Abstract
Soluble SULTs (sulfotransferases) are important in the regulation of messenger molecules and the elimination of xenobiotics. However, sulfo-conjugation of various substrates can also lead to the formation of reactive metabolites that may induce cancer and cause other damage. The aim of the present study was to identify the SULT forms expressed in the human gastrointestinal tract, especially the colon and rectum (common sites for cancer), and to determine their cellular localization. Normal colonic or rectal tissue, resected with tumours, was obtained from 39 subjects. For comparison, we additionally studied one to four samples from stomach, jejunum, ileum, cecum and liver. SULTs were detected by immunoblotting, immunohistochemistry and measurement of enzyme activities. SULT1A1, 1A3 and 1B1 were found in all parts of the gastrointestinal tract, often exceeding levels in liver (where these forms were present at high, undetectable and low levels respectively). They were predominantly localized in differentiated enterocytes. SULT1E1 and 2A1 were only detected in liver, jejunum, ileum and cecum. SULT1C1 was readily found in stomach, but was negligible elsewhere. SULT1A2 was present at low levels in individual samples. The remaining forms were not detected with the limitation that only high levels could be recognized with the antisera used. In conclusion, SULTs are abundant in the gastrointestinal tract of man. We suspect that they are involved in the presystemic elimination of bioactive food-borne components, including aglycones released by gut microbiota, as well as the bioactivation of some procarcinogens.
Keywords: colon, intestinal biotransformation, stomach, sulfotransferase (SULT)
Abbreviations: DHEA, dehydroepiandrosterone; IPTG, isopropyl β-D-thiogalactoside; PAPS, 3′-phosphoadenosine 5′-phosphosulfate; SULT, sulfotransferase; UGT, UDP-glucuronosyltransferase
INTRODUCTION
Many drugs, secondary plant metabolites and other xenobiotics enter human organisms via the gastrointestinal tract. Absorbed xenobiotics often have to be metabolized before they can be excreted. Biotransformation of some compounds may already occur in the intestinal mucosa. Lipophilic xenobiotics are usually absorbed in the small intestine and are often metabolized in two phases, the introduction of a functional group in phase-I and the usage of this functional group for the conjugation with an anionic moiety in phase-II. The situation is somewhat different in the large bowel, where bacteria may convert xenobiotics (e.g. glycosylated phytochemicals) and xenobiotic metabolites (e.g. glucuronides excreted with bile) into products that may be better absorbed than the educts. In general, these metabolites already contain hydroxy or other nucleophilic functional groups. Thus they may undergo conjugation reactions in the mucosa without need of prior phase-I metabolism. UGTs (UDP-glucuronosyltransferases) and SULTs (sulfotransferases) represent the major classes of conjugating enzymes for nucleophilic substrates.
Biotransformation of xenobiotics is usually mediated by enzymes with very broad substrate tolerance and involves some risk of the formation of toxic metabolites, in particular of chemically reactive intermediates, that may induce mutations and then lead to degenerative alterations, such as neoplasias. Indeed, stomach cancer is relatively common and colorectal cancer even is one of most prevalent neoplasias [1]. Among the phase-II enzymes, SULTs particularly are often involved in the formation of reactive intermediates [2,3]. They transfer the sulfo group from PAPS (3′-phosphoadenosine-5′ phosphosulfate) to acceptor molecules. The sulfate moiety, resulting from O-sulfonation, is a good leaving-group in certain chemical linkages. Such sulfo-conjugates are electrophilically reactive. Thus various aromatic amines, alkenylbenzenes and hydroxymethylated polycyclic aromatic hydrocarbons are examples for SULT-dependent rodent carcinogens [2]. We have expressed individual human and rodent SULTs in target cells of standard mutagenicity tests. Using these tools, we demonstrated for more than 100 chemicals a bioactivation by human SULTs to genotoxic metabolites directly or after phase-I metabolism [3]. The list of these chemicals comprises heat-induced food constituents (heterocyclic aromatic amines, 5-hydroxymethylfurfural), drugs (oxamniquine, cyproterone acetate), plant metabolites (safrole, aristolochic acids), environmental contaminants (alkylated polycyclic hydrocarbons, 3-nitrobenzanthrone) and industrial chemicals (2-nitropropane, nitro- and aminoarenes). When used at low concentrations, each promutagen was only activated by one or two human SULT forms, but different forms were involved in the activation of different compounds. In other cases, sulfo-conjugation can prevent the activation of promutagens [3]. SULTs are also important in the regulation of the levels of various hormones, such as steroids, catecholamines and iodothyronines [4–6]. The corresponding SULTs show high affinities for their endogenous substrates and could form an efficient defence system against food-borne hormones, if expressed in the gut.
For these reasons, it would be interesting to know which SULT forms are expressed in various sections and cells of the human alimentary tract. However, most studies on gastrointestinal SULTs focused on the determination of the conjugation of a specific substrate or the detection of an individual SULT protein in a specific section of the gastrointestinal tract [7–11]. Dooley et al. [12] attempted to study systematically the expression of the various SULTs in human tissues using reverse transcription PCR. However, this study focussed on skin tissues, with less intense investigations of other tissues for comparison. Moreover, mRNA and protein levels may not correlate in many cases. In the present study we aimed to identify the SULTs as proteins and/or by way of their enzyme activities in various sections of the human alimentary tract. The expression in colon and rectum was of particular interest, as these are common tumour sites.
A total of 11 human SULT forms have been characterized at the gene, message and protein levels. Two additional possible SULT genes (SULT1C3 and 6B1) were identified in the human genome [13]. We have constructed the putative SULT1C3 cDNA and demonstrated that it encodes a protein with SULT activity (W. Meinl and H. Glatt, unpublished work). Several nomenclatures have been proposed for SULTs. In the present study we use the same nomenclature as in our previous papers (e.g. [3] amd [19]). The names are identical with the names proposed by Blanchard et al. [14] for all forms except the SULT1C subfamily. We are using the names introduced by Freimuth et al. [15], who detected two members of this subfamily. Blanchard et al. [14] suggested the designations SULT1C2 for Freimuth's SULT1C1, and SULT1C4 for Freimuth's SULT1C2.
EXPERIMENTAL
Tissue samples
Tissue samples were obtained from Caucasian patients who underwent clinically indicated surgery. This material was made anonymous and used following the recommendations of the central ethical committee of the German Medical Association. SULT expression was studied in histologically normal tissue resected together with diseased tissue. Rectal tissue was from 11 patients (3 male, 8 female; aged 37–75 years) with rectal tumours. Colon tissue was from 24 patients (13 male, 11 female; aged 50–76 years) with colon tumours and from four patients (three male, one female; aged 36–74 years) with sigma diverticulitis. Cecal tissue was from a male patient (aged 68 years) with a rectal tumour. Ileal tissue was from three patients (two male, one female; aged 59–75 years) with rectal tumours and from one male patient aged 76 years with a colon tumour. Jejunal tissue was from a 33 year old male patient suffering from a pancreas carcinoma. Stomach and liver tissues were biopsies from male patients; no pathological changes were found in these biopsies.
None of the tumour patients had received radio- or chemo-therapy prior to the surgery. Treatment with osmotic laxatives preceded all intestinal surgeries, which were conducted under anaesthesia (midazolam for premedication; propafenol, sufetanil, rocuronium and isofluvane/O2/air for the surgery). There is no indication that any of these agents might affect the expression of xenobiotic-metabolizing enzymes. Some patients were treated with additional drugs. However, the number of patients treated with a given drug was too small for studying a possible influence on SULT expression.
After resection, gut samples were opened and washed with ice-cold mucosa buffer, which contained 40 mM Tris/HCl (pH 7.8), 120 mM KCl, 200 g/l glycerol and the protease inhibitor aprotinin (60000 kallikrein inhibitory units per litre; purchased under the trade name Trasylol from Bayer). Tissue samples were frozen in liquid nitrogen and stored at −80 °C until further use. For histological evaluation and immunohistochemistry, tissue specimens were fixed in phosphate-buffered formaldehyde (4%) and embedded in paraffin.
SULT1A1, 1A3 and 1B1 standards from inclusion bodies
The plasmid hST1B2/pKK233-2 [16] (hST1B2 is another designation for human SULT1B1) served as a template in the PCR procedure modifying the 5′- and 3′-flanking regions for subcloning SULT1B1 cDNA into pET24a(+) (Novagen) using the restriction sites for NdeI and HindIII. Restriction sites were introduced using the following primers from BioTeZ: 5′-GAATTCCATATGCTTTCCCCAAAAG-3′ and 5′-GAATTCAAGCTTTAAATCTCTGTGCGG-3′. After an initial denaturing step for 5 min at 96 °C, amplification was carried out for 30 cycles of 1 min at 94 °C, 1 min at 60 °C and 2 min at 72 °C, followed by a final elongation step for 10 min at 72 °C. The reaction mixture (100 μl) contained 4 units of Combi-Pol polymerase, 20 pmol of each primer, 50 ng of template, 2 mM dNTPs, 1.5 mM MgCl2 and Combi-Pol reaction buffer diluted according to the manufacturer's protocol. All PCR reagents were from InViTek.
For construction of pET-SULT1A1 and pET-SULT1A3, cDNAs were subcloned from pKK233-2-derived plasmids that contained either human SULT1A1 (also termed hP-PST) [17] or SULT1A3 (also termed hM-PST) [18] cDNA, into pET28b(+) (Novagen) using the restriction sites for NcoI and HindIII.
Escherichia coli BL21 (DE3; Novagen) was transformed with pET-SULT plasmids to yield E. coli BL21-SULT1A1, -SULT1A3 and -SULT1B1 strains. Overexpression of SULT in these strains was achieved by induction with IPTG (isopropyl β-D-thiogalactoside) as described in the pET system manual (Novagen). More than 90% of the soluble protein was SULT as determined by SDS/PAGE and staining with Coomassie Blue. When inclusion bodies were used, a single band (representing SULT) was detected even when excessive protein levels were loaded.
For preparation of inclusion bodies, bacteria harvested from 50 ml of culture were lysed in 1.25 ml of 40 mM Tris/HCl (pH 8) containing 4% (w/v) saccharose, 1 mM EDTA and 2 mg/ml lysozyme, and incubated on ice for 30 min. DNAseI was added to a final concentration of 90 μg/ml as well as MgCl2 and MnCl2 at a final concentration of 20 mM and 2 mM respectively. Incubation on ice was continued for another 30 min. Then 3 ml of a detergent buffer [1% (w/v) deoxycholic acid, 1% (w/v) Nonidet P40, 2 mM EDTA, 200 mM NaCl and 20 mM Tris/HCl (pH 7.5)] was added. The mixture was vigorously shaken and centrifuged at 10000 g at 4 °C for 10 min. The insoluble fraction was resuspended in 3 ml of a second detergent buffer [0.5% (w/v) Triton X-100, 1 mM EDTA and 20 mM Tris/HCl (pH 7.4)] and centrifuged (10000 g at 4 °C for 10 min). The washing steps were repeated, then the insoluble fraction was resuspended in 3 ml of 50 mM Tris/HCl (pH 7.5) containing 1 mM EDTA, and centrifuged (10000 g at 4 °C for 10 min). This step was repeated twice, then the inclusion bodies were resuspended in 1 ml of 0.1% SDS and stored at −80 °C.
SULT standards from cytosolic preparations of recombinant Salmonella Typhimurium strains
We have expressed all human SULTs in Salmonella Typhimurium TA1538 at levels amounting to 0.5–10% of the cytosolic protein [19]. SULTs in these preparations are more stable and easier to handle than purified enzymes and inclusion bodies, and therefore were normally used as standards in immunoblotting.
Preparation of the cytosolic fraction from human tissues and bacteria
Surgical specimens were thawed from −80 °C on ice overnight. Opened intestinal samples were placed on an ice-cooled glass plate, rinsed with ice-cold PBS, and mucosa was scraped off with a rubber scalpel. Mucosa and liver were homogenized using a Potter–Elvehjem tissue grinder. The homogenization medium (3–5 ml per g of tissue) contained 10 mM sodium phosphate buffer (pH 7.4), 150 mM KCl, 1 mM EDTA and Complete Protease Inhibitor Cocktail diluted according to the manufacturer's protocol (Boehringer Mannheim). The same buffer (1 ml) was used for sonication of bacteria harvested from 100 ml overnight culture. The homogenates were centrifuged at 100000 g for 1 h at 4 °C. The resulting supernatant was termed the cytosolic fraction. Protein concentrations were determined according to the method of Lowry using BSA as a standard.
Antisera
Antisera raised in rabbits against human SULT1B1 (previously termed ST1B2) [16] and human SULT2A1 (previously termed DHEA-ST) [20] were kindly provided by C. Falany (Department of Pharmacology, University of Alabama at Birmingham, Birmingham, AL, U.S.A.) and are designated as ‘anti-1B1-serum’ and ‘anti-2A1-serum’ respectively. The antisera named ‘anti-r1E1-serum’ and ‘anti-1C1-serum’ were raised in a rabbit against rat liver oestrogen SULT [21] and in a sheep against human SULT1C1 [22] (where it was named SULT1C2) respectively. Both antisera were gifts of M. Coughtrie (Division of Pathology and Neuroscience, Ninewells Hospital and Medical School, University of Dundee, Dundee, U.K.).
For generation of anti-1A1-serum, two female New Zealand White rabbits (Tierzucht Schönwalde) were immunized with 250 μg of human SULT1A1 inclusion bodies dissolved in PBS with 1% (w/v) SDS and mixed with complete Freund's adjuvant. Immunization was repeated after 4 weeks using incomplete Freund's adjuvant. After six additional immunizations at 4-week intervals, sera were collected. Antisera from both rabbits were probed with various human SULT forms by immunoblot analysis, and the one chosen for further experiments was termed ‘anti-1A1-serum’.
In some cases immunoabsorption was used to increase the specificity of antisera. Anti-1A1- and anti-1C1-antisera were incubated with SULT1B1–Sepharose to remove antibodies binding to SULT1B1. Likewise, anti-1B1-serum was incubated with SULT1A1–Sepharose to enhance its specificity. For generation of SULT1A1– and SULT1B1–Sepharose, IPTG-induced E. coli BL21-1A1 and -1B1 were sonicated in 50 mM NaHCO3 (pH 8.3) and the 100000 g supernatant was coupled to CNBr–Sepharose (Amersham Pharmacia Biotech) according to the manufacturer's protocol. Antisera were incubated with SULT-sepharose at 4 °C overnight. These pretreated antisera showed somewhat lower immunoreactivities in immunoblot analyses than the native antisera. Since cross-reacting SULTs could normally be distinguished due to their differing electrophoretic mobilities, the native antisera were used in immunoblot analyses, unless specified otherwise. However, the pretreated antisera with enhanced specificity were consistently used for immunohistochemistry. Moreover, negative control antisera were used in immunohistochemistry; to this end, specific antibodies were removed from anti-1A1- and anti-1B1-antisera by incubation with SULT1A1– and SULT1B1–Sepharose respectively.
Immunodetection of SULT forms in cytosolic fractions and characterization of antisera
Cytosolic fractions were separated using SDS/PAGE (11% gels) according to the method of Laemmli [22a]. Cytosolic fractions of Salmonella Typhimurium TA1538 expressing various human SULTs [19] as well as purified SULT inclusion bodies were included as standards. After electrophoresis, proteins were transferred to Hybond ECL membrane (Amersham Pharmacia Biotech) and probed with primary antisera diluted in Tris-buffered saline [50 mM Tris/HCl (pH 7.6) and 150 mM sodium chloride] containing 0.1% Tween 20 and 1% BSA. For anti-1A1-serum, BSA was replaced with non-fat dried skimmed milk powder (1%) as blocking reagent. Anti-1A1- and anti-1B1-antisera were diluted 1:10000 in blocking reagent; for all other antisera, the dilution was 1:2000. Goat anti-rabbit or donkey anti-sheep IgG–peroxidase conjugate (Sigma), at a dilution of 1:2000, was used as secondary antibody. The immunoreactive bands were visualized using the ECL system together with Hyperfilm ECL (Amersham Pharmacia Biotech).
In order to remove bound antibodies for reprobing with other antisera, membranes were incubated with 62.5 mM Tris/HCl (pH 6.7) containing 100 mM 2-mercaptoethanol and 2% SDS at 50 °C for 1 h. Afterwards they were washed twice with Tris-buffered saline containing 0.1% Tween 20 for 15 min before new blocking reagent was applied. Then membranes were probed with the next antiserum as described above. All blots contained various controls (cDNA-expressed SULTs), whose reactivity with the various antisera is known. Incomplete removal of an antibody or washing-out of antigen before reprobing would have led to unspecific or insufficient signals respectively; of these controls, this was never the case under the conditions used. Likewise, comparable results were obtained when we repeated the analysis with the antiserum used for the initial probing at the end of the reprobing series.
Immunohistochemistry
Slices of 4 μm were dewaxed in toluene and rehydrated in alcohol, and then washed sequentially in water and PBS containing 0.1% Tween 20. They were then treated with 0.3% H2O2 for 10 min and incubated with PBS containing 0.1% Tween 20 and 1% BSA (or non-fat skimmed milk powder in case of anti-1A1-serum) for 30 min at room temperature (25 °C). Antisera raised against SULT1B1 (1:1500 diluted in PBS containing 0.1% Tween 20 and 1% BSA), SULT2A1 (1:800 in PBS containing 0.1% Tween 20 and 1% BSA) or SULT1A1 (1:1500 in PBS containing 0.1% Tween 20 and 1% non-fat skimmed milk powder) were applied at 4 °C overnight. Unbound antibody was removed by washing with PBS containing 0.1% Tween 20. Bound antibodies were detected using the Vectastain ABC kit (Vector Laboratories) in combination with the diaminobenzidine chromogen system (Immunotech). Nuclei were counterstained with Methyl Green (Sigma), 0.8% in 20% ethanol.
Enzyme activity measurements
Enzyme activity of SULT2A1 and 1E1 was determined by the turnover of [3H]DHEA {[1,2,6,7-3H(N)]dehydroepiandrosterone; 2.2 TBq/mmol, NEN DuPont} and [6,7-3H(N)]17β-oestradiol (1.48 TBq/mmol) respectively, as described previously [23]. The reaction mixture (250 μl) contained 5–200 μg of cytosolic protein, 7 mM MgCl2, 20 μM PAPS, 3 μM [3H]DHEA or 20 nM [3H]oestradiol, and 50 mM Tris/HCl (pH 7.4). Preliminary experiments using varying amounts of cytosolic fraction of each tissue were performed to determine the linear range. Blanks contained water instead of PAPS. After 20 min at 37 °C, 250 μl of 100 mM Tris/HCl (pH 8.7) and 3 ml of chloroform were added to terminate the reaction. Following a single extraction, 250 μl of the aqueous layer was subjected to liquid scintillation counting.
Genotyping of SULT1A1
Genomic DNA was prepared from mucosa samples using the Qiagen Tissue kit. A polymorphism, encoding an Arg/His exchange in codon 213, was analysed by PCR and enzymatic restriction of PCR products, as described previously [24].
RESULTS
Characterization of antisera
Cytosolic fractions were prepared from Salmonella Typhimurium strains expressing the individual human SULTs. Various levels of each preparation were electrophoresed, blotted and probed with the various antisera (Supplementary Figure 1 at http://www.BiochemJ.org/bj/404/bj4040207add.htm). Each antiserum recognized several SULT forms with various sensitivities. SULT1A1, 1A3, 1B1, 1C1 and 2A1 showed strong signals with at least one antiserum even at the lowest amount of bacterial cytosol used (1 μg of total protein, corresponding to 10–60 ng of SULT protein). Somewhat higher amounts of protein were required for detecting SULT1A2 and 1E1. The remaining forms were only recognized when relatively high SULT levels were used. Semiquantitative data on the reactivity of each antiserum with each SULT form are presented in Supplementary Table 1 (at http://www.BiochemJ.org/bj/404/bj4040207add.htm).
Tissue and cellular distribution of SULT1A forms
Immunoblot analyses
SULT1A1 differed in its electrophoretic mobility (32 kDa) from all other forms except SULT4A1. Anti-r1E1-serum as well as anti-1A1-serum readily detected SULT1A1 standard but showed negligible immunoreactivity with SULT4A1. Therefore the identification of SULT1A1 protein was unambiguous. It was well-detected in all samples from gastrointestinal tissues using anti-r1E1-serum (Figure 1, top panel) and anti-1A1-serum (Figure 2, lower panel). Using cDNA-expressed SULT1A1 as a standard, tissue levels of this protein were estimated (Table 1). They were particularly high in ileum and liver. The next highest levels were observed in large bowel. The differences between cecum, colon and rectum were smaller than those among different colonic or rectal samples.
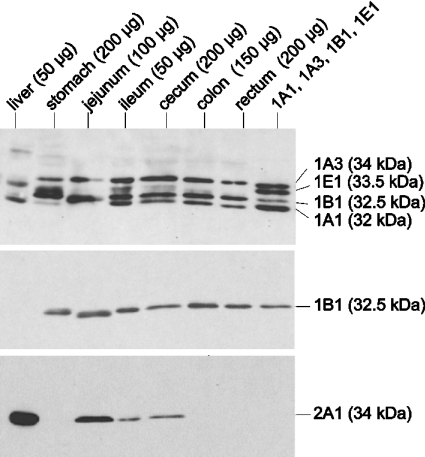
Figure 1. Immunodetection of SULTs in various parts of the gastrointestinal tract using antisera raised against rat SULT1E1 (top panel), human SULT1B1 (middle panel) and human SULT2A1 (bottom panel).
Cytosolic fractions were prepared from hepatic and gastrointestinal tissues. The amount of cytosolic protein used is indicated in parentheses. Cytosolic protein of Salmonella Typhimurium TA1538-SULT1E1 (0.5 μg) and inclusion bodies of SULT1A1 (60 ng), SULT1A3 (60 ng) and SULT1B1 (80 ng) were mixed (lane headed ‘1A1, 1A3, 1B1, 1E1’) to imitate their expression in ileum. The samples were electrophoresed and transferred to a nitrocellulose membrane. The blot was first probed with anti-r1E1-serum. After removing the antibody, the blot was reprobed with anti-1B1-serum. Antibodies were again removed and the blot was reprobed with anti-2A1-serum. Native antisera were used in these experiments.
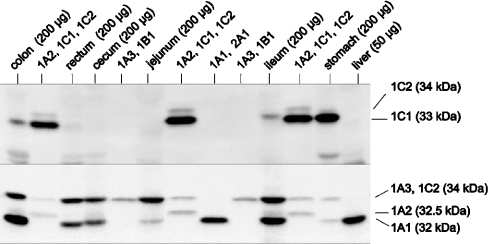
Figure 2. Immunodetection of SULTs in various parts of the gastrointestinal tract and liver using antisera raised against human SULT1C1 (upper panel) and SULT1A1 (lower panel).
Cytosolic fractions of Salmonella Typhimurium TA1538-SULT1A1 (3 μg of protein), 1A2 (5 μg), 1A3 (3 μg), 1B1 (1 μg), 1C1 (0.5 μg), 1C2 (5 μg) and human tissues (amount of protein indicated in the Figure) were electrophoresed and transferred to a nitrocellulose membrane. The blot was probed with anti-1C1-serum. Then antibodies were removed and the blot was reprobed with anti-1A1-serum. Both antisera had been pre-incubated with SULT1B1–Sepharose to abolish cross-reactivity with SULT1B1.
Table 1. Summary on SULT forms detected in cytosolic fractions from liver and various sections of the gastrointestinal tract.
SULT levels were determined semi-quantitatively using cDNA-expressed proteins as standards. This method may involve inaccuracies in the absolute value (we estimate by a factor of up to three), but should be more accurate for comparison of the same protein in different samples (within rows). Values are means of n samples. An extended version of this Table is presented in Supplementary Table 3 (at http://www.BiochemJ.org/bj/404/bj4040207add.htm). −, below the limit of detection.
| ng SULT/mg of cytosolic protein | |||||||
|---|---|---|---|---|---|---|---|
| Rectum (n=11) | Colon (n=28) | Cecum (n=1) | Jejunum (n=1) | Ileum (n=4) | Stomach (n=1) | Liver (n=1) | |
| SULT form | |||||||
| 1A1 | 210 | 180 | 150 | 75 | 1200 | 75 | 800 |
| 1A2 | − | −* | 75 | − | −* | − | 75 |
| 1A3 | 310 | 320 | 300 | 300 | 1500 | 150 | − |
| 1B1 | 130 | 120 | 100 | 50 | 420 | 50 | 25 |
| 1C1 | − | −* | − | − | −* | 25 | − |
| 1E1 | −† | −† | 6† | 12† | 50† | −† | 100† |
| 2A1 | −† | −† | 25† | 200† | 150† | −† | 1000† |
*Some samples showed faint signals close to the limit of detection.
†Levels of SULT1E1 and 2A1 protein were paralleled by results from enzyme activity assays using characteristic substrates for these forms, oestradiol (20 nM) and DHEA (3 μM) respectively (see Supplementary Table 4 at http://www.BiochemJ.org/bj/404/bj4040207add.htm).
The SULT1A1 level varied more strongly among the 28 colon samples studied (11-fold) than among the 11 rectal samples (4-fold). SULT1A1 shows a common genetic polymorphism involving an Arg/His exchange in codon 213. Using platelets as an enzyme source, Raftogianis et al. [25] found 7.7- and 6.4-fold higher SULT1A1 activity in *Arg/*Arg and *Arg/*His subjects than in *His/*His subjects. In the present study, the *His/*His genotype was associated with the lowest mean SULT1A1 protein level in colon as well as rectum, 88 and 58% compared with the *Arg/*Arg genotype (See Supplementary Table 2 at http://www.BiochemJ.org/bj/404/bj4040207add.htm). However, the highest mean levels were observed in the *Arg/*His rather than the *Arg/*Arg genotype, and the levels of the individual samples strongly overlapped between all genotypes. Thus the Arg/His polymorphism does not appear to be the major factor determining SULT1A1 expression in large bowel. The expression was not associated with the sex of the individual, the only other factor analysed (results not shown).
SULT1A2 co-migrated with SULT1B1 (32.5 kDa), but not with any other SULT form. Both antisera that detected SULT1A2 with good sensitivity (anti-1A1-serum and anti-r1E1-serum) cross-reacted with SULT1B1. Pre-incubation with SULT1B1–Sepharose abolished this cross-reactivity. Using the pretreated antiserum, a weak signal at 32.5 kDa was obtained with some samples from colon, cecum and ileum, but not with SULT1B1 standard (Figure 2, lower panel). It was also seen in liver when the amount of protein was increased to 200 μg (results not shown), in agreement with previous results with three other liver samples [19].
SULT1A3 co-migrated (34 kDa) with several other SULTs (1C2, 1C3 and 2A1). Using anti-r1E1-serum (Figure 1, top panel) and anti-1A1-serum (Figure 2, lower panel), a protein co-migrating with these SULTs was observed in rectum, colon, cecum, ileum, jejunum and stomach, but not in liver. Since the immunoreactivity of both antisera was high for SULT1A3 but low for the other 34 kDa forms, it was likely that the signal represented SULT1A3. This notion was corroborated by results of analyses using antisera that preferentially detect the other forms. Thus anti-1C1-serum, which recognized the SULT1C2 and 1C3 standards, failed to produce any 34 kDa signal in gastrointestinal samples (Figure 2, upper panel). The remaining 34 kDa form, SULT2A1, showed much higher expression in liver than in any gastrointestinal sample (see below). This high hepatic level of SULT2A1 did not produce any signal with the anti-r1E1- and anti-1A1-antisera recognizing a 34 kDa protein in the extrahepatic samples. Thus the latter signals appeared to be exclusively due to SULT1A3. Expression of SULT1A3 was very high in all ileal samples studied (Table 1). Levels amounting to approximately one-fifth of those in ileum were detected in the remaining intestinal sections (jejunum, cecum, colon and rectum). However, an 8.8-fold variation was observed among the colonic samples (see Supplementary Table 3 at http://www.BiochemJ.org/bj/404/bj4040207add.htm). This was not associated with gender (results not shown). The variation was less in rectum and small intestine (3.7- and 1.4-fold, but in smaller numbers of samples). The gastric sample showed a lower SULT1A3 level than the intestinal samples. SULT1A3 was not detected in liver.
Immunohistochemical localization
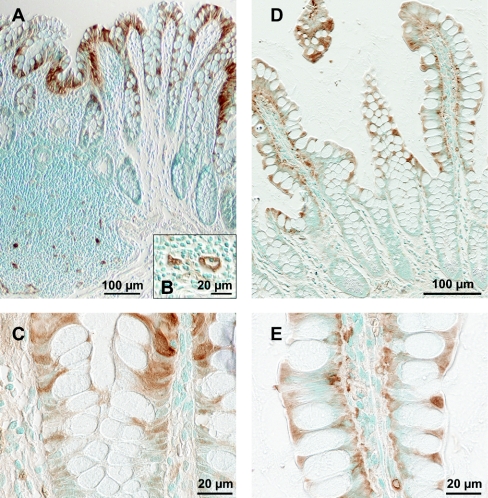
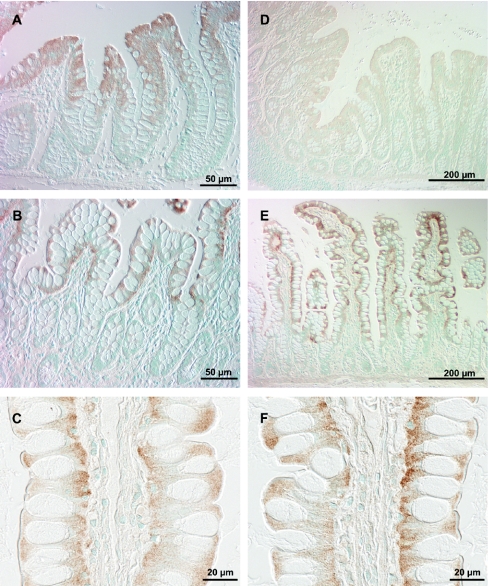
Anti-1A1-serum pretreated with SULT1B1–Sepharose (to increase the specificity) showed strong signals in both colon and ileum. No staining was observed when the SULT1A-specific antibodies were removed from the antiserum by pre-incubation with SULT1A1–Sepharose. In colon, SULT1A protein was detected in mature enterocytes (luminal half of the crypts) and endothelial cells (Figures 3A–3C). Immunostaining of the enterocytes was restricted to the nuclei in the middle of the crypt but extended to the cytoplasm and clearly intensified in more luminal cells (Figures 3A and 3C). Endothelial staining was particularly strong in the capillaries of the lymphoid follicles (Figure 3B). In ileum, staining was observed in mature enterocytes, starting at the crypt villus border and in endothelial cells (Figures 3D and 3E). Anti-1A1-serum does not distinguish between the different SULT1A forms. Since the predominant amount of the antigen is localized in the mature enterocytes and since SULT1A1 as well as SULT1A3 are abundant in colon and ileum (as shown in the immunoblot analyses), both forms must be present in these cells. The data do not make it possible to state which SULT1A forms are expressed in endothelial cells.
Figure 3. Immunodetection of SULT1A forms in colonic (A–C) and ileal (D and E) mucosa.
Formalin-fixed, paraffin-embedded slices were incubated with anti-1A1-serum (pre-incubated with SULT1B1–Sepharose) at 4 °C overnight. Bound antibodies are indicated by brown staining using the Vectastain ABC kit with diaminobenzidine as substrate. Nuclei were stained with Methyl Green.
Tissue distribution and cellular localization of SULT1B1
Immunoblot analyses
SULT1B1 co-migrated with SULT1A2 (32.5 kDa) but not with any other form. Anti-1B1-serum detected SULT1B1 with high sensitivity, but was inactive towards SULT1A2. Using this antiserum, SULT1B1 protein was detected in stomach, jejunum, ileum, cecum, colon and rectum (Figure 1, middle panel). In that blot, the immunoreactive protein in the jejunum sample migrated slightly faster than that in the neighbouring lanes. However, this difference disappeared in a repeat experiment, suggesting that this was due to an accidental technical, rather than biological, reason. SULT1B1 was not detected in liver when 50 μg of cytosolic liver protein was loaded (Figure 1, middle panel). This amount was used in order to avoid an excessive signal with anti-2A1-serum (Figure 1, bottom panel). When the amount of hepatic protein was increased to 200 μg, the presence of SULT1B1 was clearly manifested (results not shown) in agreement with previous results with three other liver samples [19].
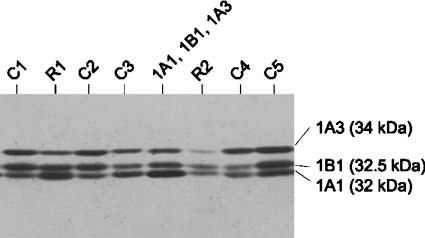
Anti-r1E1-serum clearly recognized SULT1B1 in addition to the SULT1A forms and SULT1E1. This antiserum was useful in comparing the expression levels of several SULT forms in human tissues (Figure 1, top panel). A 9-fold variation in the SULT1B1 level, which was not associated with gender (results not shown), was observed among colonic samples, whereas variability was less (nearly 5-fold) in rectum and small intestine (Supplementary Table 3.) As described in preceding sections, colon also showed the highest variation in the levels of SULT1A1 and 1A3. However, the variation of these three SULT forms was not concurrent. As shown in Figure 4, the SULT1A1 signal was stronger in some colonic samples than the SULT1A3 and 1B1 signals (e.g. C3). In other samples, the SULT1A3 (C4) or 1B1 (C1) signal was strongest.
Figure 4. Variation of SULT expression between different colonic and rectal samples.
Cytosolic protein (200 μg) of five colon (C1–C5) and two rectum (R1 and R2) samples were electrophoresed and transferred on to nitrocellulose membrane. The membrane was probed with anti-r1E1-serum. Cytosolic fractions of Salmonella Typhimurium TA1538-SULT1A1 (5 μg), TA1538-SULT1A3 (0.5 μg) and TA1538-SULT1B1 (1.5 μg) were used as standards.
Immunohistochemical localization
Anti-1B1-serum pre-incubated with SULT1A1–Sepharose (to increase the specificity) stained mature enterocytes in colon (upper third of the crypt; Figure 5A) and ileum (starting at the transition from crypt to villus; Figures 5B and 5C). Staining was cytoplasmic. In addition, a small number of cells of the lamina propria, possibly leucocytes, were stained. Staining was abolished when the anti-SULT1B1-serum was pre-incubated with SULT1B1–Sepharose to remove the anti-SULT1B1-antibodies.
Figure 5. Immunodetection of SULT1B1 (left-hand panels) and SULT2A1 (right-hand panels) in colonic mucosa (upper row) and ileal mucosa (middle and lower rows).
Formalin-fixed, paraffin-embedded slices of colon and ileum were incubated with anti-1B1-serum (pre-incubated with SULT1A1–Sepharose) and anti-2A1-serum respectively, at 4 °C overnight. Bound antibodies are indicated by brown staining using the Vectastain ABC kit with diaminobenzidine as substrate. Nuclei were stained with Methyl Green.
Tissue distribution of SULT1C forms
SULT1C1 differed in its electrophoretic mobility (33 kDa) from the other SULT forms. However, the difference was small with regard to SULT1B1 (32.5 kDa). Using anti-1C1-serum pretreated with SULT1B1–Sepharose, SULT1C1 protein was readily detected in stomach mucosa, but was not found in rectum, jejunum and cecum (Figure 2, upper panel). Low levels close to the limit of detection appeared to be present in a few samples of colon and ileum.
SULT1C2 and 1C3 proteins co-migrated with SULT1A3 and 2A1 (34 kDa) and were detected by all five antisera used, including anti-1C1- and anti-1B1-antisera. The latter antisera did not produce a 34 kDa signal on blots with the various tissue samples. Thus SULT1C2 and 1C3 were not found in the samples, but the limit of detection was unsatisfactory (400 ng per mg of cytosolic protein) due to moderate immunoreactivity of all antisera available.
Tissue distribution of SULT1E1
Immunoblot analyses
SULT1E1 protein was best detected using anti-r1E1-serum. This antiserum also recognized several other human SULTs which differed in their electrophoretic mobility from SULT1E1. A band with the characteristic mobility of SULT1E1 (33.5 kDa) was clearly detected in liver and ileum (Figure 1, top panel). Faint signals at the same position were observed in jejunum and cecum. Marginally faster-moving proteins were detected in stomach and, very weakly, in some colon samples. Clarification was required whether this band represented SULT1E1 or another form, with SULT1C1 being a candidate.
Enzyme activity
Sulfation of oestradiol at a low substrate concentration (20 nM) is a good marker for the presence of SULT1E1 enzyme [26]. Substantial oestradiol SULT activities were observed in liver and ileum, followed by jejunum and cecum (see Supplementary Table 4 at http://www.BiochemJ.org/bj/404/bj4040207add.htm). These findings agree with the results of the immunoblot analysis. Sulfation of oestradiol, at a very low rate, was also detected in colonic, rectal and gastric samples. At such low rates it is important to examine the specificity of the reaction, as other SULTs can also catalyse the sulfation of oestradiol, although they only reach their maximal activity when the concentration of oestradiol is increased by several orders of magnitude, e.g. to 6 μM for SULT1A1 [26]. In order to estimate the contribution of non-SULT1E1 forms to the sulfation of oestradiol in colonic cytosol, we mixed bacterially expressed SULT1A1, 1A3 and 1B1 at the same levels as found in a pooled colon preparation. The oestradiol SULT activity of this SULT pool was similar to that of colon cytosol (Supplementary Table 4). Thus the activity cannot be used to demonstrate the presence SULT1E1 in colon. Likewise, the oestradiol SULT activity observed in rectum and stomach samples can readily be explained by the presence of forms other than SULT1E1.
Tissue distribution and cellular localization of SULT2A1
Immunoblot analyses
SULT2A1 co-migrated (34 kDa) with several other SULTs (1A3, 1C2 and 1C3). Anti-2A1-serum efficiently detected SULT2A1, did not give any signal with SULT1A3, and only showed very weak immunoreactivity with SULT1C2 and 1C3. Moreover, the latter forms were not detected in the tissues investigated. Therefore SULT2A1 protein could be unequivocally identified in immunoblot analyses. High levels of SULT2A1 were detected in liver (Figure 1, bottom panel). Lower levels were found in small intestine (jejunum and ileum). Expression was very low in cecum and not detected in the remaining tissues (stomach, colon and rectum).
Enzyme activity
DHEA is an excellent substrate for SULT2A1 [26]. The only other human SULT forms that conjugate DHEA are SULT2B1a and 2B1b [27]. However, these forms show much lower activity with DHEA than SULT2A1 and were not detected in the tissues investigated (see below). DHEA SULT activity in tissue samples (Supplementary Table 4) correlated with the results of the immunoblot analyses, in the following order: liver > ileum, jejunum>cecum>rectum, colon, stomach (below limit of detection).
Immunohistochemistry
No immunohistochemical staining was observed in colon mucosa using anti-2A1-serum (Figure 5D), which agrees with the immunoblotting and enzyme activity results obtained with subcellular preparations. In ileum, mature enterocytes but no other cells, were stained (Figures 5E and 5F). Staining primarily occurred in the cytoplasm, but the ileal enterocytes are so compact that a minor staining in nuclei could not be ruled out.
Tissue distribution of the remaining SULT forms (2B1a, 2B1b and 4A1)
The electrophoretic mobilities of SULT2B1a (40 kDa) and 2B1b (42 kDa) were unique among the human SULT members. Anti-2A1-serum detected these forms, expressed in bacteria, with moderate sensitivity (limit of detection ∼100 and 400 ng/mg of protein respectively). Neither form was detected in hepatic or gastrointestinal samples. Recently, it was reported that the subcellular localization of SULT2B1b may vary between tissues; it was primarily detected in cytosol in prostate, but in the nuclei in placenta [28]. Since we only used the cytosolic fraction, we would have overlooked nuclear enzyme in our immunoblot analysis. However, we found no staining of nuclei (or other cellular compartments) in our immunohistochemical analyses of colon with anti-2A1-serum, which cross-reacts with SULT2B1 forms. Ileal enterocytes were stained with anti-2A1-serum, but staining was primarily observed in the cytoplasm. No appropriate antiserum was available for detecting SULT4A1. Others have detected SULT4A1 mRNA or protein in brain, but not in any other tissue [29].
DISCUSSION
Overlap of the substrate specificity [26,30] limits the possibility of elucidating the expression pattern of the various SULTs solely based on activities. Therefore we focussed on the identification of SULT forms by immunoreactivity. We put much effort into a good electrophoretic resolution of SULT forms and the characterization of a series of antisera. In general, we were able to unambiguously identify the SULT proteins detected in immunoblots. In individual cases, the results were further corroborated using enzyme reactions with substrates (oestradiol and DHEA) that are characteristic for SULT1E1 and 2A1 respectively.
We detected seven SULT forms in the alimentary tract. SULT1A1, 1A3 and 1B1 were found in all parts of the alimentary tract. They showed their highest levels in ileum. The ileal level of SULT1A1 was slightly above the level of the hepatic sample used. SULT1A3 was present in ileum at a somewhat higher level than SULT1A1, but was absent in liver (with a detection limit of 5% of the ileal level). Likewise, expression of SULT1B1 was 17 times higher in ileum than in liver. All three forms were also present at relatively high levels in large bowel. Differences in the levels were minor between cecum, colon and rectum, but substantial between colo-rectal samples from different subjects. A genetic polymorphism that affects the expression of SULT1A1 in platelets did not appear to be an important factor for the variation of SULT1A1 in gut. At present, we can only speculate that dietary habits or microbial colonization may modulate the expression of SULTs. Other SULT forms (1A2, 1C1, 1E1 and 2A1) were only detected in some parts of the gastrointestinal tract. SULT1A2 appears to be much less abundant in the human organism than the other SULT1A forms. Others detected SULT1A2 at the mRNA level [12], but not at the protein level, despite an intensive search [31]. Under our electrophoretic conditions, SULT1A2 separates from SULT1A1, although these proteins differ in only 12 amino acid residues. While SULT1A2 was detected in some gastrointestinal samples and in liver, its level was always much lower than that of SULT1A1.
SULT1C1 appears to be primarily expressed in various tissues at the foetal stage [22,32,33]. In addition, its mRNA was found in adult kidney, thyroid gland and stomach [32]. Now, we unambiguously detected SULT1C1 protein in stomach. A low level, at the limit of detection, appeared to be present in some colonic and ileal samples, whereas no signals were obtained in the remaining tissues investigated. From previous immunoblot analyses it is known that SULT1E1 and 2A1 are expressed in jejunum [8] and liver [34,35]. We confirmed these findings. In addition, we demonstrated the presence of SULT1E1 and 2A1 protein and enzyme activity in ileum and, at a low level, in cecum. Both forms were absent in stomach, colon and rectum. The negative results for SULT2A1 in all three tissues and for SULT1E1 in stomach agree with negative results reported for the mRNA by Dooley et al. [12]. However, these authors detected SULT1E1 mRNA in colon and colorectal tissue. Perhaps, SULT1E1 protein is degraded so rapidly in colon and rectum that it does not reach a detectable level, or it is expressed at sites other than the mucosa (in the present study only the mucosa was used, whereas Dooley et al. [12] did not specify their ‘colon’ and ‘colorectal’ tissue). In contrast with the present study and Dooley et al. [12], Tashiro et al. [36] detected SULT2A1 mRNA, SULT2A1 protein and DHEA activity in gastric mucosa. Their enzyme assay, involving a chromatographic step to reduce background signals, was more sensitive than our assay. They determined activities of 0.05–0.93 pmol·mg−1·min−1, which is much lower than those we found in other parts of the intestine (up to 121 pmol·mg−1·min−1 in an ileal sample). In that study [36], SULT2A1 immunoreactivity and mRNA hybridization was localized in parietal cells. In contrast, in the present study, ileal expression of SULT2A1 was exclusively observed in mature enterocytes.
In additional experiments, we studied the cellular localization of SULTs in ileal and colonic mucosa. The results indicate that SULT1A1, 1A3, 1B1 (in both tissues) as well as 2A1 (detected only in ileum) are primarily expressed in differentiated enterocytes. This localization supports the idea that the enzymes are part of the metabolic barrier of gut. In addition, we detected SULT1A protein(s) in endothelial cells. Localization of SULT1A in enterocytes of the luminal surface of colon has been described [37], but not staining of intestinal capillaries. SULT activity towards phenols (typical substrates of SULT1A forms) has been reported for human aortic endothelial cells [38] and bovine brain microvessel endothelial cells [39]. It has been suggested that SULTs in brain vessels may form a metabolic component of the blood–brain barrier [39]. Thus they may have a similar barrier function in gut endothelial cells. Xenobiotics (or their phase-I metabolites) containing a nucleophilic group are usually substrates not only for SULTs, but also for UGTs. Expression of both enzyme classes shows common characteristics in the human alimentary tract, as shown in the present study for SULTs and in that of Strassburg et al. [40] for UGTs. Various SULTs and UGTs are highly expressed in the gastrointestinal tract, including some forms that are not found in liver. Moreover, the pattern of the forms expressed differs between various parts of the gastrointestinal tract. However, sulfo-conjugation involves a much higher risk for the formation of reactive intermediates than glucuronidation [3]. Nevertheless, it is difficult to assess whether SULTs in the human alimentary tract represent a risk factor for carcinogenesis. It is important to notice that SULTs were only detected in mature enterocytes, but not in the stem cells of the epithelium. If a reactive sulfo-conjugate remained in the SULT-proficient cells, then the specific expression of SULTs in terminally differentiated cells might be a protective measure rather than a risk factor for carcinogenesis. Unfortunately, the animal species that are normally used in carcinogenicity studies, mouse and rat, only show low expression of most SULT forms in gut. For example, we only detected SULT1B1 and an unidentified form (putatively a member of the SULT1C subfamily, based on its immunoreactivity) in rat colon [41]. Moreover, the level of SULT1B1 in colon mucosa only amounted to 30% of the hepatic level in the rat [41], whereas the level of corresponding human enzyme was 5 times higher in colon mucosa than in liver (the present study). Likewise, SULT activity towards the promutagen 1-hydroxymethylpyrene was nearly 1000 times higher in the cytosolic fraction from liver compared with colon mucosa in the rat, whereas this variation was only 8-fold for corresponding preparations from humans [42]. The low SULT protein and activity levels in the gastrointestinal tract of the rat are reflected in low mRNA levels for many SULT forms in gastrointestinal and other extrahepatic tissues compared with liver [43]. In agreement with this tissue distribution, liver was the target organ of all SULT-dependent carcinogens identified in mice and rats [2]. We have just succeeded in constructing transgenic mouse models that express human SULT1A1 and 1B1 in colon at levels similar to those found in man (W. Meinl, G. Dobbernack, H. Himmelbauer and H. Glatt, unpublished work). We will use these models to study the role of these enzymes in the activation and inactivation of carcinogens and other toxic compounds, in particular those affecting the alimentary tract.
Online data
Acknowledgments
The authors thank Elisabeth Meyer and Sabine Braune for excellent technical assistance and Professor Frank Dombrowski (Institute of Pathology, Ernst-Moritz-Ardnt University, Greifswald, Germany), for histopathological evaluation of samples. This work was financially supported by Deutsche Forschungsgemeinschaft (INK 26) and Bundesministerium für Bildung und Forschung (grant BIO/0313028A).
References
- 1.World Cancer Research Fund. Food. Nutrition and the Prevention of Cancer: a Global Perspective, Washington; 1997. [Google Scholar]
- 2.Miller J. A., Surh Y.-J. Sulfonation in chemical carcinogenes. Handbook of Pharmacology, Vol. 112. In: Kauffman F. C., editor. Conjugation-Deconjugation Reactions in Drug Metabolism and Toxicity. Berlin: Springer-Verlag; 1994. pp. 429–458. [Google Scholar]
- 3.Glatt H. R. Activation and inactivation of carcinogens by human sulfotransferases. In: Pacifici G. M., Coughtrie M. W. H., editors. Human Sulphotransferases. London: Taylor & Francis; 2005. pp. 281–306. [Google Scholar]
- 4.Visser T. J. Pathways of thyroid hormone metabolism. Acta Med. Austriaca. 1996;23:10–16. [PubMed] [Google Scholar]
- 5.Eisenhofer G., Coughtrie M. W. H., Goldstein D. S. Dopamine sulphate: an enigma resolved. Clin. Exp. Pharmacol. Physiol. 1999;26:S41–S53. [PubMed] [Google Scholar]
- 6.Strott C. A. Sulfonation and molecular action. Endocr. Rev. 2002;23:703–732. doi: 10.1210/er.2001-0040. [DOI] [PubMed] [Google Scholar]
- 7.Sundaram R. S., Szumlanski C., Otterness D., van Loon J. A., Weinshilboum R. M. Human intestinal phenol sulfotransferase: assay conditions, activity levels and partial purification of the thermolabile form. Drug Metab. Dispos. 1989;17:255–264. [PubMed] [Google Scholar]
- 8.Her C., Szumlanski C., Aksoy I. A., Weinshilboum R. M. Human jejunal estrogen sulfotransferase and dehydroepiandrosterone sulfotransferase: immunochemical characterization of individual variation. Drug Metab. Dispos. 1996;24:1328–1335. [PubMed] [Google Scholar]
- 9.Harris R. M., Picton R., Singh S., Waring R. H. Activity of phenolsulfo-transferases in the human gastrointestinal tract. Life Sci. 2000;67:2051–2057. doi: 10.1016/s0024-3205(00)00791-8. [DOI] [PubMed] [Google Scholar]
- 10.Chen G., Zhang D., Jing N., Yin S., Falany C. N., Radominska-Pandya A. Human gastrointestinal sulfotransferases: identification and distribution. Toxicol. Appl. Pharmacol. 2003;187:186–197. doi: 10.1016/s0041-008x(02)00073-x. [DOI] [PubMed] [Google Scholar]
- 11.Pacifici G. M. Inhibition of human liver and duodenum sulfotransferases by drugs and dietary chemicals: a review of the literature. Int. J. Clin. Pharmacol. Ther. 2004;42:488–495. doi: 10.5414/cpp42488. [DOI] [PubMed] [Google Scholar]
- 12.Dooley T. P., Haldeman-Cahill R., Joiner J., Wilborn T. W. Expression profiling of human sulfotransferase and sulfatase gene superfamilies in epithelial tissues and cultured cells. Biochem. Biophys. Res. Commun. 2000;277:236–245. doi: 10.1006/bbrc.2000.3643. [DOI] [PubMed] [Google Scholar]
- 13.Freimuth R. R., Wiepert M., Chute C. G., Wieben E. D., Weinshilboum R. M. Human cytosolic sulfotransferase database mining: identification of seven novel genes and pseudogenes. Pharmacogenomics J. 2004;4:54–65. doi: 10.1038/sj.tpj.6500223. [DOI] [PubMed] [Google Scholar]
- 14.Blanchard R. L., Freimuth R. R., Buck J., Weinshilboum R. M., Coughtrie M. H. W. A proposed nomenclature system for the cytosolic sulfotransferase (SULT) superfamily. Pharmacogenetics. 2004;14:199–211. doi: 10.1097/00008571-200403000-00009. [DOI] [PubMed] [Google Scholar]
- 15.Freimuth R. R., Raftogianis R. B., Wood T. C., Moon E., Kim U. J., Xu J., Siciliano M. J., Weinshilboum R. M. Human sulfotransferases SULT1C1 and SULT1C2: cDNA characterization, gene cloning, and chromosomal localization. Genomics. 2000;65:157–165. doi: 10.1006/geno.2000.6150. [DOI] [PubMed] [Google Scholar]
- 16.Wang J., Falany J. L., Falany C. N. Expression and characterization of a novel thyroid hormone-sulfating form of cytosolic sulfotransferase from human liver. Mol. Pharmacol. 1998;53:274–282. doi: 10.1124/mol.53.2.274. [DOI] [PubMed] [Google Scholar]
- 17.Falany C. N., Wheeler J., Oh T. S., Falany J. L. Steroid sulfation by expressed human cytosolic sulfotransferases. J. Steroid Biochem. Mol. Biol. 1994;48:369–375. doi: 10.1016/0960-0760(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 18.Ganguly T. C., Krasnykh V., Falany C. N. Bacterial expression and kinetic characterization of the human monoamine-sulfating form of phenol sulfotransferase. Drug Metab. Dispos. 1995;23:945–950. [PubMed] [Google Scholar]
- 19.Meinl W., Pabel U., Osterloh-Quiroz M., Hengstler J. G., Glatt H. R. Human sulfotransferases are involved in the activation of aristolochic acids and are expressed in renal target tissue. Int. J. Cancer. 2006;118:1090–1097. doi: 10.1002/ijc.21480. [DOI] [PubMed] [Google Scholar]
- 20.Comer K. A., Falany C. N. Immunological characterization of dehydroepiandrosterone sulfotransferase from human liver and adrenal. Mol. Pharmacol. 1992;41:645–651. [PubMed] [Google Scholar]
- 21.Forbes-Bamforth K. J., Coughtrie M. W. H. Identification of a new adult human liver sulfotransferase with specificity for endogenous and xenobiotic estrogens. Biochem. Biophys. Res. Commun. 1994;198:707–711. doi: 10.1006/bbrc.1994.1102. [DOI] [PubMed] [Google Scholar]
- 22.Stanley E. L., Hume R., Coughtrie M. W. H. Expression profiling of human fetal cytosolic sulfotransferases involved in steroid and thyroid hormone metabolism and in detoxification. Mol. Cell. Endocrinol. 2005;240:32–42. doi: 10.1016/j.mce.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 22a.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Forbes K. J., Hagen M., Glatt H. R., Hume R., Coughtrie M. W. H. Human fetal adrenal hydroxysteroid sulphotransferase: cDNA cloning, stable expression in V79 cells and functional characterisation of the expressed enzyme. Mol. Cell. Endocrinol. 1995;112:53–60. doi: 10.1016/0303-7207(95)03585-u. [DOI] [PubMed] [Google Scholar]
- 24.Engelke C. E., Meinl W., Boeing H., Glatt H. R. Association between functional genetic polymorphisms of human sulfotransferases 1A1 and 1A2. Pharmacogenetics. 2000;10:163–169. doi: 10.1097/00008571-200003000-00008. [DOI] [PubMed] [Google Scholar]
- 25.Raftogianis R. B., Wood T. C., Otterness D. M., van Loon J. A., Weinshilboum R. M. Phenol sulfotransferase pharmacogenetics in humans: association of common SULT1A1 alleles with TS PST phenotype. Biochem. Biophys. Res. Commun. 1997;239:298–304. doi: 10.1006/bbrc.1997.7466. [DOI] [PubMed] [Google Scholar]
- 26.Falany C. N. Sulfation and sulfotransferases: 3. Enzymology of human cytosolic sulfotransferases. FASEB J. 1997;11:206–216. doi: 10.1096/fasebj.11.4.9068609. [DOI] [PubMed] [Google Scholar]
- 27.Meloche C. A., Falany C. N. Expression and characterization of the human 3β-hydroxysteroid sulfotransferases (SULT2B1a and SULT2B1b) J. Steroid Biochem. Mol. Biol. 2001;77:261–269. doi: 10.1016/s0960-0760(01)00064-4. [DOI] [PubMed] [Google Scholar]
- 28.He D., Meloche C. A., Dumas N. A., Frost A. R., Falany C. N. Different subcellular localization of sulphotransferase 2B1b in human placenta and prostate. Biochem. J. 2004;379:533–540. doi: 10.1042/BJ20031524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Falany C. N., Xie X., Wang J., Ferrer J., Falany J. L. Molecular cloning and expression of novel sulphotransferase-like cDNAs from human and rat brain. Biochem. J. 2000;346:857–864. [PMC free article] [PubMed] [Google Scholar]
- 30.Fujita K., Nagata K., Yamazaki T., Watanabe E., Shimada M., Yamazoe Y. Enzymatic characterization of human cytosolic sulfotransferases; identification of ST1B2 as a thyroid hormone sulfotransferase. Biol. Pharm. Bull. 1999;22:446–452. doi: 10.1248/bpb.22.446. [DOI] [PubMed] [Google Scholar]
- 31.Nowell S., Green B., Tang Y. M., Wiese R., Kadlubar F. F. Examination of human tissue cytosols for expression of sulfotransferase isoform 1A2 (SULT1A2) using a SULT1A2-specific antibody. Mol. Pharmacol. 2005;67:394–399. doi: 10.1124/mol.104.006171. [DOI] [PubMed] [Google Scholar]
- 32.Her C., Kaur G. P., Athwal R. S., Weinshilboum R. M. Human sulfotransferase SULT1C1: cDNA cloning, tissue-specific expression, and chromosomal localization. Genomics. 1997;41:467–470. doi: 10.1006/geno.1997.4683. [DOI] [PubMed] [Google Scholar]
- 33.Sakakibara Y., Yanagisawa K., Katafuchi J., Ringer D. P., Takami Y., Nakayama T., Suiko M., Liu M. C. Molecular cloning, expression, and characterization of novel human SULT1C sulfotransferases that catalyze the sulfonation of N-hydroxy-2-acetylaminofluorene. J. Biol. Chem. 1998;273:33929–33935. doi: 10.1074/jbc.273.51.33929. [DOI] [PubMed] [Google Scholar]
- 34.Song W. C., Qian Y., Li A. P. Estrogen sulfotransferase expression in the human liver: marked interindividual variation and lack of gender specificity. J. Pharmacol. Exp. Ther. 1998;284:1197–1202. [PubMed] [Google Scholar]
- 35.Aksoy I. A., Sochorova V., Weinshilboum R. M. Human liver dehydroepiandrosterone sulfotransferase: nature and extent of individual variation. Clin. Pharmacol. Ther. 1993;54:498–506. doi: 10.1038/clpt.1993.181. [DOI] [PubMed] [Google Scholar]
- 36.Tashiro A., Sasano H., Nishikawa T., Yabuki N., Muramatsu Y., Coughtrie M. W., Nagura H., Hongo M. Expression and activity of dehydroepiandrosterone sulfotransferase in human gastric mucosa. J. Steroid Biochem. Mol. Biol. 2000;72:149–154. doi: 10.1016/s0960-0760(00)00020-0. [DOI] [PubMed] [Google Scholar]
- 37.Windmill K. F., Christiansen A., Teusner J. T., Bhasker C. R., Birkett D. J., Zhu X. Y., McManus M. E. Localisation of aryl sulfotransferase expression in human tissues using hybridisation histochemistry and immunohistochemistry. Chem.-Biol. Interact. 1998;109:341–346. doi: 10.1016/s0009-2797(97)00144-0. [DOI] [PubMed] [Google Scholar]
- 38.Yang H. Y., Namkung M. J., Nelson W. L., Juchau M. R. Phase II biotransformation of carcinogens/atherogens in cultured aortic tissues and cells: I. Sulfation of 3-hydroxybenzo[a]pyrene. Drug Metab. Dispos. 1986;14:287–292. [PubMed] [Google Scholar]
- 39.Baranczyk-Kuzma A., Audus K. L., Borchardt R. T. Substrate specificity of phenol sulfotransferase from primary cultures of bovine brain microvessel endothelium. Neurochem. Res. 1989;14:689–691. doi: 10.1007/BF00964880. [DOI] [PubMed] [Google Scholar]
- 40.Strassburg C. P., Nguyen N., Manns M. P., Tukey R. H. UDP-glucuronosyltransferase activity in human liver and colon. Gastroenterology. 1999;116:149–160. doi: 10.1016/s0016-5085(99)70239-8. [DOI] [PubMed] [Google Scholar]
- 41.Teubner W., Meinl W., Glatt H. R. Stable expression of rat sulfotransferase 1B1 in V79 cells: activation of benzylic alcohols to mutagens. Carcinogenesis. 2002;23:1877–1884. doi: 10.1093/carcin/23.11.1877. [DOI] [PubMed] [Google Scholar]
- 42.Glatt H. R., Meinl W., Kuhlow A., Ma L. Metabolic formation, distribution and toxicological effects of reactive sulphuric acid esters. Nova Acta Leopoldina NF87. 2003;329:151–161. [Google Scholar]
- 43.Dunn R. T., Klaassen C. D. Tissue-specific expression of rat sulfotransferase messenger RNAs. Drug Metab. Dispos. 1998;26:598–604. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.