:127–138. doi: 10.1152/physiolgenomics.1999.1.3.127. [DOI] [PubMed] [Google Scholar]
- 12.Ohkura N., Hijikuro M., Yamamoto A., Miki K. Molecular cloning of a novel thyroid/steroid receptor superfamily gene from cultured rat neuronal cells. Biochem. Biophys. Res. Commun. 1994;205:1959–1965. doi: 10.1006/bbrc.1994.2900. [DOI] [PubMed] [Google Scholar]
- 13.Sirover M. A. New nuclear functions of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in mammalian cells. J. Cell. Biochem. 2005;95:45–52. doi: 10.1002/jcb.20399. [DOI] [PubMed] [Google Scholar]
- 14.Mazzola J. L., Sirover M. A. Subcellular localization of human glyceraldehyde-3-phosphate dehydrogenase is independent of its glycolytic function. Biochim. Biophys. Acta. 2003;1622:50–56. doi: 10.1016/s0304-4165(03)00117-x. [DOI] [PubMed] [Google Scholar]
- 15.Mazzola J. L., Sirover M. A. Subcellular alteration of glyceraldehyde-3-phosphate dehydrogenase in Alzheimer's disease fibroblasts. J. Neurosci. Res. 2003;71:279–285. doi: 10.1002/jnr.10484. [DOI] [PubMed] [Google Scholar]
- 16.Brown V. M., Krynetski E. Y., Krynetskaia N. F., Grieger D., Mukatira S. T., Murti K. G., Slaughter C. A., Park H. W., Evans W. E. A novel CRM1-mediated nuclear export signal governs nuclear accumulation of glyceraldehyde-3-phosphate dehydrogenase following genotoxic stress. J. Biol. Chem. 2004;279:5984–5992. doi: 10.1074/jbc.M307071200. [DOI] [PubMed] [Google Scholar]
- 17.Glaser P. E., Gross R. W. Rapid plasmenylethanolamine-selective fusion of membrane bilayers catalyzed by an isoform of glyceraldehyde-3-phosphate dehydrogenase: discrimination between glycolytic and fusogenic roles of individual isoforms. Biochemistry. 1995;34:12193–12203. doi: 10.1021/bi00038a013. [DOI] [PubMed] [Google Scholar]
- 18.Glaser P. E., Han X., Gross R. W. Tubulin is the endogenous inhibitor of the glyceraldehyde 3-phosphate dehydrogenase isoform that catalyzes membrane fusion: implications for the coordinated regulation of glycolysis and membrane fusion. Proc. Natl. Acad. Sci. U.S.A. 2002;99:14104–14109. doi: 10.1073/pnas.222542999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hessler R. J., Blackwood R. A., Brock T. G., Francis J. W., Harsh D. M., Smolen J. E. Identification of glyceraldehyde-3-phosphate dehydrogenase as a Ca2+-dependent fusogen in human neutrophil cytosol. J. Leukoc. Biol. 1998;63:331–336. doi: 10.1002/jlb.63.3.331. [DOI] [PubMed] [Google Scholar]
- 20.Robbins A. R., Ward R. D., Oliver C. A mutation in glyceraldehyde-3-phosphate dehydrogenase alters endocytosis in CHO cells. J. Cell Biol. 1995;130:1093–1104. doi: 10.1083/jcb.130.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumagai H., Sakai H. A porcine brain protein (35 K protein) which bundles microtubules and its identification as glyceraldehyde 3-phosphate dehydrogenase. J. Biochem. (Tokyo) 1983;93:1259–1269. doi: 10.1093/oxfordjournals.jbchem.a134260. [DOI] [PubMed] [Google Scholar]
- 22.Huitorel P., Pantaloni D. Bundling of microtubules by glyceraldehyde-3-phosphate dehydrogenase and its modulation by ATP. Eur. J. Biochem. 1985;150:265–269. doi: 10.1111/j.1432-1033.1985.tb09016.x. [DOI] [PubMed] [Google Scholar]
- 23.Duclos-Vallee J. C., Capel F., Mabit H., Petit M. A. Phosphorylation of the hepatitis B virus core protein by glyceraldehyde-3-phosphate dehydrogenase protein kinase activity. J. Gen. Virol. 1998;79:1665–1670. doi: 10.1099/0022-1317-79-7-1665. [DOI] [PubMed] [Google Scholar]
- 24.Engel M., Seifert M., Theisinger B., Seyfert U., Welter C. Glyceraldehyde-3-phosphate dehydrogenase and Nm23-H1/nucleoside diphosphate kinase A. Two old enzymes combine for the novel Nm23 protein phosphotransferase function. J. Biol. Chem. 1998;273:20058–20065. doi: 10.1074/jbc.273.32.20058. [DOI] [PubMed] [Google Scholar]
- 25.Singh R., Green M. R. Sequence-specific binding of transfer RNA by glyceraldehyde-3-phosphate dehydrogenase. Science. 1993;259:365–368. doi: 10.1126/science.8420004. [DOI] [PubMed] [Google Scholar]
- 26.Zang W. Q., Fieno A. M., Grant R. A., Yen T. S. Identification of glyceraldehyde-3-phosphate dehydrogenase as a cellular protein that binds to the hepatitis B virus posttranscriptional regulatory element. Virology. 1998;248:46–52. doi: 10.1006/viro.1998.9255. [DOI] [PubMed] [Google Scholar]
- 27.Meyer-Siegler K., Mauro D. J., Seal G., Wurzer J., deRiel J. K., Sirover M. A. A human nuclear uracil DNA glycosylase is the 37-kDa subunit of glyceraldehyde-3-phosphate dehydrogenase. Proc. Natl. Acad. Sci. U.S.A. 1991;88:8460–8464. doi: 10.1073/pnas.88.19.8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baxi M. D., Vishwanatha J. K. Uracil DNA-glycosylase/glyceraldehyde-3-phosphate dehydrogenase is an Ap4A binding protein. Biochemistry. 1995;34:9700–9707. doi: 10.1021/bi00030a007. [DOI] [PubMed] [Google Scholar]
- 29.McNulty S. E., Toscano W. A., Jr Transcriptional regulation of glyceraldehyde-3-phosphate dehydrogenase by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Biochem. Biophys. Res. Commun. 1995;212:165–171. doi: 10.1006/bbrc.1995.1951. [DOI] [PubMed] [Google Scholar]
- 30.Krynetski E. Y., Krynetskaia N. F., Gallo A. E., Murti K. G., Evans W. E. A novel protein complex distinct from mismatch repair binds thioguanylated DNA. Mol. Pharmacol. 2001;59:367–374. doi: 10.1124/mol.59.2.367. [DOI] [PubMed] [Google Scholar]
- 31.Ryazanov A. G. Glyceraldehyde-3-phosphate dehydrogenase is one of the three major RNA-binding proteins of rabbit reticulocytes. FEBS Lett. 1985;192:131–134. doi: 10.1016/0014-5793(85)80058-2. [DOI] [PubMed] [Google Scholar]
- 32.Nagy E., Rigby W. F. Glyceraldehyde-3-phosphate dehydrogenase selectively binds AU-rich RNA in the NAD+-binding region (Rossmann fold) J. Biol. Chem. 1995;270:2755–2763. doi: 10.1074/jbc.270.6.2755. [DOI] [PubMed] [Google Scholar]
- 33.De B. P., Gupta S., Zhao H., Drazba J. A., Banerjee A. K. Specific interaction in vitro and in vivo of glyceraldehyde-3-phosphate dehydrogenase and LA protein with cis-acting RNAs of human parainfluenza virus type 3. J. Biol. Chem. 1996;271:24728–24735. doi: 10.1074/jbc.271.40.24728. [DOI] [PubMed] [Google Scholar]
- 34.Schultz D. E., Hardin C. C., Lemon S. M. Specific interaction of glyceraldehyde 3-phosphate dehydrogenase with the 5′-nontranslated RNA of hepatitis A virus. J. Biol. Chem. 1996;271:14134–14142. doi: 10.1074/jbc.271.24.14134. [DOI] [PubMed] [Google Scholar]
- 35.Yi M., Schultz D. E., Lemon S. M. Functional significance of the interaction of hepatitis A virus RNA with glyceraldehyde 3-phosphate dehydrogenase (GAPDH): opposing effects of GAPDH and polypyrimidine tract binding protein on internal ribosome entry site function. J. Virol. 2000;74:6459–6468. doi: 10.1128/jvi.74.14.6459-6468.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin S. S., Chang S. C., Wang Y. H., Sun C. Y., Chang M. F. Specific interaction between the hepatitis delta virus RNA and glyceraldehyde 3-phosphate dehydrogenase: an enhancement on ribozyme catalysis. Virology. 2000;271:46–57. doi: 10.1006/viro.2000.0302. [DOI] [PubMed] [Google Scholar]
- 37.Dollenmaier G., Weitz M. Interaction of glyceraldehyde-3-phosphate dehydrogenase with secondary and tertiary RNA structural elements of the hepatitis A virus 3′ translated and non-translated regions. J. Gen. Virol. 2003;84:403–414. doi: 10.1099/vir.0.18501-0. [DOI] [PubMed] [Google Scholar]
- 38.Berry M. D., Boulton A. A. Glyceraldehyde-3-phosphate dehydrogenase and apoptosis. J. Neurosci. Res. 2000;60:150–154. doi: 10.1002/(SICI)1097-4547(20000415)60:2<150::AID-JNR3>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 39.Saunders P. A., Chen R. W., Chuang D. M. Nuclear translocation of glyceraldehyde-3-phosphate dehydrogenase isoforms during neuronal apoptosis. J. Neurochem. 1999;72:925–932. doi: 10.1046/j.1471-4159.1999.0720925.x. [DOI] [PubMed] [Google Scholar]
- 40.Ishitani R., Tanaka M., Sunaga K., Katsube N., Chuang D. M. Nuclear localization of overexpressed glyceraldehyde-3-phosphate dehydrogenase in cultured cerebellar neurons undergoing apoptosis. Mol. Pharmacol. 1998;53:701–707. doi: 10.1124/mol.53.4.701. [DOI] [PubMed] [Google Scholar]
- 41.Ishitani R., Sunaga K., Tanaka M., Aishita H., Chuang D. M. Overexpression of glyceraldehyde-3-phosphate dehydrogenase is involved in low K+-induced apoptosis but not necrosis of cultured cerebellar granule cells. Mol. Pharmacol. 1997;51:542–550. doi: 10.1124/mol.51.4.542. [DOI] [PubMed] [Google Scholar]
- 42.Ishitani R. A role for GAPDH in neuronal apoptosis. Seikagaku. 1997;69:1107–1111. [PubMed] [Google Scholar]
- 43.Ishitani R., Kimura M., Sunaga K., Katsube N., Tanaka M., Chuang D. M. An antisense oligodeoxynucleotide to glyceraldehyde-3-phosphate dehydrogenase blocks age-induced apoptosis of mature cerebrocortical neurons in culture. J. Pharmacol. Exp. Ther. 1996;278:447–454. [PubMed] [Google Scholar]
- 44.Saunders P. A., Chalecka-Franaszek E., Chuang D. M. Subcellular distribution of glyceraldehyde-3-phosphate dehydrogenase in cerebellar granule cells undergoing cytosine arabinoside-induced apoptosis. J. Neurochem. 1997;69:1820–1828. doi: 10.1046/j.1471-4159.1997.69051820.x. [DOI] [PubMed] [Google Scholar]
- 45.Ishitani R., Chuang D. M. Glyceraldehyde-3-phosphate dehydrogenase antisense oligodeoxynucleotides protect against cytosine arabinonucleoside-induced apoptosis in cultured cerebellar neurons. Proc. Natl. Acad. Sci. U.S.A. 1996;93:9937–9941. doi: 10.1073/pnas.93.18.9937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tajima H., Tsuchiya K., Yamada M., Kondo K., Katsube N., Ishitani R. Over-expression of GAPDH induces apoptosis in COS-7 cells transfected with cloned GAPDH cDNAs. Neuroreport. 1999;10:2029–2033. doi: 10.1097/00001756-199907130-00007. [DOI] [PubMed] [Google Scholar]
- 47.Zheng L., Roeder R. G., Luo Y. S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell. 2003;114:255–266. doi: 10.1016/s0092-8674(03)00552-x. [DOI] [PubMed] [Google Scholar]
- 48.Kimura M., Suzuki H., Ishihama A. Formation of a carboxy-terminal domain phosphatase (Fcp1)/TFIIF/RNA polymerase II (pol II) complex in Schizosaccharomyces pombe involves direct interaction between Fcp1 and the Rpb4 subunit of pol II. Mol. Cell. Biol. 2002;22:1577–1588. doi: 10.1128/mcb.22.5.1577-1588.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mitsuzawa H., Kimura M., Kanda E., Ishihama A. Glyceraldehyde-3-phosphate dehydrogenase and actin associate with RNA polymerase II and interact with its Rpb7 subunit. FEBS Lett. 2005;579:48–52. doi: 10.1016/j.febslet.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 50.Carlile G. W., Tatton W. G., Borden K. L. Demonstration of a RNA-dependent nuclear interaction between the promyelocytic leukaemia protein and glyceraldehyde-3-phosphate dehydrogenase. Biochem. J. 1998;335:691–696. doi: 10.1042/bj3350691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J., Shiels C., Sasieni P., Wu P. J., Islam S. A., Freemont P. S., Sheer D. Promyelocytic leukemia nuclear bodies associate with transcriptionally active genomic regions. J. Cell Biol. 2004;164:515–526. doi: 10.1083/jcb.200305142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sambrook J., Russell D. W. New York: Cold Spring Harbor Laboratory Press; 2001. Molecular Cloning: a Laboratory Manual. [Google Scholar]
- 53.Lee J., Rhee B. K., Bae G. Y., Han Y. M., Kim J. Stimulation of Oct-4 activity by Ewing's sarcoma protein. Stem Cells. 2005;23:738–751. doi: 10.1634/stemcells.2004-0375. [DOI] [PubMed] [Google Scholar]
- 54.Kim S. Y., Kim Y. S., Bahk Y. Y. Proteome changes induced by expression of tumor suppressor PTEN. Mol. Cells. 2003;15:396–405. [PubMed] [Google Scholar]
- 55.Lee H. J., Kim S., Pelletier J., Kim J. Stimulation of hTAFII68 (NTD)-mediated transactivation by v-Src. FEBS Lett. 2004;564:188–198. doi: 10.1016/S0014-5793(04)00314-X. [DOI] [PubMed] [Google Scholar]
- 56.Kim J., Lee J. M., Branton P. E., Pelletier J. Modification of EWS/WT1 functional properties by phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 1999;96:14300–14305. doi: 10.1073/pnas.96.25.14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dignam J. D. Preparation of extracts from higher eukaryotes. Methods Enzymol. 1990;182:194–203. doi: 10.1016/0076-6879(90)82017-v. [DOI] [PubMed] [Google Scholar]
- 58.Kim J., Lee J. M., Branton P. E., Pelletier J. Modulation of EWS/WT1 activity by the v-Src protein tyrosine kinase. FEBS Lett. 2000;474:121–128. doi: 10.1016/s0014-5793(00)01590-8. [DOI] [PubMed] [Google Scholar]
- 59.Geetha T., Kenchappa R. S., Wooten M. W., Carter B. D. TRAF6-mediated ubiquitination regulates nuclear translocation of NRIF, the p75 receptor interactor. EMBO J. 2005;24:3859–3868. doi: 10.1038/sj.emboj.7600845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilson T. E., Fahrner T. J., Johnston M., Milbrandt J. Identification of the DNA binding site for NGFI-B by genetic selection in yeast. Science. 1991;252:1296–1300. doi: 10.1126/science.1925541. [DOI] [PubMed] [Google Scholar]
- 61.Hisaoka M., Hashimoto H. Extraskeletal myxoid chondrosarcoma: updated clinicopathological and molecular genetic characteristics. Pathol. Int. 2005;55:453–463. doi: 10.1111/j.1440-1827.2005.01853.x. [DOI] [PubMed] [Google Scholar]
- 62.Graven K. K., Troxler R. F., Kornfeld H., Panchenko M. V., Farber H. W. Regulation of endothelial cell glyceraldehyde-3-phosphate dehydrogenase expression by hypoxia. J. Biol. Chem. 1994;269:24446–24453. [PubMed] [Google Scholar]
- 63.Sawa A., Khan A. A., Hester L. D., Snyder S. H. Glyceraldehyde-3-phosphate dehydrogenase: nuclear translocation participates in neuronal and nonneuronal cell death. Proc. Natl. Acad. Sci. U.S.A. 1997;94:11669–11674. doi: 10.1073/pnas.94.21.11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wansa K. D., Harris J. M., Yan G., Ordentlich P., Muscat G. E. The AF-1 domain of the orphan nuclear receptor NOR-1 mediates trans-activation, coactivator recruitment, and activation by the purine anti-metabolite 6-mercaptopurine. J. Biol. Chem. 2003;278:24776–24790. doi: 10.1074/jbc.M300088200. [DOI] [PubMed] [Google Scholar]
- 65.Ordentlich P., Yan Y., Zhou S., Heyman R. A. Identification of the antineoplastic agent 6-mercaptopurine as an activator of the orphan nuclear hormone receptor Nurr1. J. Biol. Chem. 2003;278:24791–24799. doi: 10.1074/jbc.M302167200. [DOI] [PubMed] [Google Scholar]
- 66.Schmitz H. D. Reversible nuclear translocation of glyceraldehyde-3-phosphate dehydrogenase upon serum depletion. Eur. J. Cell Biol. 2001;80:419–427. doi: 10.1078/0171-9335-00174. [DOI] [PubMed] [Google Scholar]
- 67.Filion C., Labelle Y. The oncogenic fusion protein EWS/NOR-1 induces transformation of CFK2 chondrogenic cells. Exp. Cell Res. 2004;297:585–592. doi: 10.1016/j.yexcr.2004.03.040. [DOI] [PubMed] [Google Scholar]