Abstract
Cruciferin (a 12 S globulin) is the most abundant storage protein in the seeds of Arabidopsis thaliana (thale cress) and other crucifers, sharing structural similarity with the cupin superfamily of proteins. Cruciferin is synthesized as a precursor in the rough endoplasmic reticulum. Subunit assembly is accompanied by structural rearrangements involving proteolysis and disulfide-bond formation prior to deposition in protein storage vacuoles. The A. thaliana cv. Columbia genome contains four cruciferin loci, two of which, on the basis of cDNA analysis, give rise to three alternatively spliced variants. Using MS, we confirmed the presence of four variants encoded by genes At4g28520.1, At5g44120.3, At1g03880.1 and At1g3890.1 in A. thaliana seeds. Two-dimensional gel electrophoresis, along with immunological detection using anti-cruciferin antiserum and antibodies against phosphorylated amino acid residues, revealed that cruciferin was the major phosphorylated protein in Arabidopsis seeds and that polymorphism far exceeded that predicted on the basis of known isoforms. The latter may be attributed, at least in part, to phosphorylation site heterogeneity. A total of 20 phosphorylation sites, comprising nine serine, eight threonine and three tyrosine residues, were identified by MS. Most of these are located on the IE (interchain disulfide-containing) face of the globulin trimer, which is involved in hexamer formation. The implications of these findings for cruciferin processing, assembly and mobilization are discussed. In addition, the protein phosphatase 2C-impaired mutant, abi1-1, was found to exhibit increased levels of cruciferin phosphorylation, suggesting either that cruciferin may be an in vivo target for this enzyme or that abi1-1 regulates the protein kinase/phosphatase system required for cruciferin phosphorylation.
Keywords: Arabidopsis thaliana (thale cress) abi1 mutant, cruciferin, mass spectrometry (MS), protein phosphorylation, seed storage proteins, two-dimensional electrophoresis
Abbreviations: ABA, abscisic acid; ASB-14, amidosulfobetaine-14; CRA1, cruciferin precursor; CRB, cruciferin B; CRC, cruciferin C; 1- and 2-DE, one- and two-dimensional gel electrophoresis; DTT, dithiothreitol; IA, intrachain disulfide-containing; IE, interchain disulfide-containing; IPG, immobilized pH gradient; MS/MS, tandem MS; PP2C, protein phosphatase 2C; PSV, protein storage vacuoles; TAIR, The Arabidopsis Information Resource; TOF, time-of-flight; VPE, vacuolar processing enzymes
INTRODUCTION
During their development, plant seeds accumulate large amounts of proteins. These proteins provide the main source of nitrogen for seed germination and for early seedling growth. In Arabidopsis thaliana (thale cress) and other crucifers, the main seed proteins are the 12 S globulins (cruciferin) and the 2 S albumins (napin), which are localized to PSVs (protein storage vacuoles), and the oleosins, which are integral proteins of the oil-body membrane. Salt-soluble globulins are widely distributed in dicotyledons, monocotyledons and gymnosperms, and comprise the trimeric and predominantly glycosylated 7 S vicilins and the hexameric and generally non-glycosylated 11–12 S legumins [1]. The three-dimensional structures of several vicilin-type proteins, namely phaseolin [2], canavalin [3], β-conglycinin [4] and germin [5], and the legumin-type proglycinin A1aB1b homotrimer [6] and glycinin A3B4 homohexamer [7], have revealed that seed storage globulins share striking similarities in secondary and tertiary structure with the cupin superfamily of prokaryotic and eukaryotic proteins [8,9]. Proteins in this family have been ascribed a wide variety of functions [8,9]. The characteristic cupin domain contains two conserved β-barrel motifs separated by a lesser-conserved region comprising two β-strands with an intervening loop of variable length. This compact ‘jelly-roll’ β-barrel structure is thought to confer on the cupin domain a high degree of thermal stability and resistance to protease digestion.
Temporal and spatial factors influence the accumulation of seed storage proteins in the cotyledon and axis during seed development [10–12]. The latter is exemplified by the observation that antisense suppression of napin expression causes a corresponding increase in cruciferin synthesis, thus neutralizing the overall impact on total seed protein content [13]. Cruciferin is synthesized in the rough endoplasmic reticulum as a ∼50 kDa precursor and is then transported to the PSV by either a Golgi-dependent [14] or Golgi-independent process [15–17]. Regardless of the translocation pathway used, the precursor is processed into a ∼30 kDa acidic α-peptide and a ∼20 kDa basic β-peptide, which are linked together by a single interchain disulfide bond. The processing is accompanied by the organization of the mature peptides into a hexameric complex [18–20]. Four VPEs (vacuolar processing enzymes), which are cysteine-type proteinases with specificity toward conserved Asn-Gly motifs in both napin and cruciferin, have been identified in A. thaliana, including the vegetative α-VPE [21] and γ-VPE [22] and the seed-specific β-VPE [21] and δ-VPE [23]. Recent studies have shown that the β-VPE is essential for proper processing of seed storage proteins, but that the vegetative-type VPEs, aspartic proteinases and possibly other proteolytic enzymes can partially compensate for the loss of β-VPE activity, resulting in similar patterns of seed protein processing and deposition [23,24].
Phosphorylation is the most frequent type of protein post-translational modification, and is often associated with the molecular activation/de-activation of signal-transduction pathways. The A. thaliana genome encodes more than 1300 types of protein kinases and phosphatases that perform such functions. The mapping of phosphorylation sites in proteins remains a significant challenge, due to the limited abundance of most phosphorylated proteins, low phosphorylation stoichiometry and phosphorylation-site heterogeneity. Considerable effort has been devoted to developing strategies for the enrichment and detection of phosphorylated proteins and peptides, and a variety of MS techniques, employing both electrospray ionization and MALDI (matrixassisted laser desorption–ionization) have been used to identify sites of stable and labile (i.e. histidine) phosphorylation [25–27]. Such methods were recently applied to the characterization of heat-stable phosphorylated proteins in mature A. thaliana seeds [28].
The present study was undertaken with the goal of identifying proteins that are phosphorylated during A. thaliana seed germination. In the process, we discovered that the most abundant phosphorylated seed protein was cruciferin, and that multiple phosphorylation sites could be identified using LC–MS/MS (liquid chromatography–tandem MS). Since PP2C (protein phosphatase 2C), which is encoded by ABI1, has been implicated in the regulation of ABA (abscisic acid)-dependent kinase signalling during seed development [29, 30], we also examined protein phosphorylation in the abi1-1 mutant, in which PP2C activity is significantly reduced. We found that cruciferin is hyperphosphorylated in this mutant and is, therefore, a probable target for the ABI1 protein phosphatase.
EXPERIMENTAL
Seed protein sample preparation
All reagents were obtained from Sigma–Aldrich (Oakville, ON, Canada) unless otherwise noted. A 50 mg portion of dried A. thaliana seeds (Landsberg erecta, wild-type or abi1-1 mutant) were ground in 15% (w/v) trichloroacetic acid/acetone and 0.07% 2-mercaptoethanol with ceramic beads in a FastPrep® FP120 Cell Disrupter (Qbiogene, Montreal, QB, Canada), incubated at −20 °C for 3 h, then centrifuged at 20000 g at 4 °C for 30 min. The pellets were washed with 80% (v/v) cold acetone and air-dried overnight at 4 °C. For 1-DE (one-dimensional gel electrophoresis) the dried tissues were extracted with 1 ml of an extraction buffer consisting of 50 mM Tris/HCl (pH 8.8), 6 M urea, 2% (w/v) SDS and 5 mM DTT (dithiothreitol) at room temperature (22 °C) for 1 h with stirring. After addition of iodoacetamide to a final concentration of 10 mM, the extraction was allowed to continue for an additional 1 h before centrifuging at 20000 g for 5 min at room temperature to remove tissue debris. For 2-DE (two-dimensional gel electrophoresis) the dried tissues were extracted with 1 ml of an extraction buffer consisting of 10 mM Tris base, 7 M urea, 2 M thiourea, 1% (w/v) ASB-14 (amidosulfobetaine-14), 1% Triton X-100, 50 units/ml DNase I and 5 units/ml RNase A at room temperature for 1 h with stirring. The extract was centrifuged at 20000 g for 30 min, and the supernatant removed and further centrifuged at 35000 g for 30 min. The proteins were precipitated by the sequential addition, and mixing, of 4 ml of methanol, 1 ml of chloroform and 3 ml of distilled water, and the mixture was centrifuged at 20000 g for 5 min. The white layer of precipitated proteins located between the upper and lower phases was removed, washed with methanol, centrifuged at 20000 g for 5 min, air-dried and resuspended in a rehydration buffer consisting of 10 mM Tris base, 7 M urea, 2 M thiourea, 1% ASB-14, 1% Triton X-100, 0.5% Biolytes pH 3-10 (Bio-Rad, Hercules, CA, U.S.A.) and 2 mM tributylphosphine. Protein concentration was determined using the DC RC Protein Assay kit (Bio-Rad) with BSA as a standard.
1- and 2-DE
Seed protein samples prepared for 1-DE were mixed with 10% (v/v) glycerol and Bromophenol Blue. Electrophoresis was performed using 10–20% (w/v) linear-gradient Criterion Tris/HCl gels (Bio-Rad) at a constant voltage of 150 V until the dye was 1 cm from the bottom of the gel. The gels were either stained with Bio-Safe Coomassie Blue (Bio-Rad) or transferred to membranes for immunoblotting.
A 1 mg portion of each seed protein sample prepared for 2-DE was applied to ReadyStrip™ IPG (immobilized pH gradient) Strips (pH 3–10 non-linear, 17 cm; Bio-Rad). The strips were passively rehydrated overnight at room temperature and focused using a Protein IEF (isoelectric focusing) Cell (Bio-Rad) at a maximum current of 40 μA/strip and a final voltage of 10000 V for a total of 80000 V·h (rapid ramp). SDS/PAGE was subsequently performed using an Ettan DALTSix electrophoresis cell (GE Healthcare, Baie d’Urfé, QB, Canada) running 1.5-mm-thick 10% (w/v) polyacrylamide gels. Prior to SDS/PAGE, IPG strips were treated sequentially with 2% DTT and 2.5% iodoacetamide in 6 M urea, 0.375 M Tris-HCl (pH 8.8), 2% SDS and 20% (v/v) glycerol for 10 min to reduce and alkylate proteins. Electrophoresis was carried out at 2.5 W/gel for 1 h and then at 17 W/gel until the Bromophenol Blue dye was 1 cm from the bottom of the gel. Precision Plus Protein™ Dual Color Standards (Bio-Rad) were used as molecular-mass markers.
AgNO3 staining was carried out using a Hoefer Processor Plus unit (GE Healthcare) and a modified version of the Vorum protocol [31]. Briefly, gels were first fixed with 12% (v/v) acetic acid and 0.05% formalin [35% (v/v) formaldehyde] in 50% (v/v) methanol overnight, washed with 35% (v/v) ethanol three times for 20 min, sensitized in 0.02% Na2S2O3 for 2 min, washed three times with doubly distilled water for 5 min, stained with 0.2% AgNO3 and 0.076% formalin in doubly distilled water for 20 min, washed twice with doubly distilled water for 1 min, and developed with 6% Na2CO3, 0.05% formalin and 0.0004% Na2S2O3 in doubly distilled water. The developing process was quenched with 12% acetic acid and 50% (w/v) methanol. The volume of solvent used in each step was 350 ml. To reduce the staining intensity for gels loaded with 1 mg of seed protein, the amount of Na2S2O3 in all steps was reduced to 80% of that normally used.
Proteins in gels stained with Bio-Safe Coomassie Blue or AgNO3 were quantified using an ImageScanner (GE Healthcare).
Production of an anti-cruciferin serum
Oligonucleotides BnCRC 1693F (5′-AAAGGATCCATGAGGCAGTCGCTAGG-3′) and BnCRC 1693R (5′-CCTGCGGCCGCAGCCTCGACTATCTCCTC-3′) were used to amplify the cruciferin precursor (i.e. the mature protein minus the signal peptide) from a Brassica napus (rape) cDNA which, on the basis of expressed-sequence-tag analysis, represents a highly abundant cruciferin mRNA that shares a high degree of identity with cruciferins encoded by A. thaliana. The fragment was digested with EcoRI and NotI, cloned into pET28a (Novagen, San Diego CA, U.S.A.) and introduced into the Epicurian Coli® BL21-RP Codon Plus strain (Stratagene, La Jolla, CA, U.S.A.). Bacterial cultures were grown overnight in Luria–Bertani broth at 37 °C, diluted 1:10 with fresh medium and incubated at 30 °C with vigorous shaking until the attenuance (D600) reached 0.3. Protein expression was induced with 1 mM isopropyl β-D-thiogalactoside for 24 h. Recombinant histidine-tagged cruciferin was purified from the inclusion bodies using the His•Bind® purification kit (Novagen) according to the manufacturer's protocol. Essentially, bacterial cell pellets were re-suspended in 1×binding buffer and disrupted by treatment with lysozyme and Triton X-100, followed by sonication. The debris was removed by centrifugation at 10000 g for 10 min and the pellet was re-suspended in binding buffer containing 8 M urea, incubated at 4 °C for 45 min, centrifuged, and the supernatant loaded on to an equilibrated His•Bind resin column. The histidine-tagged cruciferin was eluted with imidazole after several washes and precipitated with 10% trichloroacetic acid. After washing with acetone, the purified cruciferin (∼100 μg) was dried and re-suspended in 0.5 ml of TitreMax adjuvant (Sigma–Aldrich). Following the initial immunization, rabbits (New Zealand White) were boosted twice at 2-week intervals before collection of the antiserum.
Immunoblotting
Following electrophoresis, proteins were transferred from 1-DE or 2-DE gels to Immuno-Blot PVDF membranes using a Trans-Blot Plus Electrophoretic Transfer Cell (Bio-Rad). The transfer was carried out in Dunn carbonate buffer (10 mM NaHCO3/3 mM Na2CO3, pH 9.9) using high-intensity-field conditions, according to the manufacture's instructions. For detection of phosphorylated proteins, the membranes were blocked with 3% (w/v) BSA in 1×TBST buffer (20 mM Tris, 14 mM NaCl and 0.3% Tween-20, pH 7.6) for 1 h and then probed with the primary antibody in TBST for 1 h. The monoclonal antibodies (Calbiochem, San Diego CA, U.S.A.) and total concentrations used were as follows: anti-phosphoserine (1C8, 4A3, 4A9, 16B4 at 0.28 μg/ml), antiphosphothreonine (1E11, 4D11, 14B3 at 0.32 μg/ml) and anti-phosphotyrosine (2C8, 3B12, 9H8, 16F4 at 0.25 μg/ml). The membranes were washed four times for 10 min with TBST buffer and incubated with a secondary antibody (goat anti-mouse IgM for anti-phosphoserine, goat antimouse IgG for anti-phosphothreonine and anti-phosphotyrosine conjugated to horseradish peroxidase; Calbiochem) diluted 1:20000 in TBST buffer containing 5% (w/v) non-fat dried milk powder for 1 h. The membranes were washed four times for 10 min in TBST buffer and developed using the ECL Plus Western Blotting system (GE Healthcare). For detection of cruciferin, the membranes were blocked with 5% non-fat dried milk powder, probed with primary polyclonal antiserum against cruciferin at a dilution of 1:20000 and then with secondary antibody (goat anti-rabbit IgG conjugated to horseradish peroxidase; Calbiochem) at a dilution of 1:200000.
In-gel trypsin digestion and MS
Immunoreactive seed protein bands (1-DE) and spots (2-DE) were excised from the gels using a Proteome Works 2-D spot cutter (Bio-Rad) and placed in a 96-well microtitre plate. The proteins were then automatically destained, reduced with dithiothreitol, alkylated with iodoacetamide and digested with porcine trypsin (Promega, Madison, WI, U.S.A.) using a MassPREP protein digest station (Micromass, Wynthenshaw, Manchester, U.K.) according to protocols recommended by the manufacturer. Tryptic peptides were extracted three times with a 50:50 (v/v) mixture of acetonitrile and aq. 1% trifluoroacetic acid, dried under vacuum, and re-dissolved in 10 μl of 0.1% trifluoroacetic acid. The plate was sealed using an ALPS 300™ heat sealer (ABgene) and 6.4 μl of each digest was analysed by LC–MS/MS, using a capLC ternary nanoHPLC system interfaced to a Q-ToF Ultima Global hybrid tandem mass spectrometer fitted with a Z-spray nanoelectrospray ion source (Waters, Mississauga, ON, Canada). Solvents A and C were 0.2% formic acid in water, whereas solvent B consisted of 0.2% formic acid in acetonitrile. The peptide-digest sample was loaded on to a C18 trapping column (Symmetry 300; 0.35 mm×5 mm Opti-pak; Waters) and washed for 3 min using solvent C at a flow rate of 30 μl/min. The flow path was then switched using a ten-port rotary valve, and the sample eluted on to a C18 analytical column (PepMap; 75 μm×15 cm, 3 μm particle size; Dionex, Oakville ON, Canada). Separations were performed using the following gradient programme at a flow rate of 220 nl/min after splitting: 5:95 (%, A:B) for 3.0 min, changing to 8:92 at 4.0 min, 42:58 at 50.0 min, and 90:10 at 50.5 min, holding until 55.5 min, and then reverting to 5:95% A:B from 56.0 to 60.0 min. LC–MS/MS analysis was carried out using data-dependent acquisition, during which peptide precursor ions were detected by scanning from mass-to-charge (m/z) 400 to 1200 in TOF (time-of-flight) MS mode. Multiply charged ions rising above a predetermined threshold intensity were automatically selected for TOF MS/MS analysis, and product-ion spectra were acquired over the m/z range 50–1990. LC–MS/MS data were processed using PeptideAuto software (Waters) and searched against the Arabidopsis database at TAIR (The Arabidopsis Information Resource; http://www.arabidopsis.org/) using MASCOT Daemon (Matrix Science, Boston, MA, U.S.A.). Searches were performed using carbamidomethylation of cysteine as the fixed modification, oxidation of methionine and phosphorylation of serine, threonine or tyrosine as variable modifications, and mass tolerances of 0.5 Da for both MS and MS/MS data, allowing for one missed cleavage during trypsin digestion. Peptide matches were considered unambiguous if ion scores were significant (P<0.05) and protein identifications included at least one top-ranking peptide match.
RESULTS
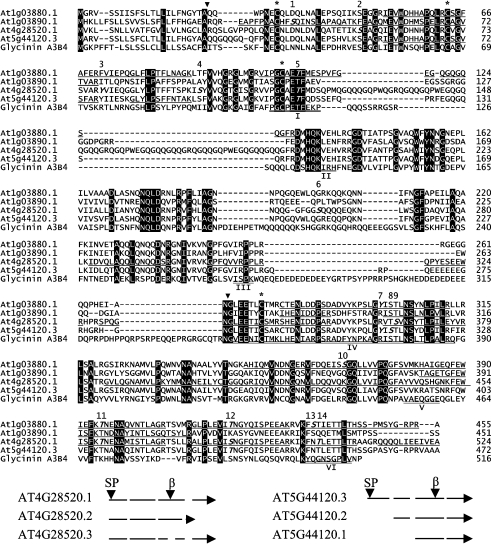
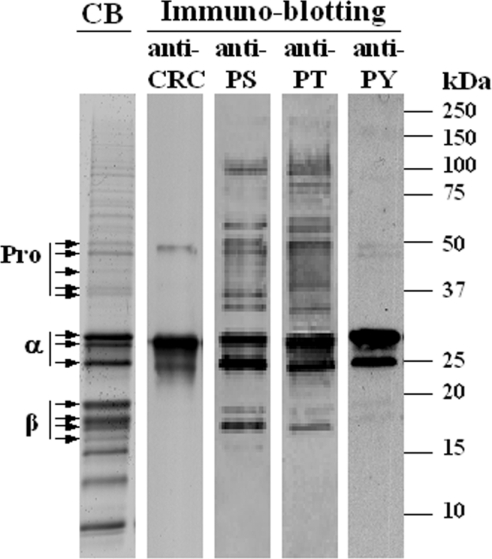
The A. thaliana cv. Columbia genome contains four loci that encode cruciferins: At5g44120 [CRA1 (cruciferin precursor)], At1g03880 [CRB (cruciferin B)], At4g28520 [CRC (cruciferin C)] and At1g03890 (putative cruciferin) [10]. The majority of the transcripts arise from At4g28520, At5g44120 and, to a lesser extent, from At1g03880 [32]. At1g03890 is located in tandem with the locus encoding CRB and was once believed to be a pseudogene; however, a cDNA has recently been assigned to it (http://signal.salk.edu/). Expressed-sequence-tag analysis, using data available from TAIR revealed three splice variants for both At4g28520 and At5g44120, resulting in a potential repertoire of eight cruciferin proteins (Figure 1). The putative signal peptidase and Asn–Gly cleavage sites shown in Figure 1 are consistent with Edman N-terminal sequencing of accumulated cruciferin precursors and β-subunits [24]. The protein derived from At4g28520.3 contains an internal deletion that eliminates the conserved Asn–Gly processing site. At5g44120.1 and At5g44120.2 yield proteins truncated at the extreme N-terminus; however, alternative signal peptides were not detected using either the SignalP 3.0 Server (http://www.cbs.dtu.dk/services/SignalP/) or Signal Peptide Prediction (http://www.bioinformatics.leeds.ac.uk/prot_analysis/Signal.html). To determine the distribution of cruciferins on 1-DE gels, LC–MS/MS analysis was performed on the major protein bands that cross-reacted with the cruciferin antiserum (Figure 2). The bulk of the cruciferin was processed into α- (25–30 kDa) and β- (17–20 kDa) subunits. A small amount of unprocessed cruciferin (40–50 kDa), signified by the detection of both α- and β-subunit peptides, was also observed. Peptides derived from the remaining lower-molecular-mass major bands corresponded to various types of oleosins and napins (results not shown). The polyclonal antiserum generated against an unprocessed pro-cruciferin expressed in Escherichia coli exhibited very strong affinity toward the α-subunits, but little affinity for the β-subunits.
Figure 1. Alignment of protein sequences of four translated 12 S globulin cruciferins from A. thaliana and 11 S globulin glycinin A3B4 from G. max using Clustal V (top), and a diagram of alternative splicing of loci At4g28520 and At5g44120 (bottom).
Top: alignment of protein sequences. Residues that are identical in all cases are indicated in white on a black background. Locations of putative signal peptidase (SP) and Asn↓Gly (β) cleavage sites are indicated by a ▼ symbol. Cysteine residues involved in inter- and intra-chain disulfide bond formation are labelled with an asterisk (*). Phosphorylation sites identified in cruciferins are in bold italics and the locations are numbered. Phosphorylation of Thr116 and Ser120 in AT4G28520.1, and Tyr292 and Tyr299 in AT1G03880.1 could not be distinguished using MS/MS (see Table 1). Phosphorylation of Thr116 (location number 5) and Tyr299 (location number 7) are tentative assignments based on phosphorylation of the corresponding residues in aligned sequences. Unique tryptic peptides identified by MS in AT1G03880.1, AT1G03890.1 and AT5G44120.3, as well as the tryptic peptides in AT4G28520.1 in the divergent region of this locus are underlined. The binding regions involved in trimer interaction are double underlined and numbered according to those in glycinin A3B4 [7]. Bottom: alternative splicing diagram. Introns are shown as broken lines, and arrows indicate the direction of transcription.
Figure 2. 1-DE SDS/PAGE and immunoblotting of A. thaliana cv. Lansberg erecta seed proteins.
Extracted proteins (20 μg) were separated on 10–20% linear-gradient Criterion gels and either stained with Coomassie Blue (CB) or subjected to immunoblotting with anti-cruciferin antiserum (anti-CRC) or anti-phosphoserine (anti-PS), anti-phosphothreonine (anti-PT) and anti-phosphotyrosine (anti-PY) monoclonal antibodies. Molecular-mass markers (kDa) and protein bands corresponding to unprocessed cruciferin (Pro) and to α- and β-subunits, as confirmed by MS, are labelled accordingly.
LC–MS/MS analysis of the 2-DE protein spots that cross-reacted with cruciferin antiserum was used to investigate which of the potential gene models were expressed. As shown in Figure 1, tryptic peptides uniquely coded by At1g03880.1, At1g03890.1 and At5g44120.3, which do not exist in any other cruciferin variants, were identified and so verified the translation of these three gene models. Of the three alternatively spliced transcripts for At5g44120, only At5g44120.3 encodes a protein with a signal peptide. Alternative signal peptides were not predicted to reside at the N-terminus of the truncated isoforms encoded by the At5g44120.1 and At5g44120.2 transcripts (Figure 1), and such proteins are unlikely to be directed to the seed PSV. Proteins derived from both At4g28520.2 and At4g28520.3 have a deletion in the β-subunit. The identification of tryptic peptides coded by At4g28520.1 and spanning the variant region (Figure 1) makes a strong argument that At4g28520.1 is the major translated gene from this locus. Morevover, the sole distinctive tryptic peptides, namely G385VLQECDGAS394 and I283DVYK287, that would distinguish the products of At4g28520.2 and At4g28520.3 from At4g28520.1 respectively, were not found in any of the samples analysed. Furthermore, the protein derived from At4g28520.3 would lack the internal processing site necessary to form α- and β-subunits (Figure 1) [24]. On the basis of sequence coverage obtained from MS analysis, the relative contributions of the four identified cruciferin isoforms to total seed protein content are in the order AT4G28520.1>AT5G44120.3>AT1G03880.1> AT1G3890.1, with AT1G03890.1 present at much lower levels than the other three.
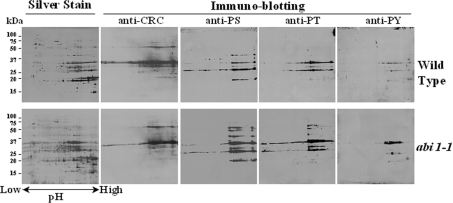
Immunoblotting with monoclonal antibodies against phosphorylated forms of serine, threonine and tyrosine indicated that cruciferin is the major phosphorylated protein in A. thaliana seeds, with phosphorylated forms of each type of hydroxyamino acid being detected. Both subunits reacted with antibodies to phosphorylated amino acid residues, although phosphorylation of α-subunits was more apparent than for β-subunits (Figures 2 and 3). Subsequently 2-DE, in conjunction with immunoblotting, revealed that the heterogeneity of cruciferin isoforms far exceeded that predicted from the limited number of expressed genes and mRNA variants described above (Figure 3).
Figure 3. 2-DE gel electrophoresis and immunoblotting of seed proteins from A. thaliana cv. Lansberg erecta wild-type or the abi1-1 mutant.
Extracted proteins (1 mg) were separated by 2-DE and visualized with silver stain (indicated) or subjected to immunoblotting with anti-cruciferin antiserum (anti-CRC) or anti-phosphoserine (anti-PS), anti-phosphothreonine (anti-PT) and anti-phosphotyrosine (anti-PY) monoclonal antibodies. Molecular-mass markers (kDa) are shown in the left hand margin.
The PP2C encoded by ABI1 has been implicated in the regulation of kinase signalling during seed development [29,30], and the abi1-1 mutant is known to exhibit pleiotropic effects. However, specific ABI1 targets have not yet been identified [33]. If cruciferin is a target for ABI1, loss of PP2C activity in the abi1-1 mutant should increase the extent of cruciferin phosphorylation. Indeed, the abi1-1 mutant exhibits hyperphosphorylation compared with the wild-type. Serine and threonine are the most frequently phosphorylated amino acids, representing more than 95% of all phosphorylated residues. This distribution is in agreement with our observations for A. thaliana seed proteins. As shown in Figure 3, proteins having phosphoserine and phosphothreonine were the most abundant forms, with proteins possessing phosphotyrosine being present at an appreciably lower level. More proteins possessing phosphoserine and phosphothreonine residues were observed with the abi1-1 mutant, whereas the number of proteins containing phosphotyrosine residues was similar in mutant and wild-type.
A total of 314 and 183 protein spots reacting with the phosphorylation-specific antibodies were excised from the abi1-1 mutant and wild-type samples, respectively, and analysed by MS. It was found that 93 protein spots (30%) from abi1-1 mutant and 17 protein spots (9%) from wild-type seeds were phosphorylated. The other proteins in which phosphorylation sites could be detected, using the same experimental conditions, were limited to glyceraldehyde-3-phosphate dehydrogenase (AT1G13440.1), two cupin family proteins (AT3G22640.1 and AT2G28490.1) and an unknown protein (AT5G63100.1), suggesting that cruciferin was the predominant phosphorylated protein in A. thaliana seeds. LC–MS/MS analysis identified a total of 19 phosphopeptides encompassing 20 phosphorylation sites in cruciferin, of which eight resided in the α-subunits and 11 in the β-subunits (Table 1). The peptide ADVYKPQLGYISTLNSYDLPILR may be phosphorylated at either Tyr10 or Ser12. MS/MS analysis was unable to confirm whether phosphorylation occurred at Thr8 or Ser12 in peptide VVPGCAETFMDSQPMQGQQQGQPWQGR, or at Tyr14 or Tyr20 in peptide CTENLDDPSDADVYKPSLGYISTLNSYNLPILR. On the basis of sequence alignments, however, it seems most likely that these two peptides would be phosphorylated at Thr8 and Tyr20 respectively, as indicated in the Figure 1. Thus, of the 20 phosphorylation sites identified, nine were located on serine, eight on threonine, and three on tyrosine residues. All four identified isoforms of cruciferin were found to be phosphorylated. Furthermore, all 20 sites were found in cruciferins isolated from abi1-1 mutant seeds, whereas only six were identified in wild-type samples (Table 1), suggesting that cruciferin is indeed a target for ABI1. These results show that cruciferin possesses multiple phosphorylation sites and exhibits complex phosphorylation-site heterogeneity, with the potential to generate a remarkable number of variants. Indeed, the diversity of cruciferin isoforms observed on 2-DE gels vastly exceeded that predicted from the various gene models, showing wide variations in pI values (Figure 3).
Table 1. Phosphorylation sites identified within A. thaliana wild-type and abi1-1 mutant cruciferins by electrospray ionization MS/MS.
pS, phosphoserine; pT, phosphothreonine; pY, phosphotyrosine.
| Peptide and phosphorylation site(s) | Locus* | Subunit | Source |
|---|---|---|---|
| EAPFPNACHFpSQINSLAPAQATK | At1g03890.1 | α | abi1-1 |
| pSEAGQIEYWDHNHPQLR | At4g28520.1 | α | abi1-1 |
| VFHLAGNNQQGGFGGpSQQQQEQK | At4g28520.1 | α | abi1-1 |
| VVPGCAEpTFMDpSQPMQGQQQGQPWQGR† | At4g28520.1 | α | abi1-1 |
| IpSYVVQGTGISGR | At4g28520.1 | α | WT/abi1-1 |
| VIPGCAEpTFMESPVFGEGQGQGQSQGFR | At1g03880.1 | α | WT/abi1-1 |
| VIPGCAEpTFQDSSEFQPR | At5g44120.3 | α | WT/abi1-1 |
| pYVIEQGGLYLPTFFTSPK | At4g28520.1 | α | WT/abi1-1 |
| FpSTIETTLTHSSPMSYGRPR | At1g03880.1 | β | WT/abi1-1 |
| VFDQEISpSGQLLVVPQGFSVMK | At1g03880.1 | β | WT/abi1-1 |
| ADVYKPQLGYIpSTLNSYDLPILR | At5g44120.3 | β | abi1-1 |
| ADVYKPQLGpYISTLNSYDLPILR | At5g44120.3 | β | abi1-1 |
| ALPLEVIpSNGFQISPEEAR | At4g28520.1 | β | abi1-1 |
| CTENLDDPSDADVpYKPSLGpYISTLNSYNLPILR‡ | At1g03880.1 | β | abi1-1 |
| FNpTLETTLTR | At4g28520.1 | β | abi1-1 |
| GLPLEVIpTNGFQISPEEAR | At5g44120.3 | β | abi1-1 |
| GLPLEVIpTNGYQISPEEAK | At1g03880.1 | β | abi1-1 |
| pTNENAMISTLAGR | At4g28520.1 | β | abi1-1 |
| pTNENAQVNTLAGR | At1g03880.1 | β | abi1-1 |
| VTpSVNSYTLPILEYVR | At4g28520.1 | β | abi1-1 |
* Denotes locus and gene model corresponding to cruciferin from which the tryptic peptide was generated.
† Phosphorylation at Thr8 or Ser12 is not distinguishable in the present study.
‡ Phosphorylation at Tyr14 or Tyr20 is not distinguishable in the present study.
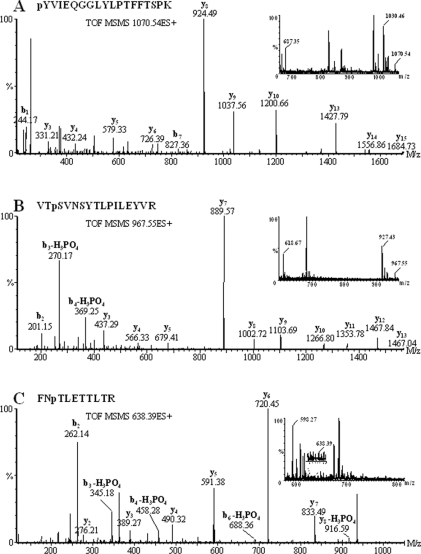
Examples of how specific phosphorylated serine, threonine and tyrosine residues were identified are shown in Figure 4. Figure 4(A) shows the product-ion MS/MS spectrum for the doubly charged precursor ion of the phosphopeptide pYVIEQGGLYLPTFFTSPK (m/z 1070.54). The corresponding molecular mass of the uncharged peptide (2139.08 Da) closely matches the predicted monoisotopic mass of non-phosphorylated form of this peptide (2059.07 Da) plus that of a single phosphate group (79.97 Da). The product-ion MS/MS spectrum shows a series of well-defined y- and b-type peptide fragment ions. In such low-energy collision-induced dissociation experiments, phosphate groups usually remain associated with the tyrosine moiety. The observed m/z values of phosphotyrosine-containing b1 (244.17) and b7 (827.36) ions closely match the expected values of 244.04 and 827.33, confirming the position of the phosphate moiety.
Figure 4. Product-ion MS/MS spectra of three cruciferin phosphopeptides.
Peaks corresponding to sequential loss of intact amino acid residues from the C or N terminus of the peptide are labelled as b- or y-type ions respectively. Peaks arising from neutral loss of H3PO4 (which confirms phosphorylation) are also indicated. Inserts show MS spectra generated by combining survey scans during chromatographic elution of both phosphorylated and non-phosphorylated forms of the intact phosphopeptide from which the corresponding MS/MS spectra were derived. The observed m/z value for each peak is indicated.
Figure 4(B) shows similar data for a doubly charged precursor ion of m/z 967.55 (1933.08 Da), which corresponds to the phosphopeptide VTpSVNSYTLPILEYVR (where pS is phosphoserine). Loss of phosphate as H3PO4 (97.98 Da) is a dominant feature in the product-ion MS/MS spectra of phosphoserine- and phosphothreonine-containing peptides. The observed m/z values of peaks corresponding to b3-H3PO4 (270.17) and b4-H3PO4 (369.25) match closely the predicted values of 270.14 and 369.21 based upon phosphorylation at the Ser3 residue. Unambiguous assignment of the b2 ion (m/z 201.15) excludes the possibility of alternative phosphorylation at the adjacent Thr2 residue. Similarly, the doubly charged precursor ion of m/z 638.39 (1274.78 Da) shown in Figure 4(C) corresponds to the phosphopeptide FNpTLETTLTR (1274.59 Da). In this case, peaks corresponding to b3-H3PO4 (m/z 345.16), b4-H3PO4 (458.24), b6-H3PO4 (688.33), and y8-H3PO4 (916.51) are consistent with phosphorylation at the Thr3 residue.
Although the factors governing peptide ionization/detection efficiencies are not well understood, and vary from one peptide to another, it has been shown that the responses obtained for phosphorylated and non-phosphorylated forms of a given peptide using MS can be used as a semiquantitative indicator of phosphorylation stoichiometry [34]. For example, by integrating the appropriate MS survey scans (insets to Figures 4A, 4B and 4C) it is possible to determine peak area ratios for doubly charged ions corresponding to phosphorylated and non-phosphylated forms of the same peptide. These values cannot be taken as direct measurements of phosphorylation stoichiometry, owing to differences in chargestate distribution and response factor for each form of the peptide. However, changes in the ratio of phosphorylated to non-phosphorylated peptide peak intensity could be used to study the dynamics of phosphorylation under different environmental or biological conditions. Hence, LC–MS/MS analysis has the potential to provide site-specific information as to the stoichiometry of cruciferin phosphorylation, in addition to confirming the locations of these sites.
DISCUSSION
Expression and heterogeneity of cruciferin
The genome of A. thaliana cv. Columbia contains four cruciferin loci from which eight types of cDNA generated by alternative mRNA splicing (gene models) have been identified. A fifth locus is present in the cv. Landsberg genome as a tandem duplication of At5g44120, and several amino acid substitutions arising from single nucleotide polymorphisms between alleles present in the two ecotypes exist. CRA1 is equivalent to AT5G44120.3, with amino acid substitutions Glu167→Gln/Val356→Glu, and CRB is equivalent to AT1G03880.1, with the Ala383→Arg substitution, while CRC is identical to AT4G28520.1. Our analysis revealed that three of the eight potential transcripts (At4g28520.1, At5g44120.3 and At1g3880.1) gave rise to the bulk of the cruciferin protein in seeds. The contribution from At1g3890.1 was much smaller, which is in keeping with the low abundance of this transcript [32]. The At4g28520.2 and At4g28520.3 transcripts encode proteins with internal deletions; however, tryptic peptides capable of distinguishing these variants from At4g28520.1 were not detected. The protein encoded by At4g28520.3 would also lack the internal Asn↓Gly β-cleavage site, and processing at this location is thought to be required for hexamer assembly [7]. The proteins encoded by At5g44120.1 and At5g44120.2 would be truncated, with deletions of 187 and 104 amino acid residues near the N-terminus of each protein respectively. Since these deletions span the signal-peptide region, and alternative signal sequences were not detected, it is unlikely that these proteins could be directed to the endoplasmic reticulum for subsequent deposition into PSVs.
Analysis of seed proteins by high-resolution 2-DE followed by staining with anti-cruciferin antiserum and phosphorylation antibodies revealed a much higher degree of cruciferin heterogeneity than was expected on the basis of the analysis described above. This same observation has also been made for the soybean (Glycine max) glycinin and β-conglycinin (the former being a functional orthologue of cruciferin) and was attributed to one or a few amino acid substitutions. Neither cruciferin (L. Wan, A. R. S. Ross, J. Yang, D. D. Hegedus and A. R. Kermode, unpublished work) nor soybean glycinin was found to be glycosylated [35,36]. Carbonylation of seed proteins, including cruciferin, has been observed; however, this oxidative process occurs only during seed germination and has been attributed to the generation of reactive oxygen species during the recovery of metabolic activity [37]. The pI values of cruciferin precursors ranged from 5.5 to 7.9. After processing, the resultant subunits had pI ranging from 5.7 to 8.9. A wide variety of proteins are phosphorylated in the mature seed [28]. The present study revealed that all four identified cruciferin isoforms were phosphorylated and that all three types of phosphophorylated amino acids (serine, threonine and tyrosine) were present, with phosphoserine and phosphothreonine accounting for most of the phosphorylated residues.
Implications of cruciferin phosphorylation for processing, assembly and mobilization
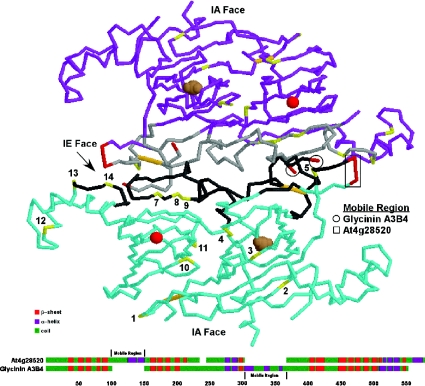
The four cruciferin proteins detected in A. thaliana seeds possess between 55 and 70 hydroxyamino acids with the potential for phosphorylation; however, alignment of these protein sequences revealed that the phosphorylated residues reside in only 14 distinct locations (Figure 1). Furthermore, hydroxyamino acids were found at the same, or immediately adjacent, locations in eight of the 14 corresponding sites in the soybean A3B4 glycinin. Consequently we were able to superimpose the locations of the phosphorylated residues on to the known three-dimensional structure of the soybean glycinin [7]. Interestingly, seven of the 14 sites mapped to regions on the interchain disulfide-containing (IE) face involved in the face-to-face interaction between trimers required for hexamer assembly (Figure 5).
Figure 5. Superimposition of A. thaliana cruciferin phosphorylation sites on the glycinin tertiary structure.
Each glycinin and cruciferin molecule consists of an α- and β-peptide linked together by intrachain (IA) and interchain (IE) disulfide bonds (orange) that denote the two faces. Although the mature proteins form hexameric structures, for the sake of clarity only two interacting glycinin A3B4 molecules (blue and purple) are depicted. The phosphorylated residues (yellow and numbered according to Figure 1) tend to cluster along the IE face and associate with the binding regions (grey) involved in subunit interactions. Magnesium (red sphere) and carbonate (brown spheres) ions with the barrel of the cupin domains are shown. A comparison of the predicted secondary structures of the cruciferin encoded by At4g28520 and glycinin A3B4 is provided below. Each protein contains a highly charged loop (red) extending from the regions indicated, and both regions are located in the same general vicinity on the three-dimensional structure. These may serve as the mobile regions preventing non-specific interactions with the IE face prior to processing and hexamer assembly.
The pro-globulin trimer is formed within the endoplasmic reticulum, with the hexamer appearing during the latter stages of localization [38,39]. Requisite for hexamer assembly is proteolytic cleavage (at the β-cleavage site) to generate α- and β-subunits, each of which contains a single cupin domain, and the subsequent linkage of these subunits by an interchain disulfide bond [40,41]. The processing appears to direct the relocalization of an acidic mobile disordered region (residues 252–325 in glycinin A3B4) from the surface of the IE face to the side of the protein. The mobile disordered region is proposed to prevent non-specific interaction between the positively charged IE face and negatively charged IA (intrachain disulfide-containing) face of the trimers prior to hexamer formation [7]. Although a long acidic region is not present in the corresponding location in any of the cruciferins, a glutamine-rich variable region is present within the α-subunit of some isoforms. Modelling of the cruciferin structure based on that of the secondary and tertiary structure of pro-glycinin [6] revealed that the glutamine-rich region also resides near the IE face in a location equivalent to that of the A3B4 mobile region and, as such, may serve to regulate hexamer formation in a similar manner.
Several hypotheses can be put forward regarding the role of phosphorylation in the assembly and/or mobilization of cruciferins. Phosphorylation sites 4 and 11 (Figure 1) are not associated with the IE face, but oppose and offset one another in the cleft between the individual α- and β-subunit cupin domains. A negative charge at one or both of these sites would clearly affect tertiary and quaternary interactions. Similarly, an increase in the overall negative charge on the IE face should serve to stabilize the interaction between the face and the positively charged glutamine-rich putative mobile region. It is plausible that removal of the phosphate residues by phosphatases, such as ABI1, accompanies the proteolytic cleavage and interchain disulfide-bond formation. Dephosphorylation of residues on the trimer-interacting face could serve to destabilize interaction with the glutamine-rich mobile region. This would promote the movement of the mobile region from the interacting face and allow hexamers to assemble.
The heterogeneity of 11S globulin may be a fundamental aspect of hexamer assembly and function in vivo, since some of the cruciferin and glycinin isoforms do not appear to possess such mobile regions. In addition, our analyses showed that only a proportion of the hydroxyamino acids at any one of the potential phosphorylation sites were modified. As has been suggested for glycinin, limiting the degree of trimer interaction through subunit heterogeneity or electrostatic repulsion on the IE face may promote disassembly and mobilization of seed storage proteins during germination [7]. Such interactions will also be affected by changes in pH and ionic strength that occur within the PSV after imbibition [42,43].
Is cruciferin an in vivo target for the ABI1 protein phosphatase?
The phytohormone ABA regulates diverse physiological processes, including seed maturation and dormancy, as well as adaptation of vegetative tissues to environmental stresses such as drought and high salinity. A series of A. thaliana mutants, designated abi, exhibit decreased sensitivity to ABA with respect to its inhibitory effects on germination and/or seedling growth and defects in molecular responses to exogenously applied ABA in vegetative tissues [44]. The ABI1 locus encodes a unique type of protein phosphatase in that the C-terminal domain possesses serine/threonine protein phosphatase activity similar to that of PP2Cs (class 2C phosphatases), whereas the N-terminal domain contains a calcium-binding EF-hand motif that generally occurs only in the heterodimeric PP2B-type serine/threonine phosphatases. A single point mutation in the abi1-1 mutant significantly reduces PP2C activity [29,30], and genome-wide gene expression profiling indicates that ABA gene regulation is impaired [45]. Although recombinant fusion proteins containing either the whole ABI1 protein or only the phosphatase domain display in vitro phosphatase activity towards 32P-labelled casein [46,47]; the in vivo targets have not yet been identified.
We have shown that the site-specific stoichiometry of cruciferin phosphorylation is altered in the abi-1 mutant, suggesting that this storage protein is an in vivo ABI1 target. This appears to be at odds with its presumed role in ABA-mediated signalling; however, it is possible that ABI1 may regulate a protein kinase/phosphatase duad responsible for cruciferin phosphorylation and dephosphorylation in an ABA-dependent manner. Testing of this hypothesis by manipulation of the ABA signalling pathway, including regulation of ABI1 activity, will be the goal of future research.
While access to the entire A. thaliana genome and improvements in proteomics techniques have greatly enhanced our ability to answer fundamental biological questions, many cellular processes occur and are regulated without changes in either gene or protein expression. Our attempts to identify the patterns of protein phosphorylation during seed germination were complicated by the discovery that the cruciferin represents the most abundant phosphoprotein in seeds. Prior depletion of cruciferin from the samples by immunoprecipitation and 32P metabolic labelling of newly phosphorylated proteins have reduced such interference and are now making these experiments possible. Meanwhile, the results of the present study make a significant contribution to our understanding of the structure of globulin-like seed storage proteins and the processes involved in their maturation and mobilization.
Acknowledgments
We thank Professor Derek Lydiate (Saskatoon Research Centre) for valuable insights regarding the possible role of phosphorylation in mobilizing seed storage proteins. This research was supported by grants from PENCE (Canadian Protein Engineering Network of Centers of Excellence), NRC-GHI (National Research Council Genomics and Health Initiative) and Genome Canada-Genome Prairie. Funding and operational support for proteomics infrastructure at the Plant Biotechnology Institute were provided by the Provincial Government of Saskatchewan and the National Research Council of Canada. This manuscript is contribution no. 48411 from the National Research Council of Canada.
References
- 1.Duranti M., Guerrieri N., Takahashi T., Cerletti P. The legumin-like storage protein of Lupinus albus seeds. Phytochemistry. 1988;27:15–23. [Google Scholar]
- 2.Lawrence M. C., Izard T., Beuchat M., Blagrove R. J., Colman P. M. Structure of phaseolin at 2.2 Å resolution. Implications for a common vicilin/legumin structure and the genetic engineering of seed storage proteins. J. Mol. Biol. 1994;238:748–776. doi: 10.1006/jmbi.1994.1333. [DOI] [PubMed] [Google Scholar]
- 3.Ko T. P., Ng J. D., McPherson A. The three-dimensional structure of canavalin from jack bean (Canavalia ensiformis) Plant Physiol. 1993;101:729–744. doi: 10.1104/pp.101.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maruyama N., Adachi M., Takahashi K., Yagasaki K., Kohno M., Takenaka Y., Okuda E., Nakagawa S., Mikami B., Utsumi S. Crystal structures of recombinant and native soybean β-conglycinin beta homotrimers. Eur. J. Biochem. 2001;268:3595–3604. doi: 10.1046/j.1432-1327.2001.02268.x. [DOI] [PubMed] [Google Scholar]
- 5.Woo E. J., Dunwell J. M., Goodenough P. W., Marvier A. C., Pickersgill R. W. Germin is a manganese containing homohexamer with oxalate oxidase and superoxide dismutase activities. Nat. Struct. Biol. 2000;7:1036–1040. doi: 10.1038/80954. [DOI] [PubMed] [Google Scholar]
- 6.Adachi M., Takenaka Y., Gidamis A. B., Mikami B., Utsumi S. Crystal structure of soybean proglycinin alaB1b homotrimer. J. Mol. Biol. 2001;305:291–305. doi: 10.1006/jmbi.2000.4310. [DOI] [PubMed] [Google Scholar]
- 7.Adachi M., Kanamori J., Masuda T., Yagasaki K., Kitamura K., Mikami B., Utsumi S. Crystal structure of soybean 11 S globulin: glycinin A3B4 homohexamer. Proc. Natl. Acad. Sci. U.S.A. 2003;100:7395–7400. doi: 10.1073/pnas.0832158100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shutov A. D., Baumlein H., Blattner F. R., Muntz K. Storage and mobilization as antagonistic functional constraints on seed storage globulin evolution. J. Exp. Bot. 2003;54:1645–1654. doi: 10.1093/jxb/erg165. [DOI] [PubMed] [Google Scholar]
- 9.Dunwell J. M., Khuri S., Gane P. J. Microbial relatives of the seed storage proteins of higher plants: conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiol. Mol. Biol. Rev. 2000;64:153–179. doi: 10.1128/mmbr.64.1.153-179.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pang P. P., Pruitt R. E., Meyerowitz E. M. Molecular cloning, genomic organization, expression and evolution of 12S seed storage genes of Arabidopsis thaliana. Plant Mol. Biol. 1988;11:805–820. doi: 10.1007/BF00019521. [DOI] [PubMed] [Google Scholar]
- 11.Sjodahl S., Gustavsson H. O., Rodin J., Lenman M., Hoglund A. S., Rask L. Cruciferin gene families are expressed coordinately but with tissue-specific differences during Brassica napus seed development. Plant. Mol. Biol. 1993;23:1165–1176. doi: 10.1007/BF00042350. [DOI] [PubMed] [Google Scholar]
- 12.Wan L. L., Xia Q., Qiu X., Selvaraj G. Early stages of seed development in Brassica napus: a seed coat-specific cysteine proteinase associated with programmed cell death of the inner integument. Plant. J. 2002;30:1–10. doi: 10.1046/j.1365-313x.2002.01262.x. [DOI] [PubMed] [Google Scholar]
- 13.Kohno Murase J., Murase M., Ichikawa H., Imamura J. Effects of an antisense napin gene on seed storage compounds in transgenic Brassica napus seeds. Plant Mol. Biol. 1994;26:1115–1124. doi: 10.1007/BF00040693. [DOI] [PubMed] [Google Scholar]
- 14.Robinson D. G., Baumer M., Hinz G., Hohl I. Vesicle transfer of storage proteins to the vacuole: The role of the Golgi apparatus and multivesicular bodies. J. Plant Physiol. 1998;152:659–667. [Google Scholar]
- 15.HaraNishimura I., Shimada T., Hatano K., Takeuchi Y., Nishimura M. Transport of storage proteins to protein storage vacuoles is mediated by large precursor-accumulating vesicles. Plant Cell. 1998;10:825–836. doi: 10.1105/tpc.10.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitsuhashi N., Hayashi Y., Koumoto Y., Shimada T., Fukasawa Akada T., Nishimura M., Hara Nishimura I. A novel membrane protein that is transported to protein storage vacuoles via precursor-accumulating vesicles. Plant Cell. 2001;13:2361–2372. doi: 10.1105/tpc.010171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimada T., Watanabe E., Tamura K., Hayashi Y., Nishimura M., Hara Nishimura I. A vacuolar sorting receptor PV72 on the membrane of vesicles that accumulate precursors of seed storage proteins (PAC vesicles) Plant Cell Physiol. 2002;43:1086–1095. doi: 10.1093/pcp/pcf152. [DOI] [PubMed] [Google Scholar]
- 18.Shewry P. R., Napier J. A., Tatham A. S. Seed storage proteins: structures and biosynthesis. Plant Cell. 1995;7:945–956. doi: 10.1105/tpc.7.7.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sjodahl S., Rodin J., Rask L. Characterization of the 12 S globulin complex of Brassica napus. Evolutionary relationship to other 11–12 S storage globulins. Eur. J. Biochem. 1991;196:617–621. doi: 10.1111/j.1432-1033.1991.tb15857.x. [DOI] [PubMed] [Google Scholar]
- 20.Inquello V., Raymond J., Azanza J. L. Disulfide interchange reactions in 11 S globulin subunits of Cruciferae seeds. Relationships to gene families. Eur. J. Biochem. 1993;217:891–895. doi: 10.1111/j.1432-1033.1993.tb18318.x. [DOI] [PubMed] [Google Scholar]
- 21.Kinoshita T., Nishimura M., Hara Nishimura I. Homologues of a vacuolar processing enzyme that are expressed in different organs in Arabidopsis thaliana. Plant Mol. Biol. 1995;29:81–89. doi: 10.1007/BF00019120. [DOI] [PubMed] [Google Scholar]
- 22.Kinoshita T., Nishimura M., Hara Nishimura I. The sequence and expression of the γ-VPE gene, one member of a family of three genes for vacuolar processing enzymes in Arabidopsis thaliana. Plant Cell Physiol. 1995;36:1555–1562. [PubMed] [Google Scholar]
- 23.Gruis D. F., Selinger D. A., Curran J. M., Jung R. Redundant proteolytic mechanisms process seed storage proteins in the absence of seed-type members of the vacuolar processing enzyme family of cysteine proteases. Plant Cell. 2002;14:2863–2882. doi: 10.1105/tpc.005009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimada T., Yamada K., Kataoka M., Nakaune S., Koumoto Y., Kuroyanagi M., Tabata S., Kato T., Shinozaki K., Seki M., et al. Vacuolar processing enzymes are essential for proper processing of seed storage proteins in Arabidopsis thaliana. J. Biol. Chem. 2003;278:32292–32299. doi: 10.1074/jbc.M305740200. [DOI] [PubMed] [Google Scholar]
- 25.McLachlin D. T., Chait B. T. Analysis of phosphorylated proteins and peptides by mass spectrometry. Curr. Opin. Chem. Biol. 2001;5:591–602. doi: 10.1016/s1367-5931(00)00250-7. [DOI] [PubMed] [Google Scholar]
- 26.Beausoleil S. A., Jedrychowski M., Schwartz D., Elias J. E., Villen J., Li J. X., Cohn M. A., Cantley L. C., Gygi S. P. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. U.S.A. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Napper S., Kindrachuk J., Olson D. J. H., Ambrose S. J., Dereniwsky C., Ross A. R. S. Selective extraction and characterization of a histidine-phosphorylated peptide using immobilized copper(II) ion affinity chromatography and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Chem. 2003;75:1741–1747. doi: 10.1021/ac026340f. [DOI] [PubMed] [Google Scholar]
- 28.Irar S., Oliveira E., Pages M., Goday A. Towards the identification of late-embryogenic-abundant phosphoproteome in Arabidopsis by 2-DE and MS. Proteomics. 2006;6:S175–S185. doi: 10.1002/pmic.200500387. [DOI] [PubMed] [Google Scholar]
- 29.Leung J., Bouvier Durand M., Morris P. C., Guerrier D., Chefdor F., Giraudat J. Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science. 1994;264:1448–1452. doi: 10.1126/science.7910981. [DOI] [PubMed] [Google Scholar]
- 30.Meyer K., Leube M. P., Grill E. A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science. 1994;264:1452–1455. doi: 10.1126/science.8197457. [DOI] [PubMed] [Google Scholar]
- 31.Mortz E., Krogh T. N., Vorum H., Görg A. Improved silver staining protocols for high sensitivity protein identification using matrix-assisted laser desorption/ionization-time of flight analysis. Proteomics. 2001;1:1359–1363. doi: 10.1002/1615-9861(200111)1:11<1359::AID-PROT1359>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 32.White J. A., Todd T., Newman T., Focks N., Girke T., de Ilarduya O. M., Jaworski J. G., Ohlrogge J. B., Benning C. A new set of Arabidopsis expressed sequence tags from developing seeds. The metabolic pathway from carbohydrates to seed oil. Plant Physiol. 2000;124:1582–1594. doi: 10.1104/pp.124.4.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Y., Sanchez J. P., Lopez Molina L., Himmelbach A., Grill E., Chua N. H. The abi1-1 mutation blocks ABA signaling downstream of cADPR action. Plant J. 2003;34:307–315. doi: 10.1046/j.1365-313x.2003.01721.x. [DOI] [PubMed] [Google Scholar]
- 34.Steen H., Jebanathirajah J. A., Rush J., Morrice N., Kirschner M. W. Phosphorylation analysis by mass spectrometry. Mol. Cell. Proteomics. 2006;5:172–181. doi: 10.1074/mcp.M500135-MCP200. [DOI] [PubMed] [Google Scholar]
- 35.Maruyama N., Fukuda T., Saka S., Inui N., Kotoh J., Miyagawa M., Hayashi M., Sawada M., Moriyama T., Utsumi S. Molecular and structural analysis of electrophoretic variants of soybean seed storage proteins. Phytochemistry. 2003;64:701–708. doi: 10.1016/s0031-9422(03)00385-6. [DOI] [PubMed] [Google Scholar]
- 36.Fukuda T., Maruyama N., Kanazawa A., Abe J., Shimamoto Y., Hiemori M., Tsuji H., Tanisaka T., Utsumi S. Molecular analysis and physicochemical properties of electrophoretic variants of wild soybean Glycine soja storage proteins. J. Agric. Food Chem. 2005;53:3658–3665. doi: 10.1021/jf0479620. [DOI] [PubMed] [Google Scholar]
- 37.Job C., Rajjou L., Lovigny Y., Belghazi M., Job D. Patterns of protein oxidation in Arabidopsis seeds and during germination. Plant Physiol. 2005;138:790–802. doi: 10.1104/pp.105.062778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herman E. M., Larkins B. A. Protein storage bodies and vacuoles. Plant Cell. 1999;11:601–613. doi: 10.1105/tpc.11.4.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimada T., Fuji K., Tamura K., Kondo M., Nishimura M., Hara Nishimura I. Vacuolar sorting receptor for seed storage proteins in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 2003;100:16095–16100. doi: 10.1073/pnas.2530568100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jung R., Nam Y. W., Saalbach I., Muntz K., Nielsen N. C. Role of the sulfhydryl redox state and disulfide bonds in processing and assembly of 11 S seed globulins. Plant Cell. 1997;9:2037–2050. doi: 10.1105/tpc.9.11.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jung R., Scott M. P., Nam Y. W., Beaman T. W., Bassuner R., Saalbach I., Muntz K., Nielsen N. C. The role of proteolysis in the processing and assembly of 11 S seed globulins. Plant Cell. 1998;10:343–357. doi: 10.1105/tpc.10.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okamoto T., Yuki A., Mitsuhashi N., Minamikawa T. Asparaginyl endopeptidase (VmPE-1) and autocatalytic processing synergistically activate the vacuolar cysteine proteinase (SH-EP) Eur. J. Biochem. 1999;264:223–232. doi: 10.1046/j.1432-1327.1999.00618.x. [DOI] [PubMed] [Google Scholar]
- 43.Muntz K., Belozersky M. A., Dunaevsky Y. E., Schlereth A., Tiedemann J. Stored proteinases and the initiation of storage protein mobilization in seeds during germination and seedling growth. J. Exp. Bot. 2001;52:1741–1752. doi: 10.1093/jexbot/52.362.1741. [DOI] [PubMed] [Google Scholar]
- 44.Koornneef M., Reuling G., Karseen C. M. The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol. Plant. 1984;61:377–383. [Google Scholar]
- 45.Hoth S., Morgante M., Sanchez J. P., Hanafey M. K., Tingey S. V., Chua N. H. Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J. Cell Sci. 2002;115:4891–4900. doi: 10.1242/jcs.00175. [DOI] [PubMed] [Google Scholar]
- 46.Bertauche N., Leung J., Giraudat J. Protein phosphatase activity of abscisic acid insensitive 1 (ABI1) protein from Arabidopsis thaliana. Eur. J. Biochem. 1996;241:193–200. doi: 10.1111/j.1432-1033.1996.0193t.x. [DOI] [PubMed] [Google Scholar]
- 47.Leube M. P., Grill E., Amrhein N. ABI1 of Arabidopsis is a protein serine/threonine phosphatase highly regulated by the proton and magnesium ion concentration. FEBS Lett. 1998;424:100–104. doi: 10.1016/s0014-5793(98)00149-5. [DOI] [PubMed] [Google Scholar]