Abstract
Fractionation of 3T3-L1 adipocyte membranes revealed that PDE3B (phosphodiesterase 3B) was associated with PM (plasma membrane) and ER (endoplasmic reticulum)/Golgi fractions, that insulin-induced phosphorylation/activation of PDE3B was greater in internal membranes than PM fractions, and that there was no significant translocation of PDE3B between membrane fractions. Insulin also induced formation of large macromolecular complexes, separated during gel filtration (Superose 6 columns) of solubilized membranes, which apparently contain phosphorylated/activated PDE3B and signalling molecules potentially involved in its activation by insulin, e.g. IRS-1 (insulin receptor substrate-1), IRS-2, PI3K p85 [p85-subunit of PI3K (phosphoinositide 3-kinase)], PKB (protein kinase B), HSP-90 (heat-shock protein 90) and 14-3-3. Expression of full-length recombinant FLAG-tagged murine (M) PDE3B and M3BΔ604 (MPDE3B lacking N-terminal 604 amino acids) indicated that the N-terminal region of MPDE3B was necessary for insulin-induced activation and recruitment of PDE3B. siRNA (small interfering RNA) knock-down of PDE3B indicated that PDE3B was not required for formation of insulin-induced complexes. Wortmannin inhibited insulin-induced assembly of macromolecular complexes, as well as phosphorylation/activation of PKB and PDE3B, and their co-immunoprecipitation. Another PI3K inhibitor, LY294002, and the tyrosine kinase inhibitor, Genistein, also inhibited insulin-induced activation of PDE3B and its co-immunoprecipitation with PKB. Confocal microscopy indicated co-localization of PDE3B and PKB. Recombinant MPDE3B co-immunoprecipitated, and co-eluted during Superose 12 chromatography, to a greater extent with recombinant pPKB (phosphorylated/activated PKB) than dephospho-PKB or p-ΔPKB [pPKB lacking its PH domain (pleckstrin homology domain)]. Truncated recombinant MPDE3B proteins and pPKB did not efficiently co-immunoprecipitate, suggesting that structural determinants for their interaction reside in, or are regulated by, the N-terminal portion of MPDE3B. Recruitment of PDE3B in macromolecular complexes may be critical for regulation of specific cAMP pools and signalling pathways by insulin, e.g. lipolysis.
Keywords: confocal microscopy, gel filtration, insulin, macromolecular complex, phosphodiesterase 3B (PDE3B), phosphorylation
Abbreviations: BiP, immunoglobulin heavy-chain-binding protein; COX, cytochrome oxidase; CT domain, C-terminal domain; DMEM, Dulbecco's modified Eagle's medium; DPBS, Dulbecco's PBS; ER, endoplasmic reticulum; FBS, foetal bovine serum; HSP-90, heat-shock protein 90; IBMX, isobutylmethylxanthine; IRS, insulin receptor substrate; NC, nitrocellulose; NFDM, non-fat dry milk; NP40, Nonidet P40; NT domain, N-terminal domain; PDE, phosphodiesterase; PH domain, pleckstrin homology domain; PI3K, phosphoinositide 3-kinase; PI3K p85, p85-subunit of PI3K; PKB, protein kinase B; pPKB, phosphorylated/activated PKB; PM, plasma membrane; PP2A, protein phosphatase 2A; RD domain, regulatory domain; siRNA, small interfering RNA; WT, wild-type
INTRODUCTION
By catalysing hydrolysis of cAMP and cGMP, cyclic nucleotide PDEs (phosphodiesterases) are critical in regulating intracellular concentrations and, consequently, physiological effects of these second messengers. The diverse, complex and large PDE superfamily contains at least 11 structurally related and functionally distinct PDE gene families (PDEs 1–11) [1]. PDE3 isoforms are encoded by two similarly organized genes, PDE3A and PDE3B. In general, PDE3A isoforms are thought to be more abundant than PDE3B in platelets, heart and vascular and airway smooth muscles, with PDE3B more abundant in tissues involved in regulation of energy homoeostasis, such as adipose tissue, liver and pancreatic β-cells [2]. Indeed, studies with intact cells, as well as with PDE3B KO (knockout) mice and mice specifically overexpressing PDE3B in pancreatic β-cells, indicate an important role for PDE3B in liver, adipose tissue and β-cells in regulation of overall energy metabolism [3,4].
Intracellular concentrations of cyclic nucleotides are tightly regulated and seem to be temporally, spatially and functionally compartmentalized. Through their targeting to different intracellular locations and interactions with molecular scaffolds, cellular structural elements and regulatory partners, PDEs modulate the magnitude, intracellular diffusion and compartmentalization of cyclic nucleotide signals. Representatives of multiple PDE families are usually present in individual cells, and their different subcellular localizations may contribute to the independent regulation of specific cAMP pools in specific microdomains [5]. In cultured vascular smooth-muscle cells, subcellular location may be important in defining mechanisms by which PDE3A and PDE3B are differentially regulated, since membrane-associated PDE3B, not cytosolic PDE3A, was induced by cAMP [6]. PDE3 isoforms have been reported to be associated with the insulin receptor in human adipocytes [7], the leptin receptor in human platelets [8] and caveolae in primary rat adipocytes [9].
PKB (protein kinase B) has been implicated in insulin-induced phosphorylation and activation of membrane-associated PDE3B [10–12]. To elucidate mechanisms for activation of membrane-associated PDE3B, we assessed effects of insulin on PDE3B and several signalling molecules, e.g. IRS-1 (insulin receptor substrate-1), PI3K (phosphoinositide 3-kinase) and PKB, in subcellular fractions of 3T3-L1 adipocytes. Our results indicate that insulin phosphorylated/activated PDE3B to a greater extent in ER (endoplasmic reticulum)/Golgi fractions than in PM (plasma membrane) fractions and suggest that co-localization and association of PDE3B and PKB might be important in activation of PDE3B by insulin. Furthermore, we found that insulin induced reversible formation of, and recruitment of PDE3B into large, high-molecular-weight (mass), macromolecular complexes (HMWC-ins) in 3T3-L1 adipocyte membranes.
MATERIALS AND METHODS
Materials
3T3-L1 cells were purchased from American Type Culture Collection (Manassas, VA, U.S.A.); DMEM (Dulbecco's modified Eagle's medium; cat. no. 11995-065), RPMI 1640 and Zenon kits were from Invitrogen; FBS (foetal bovine serum) was from Gemini Bio-Products (Woodland, CA, U.S.A.); anti-FLAG antibody, anti-FLAG–agarose, FLAG peptides, IBMX (isobutylmethylxanthine), insulin and dexamethasone were from Sigma; [3H]cAMP was from New England Nuclear (Boston, MA, U.S.A.); [γ-32P]ATP (3000 Ci/mmol) and [32P]Pi (1000 mCi/mmol) were from ICN Radiochemicals (Costa Mesa, CA, U.S.A.); SuperSignal® Westpico and Westfemto chemiluminescent reagents were from Pierce (Rockford, IL, U.S.A.); polyclonal p85 PI3K, PKB1, PKB2 and recombinant phospho- and non-phospho-PKB1, p-ΔPKB [PH domain (pleckstrin homology domain) deleted pPKB (phosphorylated/activated PKB)] and crosstide peptides were from Upstate Biotechnology; anti-IRS-1, -pPKB (S-473), -PKB1, -COX (cytochrome oxidase) IV antibodies were from Cell Signaling Technology. Anti-caveolin-1, -PKARI, -BiP (immunoglobulin heavy-chain-binding protein), -GM130, -nucleoporin p62, -β-catenin and -phosphotyrosine antibodies were purchased from BD Biosciences (San Diego, CA, U.S.A.); anti-IRS-1 (p-Tyr612) antibody was from Invitrogen; anti-adenylate cyclase, -14-3-3 and -HSP-90 (heat-shock protein 90) antibodies were from Santa Cruz Biotechnology; wortmannin, LY294002 and Genistein were from Biomol; and prestained molecular size markers were from Bio-Rad. Rabbit polyclonal antibodies to mouse PDE3B (accession no. AAN52086) were generated against peptides corresponding to amino acids 1076–1095 (NASLPQADEIQVIEEADEEE) CT domain (C-terminal domain), amino acids 266–280 (VIRPRRRSSCVSLGE) RD domain (regulatory domain) and amino acids 2–16 (RKDERERDTPAMRSP) NT domain (N-terminal domain). Affinity-purified anti-PDE3B-NT and anti-PDE3B-CT antibodies were used for immunofluorescence microscopy. For Western blotting and immunoprecipitation experiments, anti-PDE3B-NT, -RD and -CT antibodies were purified using Immunopure® IgG Protein G purification kits (Pierce). Other materials were obtained as indicated and were of the highest grade available.
Differentiation of 3T3-L1 adipocytes
As previously described [13], 3T3-L1 preadipocytes were grown (150 mm culture dishes) in complete DMEM [25 ml, containing 10% (v/v) foetal calf serum, 2 mM L-glutamine, 3.7 mg/ml sodium bicarbonate, 25 mM high glucose, 100 mg/ml sodium pyruvate, 4 µg/ml pyridoxine hydrochloride, 125 µg/ml sodium phosphate, 8 µg/ml biotin and 1% penicillin/streptomycin], at 37 °C under humidified air/5% CO2. At confluency, fresh complete DMEM containing MDI (0.3 mM IBMX, 1 µM dexamethasone and 5 µg/ml insulin) was added. After 72 h, medium was changed to complete DMEM containing 5 µg/ml insulin, which was changed every 48 h. Experiments with differentiated adipocytes were usually performed 13–14 days after initiation of differentiation.
cAMP PDE assay
PDE3 activity [that portion of total activity inhibited by 1.0 μM cilostamide (a specific PDE3 inhibitor with an IC50 of ∼17–80 nM [14])] was measured by a modification of a published method ([15]; Supplementary data at http://www.BiochemJ.org/bj/404/bj4040257add.htm), using 0.1 μM [3H]cAMP (35000 c.p.m.) as the substrate.
Immunoprecipitation and immunoblotting
For immunoprecipitations, solubilized membrane, cytosol or column fractions contained 1% (v/v) NP40 (Nonidet P40). After solubilization of membrane fractions and centrifugation (100000 g, 30 min and 4 °C), protein concentrations of supernatants were usually adjusted to 3 mg/ml. For most experiments, samples were cleared by incubation [1 h, room temperature (25 °C)] with 5 μg of pre-immune IgG, and then (30 min) with 50 μl of Protein G–Sepharose (Amersham), before centrifugation (2800 g, 4 °C and 5 min). Cleared fractions were incubated overnight (4 °C) with specified antibodies, followed by incubation with fresh Protein G–Sepharose (1 h) before centrifugation (2800 g, 4 °C and 5 min). Immunoprecipitates were washed three times with buffer B (50 mM Hepes, 250 mM sucrose, 1 mM EDTA, 10 mM PPi, 5 mM NaF, 100 mM NaCl, 1 μM okadaic acid, 1 mM Na3VO4 and Roche protease inhibitor cocktail, pH 7.5), subjected to SDS/PAGE and electrotransferred to membranes in Tris-glycine buffer (25 mM Tris/base and 192 mM glycine, pH 8.3), containing 20% (v/v) methanol. Membranes were incubated (4 °C overnight) with 5% (w/v) NFDM (non-fat dry milk) in DPBS (Dulbecco's PBS) and then with the appropriate primary antibody (usually for 2 h, but sometimes longer, depending on antibody quality and sensitivity) in 0.5% NFDM in DPBS, washed (DPBS), and incubated with HRP (horseradish peroxidase)-labelled secondary antibody (Pierce). Immunoreactive proteins were reacted with SuperSignal® Westpico or Westfemto chemiluminescent reagents; signals were detected with Imagereader LAS3000 (Fuji, Stanford, CT, U.S.A.).
Fractionation of membranes on continuous 10–45% sucrose gradients
Differentiated adipocytes were incubated in serum-free DMEM (16 h, 37 °C) to reduce possible effects of growth factors in serum. After incubation without and with insulin (100 nM, 10 min), adipocytes were washed, homogenized (Dounce homogenizer) in ice-cold buffer A (buffer B containing 50 mM sucrose) and centrifuged (500 g, 20 min and 4 °C). Supernatants were centrifuged (175000 g, 30 min) to collect total membranes, which were rehomogenized in Buffer A (1 ml), layered on top of linear 10–45% sucrose gradients [11 ml, prepared using a J17 gradient maker (Joule, Inc) and confirmed with a Palm Abbe digital refractometer] and centrifuged (Sw41 Beckman rotor, 60 min), before fractionation as described previously [9]. Gradients were manually divided into 17 equal fractions (0.7 ml), collected sequentially from the top. Portions of fractions were assayed for PDE3 activity, and subjected to SDS/PAGE/Western blotting. Protein was quantified using the BCA (bicinchoninic acid) assay kit (Pierce), with BSA as the standard.
32P-phosphorylation of PDE3B
Adipocytes were incubated overnight in serum-free DMEM, then in fresh serum-free DMEM with and without [32P]Pi (200 μCi/ml) for 90 min and, in some experiments, for an additional 30 min with wortmannin. Adipocytes were then incubated for 10 min without or with 100 nM insulin. Samples of solubilized and cleared membrane proteins from subcellular fractions, sucrose gradient fractions or Superose 6 gel filtration fractions were immunoprecipitated with 20–25 μl of anti-PDE3B-RD or -CT antibodies. Immunoprecipitated proteins were separated by SDS/PAGE, gels were washed, and 32P-labelled PDE3B was detected by phosphoimager (Amersham) analysis of wet gels, after which proteins were electrophoretically transferred to NC (nitrocellulose) membranes for immunoblotting.
Superose 6 chromatography of 3T3-L1 adipocyte microsomal fractions
After incubation in serum-free complete DMEM (16 h) and then without or with 100 nM insulin (10 min), adipocytes were washed twice with ice-cold PBS, homogenized in ice-cold buffer B and centrifuged (13000 g, 20 min). Supernatants were centrifuged (175000 g, 30 min) to separate total microsome membranes and cytosolic fractions. Pellets, solubilized in buffer B containing 1% NP40, were centrifuged (100000 g, 30 min and 4 °C; SW 41 Ti rotor). Samples (1.0 ml, 3 mg of total protein) of solubilized membranes were applied to a Superose 6 HR 10/30 column (Amersham) which was equilibrated and eluted with buffer B (without sucrose) containing 150 mM NaCl and 1% NP40. Portions (20 μl) of fractions (0.5 ml) were used for immunoblotting and assay of PDE3 activity.
siRNA (small interfering RNA) knock-down of PDE3B
siRNA duplex oligonucleotides corresponding to murine (M) PDE3B mRNA (cat. no. L-043781-00) and a control non-targetting mRNA (cat. no. D-001810-10), used as a negative control, were purchased from Dharmacon. Optimal conditions for siRNA knock-down involved transfecting adipocytes with 100 nM siRNA using MBS (modified bovine serum) mammalian transfection reagent (Stratagene) in DMEM, following the manufacturer's protocols. After 10 h, fresh complete DMEM/10% (v/v) FBS was added, and adipocytes were further incubated for 46 h. Specific PDE3B knock-down was confirmed via immunoblotting, PDE3 activity assays and quantitative real-time RT (reverse transcriptase)–PCR (see Supplementary Figure 1 at http://www.BiochemJ.org/bj/404/bj4040257add.htm).
After 56 h, adipocytes were incubated for 16 h in serum-free DMEM, and then without or with insulin (100 nM) for 10 min. Total microsomes were prepared from control, untransfected adipocytes, or adipocytes transfected with MPDE3B-siRNA or non-targeting control siRNA. Solubilized membrane fractions were prepared and subjected to chromatography on Superose 6, and fractions analysed as described above.
Expression of FLAG-tagged recombinant WT (wild-type) and mutant murine (M) PDE3B proteins in Sf21 cells and 3T3-L1 adipocytes
cDNAs encoding FLAG-tagged recombinant full-length WT MPDE3B and deletion mutants (MPDE3BΔ196, MPDE3B-Δ302, MPDE3BΔ604, lacking different amounts of N-terminal sequences, i.e. 196, 302, 604 amino acids respectively) were generated, cloned into pAcSG2 and co-transfected with linearized BaculoGold™ DNA into Sf21 cells using the BaculoGold™ transfection kit (BD Biosciences) [13]. For routine preparations of recombinant proteins, T75 flasks, containing ∼1.1×107 cells, were infected with recombinant virus for 72 h. For some experiments, solubilized recombinant full-length MPDE3B (3 ml, containing 8–10 mg of protein, 1500–2000 pmol of cAMP hydrolysed·min−1·mg−1) was purified (see Supplementary data at http://www.BiochemJ.org/bj/404/bj4040257add.htm), using a FLAG–M2–agarose affinity column.
Recombinant FLAG-tagged full-length and mutant adenoviruses, adeno-MPDE3B (AdPDE3B) or adeno-MPDE3BΔ604 (AdPDE3BΔ604) respectively were generated and amplified in HEK (human embryonic kidney)-293A cells, and virus particles characterized as described earlier [16]. Adipocytes were infected (3 h) with adenovirus AdPDE3B, AdPDE3BΔ604 or control β-galactosidase adenovirus (Adβ-gal) [provided by Dr B.J. Baum, NIDCR (National Institute of Dental and Craniofacial Research), NIH (National Institutes of Health), Bethesda, MD, U.S.A.] in complete DMEM before the medium was supplemented with 10% FBS, and incubated for another 45 h. Infection efficiency was determined by immunostaining with anti-adenovirus hexon protein using Adeno-x kit (Clontech). After 48 h, cells were washed (three times, PBS) and incubated (16 h) in complete DMEM without serum, and then without and with insulin. Fractions were prepared, subjected to Superose 6 chromatography and analysed as described above.
Superose 12 chromatography of purified recombinant MPDE3B and PKB
A Superose 12 HR 10/30 (Amersham) column was equilibrated with buffer C (50 mM Hepes, 1 mM EDTA, 150 mM NaCl, 10 mM PPi, 5 mM NaF, 1 μM okadaic acid, 1 mM Na3VO4, Roche protease inhibitor cocktail and 1% NP40, pH 7.5) containing 50 μg/ml BSA, and calibrated with gel-filtration standards from Bio-Rad Laboratories. Indicated amounts of purified recombinant FLAG-tagged PDE3B and pPKB, PKB or p-ΔPKB were incubated in a final volume of 200 μl for 30 min at 30 °C. After dilution to 1.0 ml with buffer C containing 50 μg/ml BSA, diluted reaction mixtures were immediately applied to the column through a 1.0 ml loop on the AKTA FPLC system at a flow rate of 0.5 ml/min, and were eluted with buffer C containing 50 μg/ml BSA. Samples of fractions (0.5 ml), collected on Frac 950, were assayed for protein content [A280 (absorbance); AKTA monitor UPC 900], for immunoblotting of PKB and PDE3B, and sometimes for measurement of PDE3B and PKB activities (results not shown).
Co-immunoprecipitation of FLAG-tagged recombinant MPDE3B and PKB
Recombinant MPDE3B [usually 50 arbitrary (u) units (1 u=the amount of PDE3 that hydrolysed 1 pmol of cAMP/min at 30 °C)] and its mutants were incubated (30 min, 30 °C) with indicated amounts of purified recombinant PKB, pPKB or p-ΔPKB (final volume, 200 μl). Reactions were stopped by dilution of reaction mixtures to 1.0 ml with buffer C/BSA and cleared as described above (cf ‘Immunoprecipitation and immunoblotting’ subsubsection). To block non-specific binding of Sf21 material to Protein G–Sepharose, approx. 1 ml of Protein G–Sepharose beads was incubated (overnight, 4 °C) with 5 ml (∼1 mg/ml) of Sf21 cell supernatant [prepared by homogenization of cells in buffer C containing 1% NP40 and centrifugation (12000 g, 20 min)], and thereafter washed (PBS, three times). Pretreated beads were used in clearing reaction mixtures and immunoprecipitation of recombinant proteins.
Cleared reaction mixtures were immunoprecipitated by incubation with anti-FLAG–agarose (25 μl, 2 h, 4 °C), or by incubation with anti-PDE3B or anti-PKB antibodies, followed by Protein G–Sepharose (50 μl, 1 h); immunoprecipitates were collected by centrifugation (2800 g, 4 °C, 5 min). Samples (usually 40–50 μl) of reaction mixtures (input proteins) and immunoprecipitated proteins were subjected to SDS/PAGE and immunoblotting with indicated antibodies.
Immunofluorescence microscopy
Adipocytes in 2-well collagen-coated culture slides (6×104 cells/well) (BD Biosciences) were incubated overnight in serum-free DMEM, and then without or with 100 nM insulin (10 min), before washing (cold PBS) and fixation (20 min, room temperature) with 4% (w/v) paraformaldehyde in PBS containing 1 mM CaCl2 and 1 mM MgCl2 (PBSCM). Cells were washed (PBSCM), permeabilized (5 min, 0.1% saponin in PBSCM), washed again and incubated (2 h) in 1 ml of blocking solution [10% (v/v) normal goat serum, 5% (w/v) BSA in PBSCM]. After washing (PBSCM), samples were incubated (16 h, 4 °C) with primary antibody in blocking solution. As controls, samples were incubated with nonimmune IgG prior to staining with secondary antibody. Samples were washed (PBSCM) and incubated (2 h, room temperature) in blocking buffer with secondary antibodies conjugated to Alexa Fluor® 488 or 594 (Invitrogen). Specificity of affinity-purified (see Supplementary data at http://www.BiochemJ.org/bj/404/bj4040257add.htm) anti-PDE3B-NT or CT antibodies was confirmed by the absence of PDE3B reactivity after incubation of antibodies with their immunizing peptides (results not shown). Adipocytes were viewed with a Zeiss LSM510 laser scanning confocal microscope.
RESULTS
Insulin-induced phosphorylation and activation of PDE3B in ER/Golgi membrane compartments
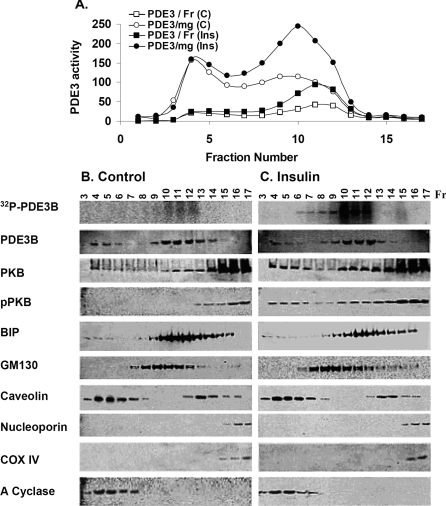
Total adipocyte membranes were isolated as described in the Materials and methods section. Based on the distribution of several molecular markers, PM, Golgi, ER and nuclear/mitochondrial fractions were well separated during fractionation on 10–45% continuous sucrose gradients (Figure 1). As shown in Figure 1(A), approx. 35–40% of the total membrane PDE3 activity is in fractions (fractions 4–7) enriched in caveolin-1 and adenylate cyclase (PM marker). In primary rat adipocytes, PDE3B was found to be associated with caveolae [9]. Approximately 60–65% of total membrane PDE3 activity is in internal membrane fractions (fractions 9–12), sometimes referred to as HDM/LDM fractions, i.e. fractions enriched in BiP (ER marker) and GM130 (Golgi marker). The same overall distribution of membrane-associated PDE3B was obtained using mouse primary adipocyte membranes [9].
Figure 1. Fractionation of 3T3-L1 adipocyte membranes on 10–45% sucrose gradients.
Adipocytes were incubated in serum-free DMEM for 16 h and then without (C) or with (Ins) 100 nM insulin (10 min, 37 °C). Some cells were incubated with [32P]Pi for 90 min before exposure to insulin. Total membranes were prepared, and 17 fractions (0.7 ml) were collected manually from continuous sucrose gradients. (A) Portions (10 μl) of fractions were assayed for PDE3 activity (expressed as pmol of cAMP hydrolysed·min−1·mg−1 or fraction−1) and protein content (results not shown). (B, C) Portions (400 μl) of fractions from adipocytes labelled with [32P]Pi incubated without (B) or with (C) insulin were analysed for [32P]PDE3B after immunoprecipitation of solubilized [32P]PDE3B with anti-PDE3B-CT antibody, separation of immunoprecipitated proteins on SDS/PAGE and phosphoimager analysis of wet gels (top row). Portions (25 μl) of fractions from adipocytes not incubated with [32P]Pi were subjected to SDS/PAGE, electrotransferred to NC membranes and immunoblotted with specific antibodies against PDE3B, PKB, pPKB, BiP (ER marker), GM130 (Golgi marker), caveolin-1, nucleoporin (nuclear marker), COX IV (mitochondria marker) and adenylate (A) cyclase (PM marker). Data shown are representative of two experiments for insulin-induced activation and [32P]PDE3B phosphorylation and Western blotting, and several experiments for activity assays.
In order to study insulin-induced phosphorylation and activation of PDE3B in the different membrane compartments, PDE3B activity and phosphorylation were measured after hormone stimulation and separation of membrane fractions by sucrose-density-gradient centrifugation. Incubation of 3T3-L1 adipocytes with insulin (100 nM, 10 min) increased PDE3 activity more than 2-fold in ER/Golgi fractions, compared with a smaller increase in caveolin-enriched PM fractions (Figure 1A). Furthermore, incubation of [32P]Pi-labelled 3T3-L1 adipocytes with insulin demonstrated that PDE3B was phosphorylated to a greater extent in ER/Golgi fractions (containing BiP and GM130) than in caveolin-enriched PM fractions (top panels, Figures 1B and 1C). There did not appear to be significant translocation of PDE3B between membrane fractions in response to insulin. Compared with unstimulated adipocytes, however, insulin stimulation resulted in enrichment of PM and ER/Golgi membranes with PKB as well as pPKB, a kinase known to be involved in insulin-induced phosphorylation and activation of PDE3B [10–12].
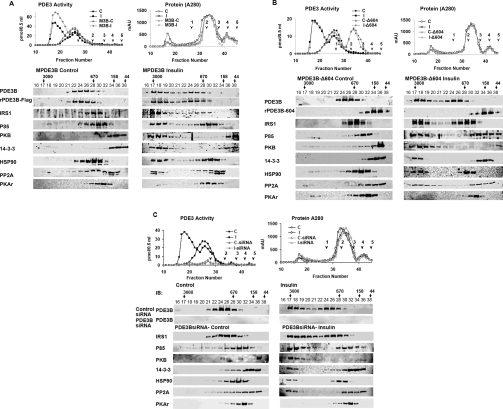
Insulin-induced formation of macromolecular complexes containing PDE3B and molecules related in its activation: effects of insulin and wortmannin
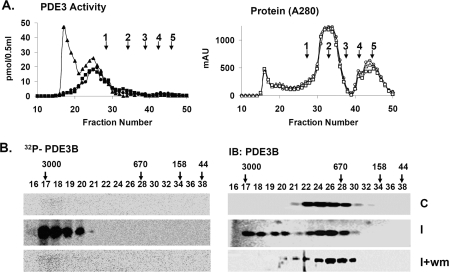
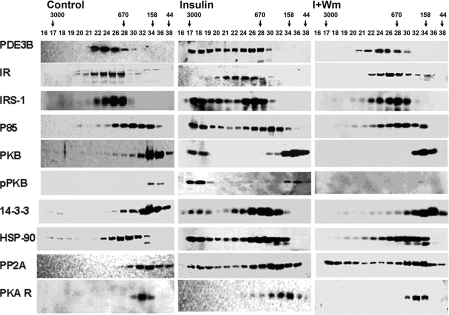
To characterize, in insulin-treated adipocytes, macromolecular or high-molecular-mass complexes (HMWC-ins) that might contain PDE3B, solubilized microsomal membranes were fractionated by gel filtration on Superose 6 (HR 10/30) columns (exclusion Mr >4×107). As seen in Figure 2(A) (left), PDE3B exhibited a molecular mass of ∼1000 kDa, much larger than its predicted monomeric molecular mass of ∼135 kDa. After incubation of adipocytes with insulin, the apparent molecular mass of ∼50% of the total PDE3B activity increased and was eluted at ≥3000 kDa, consistent with its presence in a multiprotein complex(es). Furthermore, phosphorylated PDE3B (Figure 2B) from insulin-treated adipocytes, which presumably corresponds to activated PDE3B, eluted primarily with HMWC-ins, not in fractions that contained most of the control PDE3B protein and activity. Fractions from adipocytes treated with wortmannin before insulin contained virtually no 32P-labelled/phosphorylated PDE3B (Figure 2B), as was the case for fractions originating from control cells. As seen in Figure 3 (middle), insulin treatment altered the elution patterns of several other signalling molecules, including IRS-1, IRS-2, PI3K p85 (p85-subunit of PI3K), PKB, HSP-90, 14-3-3 and PP2A (protein phosphatase 2A), to the extent that portions of these molecules co-eluted with PDE3B at ≥3000 kDa. This effect of insulin on the proteins, except for PP2A, was prevented by prior treatment of adipocytes with wortmannin (Figure 3, right). Little immunoreactive actin (results not shown) and little or no IR or PKARI co-eluted with the insulin-induced macromlecular complex(es).
Figure 2. Insulin-induced phosphorylation/activation of PDE3B: inhibition by wortmannin.
Solubilized total microsomes (3 mg of protein), prepared from adipocytes incubated for 10 min without [control (C)] or with 100 nM insulin [insulin (I)], or with insulin after incubation with 100 nM wortmannin (30 min) (I+wm) were subjected to chromatography on Superose 6. Fractions were analysed as described in the Materials and methods section. (A) (Left panel) PDE3 activity [pmol of cAMP hydrolysed·min−1·(0.5 ml)−1] (●, ▲, ■) and (right panel) protein content (AU 280 nm) (○, △, □) were measured in indicated fractions from control (●, ○), insulin (▲, △) and I+wm adipocytes (■, □). Of the PDE3 activity applied, >90% was recovered in column fractions. MW standards: 1, thyroglobulin; 2, γ-globulin; 3, ovalbumin; 4, myoglobin; 5, Vitamin B12 are indicated. Au, absorption units. (B) To detect phosphorylated [32P]PDE3B, experiments like those in (A) were performed with adipocytes labelled with [32P]Pi. PDE3B was immunoprecipitated with anti-PDE3B-CT antibody (25 μl) from samples (400 μl) of fractions, separated by SDS/PAGE, and 32P-labelled PDE3B was detected in wet gels by phosphoimager (Amersham) analysis before the same gels were used for Western immunoblotting of PDE3B, using anti-PDE3B CT antibody. The results are representative of two experiments.
Figure 3. Insulin-induced assembly of macromolecular complex(es) containing PDE3B and signalling proteins, identified during chromatography on Superose 6.
Samples (20 μl) of indicated fractions (0.5 ml) from Figure 2(A) were subjected to SDS/PAGE and immunoblotting with specific antibodies against PDE3B-CT, IR, IRS-1, PI3K p85, PKB, pPKB, 14-3-3, HSP-90, PP2A and PKARI. Results are representative of three experiments.
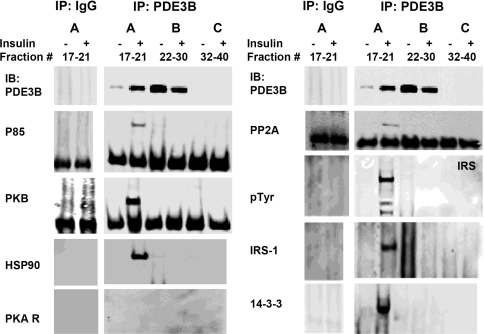
Samples of Superose 6 fractions from control or insulin-treated 3T3-L1 adipocytes (Figure 3) were pooled in three groups before immunoblotting of proteins that immunoprecipitated with anti-PDE3B CT antibodies. As seen in Figure 4, several signalling proteins co-immunoprecipitated with PDE3B (but not with non-immune rabbit IgG) from the HMWC-ins [∼3000 kDa fractions (group A)], whereas signalling proteins in pooled fractions from groups B and C from control or insulin-treated cells did not co-immunoprecipitate with PDE3B. Reactivity with anti-phosphotyrosine antibodies indicated that IRS proteins that co-immunoprecipitated with PDE3B were tyrosine-phosphorylated, and presumably ‘activated’. Taken together, our results suggest that, in insulin-treated adipocytes, PDE3B is part of a multimolecular complex (or complexes) comprising IRS-1, IRS-2, PI3K, PKB, HSP-90, 14-3-3, PP2A and additional, unidentified proteins.
Figure 4. Insulin-induced assembly of macromolecular complex(es) containing PDE3B and signalling proteins: co-immunoprecipitation with PDE3B.
Superose 6 fractions, as in Figure 3, were divided into three groups (A, B and C), and pooled for co-immunoprecipitation (group A: fractions 17–21; group B: fractions 22, 24, 26, 28 and 30; group C: fractions 32, 34, 36, 38 and 40). Pooled column fractions (150 μl from each of these fractions), were cleared, and complexes containing PDE3B were immunoprecipitated with 20 μl of anti-PDE3B CT antibodies (overnight, 4 °C). Cleared group A fractions were also immunoprecipitated with control rabbit IgG. Protein G–Sepharose-bound proteins were eluted in 100 μl of Laemmli's sample buffer. Samples (15 μl) were subjected to SDS/PAGE and immunoblotting with specific antibodies against PDE3B, PI3K p85, PKB, HSP-90 and PKARI (left panel) and PP2A, p-Tyr, IRS-1 and 14-3-3 (right panel). Some membranes used for immunoblotting lower molecular mass proteins were used also for reblotting/detecting larger proteins, including IRS-1, p-Tyr and HSP-90. Co-immunoprecipitated proteins were found largely in the higher molecular mass Superose 6 fractions (group A) after stimulation with insulin. Results were similar in three experiments. Non-specific binding of proteins was not detected in control IgG immunoprecipitates.
As seen in Figure 5, in differentiated adipocytes, adenovirus-mediated expression of recombinant full-length FLAG-tagged MPDE3B (which contains, in its N-terminal region, sites phosphorylated in response to insulin [17]) and MPDE3B-Δ604 (N-terminal truncated PDE3B in which the first 604 amino acids were deleted) demonstrated that insulin-induced activation of PDE3B and its recruitment into macromolecular complexes required the presence of the N-terminal region of PDE3B, since recombinant MPDE3B (Figure 5A), and not MPDE3B-Δ604 (Figure 5B), eluted with endogenous PDE3B, and was activated by insulin and recruited into HMWC-ins. Consistent with earlier work [18], in control adipocytes, MPDE3B-Δ604 is eluted not with endogenous ∼1000 kDa PDE3B, but with a much lower molecular mass (Figure 5B), and is not activated or recruited into macromolecular complexes by insulin. Importantly, MPDE3B-Δ604 does not interfere with the recruitment of other signalling molecules into HMWC-ins. Similarly, as seen in Figure 5(C), siRNA-induced knock-down of PDE3B (compare with Supplementary Figure 1 at http://www.BiochemJ.org/bj/404/bj4040257add.htm) does not prevent insulin-induced recruitment of signalling molecules into HMWC-ins, indicating that PDE3B is not necessary for insulin-induced complex formation, and does not serve as the central scaffold with regard to the components of the macromolecular complexes we have studied. Also, pretreatment of the cells with cilostamide, a PDE3 inhibitor, did not prevent insulin-induced complex formation (results not shown). Overexpression of 14-3-3 in adipocytes reduced insulin-induced activation of PDE3B by almost 20% (Supplementary Figure 2 at http://www.BiochemJ.org/bj/404/bj4040257add.htm), which perhaps is related to increased binding of 14-3-3 to IRS-1 after insulin stimulation; this may interfere with the ability of IRS-1 to recruit and activate PI3K [19].
Figure 5. Analysis of insulin-induced assambly of macromolecular complex(es) after overexpression of FLAG-tagged full length PDE3B (A) and mutant truncated PDE3B-Δ604 (B), or after siRNA-mediated deplation of PDE3B (C).
(A, B) Effect of adenovirus-mediated overexpression of FLAG-tagged full-length PDE3B and mutant truncated PDE3B-Δ604 on insulin-induced assembly of macromolecular complex(es). Differentiated adipocytes were infected with Adβ-gal (control) (A, B), full-length AdMPDE3B (MPDE3B) (A) or truncated AdPDE3B-Δ604 (Δ604) (B), and incubated for 10 min without [control (C), C-MPDE3B or C-Δ604] or with 100 nM insulin [insulin (I), I-MPDE3B or I-Δ604]. (A) Total microsomes (centrifugation of 13000 g supernatant at 175000 g) from adipocytes infected with AdMPDE3B or Adβ-gal were prepared and solubilized (3 mg of proteins) as described in the Materials and methods section. (B) Because Δ604 is predominantly cytosolic, total lysates (13000 g supernatant) (3 mg of protein) prepared from adipocytes infected with AdPDE3B-604 were treated with detergent, and centrifuged. (A, B) Solubilized fractions were subjected to chromatography on Superose 6 and analysed as described in the Materials and methods section. (A, B) (Left panels) PDE3 activity [pmol of cAMP hydrolysed·min−1·(0.5 ml)−1] (●, ■, ▲, ◆) and (right panels) protein content (AU 280 nm) (○, □, △, ◇) were measured in indicated fractions from (A, B) control (●, ○), insulin (■, □) and (A) AdMPDE3B- or (B) AdPDE3B-604-infected adipocyte control (▲, △), insulin (◆, ◇); of the PDE3 activity applied, >90% was recovered in column fractions. MW standards: 1, thyroglobulin; 2, γ-globulin; 3, ovalbumin; 4, myoglobin; 5, vitamin B12. Samples (20 μl) of indicated fractions (0.5 ml) were subjected to SDS/PAGE and immunoblotting with specific antibodies against the indicated proteins. Results are representative of two experiments. (C) Effect of siRNA-mediated depletion of PDE3B on insulin-induced assembly of macromolecular complex(es). Control (untreated) adipocytes, or adipocytes transfected with non-targeting control siRNA or PDE3B siRNA, were incubated for 10 min without [control (C) or siRNA-(C)] or with 100 nM insulin [insulin (I), siRNA-(I)]. Solubilized microsomal fractions were analysed as described in the Materials and methods section and Figures 5(A) and 5(B). (Left panels) PDE3 activity [pmol of cAMP hydrolysed·min−1·(0.5 ml)−1] (●, ■, ▲, ◆) and (right panels) protein content (AU 280 nm) (○, □, △, ◇) in indicated fractions from control (●, ○), insulin (■, □), and PDE3B-siRNA transfected adipocyte control (▲, △), insulin (◆, ◇); of the PDE3 activity applied, >90% was recovered in column fractions. MW standards: 1, thyroglobulin; 2, γ-globulin; 3, ovalbumin; 4, myoglobin; 5, vitamin B12. Samples (20 μl) of indicated fractions (0.5 ml) were subjected to SDS/PAGE and immunoblotting with specific antibodies against indicated proteins from the siRNA-transfected fractions. Results are representative of two experiments.
Taken together, these results suggested that, in 3T3-L1 adipocytes, activation of membrane-associated PDE3B by insulin involved reversible assembly of macromolecular complexes that contained PDE3B, PKB and other signalling molecules. We further characterized the interactions between membrane-associated PDE3B and PKB, as described below.
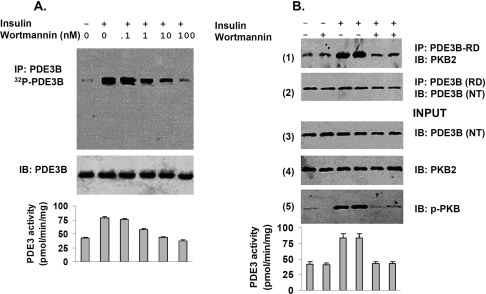
Co-immunoprecipitation of PDE3B and PKB from solubilized membranes of insulin-treated adipocytes
After incubation of 3T3-L1 adipocytes with insulin, PDE3B and PKB were co-immunoprecipitated from solubilized microsomal membranes with anti-PDE3B antibodies (Figure 6B, panel 1). As we had found in rat adipocytes and FDCP2 myeloid cells [12,20], incubation of 3T3-L1 adipocytes with wortmannin, which inhibits PI3K, blocked phosphorylation (Figure 6A, upper panel) and activation of PDE3B (Figure 6A, lower panel). As shown in Figure 6(B), wortmannin inhibited not only phosphorylation of PKB (pPKB) (panel 5) and activation of membrane-associated PDE3B (bottom), but also the interaction and co-immunoprecipitation of PDE3B and PKB/pPKB (panel 1). Another PI3K inhibitor, LY294002, and the tyrosine kinase inhibitor, Genistein (which blocked insulin receptor-mediated tyrosine phosphorylation of IRS-1), also inhibited insulin-induced activation of PKB and PDE3B and their co-immunoprecipitation (Supplementary Figure 3 at http://www.BiochemJ.org/bj/404/bj4040257add.htm).
Figure 6. Co-immunoprecipitation of PDE3B and PKB from solubilized membranes of insulin-treated adipocytes: inhibition by wortmannin.
(A) For phosphorylation of PDE3B, adipocytes were incubated with [32P]Pi as described in the Materials and methods section, and then for 10 min without or with 100 nM insulin or with insulin after incubation (30 min) with different concentrations of wortmannin (as indicated). [32P]PDE3B was immunoprecipitated from solubilized and cleared total membrane proteins (1 mg) with 25 μl of anti-PDE3B-CT antibody. Immunoisolated [32P]PDE3B was separated by SDS/PAGE and detected by phosphoimager (Amersham) analysis of the wet gels, after which proteins were electrotransferred to NC membranes for Western immunoblotting using anti-PDE3B-CT antibody. Results shown are representative of two experiments. (B) In another experiment, adipocytes not exposed to [32P]Pi were incubated (30 min) without or with 100 nM wortmannin, and then for 10 min without or with 100 nM insulin as indicated. For co-immunoprecipitation of PDE3B and PKB, solubilized, cleared, total membrane proteins (1 mg) from adipocytes were incubated with anti-PDE3B-RD antibody (25 μl) and subjected to SDS/PAGE and Western blotting with the indicated antibodies. Input proteins: 30 μg of solubilized membrane proteins. Similar results were observed in two other experiments, using anti-PKB (monoclonal) antibodies for co-immunoprecipitation of PDE3B and PKB. (A, B) Bottom panels: effects of wortmannin on insulin-stimulated PDE3 activity (expressed as pmol·min−1·mg−1) in solubilized total membranes (10 μg) prepared from adipocytes incubated without [32P]Pi, assayed as described in the Materials and methods section; values represent means±½ range.
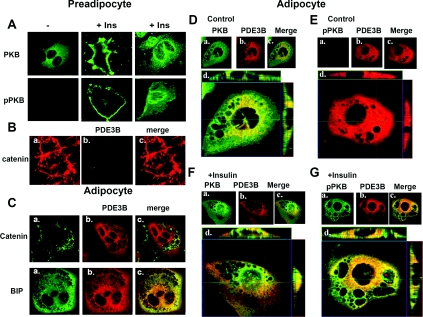
Co-localization of PKB, phospho-PKB and PDE3B by confocal microscopy
In unstimulated preadipocytes, PKB was uniformly distributed (Figure 7A, top row); and no significant staining was detected for pPKB (Figure 7A, row 2), or PDE3B or for neutral lipids (Oil Red O) (results not shown). A similar distribution of PKB in 3T3-L1 fibroblasts was previously reported [21]. After incubation of preadipocytes for 10 min with insulin, however, PKB and pPKB were significantly more concentrated in PM regions (Figure 7A, middle) and perinuclear regions (Figure 7A, right), more pronounced in some cells than in others. In preadipocytes, catenin was detected primarily at PM, at the periphery of the cell (Figure 7Ba), and PDE3B was not detected (Figure 7Bb). This configuration changed markedly with differentiation (Figure 7C), as adipocytes appeared to develop membranous invaginations (not present in preadipocytes), which stain for catenin (Figure 7Ca, top row). The ER network (BiP marker) extends extensively throughout 3T3-L1 adipocytes (Figure 7Ca, bottom row). Consistent with sucrose gradient findings (Figure 1), and our earlier studies [13], the subcellular distribution patterns of immunostaining of PDE3B (Figure 7Cb) and BiP (Figure 7Ca, bottom row) in adipocytes were almost identical, and PDE3B (red) was concentrated in ER region, and also found in the PM area (Figure 7Cb, bottom row). The overall distribution of PKB (green) was similar in both unstimulated adipocytes (Figure 7Da) and preadipocytes (Figure 7A), and little, if any, pPKB was detected in unstimulated adipocytes (Figure 7Ea). After a 10 min incubation of adipocytes with insulin (Figures 7F and 7G), more PKB and pPKB were concentrated in PM and perinuclear regions. When, after insulin treatment (Figures 7D–7G), the images of PKB or pPKB and PDE3B were superimposed, the extent of co-localization of PDE3B (red) with PKB or pPKB (green) appeared in Z-sections (Figures 7D–7G, panel d) to be greater in the ER region than in the PM region. Large amounts of green staining (PKB and pPKB) in merged images are consistent with the conclusion that substantial amounts of PKB and pPKB were not co-localized with PDE3B.
Figure 7. Subcellular localization of PDE3B, PKB and pPKB by confocal microscopy.
Preadipocytes (A, B) and adipocytes (C–G) were fixed, permeabilized with saponin and immunostained with indicated primary antibodies and Alexa Fluor® 488- or 594-conjugated secondary antibodies. In preadipocytes (A), PKB (cat. no. 2966, dilution 1:100) (green) was distributed throughout the cell. After incubation of cells with 100 nM insulin for 10 min (A), PKB and pPKB (cat. no. 4051, dilution 1:200) (green) were concentrated in PM areas and perinuclear structures. In preadipocytes, catenin (Ba) (cat. no. C-19220, dilution 1:200) was detected in PMs, at the periphery of the cells. No significant staining for PDE3B (Bb) was detected in preadipocytes using anti-PDE3B (peptide affinity-purified anti-PDE3B-NT antibody, dilution 1:200) (red) antibody. (C) Adipocytes stained for catenin (PM marker) or BiP (ER marker) (cat. no. G73320, dilution 1:200) in (a) and PDE3B (b) are merged in (c). Differentiated adipocytes developed invaginations (not present in preadipocytes), which stained for catenin. Unstimulated (D, E) or insulin-stimulated (F, G) adipocytes were stained for PKB or pPKB (a) and PDE3B (b) and are merged in (c). The extent of apparent co-localization of PDE3B (red) with PKB or pPKB (green) is greater in the ER region than in PM region. (Dd, Ed, Fd, Gd) Z-sections: reconstructed images from a stack of 18 sections (column IV) with 1 μm intervals showing areas of co-localization of PDE3B and PKB or pPKB in different planes. Centre X–Y (centre), above X–Z (top) and Y–Z (right) planes are at indicated positions. Findings are representative of several experiments.
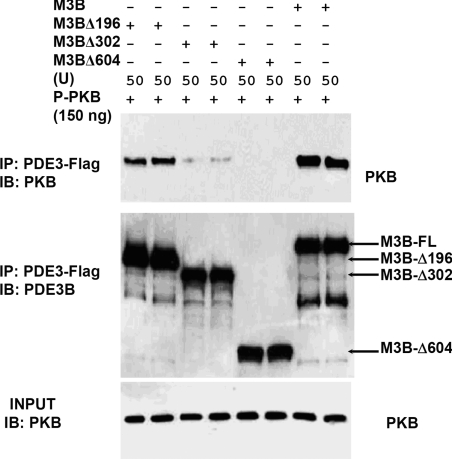
Structural determinants involved in interactions between PKB and PDE3B
To identify structural determinants involved in the interaction between PDE3B and PKB, we tested the ability of WT and truncated mutant PDE3B recombinants (expressed in Sf21 cells) to co-immunoprecipitate with PKB. As shown in Figure 8, after incubation of pPKB with WT FLAG-tagged MPDE3B, and its truncated mutants, including M3BΔ196, M3BΔ302 and M3BΔ604 (N-terminal truncated MPDE3B recombinant proteins in which the first 196, 302 or 604 amino acids were deleted respectively [13]), little, if any, M3BΔ302 and M3BΔ604 co-immunoprecipitated with pPKB using anti-FLAG antibodies. Truncated recombinant M3BΔ196 co-immunoprecipitated with pPKB, but less efficiently than WT MPDE3B, suggesting that the RD domain (1–302 amino acids) of PDE3B may be critically important for its interaction with pPKB.
Figure 8. Interaction between truncated MPDE3B recombinants and recombinant PKB and pPKB.
Recombinant FLAG-tagged WT MPDE3B (M3B), M3BΔ196, M3BΔ302 and M3BΔ604 [50 u (1 u=1 pmol of cAMP hydrolysed/min)] and recombinant pPKB (150 ng, ∼2.7 pmol, based on predicted molecular mass of ∼55000 for PKB) were incubated in buffer C containing 0.1% 2-mercaptoethanol for 30 min at 30 °C in a final volume of 200 μl. Since the different MPDE3B recombinant proteins exhibited different specific activities, all reaction mixtures were adjusted to contain similar amounts of protein by addition of protein from uninfected Sf21 cells. After dilution of reaction mixtures to 1 ml with a buffer containing 1 mg/ml BSA, samples (50 μl) were removed for analysis of Input proteins, before immunoprecipitating with 25 μl of anti-FLAG–agarose. Immunoprecipitated and input proteins were then subjected to immunoblotting with anti-PDE3B-CT and anti-PKB antibodies. Results are representative of three experiments.
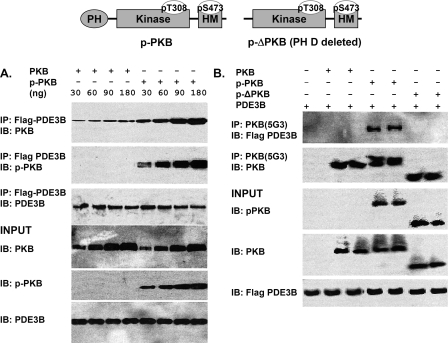
We also incubated FLAG-tagged recombinant MPDE3B with recombinant PKB, pPKB and p-ΔPKB, and immunoprecipitation was carried out with anti-FLAG or anti-PKB antibodies. As seen in Figure 9(A), more pPKB than PKB co-immunoprecipitated with FLAG–MPDE3B. As seen in Figure 9(B), after incubation of PKB, pPKB or p-ΔPKB with PDE3B, pPKB, but not PKB or p-ΔPKB, co-immunoprecipitated with MPDE3B (using anti-PKB antibodies), suggesting that activated pPKB interacted via its PH domain with MPDE3B.
Figure 9. Interactions between recombinant MPDE3B and recombinant PKB, pPKB and p-ΔPKB (PH domain deleted).
Top panel: linear structural model of pPKB, with PH-, kinase- and HM (hydrophobic motif)-domains and phosphorylated Thr308 and Ser473 residues noted. p-ΔPKB, in which the PH domain is removed, is also depicted. (A) Recombinant FLAG-tagged WT MPDE3B (M3B) (50 u) and recombinant PKB or pPKB at concentrations between 30 and 180 ng (∼0.54–3.2 pmol) were incubated (30 min, 30 °C) in buffer C (total volume, 200 μl). After addition of 800 μl of buffer C containing 1 mg/ml BSA, samples (40 μl) were taken for analysis of input proteins. Diluted mixtures were then cleared and incubated with immobilized anti-FLAG–agarose. Eluted proteins and input proteins were subjected to SDS/PAGE and immunoblotted with anti-PDE3B-CT, -PKB and -pPKB antibodies as indicated. Results were similar in two experiments. (B) Recombinant FLAG-tagged WT MPDE3B (M3B) (50 u) and 150 ng (∼2.7 pmol) of PKB, pPKB, or p-ΔPKB were incubated as in (A). After samples were taken for analysis of input proteins, diluted reaction mixtures were cleared, and immunoprecipitated with anti-PKB antibody. Immunoprecipitated proteins and input proteins were immunoblotted with anti-FLAG, -PKB and -pPKB antibodies as indicated. Results were similar in two experiments, and in two other experiments in which anti-FLAG–agarose was used for immunoprecipitation.
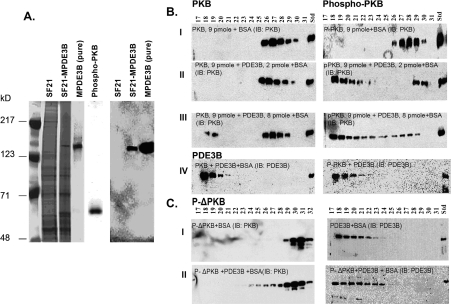
Interactions between purified recombinant MPDE3B (Figure 10A) and PKB, pPKB or p-ΔPKB were further assessed by chromatography on Superose 12 (HR 10/30) columns in buffer C containing BSA (50 μg/ml) and 1% NP40. As seen in Figures 10(B) (panel IV) and 10(C), purified recombinant MPDE3B eluted in fractions 17–21, whereas PKB and pPKB were in fractions 26–30 (Figure 10B, panel I) and p-ΔPKB in fractions 28–31 (Figure 10C, panel I). After incubation of MPDE3B with PKB, pPKB or p-ΔPKB for 30 min at 30 °C, pPKB, not PKB, interacted with and co-eluted with MPDE3B (Figure 10B, panels II and III). Some PKB did co-elute with MPDE3B when the concentration of PDE3B was increased (∼8 pmol), as seen in Figure 10(B) (panel III). Consistent with co-immunoprecipitation findings (Figure 10), p-ΔPKB did not co-elute with MPDE3B (Figure 10C, panel II), further suggesting that the PH domain of pPKB may be critically important for its interaction with PDE3B.
Figure 10. Purification of recombinant MPDE3B expressed in Sf21 cells: interactions between purified recombinant MPDE3B and recombinant PKB, pPKB and p-ΔPKB during gel filtration on Superose 12.
(A) Left panel: Sf21 cell lysates infected without (lane 2) or with (lane 3) MPDE3B-baculovirus; (lane 4) FLAG-tagged MPDE3B purified from Sf21 cell lysates as described in the Materials and methods section by anti-FLAG affinity chromatography (specific activity 0.71±0.16 μmol·min−1·mg−1, n=4) or (lane 5) (from another experiment) recombinant pPKB (Upstate Biotechnology) were subjected to SDS/PAGE. Gels were stained with Simply Blue Safe stain. Right panel: lanes 6–8: proteins from gels similar to that on the left (lanes 2–4) were electrophoretically transferred to NC membranes and immunoblotted with anti-FLAG peroxidase-conjugated antibodies for immunodetection of purified MPDE3B (lanes 7 and 8). (B, C) Purified recombinant WT PDE3B (0.25 or 1 μg, ∼2 or ∼8 pmol, based on predicted molecular mass of ∼122000) and PKB, pPKB or p-ΔPKB (500 ng, ∼9 pmol, based on predicted molecular mass of ∼55000) were incubated (30 min, 30 °C) in buffer C (total volume, 200 μl) with BSA (50 μg/ml), before dilution by addition of 800 μl of buffer C with BSA (50 μg/ml). Samples (1 ml) were then applied to Superose 12 HR 10/30 equilibrated and were eluted with buffer C and BSA (50 μg/ml). Fractions (0.5 ml) were collected on Frac 950 and portions were assayed for PDE3 and PKB activities (results not shown). Protein contents were monitored at A280 by AKTA monitor UPC 900. Portions (20 μl) of indicated fractions (0.5 ml) were subjected to SDS/PAGE, and immunoblotting with antibodies specific to PKB, pPKB and PDE3B-CT (B) and p-ΔPKB and PDE3B-CT (C). The results are representative of two experiments.
DISCUSSION
Insulin exerts profound pleiotropic effects on growth, differentiation, anabolic metabolism and energy homoeostasis, utilizing pathways shared by other growth factors and hormones. Mechanisms responsible for the specificity of insulin-signalling mechanisms remain to be elucidated. One of several explanations for divergence and specificity in biological signalling invokes spatial compartmentalization of some insulin-signalling and effector systems [22–24], including insulin-dependent regulation of caveolae- and IRS-signalling platforms, GLUT4 vesicle transport/translocation and, as suggested here, links from activated IRs to membrane-associated PDE3B in adipocytes.
Similar to results published by others [22,23], we found (F. Ahmad and V. Manganiello, unpublished work) that incubation of adipocytes with insulin resulted in rapid internalization of tyrosine-phosphorylated IR, as well as a marked increase in tyrosine-phosphorylated IRS-1, activated PI3K (which co-immunoprecipitated with IRS-1) and phosphorylated/activated PKB that was associated with intracellular membranes. Thus, although insulin did not induce significant translocation of PDE3B between PM and ER/Golgi fractions, activated signalling molecules involved in regulation of PDE3B were translocated to membrane compartments that contained PDE3B. Thus our results, as well as those of Inoue et al. [22], which demonstrated a rapid movement of IRS-1 and PI3K into microsomal fractions in insulin-stimulated 3T3-L1 adipocytes, suggest that activation of PKB could occur at internal membrane i.e. ER/Golgi compartments, as well as at PM locations. In addition, insulin also induced formation of HMWC-ins, identified during gel filtration of detergent-solubilized adipocyte membranes. Before exposure of adipocytes to insulin, PDE3B exhibited an apparent molecular mass of ∼1000 kDa; after activation by insulin (∼2-fold), the apparent molecular mass of approx. 50% of the total PDE3B activity increased and eluted at ≥3000 kDa, suggesting that PDE3B was part of these insulin-induced multimolecular complex(es).
HMWC-ins apparently contained phosphorylated/activated PDE3B and signalling molecules that are probably involved in regulation of its activity by insulin. PDE3B not recruited into HMWC-ins, or PDE3B from adipocytes not treated with insulin, exhibited little or no phosphorylation. In addition to phosphorylated PDE3B, HMWC-ins included IRS-1, PI3K p85, PKB/pPKB, HSP-90, 14-3-3 and PP2A, all of which co-eluted and co-immunoprecipitated with PDE3B after stimulation of adipocytes with insulin. Insulin-induced assembly of these proteins, except PP2A, was reversed by prior treatment of adipocytes with wortmannin. It appears that PP2A, unlike the other proteins, may not require PI3K activity (or some other enzyme inhibited by wortmannin). PP2A is apparently the principal phosphatase responsible for dephosphorylation of both PDE3B and PKB in adipocytes [25], and thus may play a central role in regulation of PDE3B activation. The separation of HMWC-ins from bulk protein by gel filtration may allow for application of proteomic/phosphoproteomic analyses of the complexes to identify critical molecules that might be further studied by expression, depletion, mutation, and real-time imaging in transfected/infected 3T3-L1 adipocytes.
After insulin stimulation, PDE3B that was recruited into HMWC-ins was highly phosphorylated. Our recent studies indicate that, in adipocytes, insulin induces multisite phosphorylation of PDE3B [17]. Whether phosphorylation of PDE3B is critical for its recruitment or for protein–protein interactions within the macromolecular complex(es) (which could also increase catalytic activity), or for both, is not certain at present. We did demonstrate, however, that insulin-induced activation of PDE3B and its recruitment into macromolecular complexes required the presence of the N-terminal region of PDE3B, which contains sites phosphorylated in response to insulin [17]. The truncated recombinant MPDE3B-Δ604, lacking the N-terminal region, was not activated or recruited into macromolecular complexes by insulin (Figure 5B). Expression of MPDE3B-Δ604, however, did not prevent insulin-induced formation of HMWC-ins. PDE3B is apparently not necessary for insulin-induced complex formation, since siRNA-induced knock-down of PDE3B or treatment of adipocytes with cilostamide, a PDE3 inhibitor, did not prevent insulin-induced complex formation (F. Ahmad and V. Manganiello, unpublished work). Although PDE3B is not the essential scaffold necessary for overall complex formation, recruitment of phosphorylated/activated PDE3B may be necessary for detailed organization of the complex and regulation of certain downstream cAMP-dependent signalling events, including, for example, insulin-induced regulation of a cAMP pool important for lipolysis. Antilipolytic effects of insulin are blocked in adipocytes from PDE3B−/− mice [4] and in primary rat adipocytes exposed to PDE3 inhibitors [26].
PKB is a likely proximate effector of PDE3B, and our studies indicated that a portion of the intracellular PKB was associated with intracellular membranes, co-eluted with PDE3B during gel filtration, and co-immunoprecipitated with endogenous PDE3B, consistent with a physical interaction between the two proteins, mediated most likely via structural determinants in the PH domain of PKB and the N-terminal regulatory region of PDE3B. This interaction was dependent on/enhanced by insulin-activation, suggesting that phosphorylation at activity-controlling or regulatory sites in either PDE3B or PKB (or both) was required.
Other interacting partners for PDE3 have been reported, e.g. PDE3B and the insulin receptor in human adipocytes [7], PDE3B and PI3Kγ in mouse heart [27], PDE3B and PDE3A with 14-3-3 [28,29], PDE3B and an unidentified ∼50 kDa protein with unknown function in rat and 3T3-L1 adipocytes [30,31], PDE3A and the leptin receptor in human platelets [8], and PDE3B and caveolae in primary rat adipocytes [9], suggesting that PDE3 regulates cAMP signalling in multiple intracellular compartments. The possibility of functional interactions with caveolae are of particular interest, since in adipocytes, caveolae-signalling platforms have been implicated in cAMP- and insulin-regulated pathways, e.g. especially lipolysis, lipogenesis and glucose uptake [26,32–36]. In FDCP2 myeloid cells [37] and human platelets [8], IL-4 (interleukin-4) and leptin may activate PDE3 and PDE3A respectively via JAK (Janus kinase)-induced tyrosine phosphorylation of IRS-2 and subsequent activation of PI3K and PKB. In platelets, a constitutive leptin receptor–PDE3A molecular complex may be involved in activation of PDE3A and its regulation of a cAMP pool that modulates platelet activation/aggregation [8].
Our studies provide an example of subcellular compartmentalization/localization of PDE3B with other signalling molecules, and how insulin-induced signals that activate PDE3B can be directed via targeting specific IRS–PI3K complexes to intracellular membranes, where activated PI3K can generate phosphoinositides that stimulate PDK (3-phosphoinositide-dependent kinase), leading to phosphorylation/activation of PKB, subsequent phosphorylation/activation of membrane-associated PDE3B, and regulation of cAMP-signalling pathways by insulin.
Online data
Acknowledgments
We thank Dr Martha Vaughan (NHLBI) for her critical reading of this paper and also acknowledge the professional skills and advice of Dr Christian A. Combs and Daniela Malide (Light Microscopy Core Facility, NHLBI) regarding microscopy-related experiments. E.D. was supported, in part, by Swedish Medical Research Council grant 3362. Part of this research was supported by the Intramural Programme of NHLBI, NIH.
References
- 1.Francis S. H., Turko I. V., Corbin J. D. Cyclic nucleotide phosphodiesterases: relating structure and function. Prog. Nucleic Acid. Res. Mol. Biol. 2001;65:1–52. doi: 10.1016/s0079-6603(00)65001-8. [DOI] [PubMed] [Google Scholar]
- 2.Shakur Y., Holst L. S., Landstrom T. R., Movsesian M., Degerman E., Manganiello V. Regulation and function of the cyclic nucleotide phosphodiesterase (PDE3) gene family. Prog. Nucleic Acid Res. Mol. Biol. 2001;66:241–277. doi: 10.1016/s0079-6603(00)66031-2. [DOI] [PubMed] [Google Scholar]
- 3.Walz H. A., Harndahl L., Wierup N., Zmuda-Trzebiatowska E., Svennelid F., Manganiello V. C., Ploug T., Sundler F., Degerman E., Ahren B., et al. Early and rapid development of insulin resistance, islet dysfunction and glucose intolerance after high-fat feeding in mice overexpressing phosphodiesterase 3B. J. Endocrinol. 2006;189:629–641. doi: 10.1677/joe.1.06522. [DOI] [PubMed] [Google Scholar]
- 4.Choi Y. H., Park S., Hockman S., Zmuda-Trzebiatowska E., Svennelid F., Haluzik M., Gavrilova O., Ahmad F., Pepin L., Napolitano M., et al. Alterations in regulation of energy homeostasis in cyclic nucleotide phosphodiesterase 3B-null mice. J. Clin. Invest. 2006;116:3240–3251. doi: 10.1172/JCI24867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Houslay M. D., Milligan G. Tailoring cAMP-signalling responses through isoform multiplicity. Trends Biochem. Sci. 1997;22:217–224. doi: 10.1016/s0968-0004(97)01050-5. [DOI] [PubMed] [Google Scholar]
- 6.Liu H., Maurice D. H. Expression of cyclic GMP-inhibited phosphodiesterases 3A and 3B (PDE3A and PDE3B) in rat tissues: differential subcellular localization and regulated expression by cyclic AMP. Br. J. Pharmacol. 1998;125:1501–1510. doi: 10.1038/sj.bjp.0702227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rondinone C. M., Carvalho E., Rahn T., Manganiello V. C., Degerman E., Smith U. P. Phosphorylation of PDE3B by phosphatidylinositol 3-kinase associated with the insulin receptor. J. Biol. Chem. 2000;275:10093–10098. doi: 10.1074/jbc.275.14.10093. [DOI] [PubMed] [Google Scholar]
- 8.Elbatarny H. S., Maurice D. H. Leptin-mediated activation of human platelets: involvement of a leptin receptor and phosphodiesterase 3A containing cellular signaling complex. Am. J. Physiol. Endocrinol. Metab. 2005;289:E695–E702. doi: 10.1152/ajpendo.00125.2005. [DOI] [PubMed] [Google Scholar]
- 9.Nilsson R., Ahmad F., Andersson U., Sward K., Manganiello V., Degerman E. Plasma membrane cyclic nucleotide phosphodiesterase 3B (PDE3B) is associated with caveolae in primary adipocytes. Cell. Signalling. 2006;18:1713–1721. doi: 10.1016/j.cellsig.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 10.Wijkander J., Landstrom T. R., Manganiello V., Belfrage P., Degerman E. Insulin-induced phosphorylation and activation of phosphodiesterase 3B in rat adipocytes: possible role for protein kinase B but not mitogen-activated protein kinase or p70 S6 kinase. Endocrinology. 1998;139:219–227. doi: 10.1210/endo.139.1.5693. [DOI] [PubMed] [Google Scholar]
- 11.Kitamura T., Kitamura Y., Kuroda S., Hino Y., Ando M., Kotani K., Konishi H., Matsuzaki H., Kikkawa U., Ogawa W., et al. Insulin-induced phosphorylation and activation of cyclic nucleotide phosphodiesterase 3B by the serine-threonine kinase Akt. Mol. Cell. Biol. 1999;19:6286–6296. doi: 10.1128/mcb.19.9.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmad F., Cong L. N., Stenson Holst L., Wang L. M., Rahn L. T., Pierce J. H., Quon M. J., Degerman E., Manganiello V. C. Cyclic nucleotide phosphodiesterase 3B is a downstream target of protein kinase B and may be involved in regulation of effects of protein kinase B on thymidine incorporation in FDCP2 cells. J. Immunol. 2000;164:4678–4688. doi: 10.4049/jimmunol.164.9.4678. [DOI] [PubMed] [Google Scholar]
- 13.Shakur Y., Takeda K., Kenan Y., Yu Z. X., Rena G., Brandt D., Houslay M. D., Degerman E., Ferrans V. J., Manganiello V. C. Membrane localization of cyclic nucleotide phosphodiesterase 3 (PDE3). Two N-terminal domains are required for the efficient targeting to, and association of, PDE3 with endoplasmic reticulum. J. Biol. Chem. 2000;275:38749–38761. doi: 10.1074/jbc.M001734200. [DOI] [PubMed] [Google Scholar]
- 14.Sudo T., Tachibana K., Toga K., Tochizawa S., Inoue Y., Kimura Y., Hidaka H. Potent effects of novel anti-platelet aggregatory cilostamide analogues on recombinant cyclic nucleotide phosphodiesterase isozyme activity. Biochem. Pharmacol. 2000;59:347–356. doi: 10.1016/s0006-2952(99)00346-9. [DOI] [PubMed] [Google Scholar]
- 15.Manganiello V., Vaughan M. An effect of insulin on cyclic adenosine 3′:5′-monophosphate phosphodiesterase activity in fat cells. J. Biol. Chem. 1973;248:7164–7170. [PubMed] [Google Scholar]
- 16.Ahmad F., Harndahl L., Tang Y., Holst L. S., Manganiello V. C. Adenovirus-mediated overexpression of murine cyclic nucleotide phosphodiesterase 3B. Methods Mol. Biol. 2005;307:93–107. doi: 10.1385/1-59259-839-0:093. [DOI] [PubMed] [Google Scholar]
- 17.Lindh R., Ahmad F., Resjo S., James P., Yang J. S., Fales H. M., Manganiello V., Degerman E. Multisite phosphorylation of adipocyte and hepatoma phosphodiesterase 3B. Biochim. Biophys. Acta. 2007;1773:584–592. doi: 10.1016/j.bbamcr.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 18.Kenan Y., Murata T., Shakur Y., Degerman E., Manganiello V. C. Functions of the N-terminal region of cyclic nucleotide phosphodiesterase 3 (PDE 3) isoforms. J. Biol. Chem. 2000;275:12331–12338. doi: 10.1074/jbc.275.16.12331. [DOI] [PubMed] [Google Scholar]
- 19.Xiang X., Yuan M., Song Y., Ruderman N., Wen R., Luo Z. 14-3-3 facilitates insulin-stimulated intracellular trafficking of insulin receptor substrate 1. Mol. Endocrinol. 2002;16:552–562. doi: 10.1210/mend.16.3.0790. [DOI] [PubMed] [Google Scholar]
- 20.Rahn T., Ridderstrale M., Tornqvist H., Manganiello V., Fredrikson G., Belfrage P., Degerman E. Essential role of phosphatidylinositol 3-kinase in insulin-induced activation and phosphorylation of the cGMP-inhibited cAMP phosphodiesterase in rat adipocytes. Studies using the selective inhibitor wortmannin. FEBS Lett. 1994;350:314–318. doi: 10.1016/0014-5793(94)00797-7. [DOI] [PubMed] [Google Scholar]
- 21.Feng J., Park J., Cron P., Hess D., Hemmings B. A. Identification of a PKB/Akt hydrophobic motif Ser-473 kinase as DNA-dependent protein kinase. J. Biol. Chem. 2004;279:41189–41196. doi: 10.1074/jbc.M406731200. [DOI] [PubMed] [Google Scholar]
- 22.Inoue G., Cheatham B., Emkey R., Kahn C. R. Dynamics of insulin signaling in 3T3-L1 adipocytes. Differential compartmentalization and trafficking of insulin receptor substrate (IRS)-1 and IRS-2. J. Biol. Chem. 1998;273:11548–11555. doi: 10.1074/jbc.273.19.11548. [DOI] [PubMed] [Google Scholar]
- 23.Saltiel A. R., Kahn C. R. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 24.Chang L., Chiang S. H., Saltiel A. R. Insulin signaling and the regulation of glucose transport. Mol. Med. 2004;10:65–71. doi: 10.2119/2005-00029.Saltiel. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Resjo S., Goransson O., Harndahl L., Zolnierowicz S., Manganiello V., Degerman E. Protein phosphatase 2A is the main phosphatase involved in the regulation of protein kinase B in rat adipocytes. Cell. Signalling. 2002;14:231–238. doi: 10.1016/s0898-6568(01)00238-8. [DOI] [PubMed] [Google Scholar]
- 26.Zmuda-Trzebiatowska E., Oknianska A., Manganiello V., Degerman E. Role of PDE3B in insulin-induced glucose uptake, GLUT-4 translocation and lipogenesis in primary rat adipocytes. Cell. Signalling. 2006;18:382–390. doi: 10.1016/j.cellsig.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 27.Patrucco E., Notte A., Barberis L., Selvetella G., Maffei A., Brancaccio M., Marengo S., Russo G., Azzolino O., Rybalkin S. D., et al. PI3Kγ modulates the cardiac response to chronic pressure overload by distinct kinase-dependent and independent effects. Cell. 2004;118:375–387. doi: 10.1016/j.cell.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 28.Onuma H., Osawa H., Yamada K., Ogura T., Tanabe F., Granner D. K., Makino H. Identification of the insulin-regulated interaction of phosphodiesterase 3B with 14-3-3 beta protein. Diabetes. 2002;51:3362–3367. doi: 10.2337/diabetes.51.12.3362. [DOI] [PubMed] [Google Scholar]
- 29.Pozuelo R. M., Campbell D. G., Morrice N. A., Mackintosh C. Phosphodiesterase 3A binds to 14-3-3 proteins in response to PMA-induced phosphorylation of Ser428. Biochem. J. 2005;392:163–172. doi: 10.1042/BJ20051103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Onuma H., Osawa H., Ogura T., Tanabe F., Nishida W., Makino H. A newly identified 50 kDa protein, which is associated with phosphodiesterase 3B, is phosphorylated by insulin in rat adipocytes. Biochem. Biophys. Res. Commun. 2005;337:976–982. doi: 10.1016/j.bbrc.2005.09.144. [DOI] [PubMed] [Google Scholar]
- 31.Chavez J. A., Gridley S., Sano H., Lane W. S., Lienhard G. E. The 47kDa Akt substrate associates with phosphodiesterase 3B and regulates its level in adipocytes. Biochem. Biophys. Res. Commun. 2006;342:1218–1222. doi: 10.1016/j.bbrc.2006.02.091. [DOI] [PubMed] [Google Scholar]
- 32.Liu P., Rudick M., Anderson R. G. Multiple functions of caveolin-1. J. Biol. Chem. 2002;277:41295–41298. doi: 10.1074/jbc.R200020200. [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto M., Toya Y., Schwencke C., Lisanti M. P., Myers M. G., Jr, Ishikawa Y. Caveolin is an activator of insulin receptor signaling. J. Biol. Chem. 1998;273:26962–26968. doi: 10.1074/jbc.273.41.26962. [DOI] [PubMed] [Google Scholar]
- 34.Cohen A. W., Razani B., Schubert W., Williams T. M., Wang X. B., Iyengar P., Brasaemle D. L., Scherer P. E., Lisanti M. P. Role of caveolin-1 in the modulation of lipolysis and lipid droplet formation. Diabetes. 2004;53:1261–1270. doi: 10.2337/diabetes.53.5.1261. [DOI] [PubMed] [Google Scholar]
- 35.Karlsson M., Thorn H., Danielsson A., Stenkula K. G., Ost A., Gustavsson J., Nystrom F. H., Stralfors P. Colocalization of insulin receptor and insulin receptor substrate-1 to caveolae in primary human adipocytes. Cholesterol depletion blocks insulin signalling for metabolic and mitogenic control. Eur. J. Biochem. 2004;271:2471–2479. doi: 10.1111/j.1432-1033.2004.04177.x. [DOI] [PubMed] [Google Scholar]
- 36.Ost A., Ortegren U., Gustavsson J., Nystrom F. H., Stralfors P. Triacylglycerol is synthesized in a specific subclass of caveolae in primary adipocytes. J. Biol. Chem. 2005;280:5–8. doi: 10.1074/jbc.C400429200. [DOI] [PubMed] [Google Scholar]
- 37.Ahmad F., Gao G., Wang L. M., Landstrom T. R., Degerman E., Pierce J. H., Manganiello V. C. IL-3 and IL-4 activate cyclic nucleotide phosphodiesterases 3 (PDE3) and 4 (PDE4) by different mechanisms in FDCP2 myeloid cells. J. Immunol. 1999;162:4864–4875. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.