Abstract
We introduce a general test of the bioenergetic importance of mtDNA (mitochondrial DNA) variants: modular kinetic analysis of oxidative phosphorylation in mitochondria from cybrid cells with constant nuclear DNA but different mtDNA. We have applied this test to the hypothesis [Ruiz-Pesini, Mishmar, Brandon, Procaccio and Wallace (2004) Science 303, 223–226] that particular mtDNA haplogroups (specific combinations of polymorphisms) that cause lowered coupling efficiency, leading to generation of less ATP and more heat, were positively selected during radiations of modern humans into colder climates. Contrary to the predictions of this hypothesis, mitochondria from Arctic haplogroups had similar or even greater coupling efficiency than mitochondria from tropical haplogroups.
Keywords: coupling efficiency, cybrid, human evolution, mitochondrial DNA (mtDNA), modular kinetic analysis, oxidative phosphorylation
Abbreviations: Δψ, mitochondrial membrane potential; ρ0, mitochondrial-DNA-less; DMEM, Dulbecco's modified Eagle's medium; FBS, foetal bovine serum; FCCP, carbonyl cyanide p-trifluoromethoxyphenylhydrazone; mtDNA, mitochondrial DNA; RFLP, restriction fragment length polymorphism; TPMP, triphenylmethylphosphonium
INTRODUCTION
Many mutations in mtDNA (mitochondrial DNA) lead to severe pathologies, such as LHON (Leber hereditary optic neuropathy) and MELAS (mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes), although the penetrance of the pathological phenotype is often puzzling [1,2]. The bioenergetic consequences of normal mtDNA variation are less well understood, although there are indications that such variation may affect bioenergetic function [3,4], disease susceptibility [5,6] and longevity [7]. Such analyses are difficult, however, because mitochondrial functions are mostly controlled by genes encoded in nuclear DNA, and it can be very difficult to be sure that small effects attributed to mtDNA variations are not confounded by different nuclear DNA backgrounds. In the present study we used cybrids with different mtDNA complements but the same nuclear DNA background to overcome this problem [8]. We then assayed bioenergetic function using the modular kinetic approach [9] to determine the effect of mtDNA sequence on mitochondrial bioenergetics. This novel application of modular kinetic analysis to cybrid mitochondria provides a powerful new tool, of general applicability, to analyse bioenergetic effects of differences in mtDNA in healthy human populations or in mitochondrial diseases.
Analysis of human mtDNA variation has identified specific combinations of polymorphisms that have been used systematically to classify mtDNAs into haplogroups and to study the origin, radiations and evolution of human populations [10,11]. More recently, it has been proposed that these polymorphisms interacted with environmental factors, leading to positive selection of adaptive variants with decreased mitochondrial coupling efficiency and increased heat production that were beneficial for humans moving out of Africa into colder Eurasia and through the Arctic to the Americas [12,13]. Specifically, haplogroups A, C, D and X, which are dominant in north-eastern Eurasian and native-American populations, are predicted to have lower oxidative phosphorylation coupling efficiency than the other haplogroups, particularly L, which is dominant in African populations, and B, which is present in central-Asian and American populations and is thought not to have been cold-selected. These haplogroups have specific polymorphisms in the mitochondrial genes MTATP6, MTCYB, MTND2 and MTND4, which code for components of the protein complexes of oxidative phosphorylation, so might cause slip or leak reactions and affect coupling efficiency. The present study tested this hypothesis experimentally in order to clarify whether, and how, oxidative phosphorylation coupling efficiency depends on genetic variants of mtDNA generated after modern humans migrated out of Africa.
EXPERIMENTAL
Subjects
Ethical approval was obtained from the Cambridge Research Ethics Committee. We recruited healthy volunteers by advertisement and obtained written informed consent from all participants. DNA was extracted from buccal swab samples using a QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). Mitochondrial haplogroups were determined by PCR–RFLP (restriction fragment length polymorphism) analysis according to criteria described in [11]. Entire mtDNAs were amplified in several overlapping fragments by PCR [14]. Each fragment was digested with restriction enzymes and resolved on agarose gels. A 15 ml sample of blood was taken from each of six volunteers who had Arctic (A, C and D) or tropical (L1, L2 and L3) haplogroups, for platelet preparation and cybrid construction.
Cells
A549.B2 ρ0 (mtDNA-less) cells derived from human lung carcinoma A549 were cultivated in DMEM (Dulbecco's modified Eagle's medium) containing 4.5 g/l glucose and 110 μg/ml sodium pyruvate, supplemented with 10% (v/v) FBS (foetal bovine serum) and 50 μg/ml uridine. Platelets from the blood samples of volunteers were fused with A549.B2 ρ0 cells as described elsewhere [15]. The resulting cybrid cell lines were routinely maintained in DMEM with 10% dialysed FBS. DNA was extracted from cultured cybrid cells as described previously [16] to confirm mitochondrial haplogroups by PCR–RFLP analysis.
Cybrid B2.3243 cells, which have the same A549 nuclear background, but harbour a pathogenic mutation at np 3243 in the tRNA-Leu(UUR) gene (MTTL1) [17,18] were kindly donated by Dr Ian Holt of this Unit. The mutation was heteroplasmic: clone Z had a high proportion of wild-type and clone X had a high proportion of mutant.
Mitochondria
Human cell mitochondria were prepared from cultured cybrid cells grown in DMEM with 10% FBS. Cells were allowed to grow up to approx. 90% confluence on three or four trays (500 cm2; Nunclon™). Following two washes with ice-cold STE buffer (comprising 0.25 M sucrose, 5 mM Tris/HCl, pH 7.4, and 2 mM EGTA), cells were scraped off the trays and harvested in STE buffer by centrifugation at 480 g (10 min, 4 °C). Cell pellets were resuspended in 5 ml of STE buffer supplemented with a protease-inhibitor cocktail (Calbiochem catalogue no. 539131) and homogenized using a glass homogenizer. After 8-fold dilution in STE buffer and removal of cell debris (3 min centrifugation at 1100 g, followed by filtration through muslin), mitochondria were pelleted by centrifugation at 12000 g (11 min, 4 °C). This procedure gave good-quality mitochondria (respiratory control ratio with α-oxoglutarate+malate of approx. 11) and yielded typically 5–8 mg of mitochondrial protein/500 cm2 tray. Protein concentration was determined by the biuret method with BSA as a standard.
Respiratory flux and membrane potential
Oxygen consumption was measured at 37 °C using a Clark-type oxygen electrode (Rank Brothers, Cambridge, U.K.) calibrated with an air-saturated medium comprising 0.115 M KCl, 10 mM KH2PO4, 3 mM Hepes, pH 7.2, 1 mM EGTA, 2 mM MgCl2 and 0.3% (w/v) defatted BSA, assumed to contain 406 nmol of atomic oxygen/ml [19]. No correction was made for consumption of oxygen by the electrode, so mitochondrial oxygen consumption at low rates will have been slightly overestimated. Simultaneously with respiratory activity, Δψ (mitochondrial membrane potential) was measured using an electrode sensitive to the lipophilic cation TPMP+ (triphenylmethylphosphonium) [20]. Mitochondria were incubated at 1.0 mg/ml (for succinate respiration) or 1.5 mg/ml (for α-oxoglutarate+malate respiration) in the presence of 80 ng/ml nigericin to collapse ΔpH and 4 μM rotenone to inhibit Complex I. The TPMP+-sensitive electrode was calibrated with sequential additions of TPMP+ up to 2 μM, then 4 mM succinate or 3.2 mM α-oxoglutarate+0.8 mM malate (with rotenone omitted) was added to initiate respiration. Experiments were terminated with 1.6 μM FCCP (carbonyl cyanide p-trifluoromethoxyphenylhydrazone), allowing correction for any small baseline drift. Δψ values were calculated from the distribution of TPMP+ across the mitochondrial inner membrane using a binding correction factor of 0.35 mg of protein/μl. Respiratory control ratios (the State 3 respiration rate with 0.8 mM ADP divided by the State 4 rate with oligomycin) with α-oxoglutarate+malate as substrate were determined in the absence of nigericin.
Modular kinetics
Mitochondrial oxidative phosphorylation can be divided conceptually into three modules. These are: (i) the reactions that produce Δψ, consisting of the substrate translocases, dehydrogenases and other enzymes and the components of the respiratory chain, here called ‘substrate oxidation’; (ii) the reactions that consume Δψ and synthesize, export and dephosphorylate ATP, consisting of the ATP synthase, the phosphate and adenine nucleotide translocases and any ATPases that may be present, called the ‘phosphorylating system’; and (iii) the reactions that consume Δψ without ATP synthesis, called the ‘proton leak’ [21]. Oxygen consumption and Δψ were measured simultaneously using mitochondria incubated with 80 ng/ml nigericin and 4 μM rotenone. Respiration was initiated by 4 mM succinate (or 3.2 mM α-oxoglutarate+0.8 mM malate, with rotenone omitted). The kinetic behaviour of a ‘Δψ-producer’ can be established by specific modulation of a ‘Δψ-consumer’ and, reciprocally, the kinetics of a consumer are revealed upon modulation of a producer [9]. To measure the kinetic response of proton leak to Δψ, the State 4 (non-phosphorylation) respiration of mitochondria in the presence of oligomycin (0.8 μg/ml; to prevent any residual ATP synthesis), which was used solely to drive the proton leak, was titrated with malonate (up to 0.5 mM). In a similar way, the State 4 respiration was titrated with FCCP (up to 0.8 μM) for measurement of the kinetic response of substrate oxidation to Δψ. State 3 (maximal rate of ATP synthesis) was obtained using an ADP-regenerating system [100 μM ATP, 20 mM glucose and 10 units/ml hexokinase (from baker's yeast; Sigma)]. Titration of State 3 respiration with malonate (up to 1.15 mM) allowed measurement of the kinetics of the Δψ-consumers (the sum of the phosphorylating system and proton leak).
Catalase assay
Cells were harvested by trypsinization, washed with ice-cold PBS, and suspended in 50 mM potassium phosphate buffer, pH 7.0, containing 0.5% Triton X-100 and 0.1 mM EDTA. The cells were disrupted by brief sonication and the supernatant was obtained by centrifugation. Catalase activity of the supernatant was measured at 37 °C in triplicate as described elsewhere [22]. One unit of catalase is defined as the activity required to decompose 1 μmol of H2O2/min. Specific activity is expressed as units per mg of protein, estimated using a BCA™ (bicinchoninic acid) protein assay kit (Pierce, Rockford, IL, U.S.A.) with BSA as a standard.
Statistics
Values are given as means±S.E.M., with n being the number of independent mitochondrial preparations (Figure 1) or cell clones (nine for Figures 2 and Figure 4; three for Figure 3). To test for significant differences in modular kinetics between haplogroups, we interpolated the mean respiration rates of mitochondria from the individual preparations or clones at the lowest individual State 4 potential (for proton leak) or State 3 potential (for the phosphorylating system) or at any potential between the highest and lowest membrane potential (for substrate oxidation). The significance of differences between means was assessed by unpaired Student's t test using Microsoft Excel X; P values<0.05 were taken to be significant. There were no significant differences between Arctic and tropical haplogroups for any of the data in Figures 2 or 4.
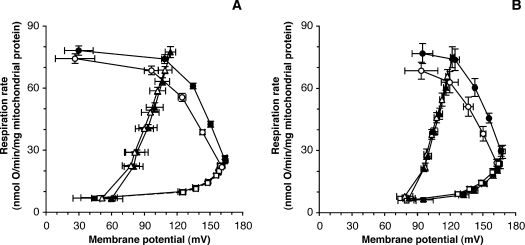
Figure 1. Modular kinetic analysis: validation of the method.
The Figures shows a modular kinetic analysis of the kinetic responses of the three modules of oxidative phosphorylation [substrate oxidation, circles; proton leak, squares; Δψ-consumers (sum of phosphorylating system and proton leak), triangles] to membrane potential, Δψ, in mitochondria isolated from cybrids, using 4 mM succinate as substrate. (A) Kinetic responses in mitochondria isolated from A, D, L1 and L2 cybrids in the absence (closed symbols) or presence (25 nM; open symbols) of the Complex III inhibitor myxothiazol. Results are means±S.E.M. for four independent mitochondrial preparations. The kinetics of substrate oxidation were significantly different at membrane potentials above 98 mV. (B) Kinetic responses in mitochondria isolated from cybrids harbouring a pathogenic mutation at np 3243 in the tRNA-Leu(UUR) gene (MTTL1). Closed symbols, clone Z (high proportion of wild-type); open symbols, clone X (high proportion of mutant). Results are means±S.E.M. for three independent mitochondrial preparations. The kinetics of substrate oxidation were significantly different at all membrane potentials.
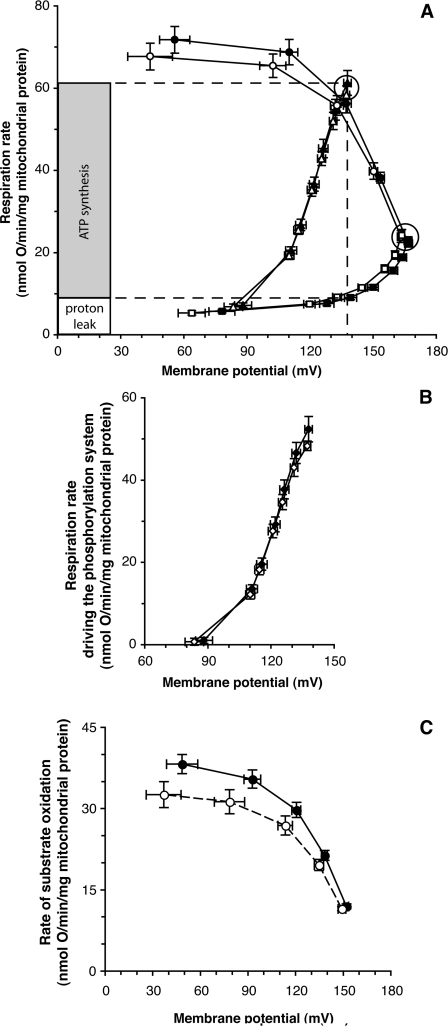
Figure 2. Modular kinetic analysis: averaged values.
The Figure shows a modular kinetic analysis of the kinetic responses of the three modules of oxidative phosphorylation to membrane potential, Δψ, in mitochondria isolated from cybrids. Closed symbols, Arctic haplogroups (A, C and D); open symbols, tropical haplogroups (L1, L2 and L3). (A) Succinate as substrate. A comparison of the kinetic responses of substrate oxidation (circles; Δψ titrated with the uncoupler FCCP), proton leak (squares; Δψ titrated with inhibitor, malonate), and Δψ-consumers (sum of phosphorylating system and proton leak, triangles; Δψ titrated with malonate) is shown. The points circled represent State 3 (maximal ATP synthesis) and State 4 (ATP synthesis) conditions. The coupling efficiency of oxidative phosphorylation (percentage of respiration used for ATP synthesis at a given Δψ) was calculated from the kinetic curves: the inset histogram shows the example of Arctic haplogroups at the Δψ of State 3. (B) Kinetic response of the phosphorylating system to Δψ, calculated from (A) by subtracting respiration driving proton leak from respiration driving the Δψ-consumers at each Δψ. (C) Kinetic response of substrate oxidation using 3.2 mM α-oxoglutarate+0.8 mM malate as substrate, with rotenone omitted. Results are means±S.E.M. for nine cell clones (three different clones for each of the three constituent haplogroups). There were three or four (A, B and C) or one (D) independent mitochondrial preparation(s) per clone.
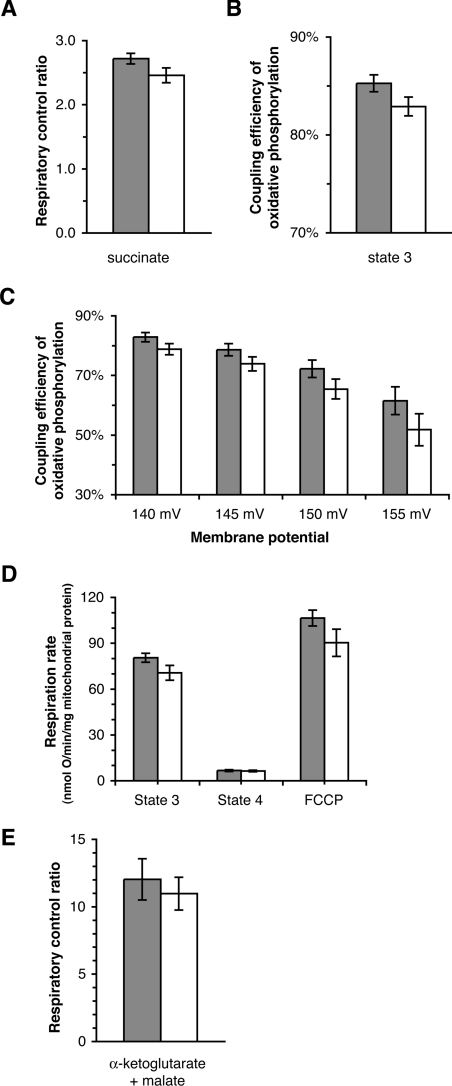
Figure 4. Coupling efficiency of oxidative phosphorylation in mitochondria isolated from cybrids.
Closed bars, Arctic haplogroups (A, C and D); open bars, tropical haplogroups (L1, L2 and L3). (A) Respiratory control ratio (State 3 respiration rate/State 4 respiration rate) using succinate as substrate. (B) Coupling efficiency using succinate as substrate at the Δψ of State 3. (C) Coupling efficiency using succinate as substrate at selected Δψ values between that at State 3 and that at State 4. Values at 145 mV or less were significantly different from values at 155 mV. (D) Respiration rates with α-oxoglutarate+malate as substrate. (E) Respiratory control ratio using α-oxoglutarate+malate as substrate. Results are means±S.E.M. for nine cell clones (three different clones for each of the three constituent haplogroups); there were three or four (A, B and C) or one (D and E) independent mitochondrial preparation(s) per clone. Values in (A, B and C) were calculated from the results shown in Figure 2(A).
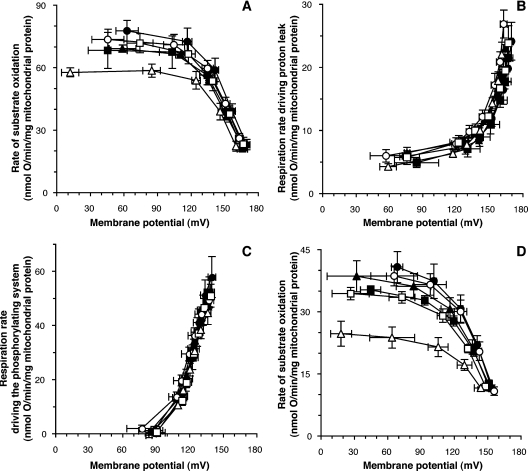
Figure 3. Modular kinetic analysis: full dataset.
The Figure shows modular kinetic analysis in mitochondria isolated from cybrids, using succinate as substrate, of (A) substrate oxidation, (B) proton leak and (C) the phosphorylating system. (D) Modular kinetic analysis of substrate oxidation using α-oxoglutarate+malate as substrate. Closed symbols, Arctic haplogroups (circles, haplogroup A; squares, haplogroup C; triangles, haplogroup D); open symbols, tropical haplogroups (circles, haplogroup L1; squares, haplogroup L2; triangles, haplogroup L3). Data are means±S.E.M. for three cell clones; there were three or four (A, B and C) or one (D) repeat(s) using independently prepared mitochondria for each clone. The kinetic response of the phosphorylating system to Δψ was calculated by subtracting respiration driving proton leak from respiration driving the Δψ-consumers at each value of Δψ.
RESULTS
Generation of cybrid cell lines
To overcome effects of different nuclear backgrounds, cybrids were used to analyse the functional effects of mtDNA. Cybrid cell lines are constructed by repopulation of ρ0 cells with exogenous mitochondria [8]. A549.B2 ρ0 cells derived from human lung carcinoma A549 were used in the present study. To obtain different populations of mitochondria, we recruited healthy human volunteers, determined their mitochondrial haplogroups and selected six with Arctic (A, C or D) or tropical (L1, L2 or L3) haplogroups. Platelets, which have no nuclei, were isolated from blood samples and fused with the ρ0 cells. The resultant cybrids had mtDNA from the different donors, but their nuclear DNA was identical. After many generations of cybrid growth (diluting platelet-derived nuclear-encoded subunits), all nuclear-encoded mitochondrial protein subunits will be specified by the host cell, but all mitochondrial-encoded subunits will be specified by the donor mtDNA. Using cybrids made it possible to investigate the effects of mtDNA variation in cells with identical nuclear DNA.
Modular kinetic analysis of oxidative phosphorylation: validation of the method
Rather than arbitrarily measuring individual reactions to look for differences caused by mtDNA variants, we applied a systems approach: top-down elasticity analysis [9]. This is better described as ‘modular kinetic analysis’, since it analyses the kinetics of the whole of oxidative phosphorylation divided into a small number of modules (in this case three: substrate oxidation, proton leak and the phosphorylating system), connected by their common substrate or product, the mitochondrial membrane potential, Δψ. Thus the analysis picks up any change in oxidative phosphorylation that is functionally important, and it is unresponsive to any changes that have no functional consequences. Comparison of the kinetic responses of each of the three modules to Δψ obtained using mitochondria isolated from different cybrids reveals any effects of mitochondrial haplogroup on the kinetics of oxidative phosphorylation. The coupling efficiency of oxidative phosphorylation (the percentage of respiration rate at a given Δψ that is used for ATP synthesis) can then be calculated from the kinetic curves. Note that any slip reactions will appear as proton leak in this analysis [23].
To validate this experimental system, we examined two models: a known chemical inhibitor and a known pathogenic mutation. We first measured the kinetics of oxidative phosphorylation in the absence or presence of a low concentration of the Complex III inhibitor myxothiazol. Figure 1(A) shows that only the kinetics of substrate oxidation were affected, as predicted, and that a change in kinetics giving rate changes of 10% was readily observed. In the second model system we measured the kinetics of oxidative phosphorylation in mitochondria isolated from cybrids harbouring a pathogenic mutation at np 3243 in the tRNA-Leu(UUR) gene (MTTL1) [17,18]. This mutation decreases expression of all mitochondrially encoded proteins, so is expected to decrease the activity of all the respiratory-chain complexes (except Complex II, whose subunits are all nuclear-encoded) and the ATP synthase. Figure 1(B) shows that our assay system reported the expected difference in the kinetics of substrate oxidation. The kinetics of the phosphorylation system were not significantly different, despite the anticipated decrease in ATP synthase activity, presumably because of low control of the ATP synthase over the phosphorylation rate. As expected, the kinetics of proton leak were not changed. Thus our test system was able to identify small changes, less than 10%, in the kinetics of different modules of oxidative phosphorylation if they were present.
Modular kinetic analysis of oxidative phosphorylation: test of the hypothesis
The electrochemical proton gradient across the mitochondrial inner membrane, formed as electrons are passed down the respiratory chain, is the primary energy source for cellular ATP synthesis. However, not all of the energy available in the redox reactions is coupled to ATP synthesis. Some is consumed by proton-slip reactions (fewer protons are pumped out of the matrix during electron flow) or proton-leak reactions (protons are pumped normally, but leak back through proton-conductance pathways that bypass the ATP synthase). As a result of these slip and leak reactions, heat is generated instead of ATP. These non-productive proton-slip and -leak pathways are physiologically important. They account for a surprisingly high proportion of cellular and organismal respiration rate, estimated to be up to 20% [24,25], so differences in their capacities could plausibly play an adaptive role that could be selected during ancient human migrations.
To test the hypothesis, we measured the kinetics and coupling efficiency of oxidative phosphorylation using isolated mitochondria from cybrid cells harbouring Arctic or tropical haplogroups. Figure 2(A) shows the averaged kinetics of the three modules of oxidative phosphorylation using succinate grouped by Arctic (closed symbols) or tropical (open symbols) mtDNA haplogroup (the results for the six individual cybrid lines are shown in Figure 3).
There was a small difference, which was not statistically significant, between the Arctic and tropical haplogroups in the rate of the substrate oxidation module at any given Δψ (Figure 2A, circles). This small difference was caused entirely by one of the three tropical haplogroup cybrids, L3, which had a lower maximum respiration rate than the other tropical haplogroup lines (Figure 3A). We also measured the kinetics of substrate oxidation using α-oxoglutarate+malate as substrate instead of succinate to check for any effects of differences in the mitochondrial Complex I genes. Once again, there was a small, but not significant, difference in the kinetics of substrate oxidation between the Arctic and tropical haplogroups (Figure 2C) that was caused by lower rates only in the L3 cybrids (Figure 3D). As there was otherwise no difference between Arctic and tropical haplogroups in the kinetics of substrate oxidation with either substrate, we assume this was an individual difference in the L3 line possibly caused by somatic mutations in mtDNA. We conclude that there is no general difference between Arctic and tropical mtDNA haplogroups in the overall kinetics of substrate oxidation.
In contrast with what the hypothesis would lead one to expect, the rate of the proton leak of mitochondria with the Arctic haplogroups was, if anything, slightly lower on average than that of the tropical ones at any value of Δψ (Figure 2A, squares) (differences were not statistically significant). For the individual haplogroups, the rate for tropical haplogroup L3 was relatively high, and the rates for Arctic haplogroups C and D were relatively low (Figure 3B), although any differences were small. This shows that mtDNA variations between the Arctic and tropical haplogroups had at most a modest effect on mitochondrial proton leak, but in the wrong direction to support the hypothesis.
Figure 2(B) shows that, on average, there was no difference in the rate of the phosphorylating system at any given value of Δψ between the Arctic and tropical haplogroups. The kinetic curves of the six individual haplogroups almost overlapped (Figure 3C). We conclude that there is no difference between Arctic and tropical mtDNA haplogroups in the overall kinetics of the phosphorylating system.
From the kinetic curves shown in Figures 2 and 3 we can analyse the coupling efficiency of oxidative phosphorylation. The classic measure is the respiratory control ratio (Figure 4A):
 |
We also independently measured the respiratory control ratio using α-oxoglutarate+malate as substrates (Figures 4D and 4E). Figures 4(A) and 4(E) show that there were small differences in average respiratory control ratio; mitochondria from the Arctic haplogroups were slightly better coupled than those from the tropical haplogroups. However, these differences were not statistically significant, and they were in the wrong direction to support the hypothesis, even if haplogroup L3, which has lower respiration rate, was omitted (results not shown).
A more precise measure of coupling efficiency is given by the percentage of respiration that is coupled to ATP synthesis, calculated as shown in Figure 2(A) at any chosen membrane potential. If this percentage is lower, then a greater proportion of the redox energy is diverted to heat production. Figure 4(B) shows that, at the value of Δψ achieved in State 3 (maximal rate of ATP synthesis), mitochondria from the Arctic haplogroups were slightly better coupled than those from the tropical haplogroups. Again, this difference was not statistically significant, and it was in the wrong direction to support the hypothesis. Mitochondria in cells normally operate at less than maximal rates. Figure 4(C) examines the coupling efficiency across the whole range of rates of ATP synthesis and values of Δψ from State 3 to State 4 (where ATP synthesis is minimal). As expected [26], coupling efficiency decreased as mitochondria moved from State 3 towards State 4. Once again, there was a small difference, but not statistically significant, between the Arctic and tropical haplogroups, and again it was in the wrong direction to support the hypothesis.
Catalase activity
Differences in respiration rates of mouse cybrid cells harbouring different mtDNA sequences can be masked by compensatory changes in mitochondrial density due to a retrograde response that correlates with threefold changes in catalase activity [27]. However, such compensatory changes would not mask changes in the modular kinetics of oxidative phosphorylation in mitochondria isolated from cybrids, and did not prevent the expected results in our pathogenic mutant model (Figure 1B). Nonetheless, to test whether catalase-associated compensation occurred in our human cybrids, we measured catalase activity to see if it depended on the mitochondrial haplogroup. Catalase specific activity did not differ between Arctic (15.4±1.0 units/mg of protein, n=9) and tropical (14.6±1.1 units/mg of protein, n=9) cybrid cells (P=0.6), suggesting that this retrograde response was not a factor in our experiments.
DISCUSSION
We conclude that the hypothesis that mitochondrial haplogroups that lower the coupling efficiency of oxidative phosphorylation were positively selected during radiations of modern humans [12,13] is not supported by our experimental test using modular kinetic analysis of cybrid mitochondria. Recent mitochondrial genetic studies have also cast doubt on the hypothesis [28,29]. The strengths of our approach to test this hypothesis are: (i) for the first time, we used a direct empirical measurement, rather than inference from genetic and physiological traits; (ii) we separated the effects of mitochondrial haplogroup from any confounding effects of nuclear DNA background; and (iii) we used a simple in vitro system with the powerful modular kinetic approach where the relevant variables could be tightly controlled and manipulated. However, our approach also has some weaknesses: (i) a small effect of mitochondrial haplogroups operating over many generations might still be too small to be detected by biochemical experiments; we estimate that our experiments had the statistical power to identify a 7.3% difference in coupling efficiency between the two haplogroups; (ii) we measured the coupling efficiency of oxidative phosphorylation using isolated mitochondria in simple defined media, but the effects of mitochondrial haplogroups might only emerge at cellular or higher levels (for example, they might involve mitochondrial–cytoplasmic feedback loops or retrograde signalling to nuclear genes, or might be tissue-specific); and (iii) interaction between nuclear and mitochondrial genomes may affect oxidative phosphorylation [30], so the effect of mitochondrial haplogroup might depend on nuclear background. A549, the parental line of the A549.B2 ρ0 cells used here, has mtDNA of European haplogroup H, which has been proposed to be relatively cold-adapted [12,13]. Perhaps the mtDNA of tropical haplogroups does not complement A549 nuclear DNA optimally and causes slightly lower coupling efficiency, masking evolutionarily relevant efficiency changes in human populations.
The novel method introduced in the present study, modular kinetic analysis using cybrids, has provided a powerful empirical test of a specific hypothesis of altered mitochondrial coupling efficiency in human populations carrying different mitochondrial haplogroups. However, it is also a more general method that can be used to investigate the bioenergetic consequences of other differences in mitochondrial haplogroups and haplotypes in humans or other species. As shown in Figure 1(B), it is also a powerful method to investigate in a precise and controlled way the bioenergetic phenotypes of hereditary or sporadic pathological mtDNA mutations that are thought to cause many mitochondrial diseases.
Acknowledgments
We thank Ms Valerie Church, Ms Julie Buckingham and Ms Helen Boysen for excellent technical assistance, and Dr Ian Holt (MRC Dunn Human Nutrition Unit) for advice and A549.B2 ρ0 and B2.3243 cells. This work was supported by the Medical Research Council (London, U.K.) and in part by a grant to T.A. from the Uehara Memorial Foundation (Tokyo, Japan).
References
- 1.Taylor R. W., Turnbull D. M. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dimauro S., Davidzon G. Mitochondrial DNA and disease. Ann. Med. 2005;37:222–232. doi: 10.1080/07853890510007368. [DOI] [PubMed] [Google Scholar]
- 3.Ruiz-Pesini E., Lapena A. C., Diez-Sanchez C., Perez-Martos A., Montoya J., Alvarez E., Diaz M., Urries A., Montoro L., Lopez-Perez M. J., Enriquez J. A. Human mtDNA haplogroups associated with high or reduced spermatozoa motility. Am. J. Hum. Genet. 2000;67:682–696. doi: 10.1086/303040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montiel-Sosa F., Ruiz-Pesini E., Enriquez J. A., Marcuello A., Diez-Sanchez C., Montoya J., Wallace D. C., Lopez-Perez M. J. Differences of sperm motility in mitochondrial DNA haplogroup U sublineages. Gene. 2006;368:21–27. doi: 10.1016/j.gene.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 5.Baudouin S. V., Saunders D., Tiangyou W., Elson J. L., Poynter J., Pyle A., Keers S., Turnbull D. M., Howell N., Chinnery P. F. Mitochondrial DNA and survival after sepsis: a prospective study. Lancet. 2005;366:2118–2121. doi: 10.1016/S0140-6736(05)67890-7. [DOI] [PubMed] [Google Scholar]
- 6.Herrnstadt C., Howell N. An evolutionary perspective on pathogenic mtDNA mutations: haplogroup associations of clinical disorders. Mitochondrion. 2004;4:791–798. doi: 10.1016/j.mito.2004.07.041. [DOI] [PubMed] [Google Scholar]
- 7.Santoro A., Salvioli S., Raule N., Capri M., Sevini F., Valensin S., Monti D., Bellizzi D., Passarino G., Rose G., et al. Mitochondrial DNA involvement in human longevity. Biochim. Biophys. Acta. 2006;1757:1388–1399. doi: 10.1016/j.bbabio.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 8.King M. P., Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 9.Brand M. D. Top-down elasticity analysis and its application to energy metabolism in isolated mitochondria and intact cells. Mol. Cell. Biochem. 1998;184:13–20. [PubMed] [Google Scholar]
- 10.Cann R. L., Stoneking M., Wilson A. C. Mitochondrial DNA and human evolution. Nature. 1987;325:31–36. doi: 10.1038/325031a0. [DOI] [PubMed] [Google Scholar]
- 11.Wallace D. C., Brown M. D., Lott M. T. Mitochondrial DNA variation in human evolution and disease. Gene. 1999;238:211–230. doi: 10.1016/s0378-1119(99)00295-4. [DOI] [PubMed] [Google Scholar]
- 12.Mishmar D., Ruiz-Pesini E., Golik P., Macaulay V., Clark A. G., Hosseini S., Brandon M., Easley K., Chen E., Brown M. D., et al. Natural selection shaped regional mtDNA variation in humans. Proc. Natl. Acad. Sci. U.S.A. 2003;100:171–176. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruiz-Pesini E., Mishmar D., Brandon M., Procaccio V., Wallace D. C. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science. 2004;303:223–226. doi: 10.1126/science.1088434. [DOI] [PubMed] [Google Scholar]
- 14.Torroni A., Petrozzi M., D'Urbano L., Sellitto D., Zeviani M., Carrara F., Carducci C., Leuzzi V., Carelli V., Barboni P., et al. Haplotype and phylogenetic analyses suggest that one European-specific mtDNA background plays a role in the expression of Leber hereditary optic neuropathy by increasing the penetrance of the primary mutations 11778 and 14484. Am. J. Hum. Genet. 1997;60:1107–1121. [PMC free article] [PubMed] [Google Scholar]
- 15.Chomyn A. Platelet-mediated transformation of human mitochondrial DNA-less cells. Methods Enzymol. 1996;264:334–339. doi: 10.1016/s0076-6879(96)64031-2. [DOI] [PubMed] [Google Scholar]
- 16.Laird P. W., Zijderveld A., Linders K., Rudnicki M. A., Jaenisch R., Berns A. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El Meziane A., Lehtinen S. K., Hance N., Nijtmans L. G., Dunbar D., Holt I. J., Jacobs H. T. A tRNA suppressor mutation in human mitochondria. Nat. Genet. 1998;18:350–353. doi: 10.1038/ng0498-350. [DOI] [PubMed] [Google Scholar]
- 18.Dunbar D. R., Moonie P. A., Jacobs H. T., Holt I. J. Different cellular backgrounds confer a marked advantage to either mutant or wild-type mitochondrial genomes. Proc. Natl. Acad. Sci. U.S.A. 1995;92:6562–6566. doi: 10.1073/pnas.92.14.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reynafarje B., Costa L. E., Lehninger A. L. O2 solubility in aqueous media determined by a kinetic method. Anal. Biochem. 1985;145:406–418. doi: 10.1016/0003-2697(85)90381-1. [DOI] [PubMed] [Google Scholar]
- 20.Brand M. D. Measurement of mitochondrial protonmotive force. In: Brown G. C., Cooper C. E., editors. Bioenergetics: A Practical Approach. Oxford: IRL Press; 1995. pp. 39–62. [Google Scholar]
- 21.Brand M. D. The proton leak across the mitochondrial inner membrane. Biochim. Biophys. Acta. 1990;1018:128–133. doi: 10.1016/0005-2728(90)90232-s. [DOI] [PubMed] [Google Scholar]
- 22.Amo T., Atomi H., Imanaka T. Unique presence of a manganese catalase in a hyperthermophilic archaeon, Pyrobaculum calidifontis VA1. J. Bacteriol. 2002;184:3305–3312. doi: 10.1128/JB.184.12.3305-3312.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brand M. D., Chien L. F., Diolez P. Experimental discrimination between proton leak and redox slip during mitochondrial electron transport. Biochem. J. 1994;297:27–29. doi: 10.1042/bj2970027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rolfe D. F. S., Newman J. M., Buckingham J. A., Clark M. G., Brand M. D. Contribution of mitochondrial proton leak to respiration rate in working skeletal muscle and liver and to SMR. Am. J. Physiol. 1999;276:C692–C699. doi: 10.1152/ajpcell.1999.276.3.C692. [DOI] [PubMed] [Google Scholar]
- 25.Brand M. D. The efficiency and plasticity of mitochondrial energy transduction. Biochem. Soc. Trans. 2005;33:897–904. doi: 10.1042/BST0330897. [DOI] [PubMed] [Google Scholar]
- 26.Brand M. D., Harper M. E., Taylor H. C. Control of the effective P/O ratio of oxidative phosphorylation in liver mitochondria and hepatocytes. Biochem. J. 1993;291:739–748. doi: 10.1042/bj2910739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moreno-Loshuertos R., Acin-Perez R., Fernandez-Silva P., Movilla N., Perez-Martos A., Rodriguez de Cordoba S., Gallardo M. E., Enriquez J. A. Differences in reactive oxygen species production explain the phenotypes associated with common mouse mitochondrial DNA variants. Nat. Genet. 2006;38:1261–1268. doi: 10.1038/ng1897. [DOI] [PubMed] [Google Scholar]
- 28.Elson J. L., Turnbull D. M., Howell N. Comparative genomics and the evolution of human mitochondrial DNA: assessing the effects of selection. Am. J. Hum. Genet. 2004;74:229–238. doi: 10.1086/381505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kivisild T., Shen P., Wall D. P., Do B., Sung R., Davis K., Passarino G., Underhill P. A., Scharfe C., Torroni A., et al. The role of selection in the evolution of human mitochondrial genomes. Genetics. 2006;172:373–387. doi: 10.1534/genetics.105.043901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mishmar D., Ruiz-Pesini E., Mondragon-Palomino M., Procaccio V., Gaut B., Wallace D. C. Adaptive selection of mitochondrial complex I subunits during primate radiation. Gene. 2006;378:11–18. doi: 10.1016/j.gene.2006.03.015. [DOI] [PubMed] [Google Scholar]