Abstract
Ligand activation of Notch leads to the release of Notch IC (the intracellular receptor domain), which translocates to the nucleus and interacts with the DNA-binding protein CSL to control expression of specific target genes. In addition to ligand-mediated activation, Notch signalling can be further modulated by interactions of Notch IC with a number of other proteins. MAML1 has previously been shown to act co-operatively with the histone acetyltransferase p300 in Notch IC-mediated transcription. In the present study we show that the N-terminal domain of MAML1 directly interacts with both p300 and histones, and the p300–MAML1 complex specifically acetylates histone H3 and H4 tails in chromatin. Furthermore, p300 acetylates MAML1 and evolutionarily conserved lysine residues in the MAML1 N-terminus are direct substrates for p300-mediated acetylation. The N-terminal domain of MAML1 contains a proline repeat motif (PXPAAPAP) that was previously shown to be present in p53 and important for the p300–p53 interaction. We show that the MAML1 proline repeat motif interacts with p300 and enhances the activity of the MAML1 N-terminus in vivo. These findings suggest that the N-terminal domain of MAML1 plays an important role in Notch-regulated transcription, by direct interactions with Notch, p300 and histones.
Keywords: acetylation, chromatin, mastermind-like protein-1 (MAML1), Notch, p300, transcription
Abbreviations: Acf-1, ATP-dependent chromatin assembly and remodelling factor 1; ANK, ankyrin; CBF1, CCAAT-binding factor 1; C/H, cysteine/histidine; CSL, CBF1/Suppressor of Hairless/Lag-1; DTT, dithiothreitol; EMSA, electrophoretic mobility-shift assay; GR, glucocoticoid receptor; HAT, histone acetyltransferase; HDAC, histone deacetylase; HES, Hairy and Enhancer of Split; Hsp90, heat-shock protein of 90 kDa; iκBα, inhibitory κBα; ISWI, imitation-SWI protein; MEF2C, MADS box transcription enhancer factor 2; MAML1, mastermind-like 1; NAP1, nucleosome assembly protein 1; NF-κB, nuclear factor κB; Notch IC, Notch intracellular receptor domain; RAM, RBP-jk (recombination signal-binding protein 1 for j-kappa)-associated molecule
INTRODUCTION
The Notch signalling pathway is an evolutionarily conserved system for cell–cell communication and plays an important role in developmental processes by influencing cellular proliferation, differentiation and apoptosis. Notch signalling controls fate decisions in many cell types, including those of the nervous system, muscles and the haematopoetic system [1–3]. Furthermore, altered Notch signalling has been associated with human cancers, especially T-cell acute lymphoblastic leukaemia (for a review, see [4]). Vertebrates contain four Notch receptors (Notch 1–4), and the receptor is a single transmembrane-spanning protein that undergoes a series of proteolytic events. Interactions with Notch ligands (Jagged1, Jagged2, Delta1 and Delta-like 1, 3 and 4) on neighboring cells result in the release of the Notch IC (intracellular domain), which translocates to the nucleus, where it controls transcription of specific target genes through its interaction with the highly conserved DNA-binding protein CSL [also known as RBP-Jk (recombination signal-binding protein 1 for j-kappa) or CBF1 in vertebrates, Suppressor of Hairless in the fruitfly Drosophila and Lag-1 in the nematode worm Caenorhabtidis elegans). In the absence of Notch IC in the nucleus, CSL binds to Notch-regulated target genes and represses transcription by interacting with the co-repressors SMRT (silencing mediator of retinoid and thyroid receptors) [5], KyoT2 [6], CIR (CBF1-interacting co-repressor) [7] and SHARP [SMRT/HDAC1 (histone deacetylase 1)-associated repressor protein] [8]. When Notch IC interacts with CSL, the formation of the co-repressor–CSL complex is disrupted, and co-activators, such as PCAF and GCN5 [9], human MAMLs (mastermind-like proteins) [10,11] and p300 [12] are recruited. The best-characterized target genes are the HES-1 (Hairy and Enhancer of Split 1) and HES-5 genes, which are mammalian homologues of the Drosophila Enhancer of Split genes [1,13]. More recently, the genes for p21, cyclin D1, HERP (homocysteine-induced endoplasmic-reticulum protein) and mitogen-activated protein kinase phosphatase 1 have been reported to be regulated by Notch (see the references cited in [3]).
The protein MAML1 was cloned on the basis of its homology with the Drosophila Mastermind [14], a neurogenic gene that has been genetically linked to Notch function [15–17]. MAML1 is a potentiator of Notch signalling for all four Notch receptors, binds to the ANK (ankyrin) repeat region of Notch IC and is believed to stabilize the interaction between Notch IC and CSL [14]. Recently, the crystal structures of the DNA-bound CSL–Notch–MAML1 complex with human proteins [18] and proteins from C. elegans [19] were reported. The structures reveal that CSL and the ANK domain in Notch form a binding pocket for a polypeptide composed of two long α-helices in the MAML1 N-terminus [18,19]. Although it is the main function of the RAM [RBP-jk (recombination signal-binding protein 1 for j-kappa)-associated molecule] domain to mediate the Notch IC interaction with CSL, the ANK domain in Notch also participates in CSL binding (see the references cited in [18]) and is crucial for assembly of a functional transcriptional activation complex (see [20] and the references cited in [18]. In addition to function as a co-activator for Notch, MAML1 was recently shown to potentiate MEF2C (MADS box transcription enhancer factor 2) transcriptional activation in myogenesis [21]. Two additional members of the MAML family have also been identified, namely MAML2 and MAML3, and all of the MAML proteins appear to function specifically in Notch signalling [10,11]. The MAML genes are expressed in all adult tissues, but have distinct expression patterns during development in mouse [10,11]. It has previously been documented that MAML1 potentiates Notch IC-mediated transcription from chromatin templates in vitro by recruiting p300 to a DNA–CSL–Notch complex [22,23]. In the present study we have continued to investigate the interplay between MAML1 and p300 in Notch-mediated transcription. Our data show that MAML1 activates transcription by directly interacting with histones and p300, and the p300–MAML1 complex specifically acetylates histone H3 and H4 tails in chromatin. Furthermore, MAML1 is acetylated by p300 and a proline repeat motif (PAPAAPAP) in MAML1 seems to be important for interaction with MAML1.
EXPERIMENTAL
Plasmids
cDNAs encoding MAML1 residues 1–300, 309–625, 499–804 and 701–1016 were amplified with PCR and subcloned into pVL1393 (BD Biosciences) after FLAG-tag sequences or subcloned into pGEX plasmids (Pharmacia). The BacVector 3000 system from Novagen was used to create baculovirus from the pVL1393-FLAG-MAML1 constructs described above. cDNA encoding full-length MAML1 was amplified with PCR from pVL1393-FLAG-MAML1 and subcloned into pCDNA. cDNAs encoding MAML1 residues 1–300 and 1–81/87–306 were subcloned into PSVSPORT (Invitrogen). cDNAs encoding p300 residues 1–672, 651–1150, 1141–1700, 1672–2414, 1647–1818 and 2042–2414 were amplified with PCR and subcloned into pET23a in front of His-tag sequences (Novagen). cDNA encoding the intracellular domain of human Notch1 (1764–2556) was subcloned from pCDNA3-hNotch1 (a gift from Dr Tom Kadesch) into pBIND (Promega).
Expression and purification of proteins
FLAG-tagged proteins were expressed in Sf9 (Spodoptera frugiperda) cells via baculovirus and purified as described in [23]. GST-tagged and His-tagged proteins were expressed in the E. coli strain BL21 and purified on glutathione–Sepharose 4B and Ni2+-nitrilotriacetate columns (Amersham Biosciences) respectively following the manufacturer's protocol. Expression of intact and tailless Xenopus laevis recombinant histones and the preparation of histone octamers was essentially done as previously described [24].
Chromatin assembly and in vitro transcription assay
The plasmid containing 12 binding sites for CSL was assembled into chromatin by using purified recombinant Drosophila Acf-1 (ATP-dependent chromatin assembly and remodelling factor 1), ISWI (imitation-SWI protein) and NAP1 (nucleosome assembly protein 1) as described previously [23]. For the transcription reactions, 70 ng of the chromatin-assembled template was preincubated with 10 ng of Notch1 IC, 50 ng of CSL, 100 ng of MAML1, 100 ng of p300 and 3 μM acetyl-CoA as indicated in Figure 1(E) below. Approx. 50 μg of HeLa nuclear extract was then added per reaction and transcription was initiated by addition of 0.4 mM NTPs. For primer-extension analysis of transcripts, a 32P-labelled probe (25000–50000 c.p.m.) extending from positions +86 to +110 of the adenovirus E4 promoter was used. Purified reverse-transcription products were analysed on 8% (w/v) polyacrylamide gels containing 7 M urea, and quantified with a PhosphorImager (Molecular Dynamics).
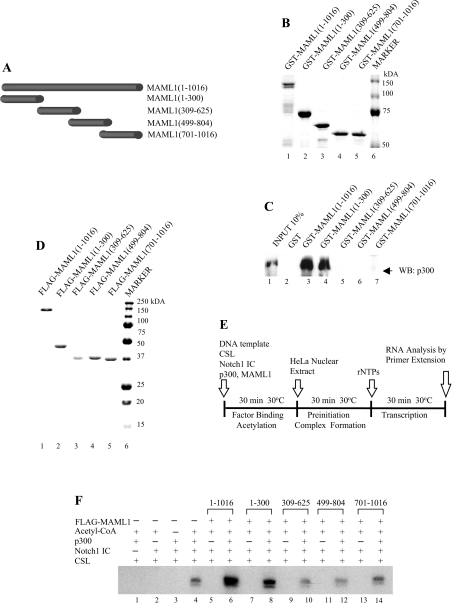
Figure 1. p300 and MAML1 act co-operatively to mediate transcriptional activation by Notch1 IC from chromatin templates.
(A) Schematic of full-length MAML1(1–1016) and its derivatives. (B) Coomassie Blue-stained SDS/polyacrylamide gel showing purified GST-tagged MAML1 proteins. (C) The MAML1 N-terminus directly interacts with p300. GST-MAML1 domains (indicated at the top), coupled to glutathione–Sepharose beads were incubated with FLAG-tagged p300 and interacting p300 was monitored by Western immunoblot (WB). The input represents 10% of the p300 used in each binding reaction. (D) Coomassie Blue-stained SDS/polyacrylamide gel showing purified FLAG-tagged MAML1 proteins. (E) Schematic of the in vitro transcription assay. (F) Transcription assay with chromatin templates. Templates were incubated with CSL, Notch1 IC, p300, MAML1 and acetyl-CoA as indicated.
Protein interaction assays
In GST–MAML1 interaction assays, approx. 2 μg of purified GST-tagged MAML1 bound to 20 μl of glutathione–Sepharose beads was incubated with 1 μg of purified FLAG-tagged Notch1 IC or 1 μg of FLAG-tagged p300 or 1 μg of His-tagged p300 in buffer A [50 mM Tris/HCl, pH 7.5, 150 mM KCl, 10% (v/v) glycerol, 0.1% Nonidet P40, 0.2 mg/ml BSA, 1 mM DTT (dithiothreitol) and 1 mM PMSF] or 200 μl of whole cell extract from HEK-293 cells transfected with p300. After centrifugation of the suspension, the supernatant was removed and the beads were washed five times with 500 μl of buffer A. The proteins were eluted from the beads, resolved by SDS/PAGE and analysed by immunoblot with a monoclonal mouse anti-FLAG M2 antibody (Sigma), a polyclonal rabbit anti-p300 antibody (Santa Cruz Biotechnology) or a monoclonal mouse anti-His antibody (Amersham Biosciences). For the MAML1 interaction assay with histones, approx. 2 μg of GST-tagged histones bound to 20 μl of glutathione–Sepharose beads was incubated with 1 μg of purified FLAG–MAML1, or 5 μg of purified FLAG-tagged MAML1 bound to 10 μl of M2–agarose was incubated with 2 μg of purified calf thymus core histones (Sigma) in buffer A (above). Beads were washed five times with 500 μl of buffer A. Eluted proteins were resolved by SDS/PAGE and analysed by immunoblot with a monoclonal mouse anti-FLAG M2 antibody (Sigma) or a polyclonal rabbit antibody recognizing acetylated histone H3 (Upstate Biotechnology).
Protein acetylation assays
For acetylation of MAML1 by p300, 0.5 μg of MAML1 proteins were incubated with 50 ng of p300 and 3 μM [3H]acetyl-CoA in a 20 μl reaction volume in buffer B [50 mM Tris/HCl, pH 8.0, 50 mM KCl, 5% (v/v) glycerol, 0.1 mM EDTA, 1 mM DTT, 1 mM PMSF and 10 mM sodium butyrate] at 30 °C for 1 h, and the reaction mixture was subjected to SDS/PAGE and autoradiography. For acetylation of MAML1 peptides, reaction mixtures containing 10 μg of peptides (JPT Peptide Technologies), 50 ng of p300 and 3 μM [3H]acetyl-CoA in a 20 μl reaction volume in buffer B were incubated at 30 °C for 1 h, then spotted on to P81 phosphocellulose filter-paper circles (Whatman) as previously described [25], and the radioactivity was measured with a liquid-scintillation counter. Standard HAT assays with chromatin templates contained 700 ng of chromatin, 500 ng of RBP-Jk, 100 ng of Notch1 IC, 1 μg of p300, 1 μg of MAML1 and 3 μM [3H]acetyl-CoA. The HAT reaction mixtures were incubated at 30 °C for 1 h and were then subjected to SDS/PAGE and autoradiography.
Transient transfections
HeLa cells were transiently transfected with the plasmids pBIND-Notch1 IC, PSVSPORT-MAML1(1-300), PSVSPORT-MAML1(1-306Δ81-87) and the reporter plasmid pG5-luc using Boehringer Mannheim's FuGENE™ 6 transfection reagent. Cells were harvested after 48 h and the levels of luciferase were measured. For the MAML1 in vivo acetylation expreriments, HEK-293 cells were transiently transfected with pCDNA-MAML1 and pCMV-p300 using Lipofectamine™ 2000 (Invitrogen). The cells were harvested 26 h after transfection and lysed in Roche lysis buffer. The KCl concentration in the whole-cell lysate was adjusted to 150 mM, and immunoprecipitation of MAML1 was performed with a rabbit antibody recognizing the MAML1 C-terminus (Chemicon) coupled to Protein G–Sepharose (GE Healthcare) following the manufacturer's protocol. For the MAML1-p300 in vivo interaction, HEK-293 cells were transiently transfected with pSVSPORT-MAML1(1-300) and pCMV-p300 using Lipofectamine™ 2000, the cells were resuspended in Roche lysis buffer after 24 h, and the KCl concentration was adjusted to 200 mM. Immunoprecipitation of MAML1 was performed with a rabbit antibody recognizing the MAML1 N-terminus (Chemicon) coupled to Protein G–Sepharose, the manufacturer's protocol being followed.
RESULTS
p300 is recruited to chromatin templates via a direct interaction with the MAML1 N-terminus
It has previously been documented that MAML1 strictly requires the presence of p300 to function from chromatin templates. In addition, p300 is dependent on the presence of MAML1 to mediate significant Notch IC transcription from chromatin templates in vitro [22,23]. It has also been shown that amino acids 1–300 and 1–74 in MAML1 shift a CSL–Notch complex in EMSA (electrophoretic mobility-shift assay) experiments, but only MAML1(1–300) potently mediates transcription from chromatin templates [22]. Consistently, MAML1(75–301) interacts with p300 in HeLa nuclear extract [22]. These observations support the model (discussed in more detail in [26]) that the MAML1 N-terminus is involved in the recruitment of p300 to promoter regions of Notch-IC-regulated genes. Since we were interested in continuing to characterize the interplay between the MAML1 N-terminus and p300, we first performed a protein–protein interaction assay to make sure that MAML1(1–300) can directly, and independently, interact with p300. GST-tagged MAML1(1–300) (and other MAML1 domains that were included) (Figure 1A) were purified (Figure 1B) by using glutathione–Sepharose beads and incubated with FLAG–p300, expressed in insect (Sf9) cells via baculovirus and affinity-purified. As presumed, and as indicated in Figure 2(C), p300 interacts strongly with the MAML1 full-length protein (1–1016) (lane 3) and the MAML1 N-terminus (1–300) (lane 4), but not with the MAML1 domains 309–625, 499–804 and 701–1016 (lanes 5–7).
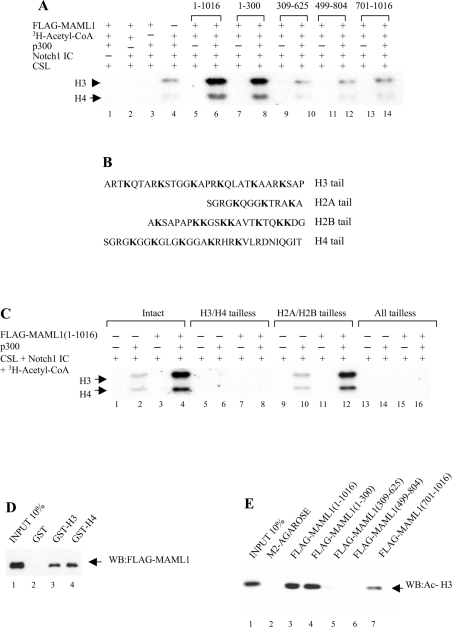
Figure 2. Substrate specificity in histone acetylation by the p300–MAML1 complex.
(A) HAT assay with chromatin templates reconstituted with recombinant intact histones. CSL, Notch1 IC, p300, MAML1 and [3H]acetyl-CoA were added to the templates as indicated. (B) Schematic of the core histones. The lysine residues in the histone tails are indicated by bold text. (C) HAT assay with chromatin templates reconstituted with recombinant intact histones (lanes 1–4) or the indicated tailless histones (lanes 5–16). Each chromatin template was incubated with CSL, Notch1 IC, p300, MAML1 and [3H]acetyl-CoA as indicated. (D) MAML1 interaction with histones. Immobilized GST-tagged histone proteins (indicated at the top) were incubated with FLAG-MAML1 and bound MAML1 was monitored by immunoblot with an antibody recognizing the FLAG-tag. The input represents 10% of the MAML1 used in each binding reaction. (E) Binding of MAML1 domains to acetylated histone H3. FLAG-tagged MAML1 proteins bound to M2–agarose were incubated with HeLa histones. Interacting acetylated histone H3 was detected in a Western blot by using an antibody recognizing acetylated histone H3 (Upstate Biotechnology). The input represents 10% of the histones used in each binding reaction. The negative control was M2–agarose without any bound FLAG-tagged protein.
To investigate further the function of the MAML1(1–300) interaction with p300 in Notch-activated transcription from chromatin templates, we expressed the MAML1 domains via baculovirus in insect cells and purified the proteins via FLAG-tags with affinity chromatography (Figure 1D). We then utilized a cell-free system with chromatin templates and reconstituted with highly purified activators (Notch) and cofactors (MAML1, p300). The CSL template, containing the E4 promoter and binding sites for CSL, was reconstituted with HeLa core histones by using purified recombinant Drosophila Acf-1, ISWI and NAP1 proteins as described by Ito et al. [27] and as previously described [23]. Assays were performed according to the protocol indicated in Figure 1(E) with HeLa nuclear extract as a source of general transcription factors and cofactors such as Mediator. As indicated in Figure 1(F), a significant level of transcription was observed when the chromatin template was incubated with CSL, Notch, p300 and acetyl-CoA (lane 4), and this activity was completely dependent on the presence of p300 (lane 2), acetyl-CoA (lane 3) and Notch1 IC (lane 1). Addition of MAML1(1–1016) increased the activity observed with CSL, Notch, p300 and acetyl-CoA approx. 5-fold (lane 6 versus lane 4), and this activity was likewise completely dependent on p300 (lane 5). These results clearly indicate a functional co-operativity between MAML1 and p300 that is also dependent upon acetyl-CoA and, presumably, related acetylation events. As further shown in Figure 1(F), the N-terminus of MAML1, amino acids 1–300, also stimulated transcription, but only in the presence of p300 (lane 8 versus lane 7). However, the MAML1 domains 309–625, 499–804 and 701–1016 failed to stimulate p300-dependent transcriptional activation by CSL and Notch (lanes 10, 12 and 14).
The p300–MAML1 complex specifically acetylates histones H3 and H4 in chromatin
p300 has previously been reported to acetylate chromatin templates containing Notch IC, CSL and MAML1 [22], and we continued to investigate in more detail the specificity in histone acetylation by the p300–MAML1 complex. The CSL template was first reconstituted with core histones purified from HeLa cells and incubated with purified factors CSL, Notch1 IC, p300, MAML1 and [3H]acetyl-CoA, as indicated in Figure 2(A). Our results indicate that there was significant acetylation of histones when the chromatin template was incubated with CSL, Notch, p300 and acetyl-CoA (lane 4), and this activity was completely dependent upon the presence of p300 (lane 2), acetyl-CoA (lane 3) and Notch1 IC (lane 1). This suggests that low levels of p300 can be recruited to the chromatin templates in the absence of MAML1. However, addition of MAML1(1–1016) strongly enhanced the histone acetylation observed with CSL, Notch, p300 and acetyl-CoA (lane 6 versus lane 4), and the increase in acetylation was totally dependent on p300 (lane 5). The N-terminal domain of MAML1, amino acids 1–300, also stimulated acetylation of chromatin templates in the presence of p300 (lane 8 versus lane 7), but the other MAML1 domains (309–625, 499–804 and 701–1016) failed to enhance p300-dependent acetylation of chromatin (lanes 10, 12 and 14). In accord with the results obtained in previous studies [22], our results show that there is a functional co-operativity between MAML1 and p300 in Notch transcriptional activation that correlates with acetylation of chromatin.
In further analysis of recombinant chromatin, the CSL template was reconstituted with recombinant intact core histones (intact) or core histones lacking the N-terminal tails (Figure 2B), H3-tailless+H4-tailless, H2A-tailless+H2B-tailless and totally tailless (all tailless). After assembly into chromatin, the CSL template was incubated with purified factors CSL, Notch1 IC, p300, MAML1 and [3H]acetyl-CoA, as indicated in Figure 2(C). There were only lower levels of histone acetylation when CSL, Notch, p300 and acetyl-CoA were incubated with chromatin templates reconstituted with intact histones (lane 2) or H2A-tailless+H2B-tailless histones (lane 10), but MAML1 strongly stimulated the p300-dependent activity (lanes 4 and 12). However, removal of the H3 and H4 tails or all four core histone tails completely abolished acetylation by the p300–MAML1 complex (lanes 5–8 and 13–16). Since we had detected only low levels of histone acetylation by p300 alone, we speculated that MAML1, in addition to recruiting p300, might directly interact with histones to facilitate histone acetylation. We had observed acetylation of the histones H3 and H4, and therefore incubated GST-tagged histone H3 and H4 with FLAG–MAML1 and monitored the protein interactions by Western blotting. As shown in Figure 2(D), both histone H3 and H4 interact strongly with MAML1 (lanes 3 and 4), and we continued to map which domain in MAML1 interacts with histone H3 by incubating FLAG–MAML1 with purified core histones. Histone H3 interaction with MAML1 was detected by Western blotting with an antibody recognizing acetylated histone H3 (Upstate Biotechnology). As indicated in Figure 2(E), there is a very significant interaction between acetylated histone H3 and MAML1(1–1016) (lane 3) and MAML1(1–300) (lane 4). We could also detect a weak interaction with MAML1(701–1016) (lane 7), but not any other MAML1 domains (lanes 5 and 6).
MAML1 interacts with the C-terminal C/H3 domain in p300
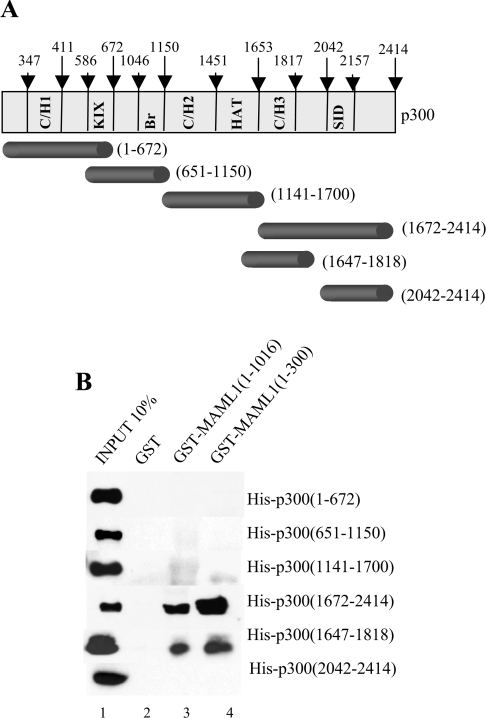
The p300 protein contains many domains that have been shown to specifically bind to different activators and co-regulators. To investigate which domain in p300 interacts with MAML1, we expressed several p300 domains (Figure 3A) in bacteria and purified the proteins via His-tags with affinity chromatography. Equal amounts of GST-tagged MAML1 proteins were incubated with recombinant p300, and the p300 domains associating with MAML1 were determined by Western blotting with an antibody recognizing the His-tag. As shown in Figure 3(B), MAML1(1–1016) and MAML1(1–300) do not interact with the p300 domains 1–672, 651–1150, 1141–1700 or 2042–2414, but show a strong binding to p300 amino acids 1672–2414 and 1647–1818 (lanes 3 and 4). p300 encloses a cysteine-and-histidine-rich domain (C/H3) between amino acids 1653–1817 that previously had been shown to interact with several activators, and our data show that the C/H3 domain in p300 is important for the MAML1 interaction.
Figure 3. MAML1 directly interacts with the C/H3 domain in p300.
(A) Schematic of subdomains in the p300 protein. (B) GST-tagged MAML1 bound to glutathione–Sepharose beads was incubated with His-tagged p300 derivatives and interacting p300 was detected by Western blotting.
p300 acetylates evolutionarily conserved lysine residues in MAML1
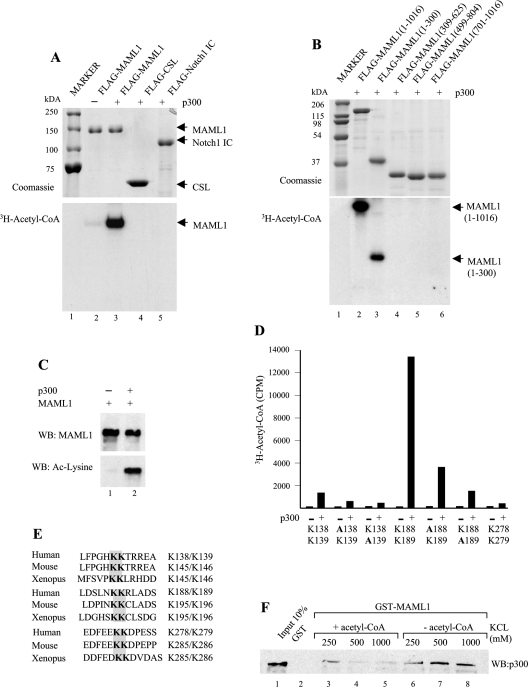
Several non-histone proteins have been reported to be acetylated by p300, and since p300 interacts with MAML1, we investigated whether p300 might acetylate factors in the Notch signalling pathway. In vitro acetylation assays were performed by incubating FLAG-tagged CSL, Notch1 IC and MAML1 with p300 and [3H]acetyl-CoA. As indicated in Figure 4(A), p300 significantly acetylates MAML1 (lane 3 versus lane 2), but not CSL or Notch1 IC (lanes 4 and 5). The data in Figure 4(B) show that p300 acetylates full-length MAML1 and the N-terminus of MAML1 (amino acids 1–300) (lanes 2 and 3), but not any other domain in MAML1 (lanes 4–6), suggesting that the MAML1 domain that interacts with p300 also harbours amino acids that can be acetylated by p300. To confirm that MAML1 can be acetylated by p300 in vivo, HEK-293 cells were co-transfected with plasmids expressing MAML1 and p300, and the whole-cell extract from transfected cells was incubated with an antibody recognizing the C-terminus of MAML1. As shown in Figure 4(C), the MAML1 antibody immunoprecipitates substantial amounts of MAML1 from the extract (upper panel), and by using an antibody recognizing acetylated lysine residues, we could show that MAML1 is also acetylated in vivo by p300 (compare lanes 1 and 2; lower panel).
Figure 4. MAML1 is acetylated in vitro and in vivo.
(A) In vitro acetylation of MAML1 relative to other components in the Notch signalling pathway. Recombinant proteins (indicated at the top and shown in the upper panel) were incubated with p300 and [3H]acetyl-CoA and reaction products were separated by SDS/PAGE and visualized by autoradiography (lower panel). (B) The acetylation site in MAML1 is located in the N-terminal domain. FLAG-tagged MAML1, full-length and derivatives (indicated at the top and shown in the upper panel) were incubated with p300 and [3H]acetyl-CoA. Reaction products were separated by SDS/PAGE and visualized by autoradiography (lower panel). (C) In vivo acetylation by MAML1. Plasmids expressing MAML1 and p300 were co-transfected into HEK-293 cells, whole-cell extracts prepared, and the MAML1 protein was immunoprecipitated with a MAML1 antibody. Proteins were separated by SDS/PAGE, and detection of the MAML1 protein and the acetylation of MAML1 were monitored by Western blotting with antibodies recognizing MAML1 and acetylated lysine residues (Cell Signalling Technology). (D) Synthetic peptides comprising MAML1 double lysine residues Lys138/Lys139, Lys188/Lys189 and Lys278/Lys279, were incubated with p300 and [3H]acetyl-CoA. Acetylation of the peptides was measured with a scintillation counter. (E) Alignment of evolutionarily conserved double lysine residues in MAML1 from human, mouse and Xenopus. (F) Acetylation of MAML1 decreases the MAML1 interaction with p300. GST-tagged MAML1 bound to Sepharose was incubated with whole cell extract from HEK-293 cells transfected with p300, in the absence or presence of 0.1 M acetyl-CoA. Beads were then washed with buffers containing 250, 500 and 1000 mM KCl respectively, and interacting p300 was monitored by immunoblot.
It has previously been documented that double lysine residues in p53 are acetylation targets for p300 [28]. Since the N-terminus of MAML1 contains three pairs of double lysine residues, Lys138/Lys139, Lys188/Lys189 and Lys278/Lys279, that might be potential targets for acetylation, we created peptides corresponding to amino acids 134–140, 186–192 and 277–283 in MAML1. The peptides were incubated with p300 and [3H]acetyl-CoA, and the levels of acetylation were measured with a liquid-scintillation counter. As shown in Figure 4(D), Lys188 and Lys189 are most strongly acetylated by p300. We also found that Lys138 and Lys139 are significantly acetylated, whereas we only detected lower acetylation levels of Lys278 and Lys279. By mutating each of the lysine residues to alanine, we found that the second lysine residue in a lysine pair (e.g. Lys139 and Lys189) were most strongly acetylated by p300. Importantly, MAML1 has previously been shown to be a conserved protein in vertebrates [29], and we noted that the positions of all of the acetylated lysine residues in MAML1 are highly conserved in vertebrates (Figure 4E). Several reports have noted that acetylation can affect protein–protein interactions in various ways (reviewed in [30]). To investigate if the p300–MAML1 interaction would be affected by acetylation of MAML1, we incubated equal amounts of GST–MAML1 with whole cell extract from HEK293 cells transfected with p300 in the absence or presence of acetyl-CoA. After pelleting and washing the Sepharose beads with buffers containing 250, 500 or 1000 mM KCl respectively, the level of p300 associated with MAML1 was determined by Western blotting. As shown in Figure 4(F), approximately the same amount of p300 interacts with MAML1 in the absence or presence of acetyl-CoA, when the washing buffer contains 250 mM KCl (lanes 3 and 6). However, when the beads containing GST–MAML1, and associating p300, are washed with buffers containing 500 or 1000 mM KCl, p300 interacts much more strongly with MAML1 in the absence of acetyl-CoA (compare lanes 7 and 8 with lanes 4 and 5). We therefore suggest that the acetylation of MAML1 might destabilize the MAML1 interaction with p300.
A proline-rich motif in MAML1 is important for p300-mediated acetylation of MAML1
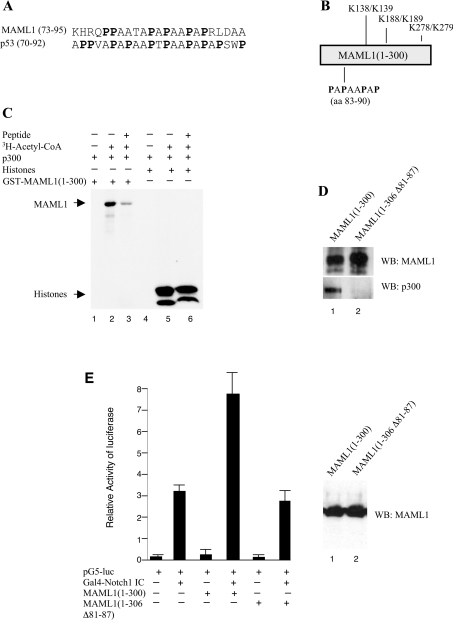
The C/H3 domain in p300 interacts with a proline motif in p53 [31], and since MAML1 also binds to the C/H3 domain, we compared the p53 proline-rich domain with the MAML1 sequence. We found that the N-terminus of MAML1 (amino acids 77–90) harbours a proline-rich motif that is indeed very similar to the proline motif in p53 (amino acids 71–92). Moreover, six amino acids (PAAPAP) are identical in the proline motifs in p53 and MAML1 (Figure 5A). The MAML1 proline motif is located approx. 50–100 amino acids in front of the double lysine residues Lys138/Lys139 and Lys188/Lys189) that are acetylated by p300 (Figure 5B). To investigate whether the proline motif in MAML1 is important for the p300 interaction, and subsequent acetylation of MAML1, we used a synthesized peptide corresponding to amino acids 73–95 in MAML1 and performed an acetylation assay as described above. As shown in Figure 5(C) (lane 2), p300 strongly acetylates MAML1, and the acetylation is dependent on acetyl-CoA (lane 1). Interestingly, the peptide almost completely inhibits the p300-mediated acetylation of MAML1 (lane 3). To confirm that the peptide would specifically inhibit p300 acetylation of MAML1, we also used purified HeLa histones in the acetylation assay. As shown in Figure 5(C), the peptide does not affect the acetylation of histones by p300 (lanes 5 and 6). Therefore we conclude that the peptide inhibits p300-mediated acetylation of MAML1, presumably by competing for p300 binding. We also investigated whether the MAML1 proline motif is important for the p300 interaction in vivo by co-expressing p300 and MAML1 proteins in HEK-293 cells. As shown in Figure 5(D) (upper panel), an antibody recognizing the MAML1 N-terminus, immunoprecipitates equal amounts of MAML1(1–300) and MAML1(1–300Δ81–87). However, the MAML1(1–300) protein pulls down substantially more p300 than does the MAML1(1–300Δ81–87) protein, which lacks the proline motif (lower panel). We further investigated whether the proline motif is important for the MAML1 N-terminus activity in vivo. HeLa cells were co-transfected with a reporter plasmid containing five GAL4 sites upstream of a luciferase gene and plasmids expressing GAL4–Notch1 IC, MAML1(1–300) or MAML1(1–306Δ81–87), as indicated in Figure 5(E). The level of transactivation by GAL4–Notch1 IC was significantly enhanced (2.8-fold) when co-transfected with MAML1(1–300). However, when GAL4–Notch1 IC was co-transfected with MAML1(1–306Δ81–87), the transactivation level was the same as for GAL4–Notch1 IC alone. As shown by a Western blot in Figure 5(E), the MAML1(1–300) and MAML1(1–306Δ81–87) proteins are equally well expressed in HeLa cells, and the different transactivation levels do not seem to be a consequence of different expression levels. Thus the proline-rich motif appears to play an important role for the transcriptional activation by the MAML1 N-terminus.
Figure 5. A proline repeat domain in MAML1 interacts with p300.
(A) Alignment of a proline repeat motif present in MAML1 and p53. (B) Schematic of the proline-rich region and double lysine residues in MAML1(1–300). (C) A proline-repeat peptide corresponding to MAML1(73–95) inhibits p300-mediated acetylation of MAML1 but not histones. GST-MAML1(1–300) or HeLa core histones were incubated with p300, acetyl-CoA and the peptide as indicated in the Figure, and reaction products were were separated by SDS/PAGE and visualized by autoradiography. (D) The MAML1 proline repeat domain is important for the p300 interaction in vivo. MAML1(1–300) or MAML1(1–300Δ81–87) were expressed together with p300 in HEK-293 cells, whole-cell extracts prepared, and the MAML1 proteins were immunoprecipitated with a MAML1 antibody. Proteins were separated by SDS/PAGE and detected with antibodies recognizing MAML1 and p300. (E) The MAML1 proline repeat domain is important for MAML1(1–300) activity in vivo. HeLa cells were co-transfected with a luciferase reporter containing five Gal4 binding sites and vectors expressing Gal4–Notch1 IC, MAML1(1-300) and MAML1(1-306Δ81-87). The bars represent mean values obtained from at least three independent experiments. For calculation of the fold stimulation value in the text, the value for the vector background was subtracted. A Western blot shows that the MAML1 proteins are equally well expressed in HeLa cells.
DISCUSSION
MAML1 has previously been suggested to potentiate Notch activity by facilitating chromatin remodelling (via p300). In the present study we have investigated in more detail the transcriptional mechanisms by the N-terminal domain of MAML. We suggest that the MAML1 N-terminus functions in chromatin remodelling through direct interactions with both p300 and histones. Furthermore, we have identified a proline-repeat motif in the MAML1 N-terminal domain that seems to be important for p300-mediated acetylation of MAML1 and for the activity of the MAML1 N-terminus in vivo.
Role of MAML1 in chromatin remodelling
MAML1 is a human homologue of the Drosophila Mastermind, and these proteins are most related in the N-terminal domain, with 35% amino acid identity and 51% similarity [14], indicating that this domain is most evolutionary conserved. The MAML1 protein comprises two acidic clusters, one located in the N-terminus and one at the end of the C-terminus, and these acidic regions are speculated to be important for protein interactions. The C-terminal acidic cluster in Mastermind does not seem to have any effect on wing formation in Drosophila, but a truncation that eliminates approximately the C-terminal third of Mastermind induces a distinct mutant phenotype when overexpressed [32]. To our knowledge, most of the proteins that have been reported to interact with MAML1 bind to the N-terminal domain in MAML1. It was demonstrated that Notch IC binds to MAML1 amino acids 1–74 in EMSA experiments [22], and in our protein-interaction assays, recombinant Notch1 IC also directly binds to MAML1(1–300) and not to any other MAML1 domain (results not shown). Evidently the MAML1 N-terminus interaction with Notch explains why the N-terminus of MAML1 is needed for Notch-mediated transcription, but also indicates that other MAML1 domains cannot significantly activate transcription on their own through interactions with other components in the transcription complex such as histones and CSL. Interestingly, the myogenesis transcription factor MEF2C interacts with MAML1 amino acids 1–70 in transient transfection assays, and co-transfection of MEF2C and Notch disrupts MAML1-enhanced myogenesis, indicating that MEF2C and Notch compete for the same binding site in MAML1 [21]. The MAML1 N-terminus also directly interacts with p300 and histones (the present study) and presumably stabilizes a p300–MAML1–histone complex to facilitate p300-mediated acetylation of histones. However, whether histone acetylation by the p300–MAML1 complex strictly requires the MAML1 interaction with histones remains to be investigated. In addition, p300 has been reported to interact with histone H3 and H4 tails, but not with histone H2A and H2B tails [33]. The p300–histone interaction might be crucial for fulfillment of the p300 co-activator function at Notch-regulated promoters, especially since our data suggest that acetylation of MAML1, which is responsible for recruitment of p300, destabilizes the MAML1 interaction with p300.
By using recombinant wild-type histones and histones lacking the tails that are targets for acetylation, we found that, in chromatin templates, histones H3 and H4 are strongly acetylated, and deletions of the H3 and H4 tails completely abolish histone acetylation. Moreover, H2A and H2B are not significantly acetylated, and deletions of the H2A and H2B tails do not affect the acetylation of H3 and H4. The acetylation pattern of recombinant chromatin by the p300–MAML1–Notch complex resembles the specificity in acetylation by p300 recruited to chromatin templates by the activator VP16, but there are some differences. In a complex with VP16, p300 potently acetylates H2A and H2B [33,34], and the p300–VP16 complex still acetylates H2A and H2B in a chromatin template containing tailless H3 and H4, even though there is a significant decrease in acetylation of H2A and H2B [33,34]. These data indicate that although p300 with its acetyltransferase activity is crucial for histone acetylation, it could be the p300-interacting proteins that determine the acetylation pattern of histones. Interestingly, it has been documented that the acetylation of specific histone tails correlates with p300-mediated and activator-dependent transcription. The dual deletion of H2A and H2B tails does not affect p300-mediated VP16 transcriptional activation, but both independent and dual deletions of H3 and H4 tails significantly reduced the transcription levels [34].
Proline-directed acetylation of MAML1 by p300
The p300 protein encloses a cysteine-and-histidine-rich domain (C/H3) between amino acids 1653 and 1817, and this domain has been reported to interact with several proteins, such as p53, E1A, TFIIB (transcription factor IIB) and Fos (see the references in [35]). In addition, p53 also interacts with the domains C/H1 and C/H2, and the bromo domain (see the references in [31]). However, we found that MAML1 only interacts with the C/H3 domain and no other domains in p300. Interestingly, a region C-terminal of the ANK repeats in Notch1 IC has previously been reported to interact with the C/H3 domain in p300 [12]. Although the structure of the CSL–Notch–MAML1 complex reveals that this region in Notch is essential for the interactions with MAML1 [18,19], it is possible that the p300 interaction with Notch IC occurs if MAML1 leaves the promoter region before p300. A proline-rich domain in p53 has been documented to interact with the C/H3 domain in p300 [31], and we found that the N-terminus of MAML1, amino acids 77–90, harbours a proline-rich motif that is very similar to the proline motif in p53 at amino acids 71–92. The proline motif in the MAML1 N-terminus seems to be the domain mainly responsible for the p300 interaction, since a peptide (containing the MAML1 N-terminal proline repeat sequence) almost completely abolished p300-mediated acetylation of MAML1. Importantly, the proline motif also appears to be important for MAML1 interaction with p300 in vivo, since a deletion of this motif strongly reduces the p300 interaction in whole cell extracts and the transactivation activity of MAML1(1–306) in cells (see Figure 5). It was recently reported that residues 15–67, N-terminal to the proline motif in human MAML1, are essential for the interaction with Notch and CSL [18] and are therefore not likely to be involved in other protein interactions. However, it is possible that domains between amino acids 90 and 300 in MAML1 might co-operate with the proline motif in binding to p300.
The double lysine residues, Lys372/Lys373 and Lys381/Lys382 in p53, have been documented to be p300 acetylation targets [28]. We found that the N-terminus of MAML1 contains three pairs of double lysine residues (Lys138/Lys139, Lys188/Lys189 and Lys278/Lys279) approx. 50–150 amino acids downstream of the proline motif in MAML1 and verified that some of these lysine residues are substrates for p300-mediated acetylation. The Lys188 and Lys189 were most strongly acetylated by p300, Lys138 and Lys139 were acetylated to a lesser extent, and Lys278 and Lys279 were not significantly acetylated. Furthermore, the second lysine residue in a lysine pair (Lys139 and Lys189) was the most important lysine residue for p300 acetylation. Interestingly, the second lysine residue in a lysine pair in p53 (Lys372 and Lys382), has also been reported to be the lysine residue most strongly acetylated by p300 [28]. MAML1(1–300) contains only three pairs of double lysine residues, but there are 11 single lysine residues in MAML1(1–300), and some of these lysine residues might also be targets for p300-mediated acetylation and be important for the total acetylation effects of MAML1.
Even though the acetylation pattern of the double lysine residues in MAML1 and p53 is very similar, the p300-mediated acetylation of these proteins might differ. Acetylation of p53 by p300 enhances the sequence-specific DNA binding activity of p53 [28]. In addition, some of the lysine residues in p53 that are targets for acetylation can also be subjected to ubiquitination. Acetylation of p53 is considered to promote protein stability, since unacetylated lysine residues are targets for ubiquitination, which leads to degradation of the p53 protein (see the references in [36]. The MAML1 N-terminus contains a cluster of basic residues in a region that has some sequence similarity to a subset of bZip DNA-binding domains, but since MAML1, so far, has not been reported to have any DNA-binding activity, we focused on investigating whether acetylation of MAML1 would affect its interaction with other proteins. Interestingly, we found that the MAML1 interaction with p300 is stronger in the absence of acetyl-CoA. We therefore conclude that one function of the acetylation might be to decrease the p300–MAML1 interaction after p300 has been recruited by MAML1 to a target gene to acetylate histones. There have been several reports that acetylation of proteins in some cases impairs the interaction with other proteins (reviewed in [30]). For example, acetylation of the molecular chaperone Hsp90 (heat-shock protein of 90 kDa) prevents its binding to the GR (glucocorticoid receptor), which ultimately leads to loss of GR transcriptional activity, since the interaction with Hsp90 is essential for GR ligand binding and nuclear translocation [37]. It has also been reported that acetylation of critical lysine residues in the protein Ku70 disrupts the interaction with the pro-apoptotic factor Bax, which allows Bax to localize to mitochondria and initiate apoptosis [38]. The transcriptional activity of NF-κB (nuclear factor κB; a heterodimer of the proteins RelA/p65 and p50) is regulated by association with IκBα (inhibitory κBα). Deacetylation of RelA/p65 by HDACs results in an increased association with IκBα that potentiates the nuclear export of NF-κB (reviewed in [30]). It is most likely that acetylation of MAML1 serves other functions in addition to affecting the interaction with p300. Since the positions of all of the acetylated double lysine residues in MAML1 are highly conserved in vertebrates, our results suggest that acetylation of MAML1 might be conserved throughout evolution and perhaps be an important modification of the protein. Further studies will focus on elucidating the specificity in timing, positioning, activation and termination of p300-mediated acetylation of MAML1, to understand how cells might use modifications of proteins, such as acetylation, to regulate responses in a complex multistep transcriptional activation pathway.
Acknowledgments
We thank Dr Robert G. Roeder (Laboratory of Biochemistry and Molecular Biology, The Rockefeller University, New York, NY, U.S.A.) for the plasmids expressing recombinant histones in bacteria, Dr James D. Griffin (Department of Medical Oncology, Dana-Farber Cancer Institute and Department of Medicine, Brigham and Women's Hospital and Harvard Medical School, Boston, MA, U.S.A.) for the pGEX-MAML1(1-1016) plasmid, and Dr Tom Kadesch (Department of Genetics, University of Pennsylvania School of Medicine, Philadelphia, PA, U.S.A.) for the pCDNA3-hNotch1 IC plasmid. This work was supported by grants from the Swedish Medical Research Council, Åke Wibergs Foundation, the Karolinska Institutets Foundation for Medical Research and the Harald and Greta Jeanssons Foundation to A.E.W.
References
- 1.Artavanis-Tsakonas S., Rand M. D., Lake R. J. Notch signalling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 2.Mumm J. S., Kopan R. Notch signalling: from the outside in. Dev. Biol. 2000;228:151–165. doi: 10.1006/dbio.2000.9960. [DOI] [PubMed] [Google Scholar]
- 3.Wu L., Griffin J. D. Modulation of Notch signalling by mastermind-like (MAML) transcriptional co-activators and their involvement in tumorigenesis. Semin. Cancer Biol. 2004;14:348–356. doi: 10.1016/j.semcancer.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 4.Pear W. S., Aster J. C. T cell acute lymphoblastic leukemia/lymphoma: a human cancer commonly associated with aberrant NOTCH1 signalling. Curr. Opin. Hematol. 2004;11:426–433. doi: 10.1097/01.moh.0000143965.90813.70. [DOI] [PubMed] [Google Scholar]
- 5.Kao H.-Y., Ordentlich P., Koyano-Nakagawa N., Tang Z., Downes M., Kintner C. R., Evans R. M., Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taniguchi Y., Furukawa T., Hun T., Han H., Honjo T. LIM protein KyoT2 negatively regulates transcription by association with the RBP-J DNA-binding protein. Mol. Cell. Biol. 1998;18:644–654. doi: 10.1128/mcb.18.1.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsieh J. J.-D., Zhou S., Chen L., Young D. B., Hayard S. D. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc. Natl. Acad. Sci. U.S.A. 1999;96:23–28. doi: 10.1073/pnas.96.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oswald F., Kostezka U., Astrahantseff K., Bourteele S., Dillinger K., Zechner U., Ludwig L., Wilda M., Hameister H., Knöchel W., et al. SHARP is a novel component of the Notch/EBP-Jk signalling pathway. EMBO J. 2002;21:5417–5426. doi: 10.1093/emboj/cdf549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurooka H., Honjo T. Functional interaction between the mouse Notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J. Biol. Chem. 2000;275:17211–17220. doi: 10.1074/jbc.M000909200. [DOI] [PubMed] [Google Scholar]
- 10.Lin S. E., Oyama T., Nagase T., Harigaya K., Kitagawa M. Identification of new human mastermind proteins defines a family that consists of positive regulators for Notch Signalling. J. Biol. Chem. 2002;277:50612–50620. doi: 10.1074/jbc.M209529200. [DOI] [PubMed] [Google Scholar]
- 11.Wu L., Sun T., Kobayashi K., Gao P., Griffin J. D. Identification of a family of Mastermind-Like transcriptional coactivators for Mammalian NOTCH receptors. Mol. Cell. Biol. 2002;22:7688–7700. doi: 10.1128/MCB.22.21.7688-7700.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oswald F., Täuber B., Dobner T., Bourteele S., Kostezka U., Adler G., Liptay S., Schmid R. M. p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol. Cell. Biol. 2001;21:7761–7774. doi: 10.1128/MCB.21.22.7761-7774.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohtsuka T., Ishibashi M., Gradwohl G., Nakanishi S., Guillemot F., Kageyama R. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 1999;18:2196–2207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu L., Aster J. C., Blacklow S. C., Lake R., Artavanis-Tsakonas S., Griffin J.D. MAML1, a human homologue of Drosophila Mastermind, is a transcriptional co-activator for NOTCH receptors. Nat. Genet. 2000;26:484–489. doi: 10.1038/82644. [DOI] [PubMed] [Google Scholar]
- 15.Bettler D., Pearson S., Yedvobnick B. The nuclear protein encoded by the Drosophila neurogenic gene mastermind is widely expressed and associates with specific chromosomal regions. Genetics. 1996;143:859–875. doi: 10.1093/genetics/143.2.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smoller D., Friedel C., Schmid A., Bettler D., Lam L., Yedvobnick B. The Drosophila neurogenic locus mastermind encodes a nuclear protein unusually rich in amino acid homopolymers. Genes Dev. 1990;4:1688–1700. doi: 10.1101/gad.4.10.1688. [DOI] [PubMed] [Google Scholar]
- 17.Yedvobnick B., Smoller D., Young P., Mills D. Molecular analysis of the neurogenic locus mastermind of Drosophila melanogaster. Genetics. 1988;118:483–497. doi: 10.1093/genetics/118.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nam Y., Sliz P., Song L., Aster J. C., Blacklow S. C. Structural basis for cooperativity in recruitment of MAML1 coactivators to Notch transcription complexes. Cell. 2006;124:973–983. doi: 10.1016/j.cell.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 19.Wilson J. J., Kovall R. A. Crystal structure of the CSL–Notch–Mastermind ternary complex bound to DNA. Cell. 2006;124:985–996. doi: 10.1016/j.cell.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 20.Lubman O. Y., Kopan R., Waksman G., Korolev S. The crystal structure of a partial mouse Notch-1 ankyrin domain: Repeats 4 through 7 preserve an ankyrin fold. Protein Sci. 2005;14:1274–1281. doi: 10.1110/ps.041184105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen H., McElhinny A. S., Cao Y., Gao P., Liu J., Bronson R., Griffin J. D., Wu L. The Notch coactivator, MAML1, functions as a novel coactivator for MEF2C-mediated transcription and is required for normal myogenesis. Genes Dev. 2006;20:675–688. doi: 10.1101/gad.1383706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fryer C. J., Lamar E., Turbachova I., Kintner C., Jones K. A. Mastermind mediates chromatin-specific transcription and turnover of the Notch enhancer complex. Genes Dev. 2002;16:1397–1411. doi: 10.1101/gad.991602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wallberg A. E., Pedersen K., Lendahl U., Roeder R. G. p300 and PCAF can act cooperatively to mediate transcriptional activation from chromatin templates by Notch intracellular domains in vitro. Mol. Cell. Biol. 2002;22:7812–7819. doi: 10.1128/MCB.22.22.7812-7819.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luger K., Rechsteiner T. J., Flaus A. J., Waye M. M. Y., Richmond T. J. Characterization of nucleosome core particles containing histone proteins made in bacteria. J. Mol. Biol. 1997;272:301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- 25.Grant P. A., Berger S. L., Workman J. L. Identification and analysis of native nucleosomal histone acetyltransferase complexes. Methods Mol. Biol. 1999;119:311–317. doi: 10.1385/1-59259-681-9:311. [DOI] [PubMed] [Google Scholar]
- 26.Lubman O. Y., Korolev S. V., Kopan R. Anchoring Notch genetics and biochemistry: structural analysis of the ankyrin domain sheds light on existing data. Mol. Cell. 2004;13:619–626. doi: 10.1016/s1097-2765(04)00120-0. [DOI] [PubMed] [Google Scholar]
- 27.Ito T., Levenstein M. E., Fyodorov D. V., Kutach A. K., Kobayashi R., Kadonaga J. T. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 1999;13:1529–1539. doi: 10.1101/gad.13.12.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu W., Roeder R. G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 29.Wu L., Kobayashi K., Sun T., Gao P., Liu J., Nakamura M., Weisberg E., Mukhopadhyay N. K., Griffin J. D. Cloning and functional characterization of the murine mastermind-like 1 (Maml1) gene. Gene. 2004;328:153–165. doi: 10.1016/j.gene.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 30.Glozak M. A., Sengupta N., Zhang X., Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 31.Dornan D., Shimizu H., Burch L., Smith A. J., Hupp T. R. The proline repeat domain of p53 binds directly to the transcriptional coactivator p300 and allosterically controls DNA-dependent acetylation of p53. Mol. Cell. Biol. 2003;23:8846–8861. doi: 10.1128/MCB.23.23.8846-8861.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Helms W., Lee H., Ammerman M., Parks A. L., Muskavitch M. A. T., Yedvobnic B. Engineered truncations in the Drosophila Mastermind protein disrupt Notch pathway function. Dev. Biol. 1999;215:358–374. doi: 10.1006/dbio.1999.9477. [DOI] [PubMed] [Google Scholar]
- 33.An W., Roeder R. G. Direct association of p300 with unmodified H3 and H4 N termini modulates p300-dependent acetylation and transcription of nucleosomal templates. J. Biol. Chem. 2003;278:1504–1510. doi: 10.1074/jbc.M209355200. [DOI] [PubMed] [Google Scholar]
- 34.An W., Palhan V. B., Karymov M., Leuba S. H., Roeder R. G. Selective requirements for histone H3 and H4 N termini in p300-dependent transcriptional activation from chromatin. Mol. Cell. 2002;9:811–821. doi: 10.1016/s1097-2765(02)00497-5. [DOI] [PubMed] [Google Scholar]
- 35.Vo N., Goodman R. H. CREB-binding protein and p300 in transcriptional regulation. J. Biol. Chem. 2001;276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- 36.Brooks C. L., Gu W. p53 ubiquitination: Mdm2 and beyond. Mol. Cell. 2006;21:307–315. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kovacs J. J., Murphy P. J. M., Gaillard S., Zhao X., Wu J.-T., Nicchitta C. V., Yoshida M., Toft D. O., Pratt W. B., Yao Y.-P. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol. Cell. 2005;18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 38.Cohen H. Y., Lavu S., Bitterman K. J., Hekking B., Imahiyerbo T. A., Miller C., Frye R., Ploegh H., Kessler B. M., Sinclair D. A. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol. Cell. 2004;13:627–638. doi: 10.1016/s1097-2765(04)00094-2. [DOI] [PubMed] [Google Scholar]